Abstract
With remarkable speed, the CRISPR–Cas9 nuclease has become the genome-editing tool of choice for essentially all genetically tractable organisms. Targeting specific DNA sequences is conceptually simple because the Cas9 nuclease can be guided by a single, short RNA (sgRNA) to introduce double-strand DNA breaks (DSBs) at precise locations. Here I contrast and highlight protocols recently developed by eight different research groups, six of which are published in GENETICS, to modify the Caenorhabditis elegans genome using CRISPR/Cas9. This reverse engineering tool levels the playing field for experimental geneticists.
Progress in science depends on new techniques, new discoveries and new ideas, probably in that order.
Sydney Brenner (Robertson 1980, from the symposium on Biology in the 1980s held at Friedrich Miescher Institut in Basel on March 20–21, 1980)
IF we are to believe Sydney Brenner then you better hold on tight because we should be in for a rush of new discoveries and ideas. New techniques based on the RNA-guided nuclease CRISPR–Cas9 (Jinek et al. 2012) have led to a recent flurry of articles describing genome editing in organisms as diverse as bacteria (Jiang et al. 2013), yeast (DiCarlo et al. 2013), worms (Friedland et al. 2013), fruit flies (Gratz et al. 2013), zebrafish (Hwang et al. 2013), mammalian cell lines (Cong et al. 2013; Mali et al. 2013), mice (Wang et al. 2013), rats (Li et al. 2013b,c), plants (Li et al. 2013a; Nekrasov et al. 2013), and food crops such as rice and wheat (Shan et al. 2013). Furthermore, Cas9 proteins can be repurposed to target effector domains to specific genomic locations to control gene expression (Qi et al. 2013; Gilbert et al. 2013). The ability to efficiently edit and perturb genomes with high precision has enormous potential for experimental design.
The challenge for researchers will be to devise the most efficient strategy to fully exploit the strengths of the system. Since each genetic model organism has its unique strengths and challenges, here I focus narrowly on how the CRISPR–Cas9 system has been adapted to Caenorhabditis elegans in nine recent studies, five of which are reported in this issue of GENETICS (Chiu et al. 2013; Cho et al. 2013; Katic and Großhans 2013; Tzur et al. 2013; Waaijers et al. 2013). A recent Commentary article covers the same ground for fruit flies (Golic 2013). I first describe how double-strand DNA breaks (DSBs) induced with transposons and TALENs (transcription activator-like effector nucleases) can be repaired from templates, because the same rules apply to CRISPR–Cas9 gene editing. I then highlight and contrast the different ways to deliver CAS9 to the germline. Finally, I look to the future and briefly discuss potential uses of Cas9 to regulate gene expression or to induce targeted changes in chromatin at particular loci.
It has been clear for >30 years from experiments primarily on yeast, worms, and flies that genomes can be edited with homologous DNA sequences (Hinnen et al. 1978). Genome editing is stimulated by homologous DNA templates with free ends (i.e., linear templates) (Orr-Weaver et al. 1981) or by generating DSBs in the genome itself, for example, by excision of a P-element transposon (Gloor et al. 1991). Genomic breaks can be repaired by nonhomologous end joining (NHEJ) resulting in stochastic changes that include small insertions and deletions (indels). Alternatively, repair can be directed by homologous sequences elsewhere in the genome (Gloor et al. 1991), homologous oligonucleotides (Banga and Boyd 1992), or extra-chromosomal transgenes (Plasterk and Groenen 1992), resulting in the incorporation of specific, predetermined sequences.
