Abstract
We report an analysis of allele-specific expression (ASE) and parent-of-origin expression in adult mouse liver using next generation sequencing (RNA-Seq) of reciprocal crosses of heterozygous F1 mice from the parental strains C57BL/6J and DBA/2J. We found a 60% overlap between genes exhibiting ASE and putative cis-acting expression quantitative trait loci (cis-eQTL) identified in an intercross between the same strains. We discuss the various biological and technical factors that contribute to the differences. We also identify genes exhibiting parental imprinting and complex expression patterns. Our study demonstrates the importance of biological replicates to limit the number of false positives with RNA-Seq data.
Keywords: RNA-Seq, allele-specific expression, genomic imprinting, linkage analysis
GENETIC variation affecting gene expression has been shown to be extremely common in natural populations and an important contributor to complex traits, both in human populations, experimental organisms, and livestock. Such variation is also likely to be important in monogenic traits by modifying the trait phenotypes resulting from structural or other mutations (Rockman and Kruglyak 2006). The loci that contribute to gene expression levels are termed expression quantitative trait loci (eQTL) and are commonly divided into cis-eQTLs and trans-eQTLs. Cis-acting elements affect gene expression only on the same DNA molecule, thus acting in an allele-specific manner. For example, a cis-eQTL might result from sequence differences in a promoter or an enhancer of the gene or sequences important for the stability of the RNA so that its turnover rate is affected. Trans-acting loci, on the other hand, act indirectly on the target gene through diffusible RNA or protein, such as a transcription factor. Such loci would be expected to affect the target gene residing on both chromosome copies in a diploid organism and, hence, the target gene would not exhibit allele-specific expression. In most studies using genetic linkage or genome-wide association analyses, cis-regulation has been assumed based on the fact that the eQTL lies in close proximity to the gene whose expression is affected. Thus, it is more precise to term such loci as “local” eQTL rather than cis (Kruglyak 2008; van Nas et al. 2010).
The identification of eQTL on a global basis became feasible ∼10 years ago with the development of gene expression microarrays (Brem et al. 2002; Schadt et al. 2003). In these studies, eQTL were mapped using linkage analysis in which the locus affecting gene expression was traced by segregation through genetic crosses in experimental organisms, human families, or livestock species (Kruglyak 2008; Le Mignon et al. 2009; van Nas et al. 2010). A weakness of such studies in the case of mammals is that the mapping resolution was poor, generally in the range of tens to hundreds of genes. With the development of genome-wide association studies in humans as well as model organisms, it became feasible to map eQTL much more finely (Cookson et al. 2009; Bennett et al. 2010; Grundberg et al. 2012; Orozco et al. 2012). Both linkage and association need large numbers of samples (hundreds) to have sufficient power.
The development of high throughput RNA sequencing, termed RNA-Seq, offers a novel approach to genome-wide expression profiling. RNA-Seq allows digital quantitation of transcript levels, improving on the semi-quantitative nature of microarrays. It can also distinguish alleles of the genes being regulated if the RNA products include sequence differences, either in the exons or introns. RNA-Seq can, of course, be performed on large numbers of samples to identify eQTLs, using either linkage or association analyses. It also offers a potentially powerful approach for identifying cis-acting eQTL (as opposed to local eQTL) through quantitation of allele-specific expression, by using a limited number of samples. However, analysis of a gene requires that the expressed sequences of two alleles differ by at least one base. Such analyses are particularly straightforward in studies of inbred strains of mice, where either sequencing or high-density genotyping has been performed for many different strains and complete haplotype information is available. When two such inbred strains are crossed to produce F1 heterozygous mice, RNA-Seq can be used to reveal imprinted genes and ASE in general. In studies of imprinted genes, reciprocal crosses can be used to discriminate parent of origin from other biases. Thus far, relatively few studies have used RNA-Seq to identify ASE genes or imprinted genes (Babak et al. 2008; Wang et al. 2008; Babak et al. 2010; Gregg et al. 2010a,b) and the results reported were sometimes inconsistent. In a recent report, DeVeale et al. (2012) have shown that it is important in such studies to consider certain technical limitations of RNA-Seq as well as biological variability.
Here, we have analyzed genes exhibiting ASE in mouse liver using RNA-Seq data from an F1 mouse design, with reciprocal crosses from inbred strains C57BL/6J (B) and DBA/2J (D). We have first examined the technical limitations of RNA-Seq using independent biological replicates. We then carried out a comparative study of cis-eQTL using RNA-Seq as opposed to expression microarrays. We previously carried out a large genetic cross using the same strains in which we identified several thousand apparent cis-eQTL (Davis et al. 2012), and we compare the two approaches. We also identify parentally imprinted genes in adult liver and examine instances of complex gene expression patterns for imprinted and ASE genes.
Materials and Methods
Mice and tissues
RNA-Seq was performed on liver mRNA from F1 male and female DBA/2J and C57BL/6J mice, purchased from The Jackson Laboratory (Bar Harbor, ME). Reciprocal F1 male and female mice were generated by breeding the parental strains in the vivarium at UCLA. For six liver RNA libraries, RNA from three mice was pooled into four independent samples of high-fat-fed B × D and D × B males and females and two samples of chow-fed B × D and D × B males. All mice were fed ad libitum and maintained on a 12-hour light/dark cycle. F1 pups were weaned at 28 days and fed a chow diet (Ralston-Purina) until 8 weeks of age, at which time half were placed on a high-fat diet (Research Diets D12266B). All F1 mice were killed at 16 weeks, with liver harvested at that time.
