Abstract
Targeted mouse mutants are instrumental for the analysis of gene function in health and disease. We recently provided proof-of-principle for the fast-track mutagenesis of the mouse genome, using transcription activator-like effector nucleases (TALENs) in one-cell embryos. Here we report a routine procedure for the efficient production of disease-related knockin and knockout mutants, using improved TALEN mRNAs that include a plasmid-coded poly(A) tail (TALEN-95A), circumventing the problematic in vitro polyadenylation step. To knock out the C9orf72 gene as a model of frontotemporal lobar degeneration, TALEN-95A mutagenesis induced sequence deletions in 41% of pups derived from microinjected embryos. Using TALENs together with mutagenic oligodeoxynucleotides, we introduced amyotrophic lateral sclerosis patient-derived missense mutations in the fused in sarcoma (Fus) gene at a rate of 6.8%. For the simple identification of TALEN-induced mutants and their progeny we validate high-resolution melt analysis (HRMA) of PCR products as a sensitive and universal genotyping tool. Furthermore, HRMA of off-target sites in mutant founder mice revealed no evidence for undesired TALEN-mediated processing of related genomic sequences. The combination of TALEN-95A mRNAs for enhanced mutagenesis and of HRMA for simplified genotyping enables the accelerated, routine production of new mouse models for the study of genetic disease mechanisms.
Keywords: TALENs, disease model, one-cell embryo, mouse mutant, nuclease, Fus, C9ORF72
GENETIC engineering of cells and organisms to create targeted mutants is a key technology for genetics and biotechnology. The ascent of the mouse as a mammalian genetic model is based on gene targeting through homologous recombination (HR) in embryonic stem (ES) cells (Capecchi 2005). Classical gene targeting via ES cells is a time- and labor-intense procedure that proceeds in the steps of vector construction, ES cell mutagenesis, chimera generation, and the transmission of mutant alleles through the germline (Hasty et al. 2000). Since the frequency of spontaneous HR in ES cells is low, it was a key finding that double-strand breaks (DSBs), created by sequence-specific nucleases, enhance local DNA repair by several orders of magnitude (Rouet et al. 1994). DSBs may be repaired through HR, using the sister chromosome as template or using gene targeting vectors that provide sequence homology regions flanking a desired genetic modification (Court et al. 2002; San Filippo et al. 2008). Alternatively, DSBs can be sealed by the nonhomologous end-joining (NHEJ) pathway that religates open ends without a repair template (Lieber 2010). By this means the DNA ends are frequently edited through the loss of multiple nucleotides, causing frameshift (knockout) mutations within coding regions. Targeted DSBs were first induced by zinc-finger nuclease (ZFN) fusion proteins that combine a DNA-binding domain made of zinc-finger motifs with the nuclease domain of FokI (Porteus and Carroll 2005). The application of ZFNs in one-cell embryos provided proof-of-principle for the direct mutagenesis of the mouse, rat, and rabbit genome in a single step (Geurts et al. 2009; Carbery et al. 2010; Meyer et al. 2010; Flisikowska et al. 2011). Nevertheless, ZFNs do not provide a universal tool since the available code for the recognition of nucleotide triplets is incomplete and multiple elements cannot be combined in a simple modular fashion. In contrast, the DNA-binding code of the transcription activator-like (TAL) proteins of Xanthomonas is based on the recognition of single nucleotides by individual peptide motifs, such that combinations of just four basic modules can be combined into domains that bind any target sequence (Boch et al. 2009; Moscou and Bogdanove 2009). Based on extensive experience with ZFNs, the TAL system could be readily adapted for gene editing by the fusion of DNA-binding modules with FokI into TAL effector nucleases (TALENs) (Cermak et al. 2011; Miller et al. 2011). Taking advantage of its modular nature, a variety of cloning protocols enable us to assemble TALEN coding regions within a short time (Cermak et al. 2011; Reyon et al. 2012).