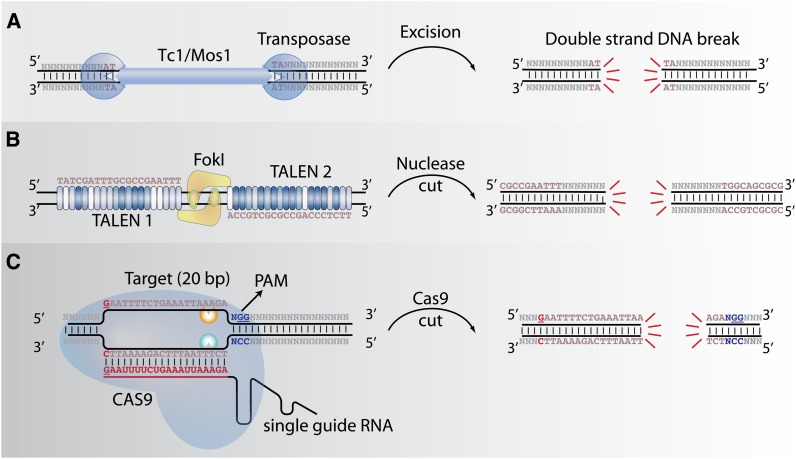
In C. elegans, genome-editing techniques have until recently primarily been based on transposons, which cause DSBs upon their excision (Figure 1A). In the absence of a repair template, gene mutations or deletions can be generated after excision of an endogenous Tc1 transposon (Zwaal et al. 1993). However, semistable, transgenic lines with extrachromosomal arrays containing many copies of injected DNA are relatively simple to generate (Mello et al. 1991) and provide templates for DSB repair after excision of the endogenous Tc1 transposon (Plasterk and Groenen 1992). The first examples of endogenous gene editing, including tagging a gene with GFP, were based on Tc1 excision (Barrett et al. 2004). This strategy has the disadvantage that Tc1 is active only in mutator strains. In these strains, several classes of transposons are active and transposition of multiple transposons lead to a heavy mutational load (Bessereau 2006). Improved gene-editing methods were developed with the Mos1 transposon from Drosophila mauritania, which is active in C. elegans (Bessereau et al. 2001). Excision of a single Mos1 transposon can be used to generate specific deletions and insertions (Robert and Bessereau 2007) as well as to insert affinity tags and GFP by repair from extrachromosomal plasmids (Gendrel et al. 2009) without inducing breaks throughout the genome. The general usefulness of Mos1 for gene editing was greatly expanded by the effort of a consortium of European labs to generate a collection of 13,000 strains carrying Mos1 elements (Bazopoulou and Tavernarakis 2009; Vallin et al. 2012). This resource enabled insertion of single copies of transgenes into specific genomic locations as well as generation of large targeted deletions (∼25 kb) using positive and negative selection markers (Frøkjær-Jensen et al. 2008, 2010, 2012). One challenge of gene editing in C. elegans is the inability to generate sustained expression in the germline from extrachromosomal transgenes (Kelly et al. 1997). Nevertheless, injected transgenes are transiently expressed in the germline (Kelly et al. 1997), and this is sufficient to mobilize Mos1 elements, particularly if the strong ubiquitous eft-3 promoter is used (Frøkjær-Jensen et al. 2008, 2012).
Figure 1.
Ways to generate site-specific double-strand DNA breaks. (A) Transposition of the class II DNA transposons Tc1 or Mos1 is catalyzed by a single enzyme (the transposase) and excision results in a DSB. (B) TALENs are composed of sequential repeats that encode the DNA targeting specificity. To generate a DSB, two TALENs are targeted to adjacent sequences separated by 14–20 bp and upon dimerization the FokI nuclease domains cleave the intervening DNA. (C) The CRISPR–Cas9 enzyme is guided to the target site by a single-guide RNA (sgRNA). The target sequence is determined by the 20 nucleotides in red followed by the PAM sequence (NGG). The DSB is generated 3 bp upstream of the PAM.
Ideally, the ability to modify the genome would not be limited to locations near endogenous or exogenous transposons. Rapid improvements in engineered DNA nucleases have now largely removed this limitation. The first synthetic technique to generate DSBs at targeted genomic regions was based on chimeric proteins consisting of a DNA binding domain (a zinc finger) and a nonspecific nuclease (FokI) (Kim et al. 1996). Such zinc finger nucleases (ZFNs) were designed to work in pairs to increase DSB specificity by requiring binding of two chimeric proteins at nearby DNA sequences for nuclease activity (Smith et al. 2000). However, the DNA sequence recognition code of zinc fingers is complicated, depends on the DNA context, and must be selected for experimentally to ensure high efficiency of each ZFN (Maeder et al. 2008).
