Abstract
Inference of population structure and individual ancestry is important both for population genetics and for association studies. With next generation sequencing technologies it is possible to obtain genetic data for all accessible genetic variations in the genome. Existing methods for admixture analysis rely on known genotypes. However, individual genotypes cannot be inferred from low-depth sequencing data without introducing errors. This article presents a new method for inferring an individual’s ancestry that takes the uncertainty introduced in next generation sequencing data into account. This is achieved by working directly with genotype likelihoods that contain all relevant information of the unobserved genotypes. Using simulations as well as publicly available sequencing data, we demonstrate that the presented method has great accuracy even for very low-depth data. At the same time, we demonstrate that applying existing methods to genotypes called from the same data can introduce severe biases. The presented method is implemented in the NGSadmix software available at http://www.popgen.dk/software.
Keywords: admixture, resequencing, population structure, association studies, NGS
ADMIXTURE occurs when isolated populations begin interbreeding and their offspring represent a mixture of alleles from different ancestral populations. Estimating the admixture proportions of an individual is a valuable tool in both population genetics and genetic epidemiology. In population genetics admixture analysis allows the researcher to classify individuals with unknown ancestry into discrete populations. This has successfully been used to describe the genetics of different populations (Rosenberg et al. 2002) and even extinct populations (Rasmussen et al. 2010). Knowing the individual admixture proportions is also useful in genetic association studies. Conventional association studies assume that the individuals are sampled from the same homogeneous population and a violation of this assumption will lead to an uncontrolled false positive rate (Marchini et al. 2004; Clayton et al. 2005). Population stratification is the presence of a systematic difference in allele frequencies between subpopulations often due to different ancestries leading to false positive findings in the association study. Various methods can be used to alleviate this problem, for example by including the admixture proportions in the association model (Price et al. 2010).
Next generation sequencing (NGS) platforms such as Illumina sequencing are used to generate large amounts of sequencing data. Although the price of sequencing is rapidly decreasing, it is still expensive to generate high-depth whole genomes for a large number of individuals. Low-depth sequencing is a much cheaper alternative that still retains most of the information in the genome (Pasaniuc et al. 2012). However, using low-depth sequencing is not unproblematic. Unlike traditional Sanger sequencing and genotyping platforms, NGS platforms do not provide genotype calls directly. Sites from pairs of homologous chromosomes are not sequenced in equal proportions but instead the chromosomes are sampled with replacement. If the sequencing depth is low, it can happen that only one of the homologous chromosomes is sequenced. On top of that there are nonnegligible errors in the sequencing data, which adds another layer of uncertainty to genotype calls. The error rates from NGS technology are high and may exceed the level of genetic variability we would expect from a biological viewpoint.
Current methods and models for finding population structure and admixture require exact knowledge of the individual genotypes. Therefore, existing methods should be applied to genotypes inferred from NGS data only if the sequencing depth is sufficiently high.
In this article we develop a new method for inferring individual ancestry based on genotype likelihoods calculated from NGS data, assuming a known number of admixing populations. This allows us to take the uncertainty in the NGS data into account and avoid inferring individual genotypes. The method is applicable not only to NGS data but also to other forms of data where the information on some or all genotypes is incomplete. The information on the genotype contained in the data must simply be condensed into genotype likelihoods and the new method can be applied.
Population structure is often estimated using the Bayesian method STRUCTURE (Pritchard et al. 2000); however, in recent years full maximum-likelihood approaches like FRAPPE (Tang et al. 2005) and ADMIXTURE (Alexander et al. 2009) have become popular especially for large data sets. This is due to efficient expectation-maximization (EM) algorithms that allow for simultaneous optimization of millions of parameters. Here we extend this maximum-likelihood framework to work on genotype likelihoods. The core of the underlying model is the same as in many previous methods (Pritchard et al. 2000; Tang et al. 2005; Alexander et al. 2009; Huelsenbeck et al. 2011) but the presented method, called NGSadmix, is based on genotype likelihoods that contain all relevant information of the unobserved genotypes.
Materials and Methods
Genotype likelihoods
The information on the unobserved genotypes contained in next generation sequencing data is best summarized in genotype likelihoods (Nielsen et al. 2011). We denote the sequencing data X = {X1, X2, … , XN} and Xi = {Xi1, Xi2, … , XiM} for N individuals and M sites. The genotype likelihoods are the likelihood of observing the sequencing data for a single individual given the unobserved genotype, defined as
Several methods for calculating the genotype likelihoods exist. The SOAPsnp model (R. Li et al. 2009) computes a mismatch matrix that is used for estimating type-specific errors, and SAMtools (Li 2011) uses a model derived from the MAQ model (Li et al. 2008). We use the simple GATK model (McKenna et al. 2010) for calculating the genotype likelihoods. The GATK model assumes independence of the reads and solely uses the observed bases overlapping a specific position along with their associated quality scores. Thus, the genotype likelihood is computed as
| (1) |
| (2) |
Here d is the depth at site j for individual i, bk is the observed base, and P is the probability of error as calculated from the quality score of bk.