Library preparation for Illumina sequencing
Library preparation was performed as recommended by the manufacturer (Illumina, Hayward, CA). Briefly, total RNA was extracted using the RNeasy Mini kit with DNase treatment (Qiagen, Valencia, CA). Poly(A) mRNA was isolated and fragmented, and first-strand cDNA was prepared using random hexamers. Following second-strand cDNA synthesis, end repair, addition of a single A base, adaptor ligation, and agarose gel isolation of ∼200-bp cDNA, PCR amplification of the ∼200 bp cDNA was performed. Liver samples were sequenced using the Illumina GAIIX sequencer to a coverage of ∼40 million single-end reads of 75 bp.
Read mapping
We first aligned reads of 75 bp to the mouse reference genome version mm9 using mrsFAST (Hach et al. 2010) allowing up to five mismatches. It is known that aligning the reads to the reference genome introduces a bias toward the reference genome, in our case a bias toward the C57BL/6J genome. Thus, we used the known genomic SNPs between DBA/2J and C57BL/6J (based on the Mouse Sequencing Consortium; Waterston et al. 2002; Keane et al. 2011), and converted the allele of those positions to “N” (base not indicated) in the reference genome; we aligned the reads to this new artificial genome (see Supporting Information, Figure S3).
The reads were then divided into two categories. The first category included the mapped reads or the set of reads that align to the genome with five or fewer mismatches. The second category was reads that failed to map to the reference genome. Many RNA-Seq reads failed to align to a genome because they spanned the exonic junctions. To overcome this problem, we mapped the unmapped reads with TopHat (Trapnell et al. 2009), which is designed to map reads to the genome by splitting the reads into smaller fragments. The reads aligned to the genome in this process were added to the map read set.
We selected reads with base modifications of the RNA located in one exon and corresponding to a known genomic SNP between DBA/2J and C57BL/6J (based on the Mouse Sequencing Consortium; Waterston et al. 2002; Keane et al. 2011). The read was required to have a base quality of ≥20.
Imprinted genes analysis
The reads of the different SNPs for the same exon were summed to improve the power of statistical analysis. An exon was considered imprinted if the two B/D expression ratios are opposite between the two reciprocal crosses and significant (P-value ≤0.05). The significance was calculated by the Fisher exact test for which the P-value was corrected for multiple testing by the Benjamini–Hochberg method. The paternal bias was defined as “B/(B + D) for DxB − B/(B + D) for BxD.”
Allele-specific expression analysis
The reads of the different SNPs for the same exon were summed to improve the power of statistical analysis. An exon was considered to have ASE if the B/D expression ratio is significantly greater than to 1.5 or less than 1/1.5 (P-value ≤0.05). The significance was calculated by a Fisher exact test for which the P-value was corrected for multiple testing by the Benjamini–Hochberg method to control the false positives. Gene set enrichment analysis was performed using DAVID (Huang da et al. 2009).
Sex and diet effects
An exon was declared as impacted by the sex (or diet) if the B/D ratio in one sex (or one diet) was significantly >1.5 or <1/1.5 with a P-value ≤0.05, whereas the B/D ratio in the other sex (or other diet) is not significant (P-value ≥0.2%). The significance was calculated by a Fisher exact test for which the P-value was corrected for multiple testing by the Benjamini–Hochberg method.
Sequenom validation of allele-specific expression
DNA was extracted from one mouse per F1 reciprocal cross and RNA was extracted from three mice per cross (independent sample from RNA-Seq RNA), pooled, and cDNA generated. DNA and cDNA were analyzed in a primer extension assay, designed to target the polymorphic nucleotide. The primer extension assay was carried out using the MassARRAY (Sequenom iPLEX Gold genotyping protocol) platform according to the manufacturer’s specifications by McGill University and the Génome Québec Innovation Centre. Primer extension products were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. The area of each peak was proportional to the transcript abundance and was measured by the MassARRAY software to generate an allelic ratio (allele 1:allele 2) calculation. The allelic ratio obtained for cDNA was normalized using that measured from genomic DNA, where the allelic ratio is expected to be 1:1 to correct for technical artifacts.
Results
Experimental design
We studied six different liver RNA samples isolated from F1 mice (each from a pool of three mice) derived from strains B and D corresponding to two reciprocal crosses. These samples differed in diet, sex, and the direction of the cross (Table S1). For analyzing parent-of-origin effects, the three samples from each cross were considered as independent biological replicates (Figure 1A). For analyzing general ASE effects, the two reciprocal crosses were used as independent biological replicates in the three sex and diet contexts (Figure 2A). RNA-Seq was performed on the Illumina platform using poly(A)-containing RNA as described in Materials and Methods.
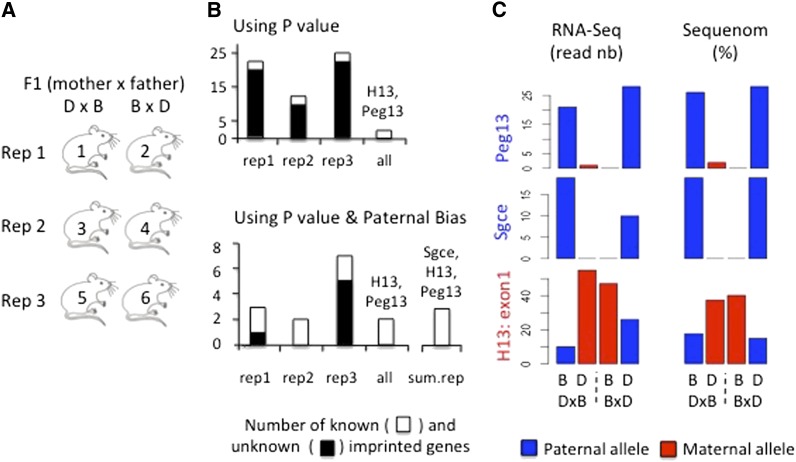
Figure 1.