TALENs are conceptually similar to ZFNs and consist of the DNA-binding domain of a TALE protein fused to the FokI nuclease (TALEN) (Figure 1B) (Christian et al. 2010). The DNA targeting code of TAL effectors is based on sequential sequence repeats in the DNA binding domain that each bind to a single (or in some cases, two) base pair (Boch et al. 2009; Moscou and Bogdanove 2009). This cipher makes it relatively easy to target each pair of TALENs simply by cloning repeats in the order corresponding to the target DNA sequence (Cermak et al. 2011).
ZFNs and TALENs have been used to introduce hereditary mutations by end joining as well as to alter the sequence of the targeted site via homologous recombination (for a review, see Gaj et al. 2013). The first successful approach to generating hereditary changes in C. elegans (and C. briggsae) with ZFNs and TALENs was based on mRNA injection and NHEJ (Wood et al. 2011). This method was recently substantially expanded to include repair templated by oligonucleotides and was shown to function in further diverged nematode species, Pristionchus pacificus, and the male/female Caenorhabditis species 9 (Lo et al. 2013). Furthermore, two FRT sites could be inserted to flank a locus and subsequently excised by FLP to delete the full intervening sequence (Lo et al. 2013). An alternative TALEN-based method allows nonhereditary, conditional knockouts in somatic cells by inducible and cell-specific expression of TALENs from extrachromosomal arrays (Cheng et al. 2013). In themselves, these TALEN-based methods will be very useful and should also facilitate development of similar methods based on CRISPR–Cas9.
The allure of the CRISPR–Cas9 system relative to these other approaches is that it is conceptually simpler and that the targeting specificity is very high. Namely, a single protein (Cas9) is used to generate the DNA break, which is targeted to a DNA sequence using a single short RNA (Figure 1C). In bacteria, Cas9 is guided to specific DNA sequences by two small RNAs, called CRISPR RNAs (crRNAs) and trans-acting crRNAs (tracrRNAs). The Cas9 nuclease has two independent nuclease domains that each cut one DNA strand, leading to a DSB (Jinek et al. 2012). For a recent review of the elegant work that led to an understanding of how several types of CRISPR–Cas9 systems work mechanistically in bacteria, see Wiedenheft et al. (2012). The two small RNAs can be engineered to comprise a single guide RNA (sgRNA) (Jinek et al. 2012), further simplifying the system. Once the sgRNA is loaded into the CAS9 protein, the complex cleaves DNA that is complementary to a 20-bp stretch (the “protospacer”) of the sgRNA. The sgRNA sequence around the protospacer is not completely arbitrary: there are requirements at its 5′ end for synthesis and at the 3′ end for DNA cleavage. The guide RNA can be synthesized in vivo using the U6 PolI promoter; U6 transcription requires a single leading G. Alternatively, the RNA can be made in vitro from the phage T7 promoter; T7 transcription requires two leading Gs. To cleave DNA, the 20-bp guide sequence must be followed by another nucleotide and then two G’s (the “PAM motif”). So, genomic target sites have the generic form GN19NGG for U6 transcription and GGN18NGG for T7 transcription, where the underlined 20-bp residues are targeted for cleavage.
Nine articles published within the last 4 months have described methods by which to generate inherited mutations in C. elegans with CRISPR–Cas9 (Friedland et al. 2013; Lo et al. 2013; Chiu et al. 2013; Cho et al. 2013; Katic and Großhans 2013; Tzur et al. 2013; Waaijers et al. 2013; Dickinson et al. 2013; Chen et al. 2013) (see summary in Table 1). The methods differ mainly in three ways: in how the CRISPR–Cas9 effector complex is delivered (DNA, RNA, or protein), in how repair is mediated (NHEJ or homology directed repair), and in how gene-editing events are identified (PCR screen or selection markers).
Table 1. Summary of CRISPR–Cas9 mediated gene editing in C. elegans.