Model
Existing methods for estimating population structure are based on genotype data from many single-nucleotide polymorphisms (SNPs) for a large number of individuals.
We assume that the variable sites are diallelic. For a variable site we observe two different alleles and have three possible genotypes. Without loss of generality we can assign our two alleles randomly and denote the two alleles as A, B. The allele frequency is the frequency with which A occurs. We identify the genotype by the counts of the B allele. So AA = 0, AB = 1, BB = 2.
For individual i at SNP j we consider the three relevant genotype likelihoods:
| (3) |
Here Xij is the sequencing data for individual i at site j, Gij is the unobserved genotype and, p(Xij|Gij = 0) is (proportional to) the probability of observing the sequencing data Xij given that individual i is genotype 0 at SNP j.
The individual admixture proportion is the proportion of an individual’s alleles that has ancestry in a postulated ancestral population. We write the proportion of individual i’s genome that originates from population k as p(k) = qik.
The model assumes K different ancestral populations, each with its own allele frequencies. We denote the allele frequencies of allele A in population k at SNP j as fjk. If the frequencies and admixture proportions are known, the probability that an allele is A for individual i at site j is . The probability for observing genotype Gij in individual i at site j, assuming Hardy–Weinberg equilibrium, is
| (4) |
Likelihood function:
When the genotypes are observed, assuming that sites are independent, the likelihood is written as
| (5) |
If the sites are not independent, then this is a composite likelihood that will still have consistent estimates. This likelihood corresponds to the likelihood used in Tang et al. (2005) and Alexander et al. (2009) and will be used when dealing with called genotypes.
When using NGS data, the genotypes are not observed and we instead work with genotype likelihoods. The above likelihood is extended by summing over all possible genotypes:
| (6) |
In the case of known genotypes the factor p(Xij|Gij) = 1 if Gij is the observed genotype and zero otherwise, and the two likelihoods are equivalent.
Estimation
We define the following maximum-likelihood estimators of the admixture proportions and the population frequencies:
| (7) |
We note that the likelihood is invariant to switching the labels in the ancestral populations; thus there are at least K! equivalent global maximums. Also note that the likelihood must be maximized under the constraints that qik, fjk ∈ [0, 1] and .
EM algorithm:
The EM algorithm iteratively optimizes the parameters. A new and better guess for the parameters is found by using the previous guess. The parameters guess for iteration n + 1 is given by
| (8) |
| (9) |
where
| (10) |
| (11) |
Here we have used the shorthand notation
A derivation of this EM algorithm from the likelihood function is found in Supporting Information, File S1. We initialize the algorithm by a randomly chosen point in the parameter space. When there is no uncertainty in the genotype data, the expression reduces to H(Xij|Qn, Fn) = Gij and it follows that for called genotypes the EM algorithm is the same as in Tang et al. (2005) and Alexander et al. (2009).
Accelerated convergence of the EM algorithm:
When the parameter space is high dimensional, the convergence of the EM algorithm can be slow. When the progress of the algorithm in the parameter space is monitored, it is clear that many small steps in the same direction could be replaced by larger steps. This is the principle of squared iterative methods (Varadhan and Roland 2008) for accelerating EM algorithms. This acceleration is similar to the approach of Alexander et al. (2009). In each iteration of the accelerated EM algorithm, the initial value of the parameters is updated twice, using the regular EM algorithm step described in the previous section. An optimal combination based on the old and the two new parameter estimates is calculated (we choose to use scheme S3 of Varadhan and Roland 2008) and this extrapolation is then again updated by a regular EM algorithm step.
Simulations
This section describes how we carried out simulations for the purpose of validating NGSadmix and compared the performance to existing methods. Each simulated scenario is based on a choice of the admixture coefficients for each individual, the joint distribution of allele frequencies in the ancestral populations, the average sequencing depth of each individual, and the sequencing error rate.
Allele frequencies in the ancestral populations:
To use a realistic joint distribution of allele frequencies for the ancestral populations, we use allele frequency estimates from two data sets. The first set of allele frequencies is based on Human Genetic Diversity Project (HGDP) population allele frequencies. The data were obtained from the University of California, Santa Cruz (UCSC) table browser (http://genome.ucsc.edu/cgi-bin/hgTables). We used the allele frequencies from three closely related populations, namely the Han Chinese, the Japanese, and the Cambodian populations. The second set is based on allele frequencies estimated from HapMap 3 (Altshuler et al. 2010b), where we used the allele frequencies from three distantly related populations: Centre d’Etude du Polymorphisme Humain collected in Utah (CEU); Yoruba in Ibadan, Nigeria (YRI); and Han Chinese in Beijing, China (CHB).