Identification of liver-imprinted genes using RNA-Seq. (A) Schematic of experimental design with three independent reciprocal crosses. In each cross, the liver of three mice per F1 were pooled and subjected to RNA-Seq analysis. (B) Number of genes that exceed threshold for the P-value (top) or the P-value and the paternal bias (bottom) in the three independent reciprocal crosses (rep1, rep2, and rep3). The thresholds are 0.05 for the P-value corrected by Benjamini–Hochberg and 50% for the paternal bias. The paternal bias was defined as B/(B + D) for DxB − B/(B + D) for BxD. When the difference is close to 1 (or −1) the gene is paternally (or maternally) expressed. Open, the known imprinted genes (excluding those found only by Gregg et al. (2010a,b); solid, the genes identified by Gregg et al. (2010a,b) or unknown. Rep1, rep2, and rep3, number found in the biological replicates 1, 2, and 3, respectively; all, imprinted genes observed in all three replicates; sum.rep, imprinted genes observed after summing the reads of the three repetitions for all the SNPs of the same exon. For this last analysis, only the stringent criteria based on both P-value and paternal bias was used. (C) Validation by Sequenom technology of the three imprinted genes observed by RNA-Seq in liver. The results obtained by RNA-Seq technology (left) are expressed as reads mapping to the SNP position. The sequenom results (right) are expressed as percentage of total mRNA sequences containing the C57BL/6J (B) vs. DBA/2J (D) base (total = 100%). Blue bars represent the paternal allele and red bars represent the maternal allele.
Figure 2.
Identification of genes with allele-specific expression using RNA-Seq. (A) Schematic of experimental design with the two reciprocal crosses used as independent biological replicates in three diet and sex contexts (HF, high fat; chow, chow diet). (B) Number of exons (right) or genes (left) not exhibiting ASE (solid), exhibiting ASE in only one sample (shaded), or in the two replicates (Keane et al. 2011). These numbers are the mean number of exons (right) or genes (left) identified in the three diet or sex studies. (C) Distribution of the expression (sum of the reads for the two alleles B and D) for exons under ASE in only one sample (shaded boxplot, example with female high fat B × D or D × B) or shared by these two replicates (open boxplot).
Analysis of imprinted genes
To study imprinted genes, we used the three independent biological replicates characterized by the same genetic background (Figure 1A) to control for random technical and biological variations. We summed the allele-specific reads across SNPs in the same exon to improve the power of the statistical test. In so doing, we identified between 12 and 25 imprinted genes with only 2 genes known to be imprinted (Figure 1B, right). These numbers decreased to between 3 and 7 genes per replicate (Figure 1B, left) when we used a more stringent criteria, combining P-value ≤0.05 and paternal bias ≥50% (see Materials and Methods). Only two imprinted genes were shared by the three replicates, corresponding to the known imprinted genes H13 and Peg13. We also summed the allelic-specific reads across SNPs in the same exon, combining all three replicates to improve the statistical power of the tests; for this analysis we applied only the stringent criteria for selecting the imprinted genes. This resulted in the identification of an additional imprinted gene, Sgce, which is also known to be imprinted. These results are summarized in Table S2. We validated the imprinted status of these three genes by an independent platform (Sequenom) (Figure 1C).
Imprinted genes are generally organized in clusters. As shown in Table 1, for the two clusters on chromosomes 6 and 15 containing Sgce and Peg13, we have four other genes considered to be expressed in liver (>10 reads for at least one allele) and known to be imprinted in other tissues (http://www.har.mrc.ac.uk/research/genomic imprinting). However, none of these four genes was found to be imprinted (Table 1).
Table 1. Expression and imprinting status in liver of two gene clusters around Sgce and Peg13 and known to be imprinted.
| Read counts for D × B | Read counts for B × D | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Gene | Knowna | Exon no. | B | Db | B/B + D | Bb | Db | B/B + D | Paternal biasc | PvalBH |
| 6 | Casd1 | Mat | 18 | 20 | 36 | 0.36 | 27 | 25 | 0.51 | −0.16 | NS |
| 6 | Sgce | Pat | 8 | 19 | 0 | 1 | 0 | 10 | 0 | 1 | 0.00014 |
| 6 | Ppp1r9a | Mat | 1 | Read count <10 | Read count <10 | ND | ND | ||||
| 6 | Pon2 | Mat | 9 | 253 | 252 | 0.5 | 161 | 168 | 0.49 | 0.01 | NS |
| 6 | Pon2 | Mat | 7 | 13 | 34 | 0.28 | 12 | 27 | 0.31 | −0.03 | NS |
| 6 | Pon2 | Mat | 6 | 40 | 33 | 0.55 | 30 | 15 | 0.67 | −0.12 | NS |
| 6 | Pon2 | Mat | 4 | 90 | 90 | 0.5 | 74 | 77 | 0.49 | 0.01 | NS |
| 6 | Asb4 | Mat | 1 | Read count <10 | Read count <10 | ND | ND | ||||
| 15 | Peg13 | Pat | unique exon | 68 | 1 | 0.98 | 0 | 86 | 0 | 0.98 | 4.1E-26 |
| 15 | Trappc9 | Mat | 10 | 9 | 8 | 0.53 | 21 | 10 | 0.68 | −0.15 | NS |
| 15 | Trappc9 | Mat | 9 | 11 | 7 | 0.61 | 17 | 11 | 0.61 | 0 | NS |
| 15 | Eif2c2 | Mat | 1 | Read count <10 | Read count <10 | ND | ND | ||||
| 15 | Slc38a4 | Pat | 16 | 907 | 0 | 1 | 605 | 1 | 1 | 0 | NS |
Genes referenced as imprinted (Gregg et al. 2010a,b, http://www.har.mrc.ac.uk/research/genomic_imprinting) and for which at least one polymorphism with one read was detected in one of our samples. Genes identified as imprinted in our study are in boldface.