| Author | Journal | Publication Date | CAS-9 expression | sgRNA expression | Selection strategy | Species | Repair mechanism | Repair template | Number of genes modified | Gene-editing frequency (P0) Average ±SEM | Gene-editing frequency (F1) Average ±SEM | Indels observed (Average ±SEM) | % Precise templated modifications | Off target mutations (sgRNAs, candidate matches, genomic loci sequenced) | Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Friedland et al. | Nature Methods | June 2013 | DNA (Peft-3) | DNA (U6 promoter) | Phenotype, fluorescent markers and PCR | C. elegans | NHEJ | N.A. | 4 (unc-119, dpy-13, klp-12, Y61A9LA.1) | N.D. | 26% ±14% (N = 7) | 4 ±1 bp (N = 26) | N.A. | 0% (4, 8, 14) | First example of CAS9–CRISPR genetic modification in C. elegans |
| Waaijers et al. | GENETICS | This issue | DNA (Phsp-16.48) | RNA (T7) and DNA (U6 promoter) | Visible phenotype screening | C. elegans | NHEJ | N.A. | 4 (lin-5, rol-1, dpy-11, unc-119) | 13% ± 3% (N =6) | 7% ± 3% (N = 6) | 11 ± 3 bp (N = 13) | N.A | N.D. | Reported toxicity with Peft-3:cas9 DNA injection. |
| Tzur et al. | GENETICS | This issue | DNA (Peft-3) | DNA (U6 promoter) | Fluorescent markers and PCR | C. elegans | Templated | dsDNA (plasmid) | 2 (klp-12, lab-1) | 25% ± 16% (N = 3) | 7% ± 5% (N = 3) | N.A. | 100% (N = 6) | N.D. | Reported small effect of lab-1 deletion on upstream gene (asfl-1) |
| Chen et al. | Nucleic Acid Research | In press | DNA (Peft-3) | DNA (U6 promoter) | Positive (hygromycin), negative (peel-1), drug selection (benomyl), visual (fluorescent array markers) | C. elegans | NHEJ and templated | dsDNA (plasmid) | 3 (ben-1, unc-4, unc-5) | 6 ± 4% (N = 2) | 32 ± 19% (N = 4) | 2.7 ± 0.5 bp (N = 16) | 100% (N = 4) | N.D. | Four 9-kb insertions fully sequenced. |
| Dickinson et al. | Nature Methods | In press | DNA (Peft-3) | DNA (U6 promoter) | Positive (unc-119), negative (peel-1), visual (fluorescent array markers) | C. elegans | Templated | dsDNA (plasmid) | 3 (nmy-2, his-72, lin-31) | 11 ± 4% (N = 7) | N.D. | 1x unc-119 duplication, 1x GFP transgene deletion | 85% (N = 13) | 0% (2, 14, 182) | Four different mutations were introduced into lin-31 simultaneously. |
| Lo et al. | GENETICS | August 2013 | mRNA (T7) | T7 mRNA (Note: tracrRNA and crRNA separate) | PCR, cel-1 mismatch assay, drug selection (benomyl) | C. elegans | NHEJ | N.A. | CAS9: 1 (GFP) | CAS9: 38% (N = 1) | CAS9: 1.2% (N = 1) | N.D. | N.A. | N.D. | Single sgRNA was non-functional. |
| Katic and Großhans | GENETICS | This issue | mRNA (T7) | mRNA (T7) | Drug selection (benomyl), phenotype | C. elegans | NHEJ and templated | dsDNA (plasmid) | 3 (ben-1, unc-36, daf-2) | 35 ± 30% (N = 2) | 0.4% (N = 1) | 134 ± 100 bp (N = 8) | 100% (N = 1) | sgRNA with two mismatches targets ben-1 at 9% frequency (P0) | 2-bp mismatch designed into sgRNA at 5′ end still permits gene targeting. |
| Chiu et al. | GENETICS | This issue | mRNA (SP6) | mRNA (SP6) | Phenotype | C. elegans | NHEJ | N.A. | 2 (dpy-11, unc-4) | 13 ± 7% (N = 2) | No data | >1800 ± 500 bp (N = 9) | N.A. | No mutations or deletions detected. | Whole-genome sequencing of two independent alleles. |
| Cho et al. | GENETICS | This issue | Purified protein | mRNA (T7) | PCR, T7E1 mismatch assay | C. elegans | NHEJ | N.A. | 2 (dpy-3, unc-1) | 45 ± 5% (N = 2) | 6 ± 3% (N = 2) | 98 ± 25 (N = 18) | N.A. | N.D. | One example of biallelic gene targeting. |
PAM, protospacer associated motif; DSB, double-strand DNA break; TALEN, TAL effector nuclease; CRISPR, clustered regularly interspaced short palindromic repea
DNA Injection