Simulation scenarios:
For each of the two sets of allele frequencies we simulated four different scenarios each with 100,000 SNPs and three ancestral populations.
The simulated scenarios A, B, and C all consist of 50 samples with 30 nonadmixed samples from three ancestral populations, 10 samples that are equally admixed from all three populations, and 10 samples that are composed of two of the ancestral populations in equal proportions; see top plot in Figure 1. The sequencing depths are different between the three scenarios. In scenario A the average depth varies between individuals. In scenario B each individual is sequenced at an average depth of 2 and in scenario C we let 25 individuals have a high average depth (20×) and 25 have a low average deoth (1×).
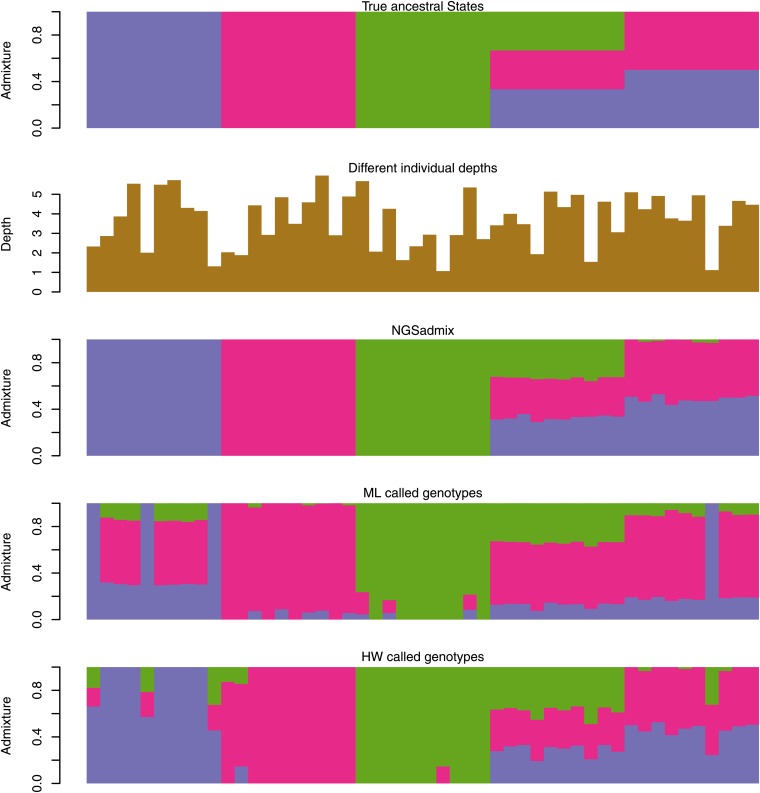
Figure 1.
Scenario A simulations based on HGDP allele frequencies. The top plot is the true admixture proportions used for simulating the 50 samples. Each bar reflects the admixture of a single individual. A single color bar means that there is no admixture, and proportions of admixture are seen as the proportion of different colors. The second plot shows the individual average sequencing depths. The third plot shows the admixture proportions estimated from genotype likelihoods using NGSadmix. The final two plots show admixture proportions estimated by ADMIXTURE from called genotypes—ML genotypes and HW genotypes.
In scenario D we examine the behavior of NGSadmix for a wider range of admixture proportions. We simulated 340 individuals with 150 nonadmixed samples, 50 samples that are equally admixed from all three ancestral populations, and 50 samples that are composed of two of the ancestral populations in equal proportions. We divided the remaining 90 samples into groups of 10 and gradually increased the admixture proportion for two of the populations from 5% to 45% by steps of 5% (see Figure S9). This scenario was simulated 100 times. For each realization the individual depths were uniformly sampled from 0.5× to 6× and the allele frequencies were randomly sampled without replacement.
Simulation of genotype likelihoods:
From the admixture coefficients and the allele frequencies in the ancestral populations, we simulate the genotypes of each individual according to the probabilities given by Equation 4. With these genotypes we generate the genotype likelihoods, using Equation 1, by simulating the sequencing errors and sequencing depth, assuming a Poisson distribution. We assume a symmetric error rate of 1% and assume that this error rate is reflected in the base quality scores.
From the simulated genotype likelihoods we remove sites with a minor allele frequency <5% estimated from the genotype likelihoods. We also remove sites with >80% missing data. For each simulated scenario we use the first 100,000 SNPs that pass these filters.