Number of reads corresponding to the two alleles B and D after summing the reads across SNPs in the same exon and across the three replicate sets DxB vs. BxD (named rep1, rep2 and rep3 as shown Figure 1A). “read count < 10” means that the gene had a total read count < 10 (whatever the allele) and therefore was considered as not or very weakly expressed in liver (as indicated by “low expression”).
The paternal bias was defined as “B/(B+D) for DxB - B/(B+D) for BxD”. PvalBH: P value corrected for multiple testing.
ND: not determined.
Analysis of ASE
To limit the bias due to the sequence alignment step performed against the C57BL/6J reference genome, which enriches captured sequences for the B allele, we applied a specific procedure that consists of changing the SNP of the reference genome with an N (see Materials and Methods). After this adjustment, we observed a ratio of “exon number with B > D/exon number with D > B” close to 1 (1.13) and we then analyzed the genes under ASE in each of the six F1 samples. We used the two reciprocal crosses as biological replicates in three diet and sex contexts, to control for random technical and biological variations. Thus, we had three replicate comparison sets (rep1, rep2, and rep3), as indicated in Figure 2A. The number of ASE genes that we found was quite similar across the six samples. On average, of 2256 genes (4147 exons with a read sum across SNPs in the same exon >10 for the two alleles), we observed 383 genes (523 exons) that exhibited significant ASE (Figure 2B). An ASE was considered significant if the associated P-value corrected for multiple testing was ≤0.05 and the expression ratio B/D or D/B was ≥1.5. The number of ASE genes reproducible across the two biological replicates was similar for all three comparisons and averaged 284 genes (397 exons). In summary, 19.5 ± 0.004% of the genes analyzed exhibit ASE in individual samples. This percentage decreased to 14.6 ± 0.004% after analyzing biological replicates (Figure 2B). This reduction in the number of ASE genes identified reflects the biological and technical variations, in part due to the expression level as shown in Figure 2B, right. The effects of sex and diet on ASE were found to be minimal and were not further analyzed: among the 284 genes under ASE, 1 and 4 genes were observed with a reproducible and significant effect of diet and sex, respectively (Table S3 and Table S4). Of the genes that were more highly expressed in C57BL/6 mice, there was a significant enrichment in “antigen processing and presentation,” “complement and coagulation cascades,” and “fatty acid metabolism” signaling pathways. Genes more highly expressed in DBA/2J were enriched for “oxidation-reduction,” “drug metabolism,” and “response to cytokine stimulus” signaling pathways (Table S5).
Within-gene differences in ASE including parent of origin
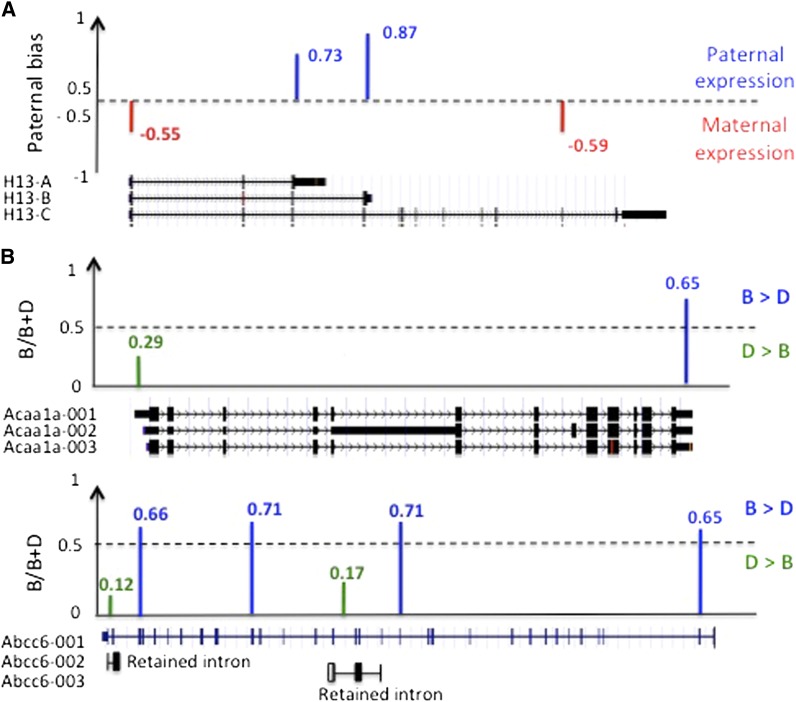
Processing of pre-mRNA is highly regulated since it results in multiple mRNA and protein products that may have distinct or even opposite functions. We therefore analyzed the imprinting and ASE profiles at the exon or intron levels to detect possible variation in expression patterns within the same gene. A striking example of this was observed in data for the imprinted H13 gene. This gene exhibited four exons for which a SNP was available and reproducible across the three biological replicates. Interestingly, we found that exons 3 and 4 were paternally expressed, whereas exons 1 and 10 were overexpressed for the maternal copy (Figure 3A). This complex profile is related to the different isoforms of this gene as shown in Figure 3A. The isoform information indicates that the long isoform C is likely overexpressed for the maternal copy, in contrast to the short isoforms A and B for which the paternal copy is likely overexpressed. Overall, among the genes observed under ASE, we found 19 genes for which two exons or one exon and one intron yielded opposite ASE patterns. Figure 3B shows two examples of within-gene differences under ASE (Acaa1a and Abcc6). These observations suggest the existence of different isoforms in the two strains used to generate the F1 mice, with genetic control elements acting in cis, possibly located at the splice sites.