We recently reported that TALEN target sites are distributed in the mouse genome at an average spacing of 14 bp, enabling genome-wide targeted mutagenesis at high precision. In particular, we provided proof-of-principle that TALENs and oligodeoxynucleotides (ODNs) can be applied in one-cell embryos to introduce targeted mutations (Wefers et al. 2013). For HR- and NHEJ-mediated gene modifications, we achieved rates of 2% and 6%, respectively, using experimental conditions that were not yet optimized. Higher rates of NHEJ-mediated nucleotide deletions (>40%) were obtained upon the microinjection of TALEN mRNAs into the cytoplasm of one-cell embryos, tolerating larger injection volumes (Sung et al. 2013). Nevertheless, for the creation of targeted mutations it is instrumental to deliver DNA templates for HR together with TALEN mRNAs directly into the pronucleus, tolerating only minimal injection volumes. To set up an efficient routine procedure for mutagenesis we enhanced the activity of TALEN mRNAs to optimize nuclease expression upon pronuclear delivery, such that one or more knockin or knockout alleles are obtained among a group of mice derived from a single microinjection experiment. Upon the establishment of a mutant by embryo manipulation, the genotyping of breeding colonies imposes a constant workload. PCR-based protocols for the detection of subtle mutations often require the digestion of PCR products and gel electrophoresis. To minimize these efforts we validated whether high-resolution melt analysis (HRMA) represents a reliable and simplified tool for the genotyping of mouse mutants. HRMA identifies mutant PCR products by their specific denaturation profile (Liew et al. 2004) and requires no restriction digestion and size separation of PCR products.
We applied this streamlined procedure to introduce targeted and knockout mutations into the Fus and C9orf72 genes to create disease models for inherited amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). Mutations disrupting the C-terminal nuclear localization sequence (NLS) of FUS have been identified in ALS patients (Kwiatkowski et al. 2009), whereas a hexanucleotide repeat expansion in the first intron of the C9orf72 gene was found in patients representing ALS, FTLD, or both diseases (DeJesus-Hernandez et al. 2011; Renton et al. 2011). In the Fus gene dominant mutations within the NLS disrupt the nuclear import of FUS and lead to its cytoplasmic deposition in the brain and spinal cord of patients (Bosco et al. 2010; Gal et al. 2011; Ito et al. 2011; Kino et al. 2011; Dormann and Haass 2013). This defect is a key to pathogenesis since mutations that severely impair nuclear import, such as the P525L replacement, lead to an early onset and rapid progression of the disease. Since FUS is involved in multiple steps of gene expression, including transcription, pre-mRNA splicing, and mRNA transport, neurodegeneration may be caused by the loss of essential nuclear functions and/or the gain of a toxic function in the cytosol. Depletion of FUS in zebrafish and fruit flies causes a motoneuron phenotype but is perinatal lethal in mice (Hicks et al. 2000; Kabashi et al. 2011; Sasayama et al. 2012). To faithfully mimic the human codon replacements R521G and P525L, we targeted the analogous positions R513 and P517 of the mouse Fus gene, using TALENs and ODNs. As a cause of C9orf72-associated pathogenesis, the intronic repeat expansion may be deleterious through RNA-mediated toxicity or by the translation of repeat sequences, causing the production and aggregation of dipeptide repeat proteins (Ash et al. 2013; Mori et al. 2013) or both (Taylor 2013). To clarify whether C9orf72 loss-of-function also contributes to the FTLD phenotype and to decipher its cellular function, we disrupted the mouse homolog of the C9orf72 gene, 3110043O21Rik, by the creation of TALEN-induced frameshift mutations.
Using our advanced TALEN mutagenesis procedure we obtained recombined Fus alleles in 6.8% and nucleotide deletions within C9orf72 in 41% of mice derived from pronuclear embryo injections, validating this approach for the expedited recapitulation of disease-associated alleles. The established Fus codon replacement and C9orf72 knockout mutants will be instrumental to studying genetic ALS and FTLD disease mechanisms.
Materials and Methods
TALEN target sites
For the selection of TALEN target sequences we used the “TALENdesigner” (www.talen-design.de) as described in Wefers et al. (2013). Selected target sites cover two recognition sequences of 15 bp preceded by a T, separated by a spacer of 14–15 bp. To minimize off-target recognition, potential sites were analyzed using the “Paired Target Finder” (https://tale-nt.cac.cornell.edu) (settings: spacer length 13–20 bp, cutoff 3.0) (Doyle et al. 2012).