The first demonstration of Cas9 activity in C. elegans was based on a relatively simple protocol to screen for random mutations after DNA injection (Friedland et al. 2013). The authors used two separate plasmids to express Cas9 from an eft-3 promoter and sgRNA from a U6 small nuclear RNA promoter. They were able to induce mutations in two genes with clear mutant phenotypes from the progeny of individual F1 animals. Importantly, they were also able to identify, by PCR screening, mutations induced in two genes with no obvious phenotypes, suggesting that the mutation frequency is high enough that selection markers are not necessarily required. A follow-up article published in this issue of GENETICS expands their protocol to include templated repair from a co-injected plasmid to interrupt the klp-12 gene, or fully replace the lab-1 gene with GFP by screening only for fluorescence (Tzur et al. 2013). Another article in this issue of GENETICS describes use of a heat-shock promoter to drive expression of CAS9 and a U6 promoter to express the sgRNA to generate random deletions in four genes with visible mutant phenotypes (Waaijers et al. 2013). Two articles expand on the DNA-based protocols by incorporating the use of positive and negative selections to edit the genome (Chen et al. 2013; Dickinson et al. 2013), similar to the protocols developed for targeted transgene insertion by Mos1 (Frøkjær-Jensen et al. 2012). Chen et al. (2013) generated templated modifications in the unc-4, unc-5, and ben-1 genes by using the antibiotic hygromycin for positive selection (Greiss and Chin 2011) and fluorescent markers together with a heat-shock-inducible toxin gene peel-1 for negative selection (Seidel et al. 2011). The results presented by Dickinson et al. (2013) stand out for having most fully realized the potential of the CRISPR–Cas9 system for à la carte genome editing. The authors tagged the nmy-2 and his-72 genes with GFP by using unc-119(+) as a positive selection marker that was inserted next to the modified gene; fluorescent co-injection markers and inducible peel-1 were used to select against extrachromosomal arrays. unc-119(+) was flanked by LoxP sites and subsequent injection of DNA encoding Cre recombinase was used to re-excise unc-119(+) to minimally perturb the genomic environment, although no effect of the selection marker was detected on the expression level of nmy-2. In their most elegant experiment, Dickinson et al. (2013) mutated the lin-31 gene simultaneously at four threonine residues to mimic phosphorylation (4T → E) or lack of phosphorylation (4T → A), leaving only a 34-bp “scar” from a LoxP site distal to the 3′-UTR. All the DNA-based injection protocols showed similar high frequencies of gene editing, with averages ranging from 7 to 32% of F1 animals (Table 1).
RNA Injection
Three groups performed gene editing using Cas9 and sgRNAs transcribed in vitro (Chiu et al. 2013; Katic and Großhans 2013) or with tracrRNA and crRNA transcribed separately in vitro (Lo et al. 2013). Chiu et al. (2013) generated random mutations in genes that cause obvious phenotypes, whereas Lo et al. (2013) targeted a single-copy GFP gene and screened for lack of fluorescence. It is important to note that Lo et al. (2013) were not able to generate mutations with a sgRNA, but did so efficiently with separate tracrRNA and crRNA. Perhaps two guide RNAs are sometimes more efficient, or perhaps the exact structure of the sgRNA scaffold is very important. Katic and Großhans (2013) showed that plasmids mixed in with the RNA are efficient templates for gene repair by selecting for suppressor reversion of a daf-2 hypomorph to wild type. Similarly, Lo et al. (2013) show that ssDNA oligonucleotides can be used as templates for repair for TALEN-based gene editing. Therefore, it is likely that oligonucleotides can also be used as repair templates for Cas9–CRISPR-based gene editing.
Protein Injection
Cho et al. (2013) demonstrated that purified CAS9 protein injected into the germline along with sgRNA synthesized in vitro can create DSBs. They targeted two genes that offer visible phenotypes (dpy-3 and unc-1) and screened for NHEJ deletions with a mismatch enzyme assay. The overall frequency of gene editing in F1 progeny was 0.4–1.2% for RNA injections and 6% for protein injections, which is considerably lower than the targeting efficiency from DNA injections. However, this is somewhat misleading because with RNA and protein injections “rescued” F1 animals were not distinguished from nontransgenic animals. In fact, the gene targeting frequency of injected animals (P0) is similar to or higher for RNA and protein injections compared to DNA injections (Table 1).