Figure 3.
RNA-Seq reveals complex expression profiles within genes. (A) Imprinting pattern of H13 in adult liver for different exons. The hash marks indicating direction of the bias are the only SNPs in this region. The reads corresponding to the paternally expressed transcripts mapped to the 3′-UTRs of the two short transcripts and therefore could not be part of the long transcript. The first SNP in exon 1 was biased toward the maternal copy, because, while all three transcripts contain exon 1, the maternally expressed long transcript was 10 times more highly expressed than the short transcripts. For more details, see Table S2. (B) ASE patterns with opposite B/D expression ratios, likely reflecting different isoform expression, within the same gene. (Top) Acaa1a with two exons 5′-UTR and 3′-UTR with opposite expression profiles. (Bottom) Abcc6 with four exons overexpressed for the C57BL/6J allele contrary to two introns, overexpressed for the DBA/2J allele. These latter could reflect the expression profiles of two short isoforms known for having retained an intron.
Comparison of ASE identified by RNA-Seq with local-eQTL identified by mapping with microarrays
As discussed above, we previously identified apparent cis-acting eQTL in a large genetic intercross between strains C57BL/6J (B) and DBA/2J (D) (Davis et al. 2012). For this study, we utilized a 10-Mb window on either side of the gene for the classification of local eQTL. A fraction of these local eQTL could represent eQTL mapping near the regulated gene acting in trans; for example, genes exhibiting some form of autoregulation could exhibit local eQTL that act in trans (Figure 4B, right). In the previous study, we detected 2382 significant local-eQTL (LOD score ≥6) among 15,480 genes, using an analysis of microarray data in liver.
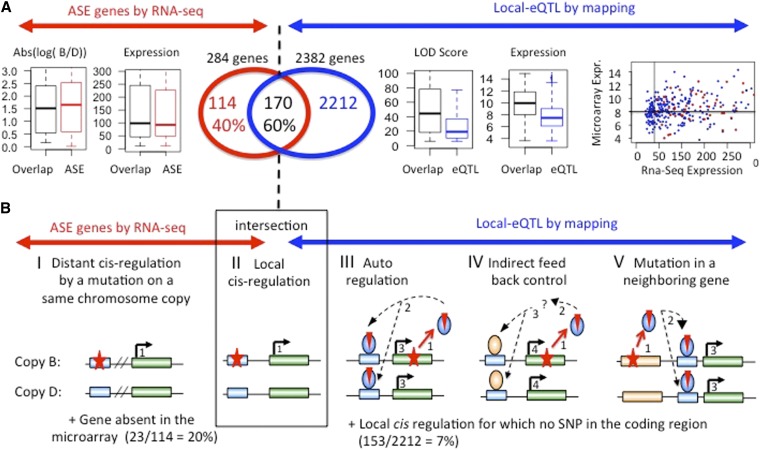
Figure 4.
Comparison of ASE identified by RNA-Seq with local-eQTL identified by mapping with microarrays. (A) Overlap between local-eQTL genes and ASE genes. Left of the hashed line: Boxplots of the expression (sum of B + D reads) and of the |log(B/D)| expression ratio for the ASE genes overlapping (black and named “overlap”) or nonoverlapping (red and named “ASE”) with eQTL genes identified by linkage analysis. Right of the hashed line: Boxplots of the LOD score and expression of the local eQTL genes overlapping (black and named “overlap”) or nonoverlapping (blue and named “eQTL”) with ASE genes. Far right: plot between the expression of all these genes in the microarray experiment (y-axis) and RNA-Seq (x-axis) experiment. (B) Possible regulatory mechanisms or technical bias corresponding to the local eQTL genes and ASE genes. The two B and D alleles are shown for each situation. Red star, mutation; nos. 1–4 and dotted arrows indicate the successive events caused by the mutation. The last event indicated by a full arrow corresponds to the allele-specific expression of the ASE gene. Green box indicates the coding regions of the ASE gene in which the causal mutation can be located (III and IV). In these two cases, the variant acts locally in trans (i.e., indirectly) on the expression level of the ASE gene. Blue box symbolizes the regulatory regions of the ASE gene in which the causal mutation can be located (I and II). In these two cases, the mutation acts in cis (sensu stricto) on the expression level of the ASE gene, i.e., no additional events occur for the regulation. In I, the mutation is distant from the gene regulated; in II, the mutation is close. Light brown box corresponds to a gene close to the ASE gene in which the causal mutation can be located (V).