Calling genotypes:
To compare NGSadmix results with admixture estimates based on called genotypes, we called genotypes from the simulated genotype likelihoods, using two different methods: maximum-likelihood (ML) genotypes where the genotype with the highest likelihood is chosen and maximum posterior probability genotypes [Hardy–Weinberg (HW) genotypes]. The posterior genotype probability is found using a prior based on an estimate of the minor allele frequency (Kim et al. 2011) under the assumption of Hardy–Weinberg equilibrium. This prior is shared for all individuals. To see the effect of using a cutoff when calling genotypes we also estimated the admixture proportions based on HW genotypes with a posterior probability >0.95 (filtered genotypes).
1000 Genomes sequencing data
Overlap with the HapMap 3 genotype data:
The HapMap 3 (Altshuler et al. 2010b) data set contains genotypes for 1.6 million SNPs in 1184 reference individuals from 11 populations. Some of these individuals have been resequenced in the 1000 Genomes Project (Altshuler et al. 2010a; Abecasis et al. 2012). This allows us to validate NGSadmix on low-coverage sequencing data by comparing our estimates with admixture coefficients estimated from the HapMap 3 genotype data. From the 9 partially overlapping populations we chose 5 populations that all had at least 20 individuals, European (CEU), Yoruban (YRI), Chinese (CHB), Mexican ancestry in the United States (MXL), and African ancestry in the United States (ASW), and chose 20 unrelated individuals from each population to constitute a 5-population scenario. Similarly we also chose 20 individuals from each of two more closely related populations, namely the Han Chinese (CHB) and the Japanese (JPT), to constitute a 2-population scenario.
Analysis of sites with known genotypes:
Using PLINK (Purcell et al. 2007), we extracted SNPs from the HapMap 3 genotype data with a joint minor allele frequency (MAF) >5%, with no more than 5% missing genotypes and without being out of Hardy–Weinberg equilibrium (P > 0.000001). Genotype likelihoods were calculated using Equation 1 from the 1000 Genomes low-coverage sequencing data for the sites overlapping the HapMap 3 genotype data. To be able to compare NGSadmix results with called genotypes based on haplotype imputation, we also performed whole-genome haplotype imputation for the two 1000 Genomes data sets. For each site we first performed a likelihood-ratio test for variability, assuming diallelic SNPs, and chose a P-value cutoff of 10−6. The likelihood function used to test for variability is described in Kim et al. (2011) and the method for finding the major and the minor allele is described in Skotte et al. (2012). For the variable sites the genotype likelihoods were calculated using Equation 1. Implementations of all methods mentioned above are available in the ANGSD software (http://www.popgen.dk/angsd). We used the calculated genotype likelihoods in the haplotype imputation software Beagle (Browning and Yu 2009). For the 100 individuals in the five-population scenario we inferred 16,536,092 polymorphic sites and we found 7,312,452 polymorphic sites in the 40 individuals in the two-population scenario. For fast imputation we separated the genome in 10-Mb regions and merged the imputed genotypes afterward. We then used the sites that overlapped with the HapMap 3 genotypes for the admixture analysis.
Analysis of inferred polymorphic sites:
To assess the performance on SNPs detected directly from the sequencing data we also inferred polymorphic sites from the 1000 Genomes low-coverage data instead of limiting our analysis to a subset of sites known to be polymorphic. For the first 10 chromosomes we chose five random contiguous 10-Mb regions for a total of 500 Mb. This was done by calling polymorphic sites across the genome, using the above-mentioned likelihood-ratio test. Using genotype likelihoods based on Equation 1 we inferred 2.9 million sites, and with SAMtool’s genotype likelihoods we obtained 2.3 million sites. Implementations of the genotype-likelihood estimators and the likelihood-ratio test are available in the ANGSD software (http://www.popgen.dk/angsd).
Implementation
The presented method for estimating individual admixture proportions based on genotype likelihoods, NGSadmix, has been implemented in C++, using POSIX threads (Pthreads). The input files are the general and widely used Beagle input files (Browning and Yu 2009). The NGSadmix software is available at http://www.popgen.dk/software.
Results
Using simulations we explore different study designs that could occur for real data and evaluate the performance of NGSadmix by comparing the estimated admixture proportions to true admixture proportions as well as estimates of admixture proportions based on genotypes called from the simulated sequencing data. We then apply the method on low-depth sequencing data from the 1000 Genomes Project, while comparing the estimates to admixture proportions inferred from HapMap 3 genotype data as well as genotypes called from the sequencing data.
Simulations
We considered two different population scenarios for our simulations: three closely related ancestral populations based on HGDP allele frequencies and three more distant populations based on HapMap 3 allele frequencies. For each of these sets of allele frequencies, we simulated four different study designs, denoted scenarios A, B, C, and D. We simulated sequencing data conditional on the admixture proportions and allele frequencies in the ancestral populations. Further details of the simulations can be found in Materials and Methods.