Experimental Considerations
Each method has strengths and weaknesses; as always, the method of choice will depend on the particular experiment. In general, RNA- and protein-based methods have the advantage that they can relatively easily be adapted to nematodes other than C. elegans because a detailed knowledge of active germline promoters is unnecessary; this has already been shown for TALENs (Wood et al. 2011; Lo et al. 2013). DNA-based methods have the advantages that positive and negative selection markers are easily incorporated into the procedure and that most labs are familiar with standard DNA injection rather than RNA injection. Random mutations mediated by NHEJ are quick to generate, but in the absence of a clear mutant phenotype require more effort to isolate, typically through PCR and assays that detect DNA mismatches. In contrast, specific gene editing determined by a co-injected plasmid requires more effort to generate the repair template but less work to isolate mutants because of selectable markers (genetic or antibiotic). Also, selectable markers add an (optional) additional step to remove the marker, but have the advantage that severe mutations are balanced and that larger regions of the genome can be modified in a single injection.
As noted by Chiu et al. (2013), the mutational spectrum generated by NHEJ repair appears to be different between DNA injections and RNA/protein injections. Indels generated by DNA injection are relatively short with most changing <10 bp. In contrast, RNA and protein injection generate large indels on the order of 100–2000 bp, with one instance of a possible chromosomal rearrangement (Chiu et al. 2013). Larger indels may complicate identification of mutations and the exact extent of indels if sequences of PCR primers near the targeted region are frequently deleted.
Off-site mutations are a concern for any genome-editing technique. However, experiments in human cells indicate that mismatches in the sgRNA potently interfere with cleavage: three interspaced or five concatenated mismatches in the 20 nucleotide targeting sequence eliminate Cas9 cleavage in most cases. The GG nucleotides in the PAM motif that follow the targeted DNA sequence are especially important: the only change that is tolerated is from NGG to NAG, with a concomitant 80% loss of cleavage efficiency (Hsu et al. 2013). In C. elegans, the most thorough characterization of off-site mutations was performed by Chiu et al. (2013), based on whole-genome sequencing of two dpy-12 alleles generated by CRISPR–Cas9 RNA injection with a single sgRNA. In this instance the authors were unable to identify any second-site mutations or deletions caused by CRISPR–Cas9 cleavage. Several other groups (Table 1) identified loci with partial homology to sgRNAs and sequenced candidates for off-site mutations, but none were detected. These experiments suggest that the CRISPR–Cas9 nuclease is not inherently mutagenic in C. elegans if care is taken to design sgRNAs to targets with minimal sequence similarity to other loci and with a keen eye to the mismatch tolerance of Cas9. Furthermore, the short lifespan of C. elegans makes it relatively easy to minimize the occurrence of background mutations by outcrossing.
Unintended mutations can also occur at the cleavage site due to error-prone repair from DNA (Nassif et al. 1994). This is not unique to the Cas9 nuclease but can occur using any of the genome-editing methods described in this review. Fidelity of oligonucleotide-based repair from a large number of genes was assayed after TALEN cleavage in C. elegans (Lo et al. 2013). Precise repair was obtained in 30% (38/128) of gene modifications and in 14% (12/85) of short deletions templated by oligonucleotides (Lo et al. 2013). Oligonucleotide-mediated repair has not been tested with CRISPR–Cas9 in C. elegans but it is possible that Cas9 with only a single active nuclease site may result in improved frequency of correct repair from oligos because repair of single-strand DNA breaks biases the repair pathway toward homology-mediated repair and disfavors NHEJ (Kim et al. 2012). CAS9 cleaves each DNA strand with a different part of the protein, and CAS9 mutants defective in one part can generate single-strand DNA nicks (Jinek et al. 2012).
From plasmid-guided repair using Cas9, Dickinson et al. (2013) observed mistakes in 14% (1/7) of transgene insertions and a gene duplication in 17% (1/6) of GFP-tagged genes. The frequency of incorrect repair is similar to that observed for Mos1-mediated insertion and deletion (Frøkjær-Jensen et al. 2008, 2010) and likely results from the repair process rather than the method used to generate a DNA break. These results indicate that several independent alleles of any modification should be generated and that molecular characterization of the sequence of the edited gene should be standard practice.