Among the 284 ASE genes that replicated among samples, 170 (∼60%) overlap with these 2382 local-eQTL genes (Figure 4A). The 170 overlapping genes tended to be expressed at higher levels and to exhibit higher LOD scores in the linkage studies compared to the 2212 other eQTL genes that did not overlap (Figure 4A, right). Of the 2212 local-eQTL genes that do not overlap, 153 (7%) did not have a SNP between D and B and, therefore, could not be analyzed by RNA-Seq. Of the 114 ASE genes that were not found in the local-eQTL gene set, 23 genes (20%) were absent from the microarrays that were used in the linkage study and, therefore, would not have been detected. These 114 nonoverlapping ASE genes had similar expression levels, P-values, and B/D ratios compared with genes that did overlap (Figure 4A, left). Thus, the 170 local e-QTL that were found by microarray and RNA-Seq can be considered as confirmed cis-eQTL. We investigated the possibility that the distance from the eQTL LOD score peak and the gene may be shorter for the eQTL set identified by RNA-Seq and linkage analysis vs. the eQTL set only identified by linkage analysis. We found that the distances were similar for both gene sets (Figure S1). This result is not surprising since the position of the maximum LOD score obtained by linkage analysis is not a reliable predictor of the position of the causal mutation underlying a QTL. Concerning the fraction of 2382 eQTL genes that have a SNP between D and B with at least 10 reads for each allele, we plotted their expression measured by microarray against their expression measured by RNA-Seq data (Figure 4A, extreme right). We have distinguished the eQTL overlapping with ASE genes from those that did not overlap (blue). In addition, we tested a set of the genes that overlapped, 95 genes with cis-eQTLs (within 2 Mb of the transcript location) and LOD scores > 6.1, and found that 90% of the ASE was in the same direction. This is shown graphically in Figure S2. The genes with a read count ≤40 were not detected as ASE, whereas they were detected as local eQTL, showing that the RNA-Seq analysis lacks power when the number of reads is limited. For the other genes with a read count >40, some are detected as ASE, whereas others were not detected, indicating that ASE analysis (sequencing) and eQTL analysis (linkage) do not reveal the same expression regulatory events, as discussed below. It is noteworthy that the probes used for linkage analysis with microarrays were removed from the analysis when they contained a SNP, to avoid artifactual differential expression.
Conclusions
Two significant conclusions have emerged from our ASE analysis of F1 heterozygous mice. First, linkage analyses and sequencing of RNA can yield different conclusions with respect to cis-eQTL. And second, RNA-Seq analysis of ASE, including parent-of-origin effects, has technical limitations that must be considered to avoid inappropriate conclusions.
During the last few years, several studies have utilized RNA-Seq data to study ASE (Babak et al. 2010) and phenomena such as RNA editing (Ju et al. 2011; Li et al. 2011; Peng et al. 2012) and imprinting (Babak et al. 2008; Wang et al. 2008; Gregg et al. 2010a,b; Wang et al. 2011). More recently, some conclusions from these earlier studies have been shown to result from systematic technical difficulties in the interpretation of RNA-Seq data and from a lack of power (DeVeale et al. 2012; Kleinman and Majewski 2012; Pickrell et al. 2012). Our study emphasizes the importance of biological replicates for analysis of ASE and imprinting. For example, in our study, 25 genes exhibited parent-of-origin imprinting in individual samples, but only two of these were shared among the three biological replicates and confirmed by a separate technology. Similarly, for ASE, 19.5% of the genes exhibited apparent ASE in one sample, but this percentage decreased to 14.6% for those exhibiting ASE in two separate biological replicates.
There are a number of possible explanations for the discrepancies of cis-eQTL identified by RNA-Seq as compared to local-eQTL identified by genetic mapping. In our comparison, a far larger number of eQTL were identified by linkage (2382 among 15,480 genes analyzed), than by RNA-Seq (284 among 2256 genes analyzed). Notably of the 2382 genes, only 7% could not be analyzed by RNA-Seq because of absence of polymorphism. It is unlikely that the majority of the linkage data represent false positives since, in a previous comparison of two separate linkage studies with the same strains, we found that 76% of local-eQTL with a LOD score of ≥6.0 replicated (van Nas et al. 2010). Many of the local-eQTL identified in the linkage studies exhibited relatively small effect sizes, and a large fraction of these local eQTL may not have been detected in the RNA-Seq studies because of a lack of power due to a limited number of reads. This emphasizes the importance of a large number of reads per sample to detect ASE of small magnitude. On the other hand, numerous local eQTL presented enough read count in RNA-Seq data to be detected (Figure 4A, extreme right). Genetic linkage or genome-wide association analyses that report local eQTL cannot distinguish, among the local eQTL, the cis-eQTL from the trans-eQTL. We observed that the 2212 local eQTL that did not overlap with ASE genes had lower LOD scores than those that overlapped (Figure 4A, right). It is likely that a certain fraction of such eQTL is trans-eQTL, i.e., acting indirectly on the genes being regulated. Several regulatory mechanisms can explain such local trans-eQTL, including autoregulation, indirect regulation by feedback, or regulation by the product of a neighboring gene (Figure 4B). This latter mechanism is all the more likely since a 10-Mb window on either side of the gene was used for determining local-eQTL. A high frequency of such regulatory mechanisms would be consistent with a relatively large number of local trans-eQTL as suggested by our results.
Approximately 60% of 284 ASE genes detected by RNA-Seq overlapped with local-eQTL. This was somewhat higher than a previous study that observed an overlap of 40% (Babak et al. 2010). We asked how likely it would be that 60% percent of the genes in the two sets would overlap by chance. Using the hypergeometric distribution (Halbritter and Tomlinson 2013), we calculated the probability that 170 or more of the 284 ASE genes (of ∼15,000 possible genes) would overlap with 2382 from the same population by chance alone as P ≤ 4.6 × 10−65. Thus, while we discuss reasons for lack of overlap, it is extremely unlikely that the percentage that did overlap occurred by chance. The 40% of ASE detected by our RNA-Seq that did not overlap had similar characteristics in terms of expression levels, P-value, and B/D ratio, as the ASE fraction that overlapped with local-eQTL. There are several possible explanations for these findings. First, 20% of the nonoverlapping fraction corresponds to genes not present on the microarray. Second, this fraction undoubtedly resulted, in part, from small differences in the genetic background and/or environmental conditions. Third, the lack of overlap could also be attributed, in part, to long-range cis-acting elements (>10 Mb). The emerging field of the chromosome conformation capture allows detection of loops that bring genes into proximity with distal regulatory elements. Whereas the regulatory chromatin loops observed are generally on a kilobase scale, a few megabase-scale loops have been reported. For example in the Drosophila genome, there is a loop of ∼10 Mb on chromosome 3R between ANT-C and BX-C and another ∼6 Mb on chromosome 2R between hbs and sns (or synj) (Sexton et al. 2012).