Scenario A: Variable depth:
Inspired by the observed average depth distribution in the 1000 Genomes data (see Figure S1), we simulated 100,000 SNPs for 50 samples with varying average depth based on HGDP frequencies, as described in Materials and Methods. The true admixture proportions are shown in the top panel of Figure 1 followed by the individual average sequencing depths. The other panels in Figure 1 show the estimated admixture coefficients using NGSadmix, the estimated admixture coefficients based on maximum-likelihood genotypes (ML), and the maximum posterior genotypes (HW). On filtered HW genotypes (HW filtered) we could not obtain convergence, and the estimates are shown in Figure S6. NGSadmix performs better than the analysis based on called genotypes no matter how the genotypes were called. The HW genotypes called by applying a prior based on the allele frequencies seem to perform better than calling genotypes based on the highest genotype likelihood (ML genotypes).
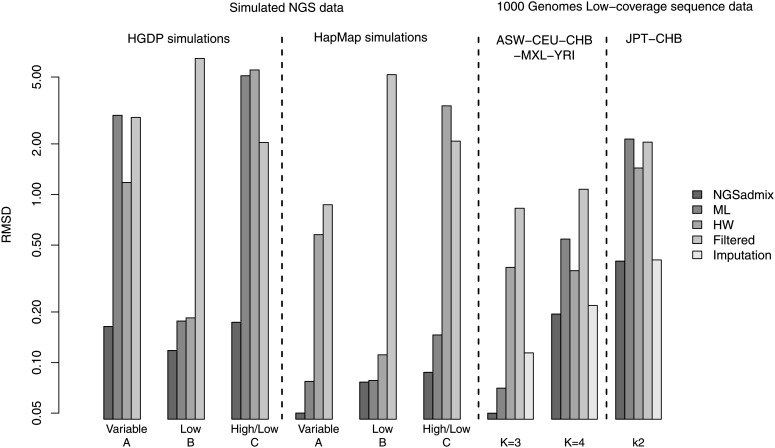
To quantify the performance beyond visual inspection we calculated the root mean square deviation (RMSD) of the estimated admixture proportions from the true admixture proportions (see leftmost group of bars in Figure 2). Using NGSadmix gives a RMSD of 0.16 while the best-performing method based on called genotypes has an RMSD of 1.18. Similarly, the largest deviation between estimated proportions and true proportions is shown in Figure S2.
Figure 2.
Root mean square deviation (RMSD) from the true admixture proportions for the different estimated admixture proportions. Left, scenario A, B, and C simulations using HGDP frequencies and then scenario A, B, and C simulations using HapMap frequencies. Right, the 1000 Genomes five-population scenario sample assuming K = 3 ancestral populations and K = 4 ancestral populations and then the 1000 Genomes two-population scenario sample assuming K = 2 ancestral populations. Note that RMSD is shown on log-scale.
The same scenario was also simulated based on allele frequencies from three distinct populations from HapMap 3. All approaches perform better since the populations are easier to distinguish (see Figure S5). Interestingly, HW genotypes do worse than the ML genotypes for the more distant populations (see Figure 2). NGSadmix still outperforms the methods based on called genotypes.
Scenario B: Low depth:
When simulating a scenario with a low and equal depth of 2× for all individuals, calling genotypes does not show the large bias clearly visible in the variable-depth scenario (see Figure S3 and Figure S4). NGSadmix still performs better with an RMSD of 0.11 while the best-performing method based on called genotypes has an RMSD of 0.18 in the HGDP-based simulations (HGDP Low in Figure 2). Genotype callers based on multiple samples such as using allele frequency priors generally generate better genotype calls than individual genotype callers (Nielsen et al. 2011). However, for this simulation the called ML genotypes based solely on the individual genotype likelihoods give a slightly better result. This is true both for the closely related populations and for the distantly related populations.
Scenario C: High and low depth:
This scenario seeks to mimic a design where a reference panel is sequenced at high depth or genotyped using SNP chips while some individuals are sequenced at very low depth (see Figure S7 and Figure S8). NGSadmix gives approximately correct admixture proportions when simulating both distant and closely related populations. However, when calling genotypes the estimated admixture proportions for the closely related populations resemble the difference in sequencing depth more than the actual ancestry. These problems are clearly reflected in the RMSD and in the maximum difference between the true and estimated admixture proportions (High/Low in Figure 2 and Figure S2). For the HapMap populations the ML genotypes give good results compared to the HW and filtered genotypes.