Although the articles described here all focused on genome editing, there are many other exciting uses for Cas9. In particular, a mutant version of CAS9, which lacks nuclease activity (CAS9*), can be used as a highly specific, modular DNA-binding protein. By fusing effector proteins to the catalytically dead CAS9* protein a DNA recognition complex with enzymatic activity that can be targeted to specific genomic regions is generated. For example, chimeric Cas9* proteins targeted to the promoter region or to the coding sequence of bacterial genes repress transcription by interfering with RNA polymerase binding and elongation, respectively (Qi et al. 2013). Similarly, CAS9* fused to transcriptional repressors or enhancers can silence or activate genes in human cell lines (Gilbert et al. 2013). CAS9* fusion to effector domains that modify histones should induce local epigenetic modifications, like TALENs have been repurposed for inducible chromatin modification at target sites in response to light (Konermann et al. 2013). If these techniques can be adapted in C. elegans, and the similar technique adapted based on TALENs (Cheng et al. 2013) suggest they can, they should allow spatial and temporal control of gene expression at the level of individual isoforms.
In summary, we are now in the exciting situation that through the efforts of many labs, several different protocols that allow robust, inheritable gene editing in C. elegans have been developed. The simplest protocols can be used to rapidly generate knockouts and protocols with more bells and whistles can be used to engineer custom modifications, including the insertion of fluorescent tags and specific mutations. These advances level the playing field for C. elegans geneticists relative to yeast, fly, and mouse geneticists, who have been able to reverse engineer knockins, knockouts, and conditional alleles based on FLP and CRE recombinases for some time. Combined with the ability to perform forward genetic screens and rapidly identify mutations by whole-genome sequencing (Hobert 2010), these advances offer opportunities for creative experimentation. Only our imaginations will limit the use Cas9 proteins to perform subtle manipulations of the C. elegans genome.
Acknowledgments
I thank Oliver Hobert, David Greenstein, and Erik Jorgensen for comments on the manuscript. C.F.J. has been funded by postdoctoral fellowships from the Lundbeck Foundation and Carlsberg Foundation to conduct research in Erik M. Jorgensen’s laboratory, who is funded by National Institutes of Health grant 5R01GM095817-03 and Howard Hughes Medical Institute.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Banga S. S., Boyd J. B., 1992. Oligonucleotide-directed site-specific mutagenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 89: 1735–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett P. L., Fleming J. T., Göbel V., 2004. Targeted gene alteration in Caenorhabditis elegans by gene conversion. Nat. Genet. 36: 1231–1237 [DOI] [PubMed] [Google Scholar]
- Bazopoulou D., Tavernarakis N., 2009. The NemaGENETAG initiative: large scale transposon insertion gene-tagging in Caenorhabditis elegans. Genetica 137: 39–46 [DOI] [PubMed] [Google Scholar]
- Bessereau, J.-L., Transposons in C. elegans (January 18, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.70.1, http://www.wormbook.org.
- Bessereau J. L., Wright A., Williams D. C., Schuske K., Davis M. W., et al. , 2001. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413: 70–74 [DOI] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., et al. , 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., et al. , 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., L. A. Fenk, and M. de Bono, 2013. Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acid Research. (in press). [Google Scholar]
- Cheng Z., Yi P., Wang X., Chai Y., Feng G., et al. , 2013. Conditional targeted genome editing using somatically 7 expressed TALENs in C. elegans. Nat. Biotechnol. (in press). [DOI] [PubMed] [Google Scholar]
- Chiu H., Schwartz H. T., Antoshechkin I., Sternberg P. W., 2013. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics 1167–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J.-S., Lee J., 2013. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 1177–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M., Cermak T., Doyle E. L., Schmidt C., Zhang F., et al. , 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J., et al. , 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41: 4336–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, D. J., J. D. Ward, D. J. Reiner, and B. Goldstein, 2013 Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nature Methods. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiácovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Hollopeter G., Taylor J., Harris T. W., et al. , 2010. Targeted gene deletions in C. elegans using transposon excision. Nat. Methods 7: 451–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F., 3rd, 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel M., Rapti G., Richmond J. E., Bessereau J.-L., 2009. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature 461: 992–996 [DOI] [PubMed] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., et al. , 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G. B., Nassif N. A., Johnson-Schlitz D. M., Preston C. R., Engels W. R., 1991. Targeted gene replacement in Drosophila via P element-induced gap repair. Science 253: 1110–1117 [DOI] [PubMed] [Google Scholar]