There are several advantages of the ASE approach using RNA-Seq compared to genetic linkage mapping: RNA-Seq requires a very limited number of samples; it identifies cis-eQTL sensu stricto (i.e., expression regulated by a mutation acting in cis); it does not depend on arbitrary genomic distance cutoffs; and it can be applied to any species. On the other hand, ASE analysis by RNA-Seq is unable to detect trans-eQTL, and only genes with RNA polymorphisms can be analyzed. Regarding reference allele bias, several methods have been used to address this in allele-specific expression studies. We followed the approach of Degner et al. (2009), which masks the bases in the reference genome in which a SNP exists, in our case, between the DBA/2J and C57BL/6J (reference) genomes. We altered all bases in the genome that had a SNP between the two strains to neutral base N. It has been shown in other genomes that mapping the RNA-Seq reads to an artificial masked genome removes the bias toward the reference genome (Degner et al. 2009). Another method is to create an enhanced reference genome that includes the alternative alleles at known polymorphic loci. This set of methods produces less bias toward the reference genome compared to the masked genome method (Rozowsky et al. 2011; Satya et al. 2012). However, these methods require more resources (time and storage) to perform the read mapping. Another method for addressing reference allele bias is to use SNP tolerant read mapping (Wu and Nacu 2010). The best method for calling allele-specific expression is an active research area, and in the next few years, studies comparing multiple methods will provide clarity on which method performs best.
Our study is the first to examine the parent-of-origin-specific gene expression in adult liver. Previous studies have suggested that ∼100 genes are imprinted in the mammalian transcriptome (Barlow 1995; Wood and Oakey 2006). Recent RNA-Seq studies focused on embryos or placental tissue and reported between 21 and 35 imprinted genes in the respective tissues (Babak et al. 2008; Wang et al. 2008, 2011). Other reports by Gregg et al. (2010a,b) identified 1300 imprinted loci in mouse embryonic and adult brain. The identification of such a large number of imprinted genes is likely due in part to technical artifacts or inadequate replication (DeVeale et al. 2012), though we cannot rule out the possibility that brain tissue may contain significantly more imprinted genes than liver. In our study using roughly the same sequencing depth per sample as other studies reported above, we observed only three genes exhibiting parent-of-origin imprinting that were shared by the three replicates. These results show the importance of using independent biological replicates and suggest a more limited number of imprinted genes in metabolic tissues in adults compared to embryos. Further analyses of imprinting in adult tissues with different developmental contexts should contribute more generally to our understanding of the maintenance of parent-of-origin effects between physiological stages. Moreover, the previous studies showed in general an arrangement in clusters, with each cluster containing a local imprinting control region from which epigenetic information spread to nearby genes (Morgan et al. 2005; Ferguson-Smith 2011; Messerschmidt 2012). Notably, we did not observe such a clustered organization. Further analyses of imprinting in adult tissues are clearly required.
Processing of pre-mRNA is highly regulated by various mechanisms, including alternative splicing of internal exons, alternative initiation of transcription, or alternative use of polyadenylation sites in the 3′-UTR (Wang et al. 2008; Cooper 2010; Nilsen and Graveley 2010). The number of transcripts is therefore higher than the number of genes: 201,816 and 90,956 transcripts for 20,476 and 23,153 genes observed in human and mouse respectively (http://www.ensembl.org). One important advantage of RNA-Seq compared with microarrays is that it captures all expressed transcripts from a tissue, known and unknown, allowing investigation of their expression patterns. For both imprinted genes and genes under ASE, we identified several examples of opposite allelic expression patterns within the same gene. One of these was a complex imprinting pattern for the H13 gene that encodes a signal peptide peptidase with two exons paternally expressed and two others maternally expressed. This pattern could be related to the different isoforms identified for this gene and caused by various poly(A) sites utilized in an allele-specific manner (Wood et al. 2008). Using uniparental partial disomies for distal chromosome 2, Wood et al. (2008) provided strong evidence that epigenetic modifications can influence alternative polyadenylation. For the genes under ASE, Abcc6 was observed with two introns overexpressed in the D strain, while the exons with polymorphisms were overexpressed in the B strain. In addition to the long, functional isoform, two short isoforms were observed for this gene, both retaining one intron and incapable of translation into the full-length protein. These results indicate that the noncoding isoforms are overexpressed in the D strain in which the protein is known to be nonfunctional (Meng et al. 2007).