Scenario D: Small admixture proportions:
In practice the contribution of a single ancestral population to an individual’s ancestry can be much lower than the proportions studied above. In this scenario we simulated individuals with sequencing depth between 0.5× and 6× and with a wide range of admixture proportions as low as 5%; see top panel in Figure S9. NGSadmix identifies even low levels of ancestral contribution quite accurately for distantly as well as closely related populations (see Figure S9 and Figure S10). Figure S9 and Figure S10 illustrate how sequencing depth can severely bias the admixture proportions estimated from called genotypes. To further describe the precision of the estimated admixture proportions, we simulated 100 realizations of this scenario. For each simulation we also estimated the admixture proportions directly from the true simulated genotypes. Figure S11 and Figure S12 show that even though some accuracy is lost, NGSadmix does not perform much worse than estimates from the true simulated genotypes. From Figure S13 we see that NGSadmix generally outperforms estimating admixture proportions from called genotypes regardless of the admixture proportions. Figure S14 and Figure S15 show that the distribution of the coefficients estimated from called genotypes strongly depends on sequencing depth with small differences between admixture proportions.
1000 Genomes sequencing data
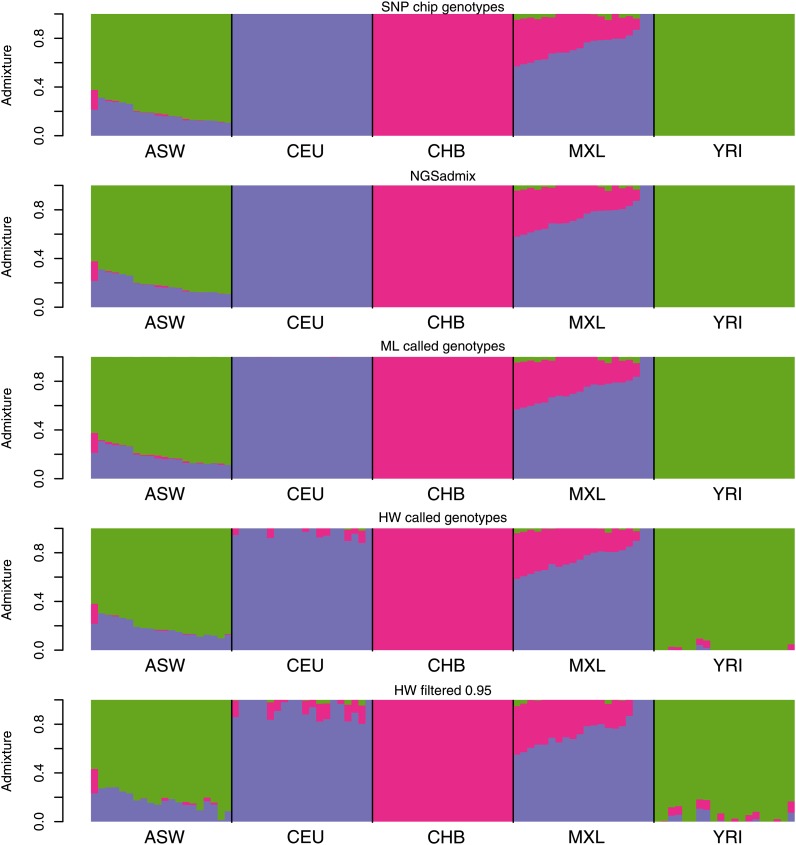
We tested the performance of our method on the 1000 Genomes low-coverage sequencing data. We selected 100 individuals from five distinct HapMap 3 populations that overlap with the 1000 Genomes low-coverage sequencing data. To evaluate the performance, we first used ADMIXTURE (Alexander et al. 2009) on publicly available HapMap 3 genotypes. Results assuming three ancestral populations are shown in Figure 3 and those for four populations are in Figure S17. For the same sites we generated genotype likelihoods based on the sequencing data. The admixture proportions estimated by NGSadmix are shown in the second panels of Figure 3 and Figure S17. The estimates based on sequencing data are almost indistinguishable from those based on HapMap 3 genotypes with the maximum observed difference in admixture proportion of 1.5% (labeled K = 3 in Figure S2). Analysis based on called genotypes also captures most of the admixture signal but for HW genotypes several of the nonadmixed individuals show a large amount of admixture (see Figure S16). Estimates based on called genotypes in general have a higher RMSD and a higher maximum deviance (labeled K = 3 in Figure 2 and Figure S2). However, estimates based on haplotype imputed genotypes performed only slightly worse than NGSadmix. Similar results are observed when assuming a higher number of populations (Figure S17).
Figure 3.
Estimated admixture proportions from both HapMap 3 genotype data (top) and NGSadmix results (bottom) for low-depth sequencing data from the 1000 Genomes.
We also tested NGSadmix on Japanese and Han Chinese since these populations are more closely related and are much harder to distinguish. The inferred population structure might not be due to two distinct homogeneous ancestral populations but might instead be the result of a more complex population history. All of the methods perform worse in this data set. As observed in the simulated data, the estimated admixture proportions correlate with the sequencing depth for ML and HW genotypes, while the admixture proportions estimated by NGSadmix and haplotype imputed genotypes correspond fairly well to those inferred using HapMap 3 genotypes (see Figure S18).