- Golic K. G., 2013. RNA-guided nucleases: a new era for engineering the genomes of model and non-model organisms. Genetics 195: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiss S., Chin J. W., 2011. Expanding the genetic code of an animal. J. Am. Chem. Soc. 133: 14196–14199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R., 1978. Transformation of yeast. Proc. Natl. Acad. Sci. USA 75: 1929–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O., 2010. The impact of whole genome sequencing on model system genetics: get ready for the ride. Genetics 184: 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., et al. , 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31: 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A., 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I., Großhans H., 2013. Targeted heritable mutation and gene conversion by Cas9-CRISPR in Caenorhabditis elegans Genetics 195: 1173–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K., Fire A., 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146: 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. G., Cha J., Chandrasegaran S., 1996. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 93: 1156–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Kim S., Kim D. H., Choi B.-S., Choi I.-Y., et al. , 2012. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res. 22: 1327–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann, S., M. D. Brigham, A. E. Trevino, P. D. Hsu, and M. Heidenreichet al, 2013 Optical control of mammalian endogenous transcription and epigenetic states. Nature 500: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Qiu Z., Shao Y., Chen Y., Guan Y., et al. , 2013b Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 31: 681–683 [DOI] [PubMed] [Google Scholar]
- Li J.-F., Norville J. E., Aach J., McCormack M., Zhang D., et al. , 2013a Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31: 688–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Teng F., Li T., Zhou Q., 2013c Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat. Biotechnol. 31: 684–686 [DOI] [PubMed] [Google Scholar]
- Lo T.-W., Pickle C. S., Lin S., Ralston E. J., Gurling M., et al. , 2013. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics 195: 331–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder M. L., Thibodeau-Beganny S., Osiak A., Wright D. A., Anthony R. M., et al. , 2008. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell 31: 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou M. J., Bogdanove A. J., 2009. A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Nassif N., Penney J., Pal S., Engels W. R., Gloor G. B., 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., Staskawicz B., Weigel D., Jones J. D. G., Kamoun S., 2013. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31: 691–693 [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J., 1981. Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA 78: 6354–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Groenen J. T., 1992. Targeted alterations of the Caenorhabditis elegans genome by transgene instructed DNA double strand break repair following Tc1 excision. EMBO J. 11: 287–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., et al. , 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V., Bessereau J.-L., 2007. Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 26: 170–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M., 1980. Biology in the 1980s, plus or minus a decade. Nature 285: 358–359 [DOI] [PubMed] [Google Scholar]
- Seidel H. S., Ailion M., Li J., van Oudenaarden A., Rockman M. V., et al. , 2011. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biol. 9: e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J., Zhang Y., Chen K., et al. , 2013. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31: 686–688 [DOI] [PubMed] [Google Scholar]
- Smith J., Bibikova M., Whitby F. G., Reddy A. R., Chandrasegaran S., et al. , 2000. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 28: 3361–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur Y. B., Friedland A. E. , Nadarajan S., Church G. M., Calarco J. A., et al. , 2013. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics 195: 1181–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallin E., Gallagher J., Granger L., Martin E., Belougne J., et al. , 2012. A genome-wide collection of Mos1 transposon insertion mutants for the C. elegans research community. PLoS ONE 7: e30482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijers, S., V. Portegijs, J. Kerver, B. B. C. G. Lemmens, M. Tijsterman et al, 2013 CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans Genetics 195: 1187–1191. [DOI] [PMC free article] [PubMed]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., et al. , 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B., Sternberg S. H., Doudna J. A., 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338 [DOI] [PubMed] [Google Scholar]
- Wood A. J., Lo T.-W., Zeitler B., Pickle C. S., Ralston E. J., et al. , 2011. Targeted genome editing across species using ZFNs and TALENs. Science 333: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaal R. R., Broeks A., van Meurs J., Groenen J. T., Plasterk R. H., 1993. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc. Natl. Acad. Sci. USA 90: 7431–7435 [DOI] [PMC free article] [PubMed] [Google Scholar]