Acknowledgments
Funding was provided by the National Institutes of Health (NIH) grants HL28481 and HL30568 to A.J.L. F.H. and E.E. are supported by National Science Foundation grants 0513612, 0731455, 0729049, 0916676, and 1065276 and NIH grants HL080079 and DA024417. S.L. was supported by Agrocampus Ouest for her mobility at UCLA for 6 months. P.F.R was a Ph.D fellow supported by INRA and the regional council of Brittany. HD07228 provided funding for L.J.M. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Communicating editor: D. Threadgill
Literature Cited
- Babak T., DeVeale B., Armour C., Raymond C., Cleary M. A., et al. , 2008. Global survey of genomic imprinting by transcriptome sequencing. Curr. Biol. 18: 1735–1741 [DOI] [PubMed] [Google Scholar]
- Babak T., Garrett-Engele P., Armour C. D., Raymond C. K., Keller M. P., et al. , 2010. Genetic validation of whole-transcriptome sequencing for mapping expression affected by cis-regulatory variation. BMC Genomics 11: 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow D. P., 1995. Gametic imprinting in mammals. Science 270: 1610–1613 [DOI] [PubMed] [Google Scholar]
- Bennett B. J., Farber C. R., Orozco L., Kang H. M., Ghazalpour A., et al. , 2010. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 20: 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R. B., Yvert G., Clinton R., Kruglyak L., 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Cookson W., Liang L., Abecasis G., Moffatt M., Lathrop M., 2009. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 10: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. N., 2010. Functional intronic polymorphisms: buried treasure awaiting discovery within our genes. Hum. Genomics 4: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. C., van Nas A., Castellani L. W., Zhao Y., Zhou Z., et al. , 2012. Systems genetics of susceptibility to obesity-induced diabetes in mice. Physiol. Genomics 44: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner J. F., Marioni J. C., Pai A. A., Pickrell J. K., Nkadori E., et al. , 2009. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25: 3207–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeale B., van der Kooy D., Babak T., 2012. Critical evaluation of imprinted gene expression by RNA-Seq: a new perspective. PLoS Genet. 8: e1002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith A. C., 2011. Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 12: 565–575 [DOI] [PubMed] [Google Scholar]
- Gregg C., Zhang J., Butler J. E., Haig D., Dulac C., 2010a Sex-specific parent-of-origin allelic expression in the mouse brain. Science 329: 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C., Zhang J., Weissbourd B., Luo S., Schroth G. P., et al. , 2010b High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science 329: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E., Small K. S., Hedman A. K., Nica A. C., Buil A., et al. , 2012. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44: 1084–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hach F., Hormozdiari F., Alkan C., Birol I., Eichler E. E., et al. , 2010. mrsFAST: a cache-oblivious algorithm for short-read mapping. Nat. Methods 7: 576–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbritter, F., and S. R. Tomlinson, 2013 Hypergeometric Distribution, edited by F. Halbritter and S. R. Tomlinson. GeneProf, Edinburgh, UK. [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Ju Y. S., Kim J. I., Kim S., Hong D., Park H., et al. , 2011. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals. Nat. Genet. 43: 745–752 [DOI] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman C. L., Majewski J., 2012. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 335: 1302. [DOI] [PubMed] [Google Scholar]
- Kruglyak L., 2008. The road to genome-wide association studies. Nat. Rev. Genet. 9: 314–318 [DOI] [PubMed] [Google Scholar]
- Le Mignon G., Desert C., Pitel F., Leroux S., Demeure O., et al. , 2009. Using transcriptome profiling to characterize QTL regions on chicken chromosome 5. BMC Genomics 10: 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Wang I. X., Li Y., Bruzel A., Richards A. L., et al. , 2011. Widespread RNA and DNA sequence differences in the human transcriptome. Science 333: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H., Vera I., Che N., Wang X., Wang S. S., et al. , 2007. Identification of Abcc6 as the major causal gene for dystrophic cardiac calcification in mice through integrative genomics. Proc. Natl. Acad. Sci. USA 104: 4530–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt D. M., 2012. Should I stay or should I go: protection and maintenance of DNA methylation at imprinted genes. Epigenetics 7: 969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H. D., Santos F., Green K., Dean W., Reik W., 2005. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14(Spec No 1): R47–R58 [DOI] [PubMed] [Google Scholar]
- Nilsen T. W., Graveley B. R., 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco L. D., Bennett B. J., Farber C. R., Ghazalpour A., Pan C., et al. , 2012. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell 151: 658–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z., Cheng Y., Tan B. C., Kang L., Tian Z., et al. , 2012. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 30: 253–260 [DOI] [PubMed] [Google Scholar]
- Pickrell J. K., Gilad Y., Pritchard J. K., 2012. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 335: 1302, author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L., 2006. Genetics of global gene expression. Nat. Rev. Genet. 7: 862–872 [DOI] [PubMed] [Google Scholar]
- Rozowsky J., Abyzov A., Wang J., Alves P., Raha D., et al. , 2011. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol. Syst. Biol. 7: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satya R. V., Zavaljevski N., Reifman J., 2012. A new strategy to reduce allelic bias in RNA-Seq readmapping. Nucleic Acids Res. 40: e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E. E., Monks S. A., Drake T. A., Lusis A. J., Che N., et al. , 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302 [DOI] [PubMed] [Google Scholar]
- Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., et al. , 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nas A., Ingram-Drake L., Sinsheimer J. S., Wang S. S., Schadt E. E., et al. , 2010. Expression quantitative trait loci: replication, tissue- and sex-specificity in mice. Genetics 185: 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Soloway P. D., Clark A. G., 2011. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics 189: 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Sun Q., McGrath S. D., Mardis E. R., Soloway P. D., et al. , 2008. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS ONE 3: e3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., et al. , 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Wood A. J., Oakey R. J., 2006. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2: e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. J., Schulz R., Woodfine K., Koltowska K., Beechey C. V., et al. , 2008. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 22: 1141–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. D., Nacu S., 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]