So far only sites that are known to be polymorphic have been analyzed. For some populations a large set of known SNPs might not be available. Therefore, we inferred SNPs from the sequencing data and based the analysis on those SNPs. We chose to call SNPs from 50 10-Mb randomly selected regions. This resulted in >2 million inferred SNPs. The results based on this analysis can be seen in Figure S19. Note that for the admixed individuals we cannot expect the exact same results from two different sets of SNPs since alleles from one population will be present in long tracts along the genome. However, we still see a very high correlation with the analysis from the SNP chip data. We estimated genotype likelihoods using both Equation 1 and SAMtools (H. Li et al. 2009). Although they gave a very different number of polymorphic sites, they gave similar admixture proportion estimates.
Convergence and computational speed
Convergence may sometimes be a problem due to the large number of parameters that are estimated simultaneously. Therefore, each scenario was run multiple times with different random starting points. Using NGSadmix on genotype likelihoods, all runs typically converge to the same maximum. When calling genotypes from the sequencing data, convergence was in some scenarios more of a problem, and here we had to run ADMIXTURE many times to determine the maximum (see Table S1).
When applying NGSadmix on 100 individuals from the 1000 Genomes project with almost 3 million SNPs, the method took on average almost 5 hr, using 10 central processing unit (CPU) threads. When running ADMIXTURE on SNP chip genotypes from the same data, the run time was on average 0.5 hr, also using 10 CPU threads. For the 100 realizations of scenario D with 340 individuals the average time for NGSadmix for the HapMap frequencies was 12.2 min (SD = 1.9), using 6 CPUs.
Discussion
Using simulations and real sequencing data, we have shown that it is possible to accurately estimate the admixture coefficients based on genotype likelihoods. This approach works regardless of depth distribution, admixture proportion, and sequencing/genotype technology, assuming that the error rates are reflected in the genotype likelihoods. The algorithm does not use any outside information from reference populations, and thus it can be applied in studies where appropriate reference panels from ancestral populations are not available. Calling genotypes can be an option if the depth is high enough to call genotypes correctly. However, having a low depth can bias the results of the admixture analysis as we have shown through simulations. In our simulations we have shown that basing the analysis on genotype likelihoods always outperforms the called genotypes. The biggest difference is seen when the populations are closely related and when depth distribution varies between individuals. The bias from calling genotypes occurs mainly when the depth differs between groups of individuals since these groups will have differential genotype errors, which is not the case when all individuals have exactly the same depth. However, even if the depth is exactly the same for all individuals, the genotype-likelihood approach is always better. This is because the genotype likelihoods contain all relevant information while the uncertainty of a genotype is lost when calling genotypes. In the presented results all individuals had an average depth of at least 0.5×. It is hard to make a precise statement for the minimum depth needed since this depends on other factors such as how different the admixing populations are, the number of individuals included in the analysis, and most importantly the depth of the other individuals. Similarly it is also hard to make a precise statement for the minimum number of SNPs needed, as this depends on the above-mentioned factors, as well as on how informative each marker is. NGSadmix suffers from the same problems as other methods in that too much missing data can give rise to convergence problems and we recommend removing sites with >80% missing data. This means that in general the depth can be very low for some individuals but not for all.
Calling genotypes jointly for multiple samples will in general give a higher accuracy of the genotype calls (Nielsen et al. 2011). Joint genotype calling can be performed based on the allele frequencies or based on the haplotype frequencies as used in haplotype imputation. However, for admixed individuals or individuals with unknown ancestry the frequencies used for the genotype calling might not represent the frequencies of the individuals’ ancestry. This is the reason we see the ML genotypes perform better than the HW genotype in most of the simulations and for the 1000 Genomes data sets. However, as we have demonstrated, even the ML genotypes can lead to very biased admixture estimates. If individuals are admixed, the genotypes should be called using an allele frequency prior that reflects their ancestry. This is precisely the prior estimated by NGSadmix by weighting the allele frequencies for each population by the individual ancestry. Thus NGSadmix can also be used to call genotypes from NGS data even if individuals are admixed. For the two 1000 Genomes data sets we also used Beagle (Browning and Yu 2009) to call genotypes though haplotype imputation, which resulted in better RMSD than that of both ML and HW genotypes but slightly worse than that of NGSadmix. However, this will probably not always be true. Given a large and fairly homogeneous sample, the haplotype imputation could potentially outperform NGSadmix, but a more heterogeneous sample with large difference in depth and populations could potentially lead to large biases.
Admixture analysis is important for many scientific fields. For example, understanding the individual’s ancestry is crucial in disease association studies. If the population structure of one’s sample is not dealt with, it will lead to an increase in the false positive rate (Marchini et al. 2004). For low-depth sequencing data the presented method can first be used to estimate the admixture proportion. These estimates can then be used as covariates in a generalized linear model framework as that also takes genotype uncertainty into account (Skotte et al. 2012). This will allow researchers to perform association on admixed samples while taking genotype uncertainty into account without inflating the false positive rates.
Admixture analysis is equally important in population genetics where is it used to infer information of the history of a sample or population. For some samples, especially ancient DNA samples, it is not possible to sequence at high depth. However, as we have shown it is possible to infer the admixture proportion even for samples sequenced at very low depth if a reference panel of high quality is available. This panel could be either individuals sequenced at high depth or individuals genotyped using a SNP chip. Genotype errors from the SNP chip can easily be incorporated in the analysis by estimating a genotype likelihood of the observed genotype given the true genotype and the error rate. This will allow samples genotyped on different platforms with different genotyping errors to be analyzed jointly without introducing a bias. Thus NGSadmix can also be very useful for traditional genotype data.
Convergence is sometimes a problem when relying on numerical optimization. NGSadmix converges almost every time for all presented scenarios. For the called genotypes, convergence can be a problem. For certain scenarios we had to run ADMIXTURE hundreds of times to determine the maximum likelihood. This is not a problem with the ADMIXTURE implementation, but it is the bias caused by calling genotypes that can produce many local maxima.
The computational speed can be a problem when dealing with next generation sequencing data. However, the presented analysis is based only on sites that are polymorphic, which means that only a small fraction of the genome is included in the analysis. Even though NGSadmix is slower than ADMIXTURE, it is still computationally feasible to estimate admixture proportions in a very large number of individuals.
Finally it should also be addressed that the performance of this model is limited by the correctness of the genotype likelihoods. If there is some error structure that is not properly accounted for in the calculation of genotype likelihoods, the method might give biased results. However, for the data analyzed from the 1000 Genomes Project we did not observe any such problem and achieved results that were virtually indistinguishable from the results from the HapMap SNP chip genotypes. In addition, changing genotype likelihood estimators did not change the results, which demonstrates the robustness of NGSadmix.
Supplementary Material
Acknowledgments
This work was supported in part by LuCamp (www.lucamp.org), Villum Foundation, and the Danish National Research Foundation.
Footnotes
Communicating editor: N. A. Rosenberg
Literature Cited
- Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., Durbin R. M., et al. , 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D. H., Novembre J., Lange K., 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19: 1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D. M., Durbin R. M., Abecasis G. R., Bentley D. R., Chakravarti A., et al. , 2010a A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D. M., Gibbs R. A., Peltonen L., Dermitzakis E., Schaffner S. F., et al. , 2010b Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B. L., Yu Z., 2009. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am. J. Hum. Genet. 85: 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. G., Walker N. M., Smyth D. J., Pask R., Cooper J. D., et al. , 2005. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 37: 1243–1246 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Andolfatto P., Huelsenbeck E. T., 2011. Structurama: Bayesian inference of population structure. Evol. Bioinform. Online 7: 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Lohmueller K. E., Albrechtsen A., Li Y., Korneliussen T., et al. , 2011. Estimation of allele frequency and association mapping using next-generation sequencing data. BMC Bioinformatics 12: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R., 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Li Y., Fang X., Yang H., Wang J., et al. , 2009. SNP detection for massively parallel whole-genome resequencing. Genome Res. 19: 1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J., Cardon L. R., Phillips M. S., Donnelly P., 2004. The effects of human population structure on large genetic association studies. Nat. Genet. 36: 512–517 [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Paul J. S., Albrechtsen A., Song Y. S., 2011. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 12: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B., Rohland N., McLaren P. J., Garimella K., Zaitlen N., et al. , 2012. Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nat. Genet. 44: 631–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Zaitlen N. A., Reich D., Patterson N., 2010. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 11: 459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M., Li Y., Lindgreen S., Pedersen J. S., Albrechtsen A., et al. , 2010. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463: 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A., Pritchard J. K., Weber J. L., Cann H. M., Kidd K. K., et al. , 2002. Genetic structure of human populations. Science 298: 2381–2385 [DOI] [PubMed] [Google Scholar]
- Skotte L., Korneliussen T. S., Albrechtsen A., 2012. Association testing for next-generation sequencing data using score statistics. Genet. Epidemiol. 36: 430–437 [DOI] [PubMed] [Google Scholar]
- Tang H., Peng J., Wang P., Risch N. J., 2005. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 28: 289–301 [DOI] [PubMed] [Google Scholar]
- Varadhan R., Roland C., 2008. Simple and globally convergent methods for accelerating the convergence of any EM algorithm. Scand. J. Stat. 35: 335–353 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.