TALEN construction and expression
Details on the construction of TALEN coding regions, expression vectors, and TALEN sequences are given in Supporting Information, File S1. For the expression of TALENs in mammalian cells we used the expression vector pCAG-TALEN-pA as described in Wefers et al. (2013). pT7-TALEN-95A was derived from pCAG-TALEN-pA by replacement of the poly(A) signal sequence with a segment of 95 adenine residues derived from a mouse Oct4 cDNA clone.
Oligodeoxynucleotides
The oligodeoxynucleotides ODNR513G (5′-TGGGTAGGGTAGTTCAGTAACACGTAATCTAACATAACTTTTTCTTTCAGGGGCGAGCACAGACAGGATGGCAGGGAGAGACCATATTAGCCTGGCTCCTGAAGTTCTGGAACTCTTCCTGTACCCAGTGTTACCCTTGT-3′) and ODNP517L (5′- TCAGTAACACGTAATCTAACATAACTTTTTCTTTCAGGGGCGAGCACAGACAGGATCGCAGGGAGAGACTATATTAGCCTGGCTCCTGAAGTTCTGGAACTCTTCCTGTACCCAGTGTTACCCTTGTTATTTTGTAAACT-3′) were synthesized and HPLC purified by Metabion (Martinsried, Germany), each having a length of 140 nt, including the targeted mutation (shown in boldface type) and a silent replacement (underlined), covering 70 bp upstream and downstream of the targeted codon.
Microinjection of one-cell embryos
The injection of TALEN mRNA and targeting molecules (ODNs) was performed as described in Wefers et al. (2012, 2013), except that injections were done only into pronuclei. Briefly, capped TALEN mRNA was prepared in a single step by in vitro transcription from pT7-TALEN-95A plasmid DNA linearized with XbaI and AleI (New England Biolabs, Frankfurt, Germany), using the mMessage mMachine T7 Ultra kit (omitting the polyadenylation step) and the MEGAclear kit (Life Technologies, Carlsbad, CA). The quality of synthesized mRNAs was controlled by agarose gel electrophoresis under denaturing conditions, using the NorthernMax-Gly system and the RNA Millenium size marker (Life Technologies). Each TALEN mRNA was then diluted in injection buffer (10 mM Tris, 0.1 mM EDTA, pH 7.2) to a working concentration of 90 ng/µl TALEN-Rik2 mRNA or 20 ng/µl TALEN-Fus15 mRNA. The targeting oligodeoxynucleotides were dissolved in water and diluted with injection buffer to a working concentration of 15 ng/µl. For microinjections, one-cell embryos were obtained by mating of (DBA/2 × C57BL/6)F1 males with superovulated FVB/N females (Charles River, Sulzbach, Germany). One-cell embryos were injected with either only TALEN-Rik2 mRNA or a mixture of TALEN-Fus15 mRNA and the targeting oligodeoxynucleotides (15 ng/µl) (ODNR513G and ODNP517L) into the larger pronucleus, but not into the cytoplasm. Test experiments showed that microinjections of Venus mRNA (90 ng/µl), using pronuclear capillaries, led to green fluorescence in all embryos, but the direct delivery of the same volume into the cytoplasm is less effective and labels only ∼10% of embryos. Injected zygotes were transferred into pseudopregnant CD1 female mice to obtain live pups. All mice showed normal development and appeared healthy. Mice were handled according to institutional guidelines approved by the animal welfare and use committee of the government of Upper Bavaria and housed in standard cages in a specific pathogen-free facility on a 12-h light/dark cycle with ad libitum access to food and water.
Isolation of genomic DNA
Genomic DNA was isolated from tail tips of founder mice and their progeny, using the Wizard Genomic DNA Purification Kit (Promega, Mannheim, Germany), following the manufacturer’s instructions.
HRMA
For the screening of TALEN-induced mutations, the TALEN target regions of C9orf72 and Fus (amplicon size 140 and 133 bp, respectively) were amplified in a 10-µl PCR reaction containing 40 ng lyophilized genomic DNA, 1 µl LC Green Plus+ Dye (Bioké, Leiden, The Netherlands), 200 nM of each dNTP, 250 nM each forward and reverse primers (Table S3), and 0.2 µl Phire Hot Start II DNA Polymerase (Thermo Scientific, Dreieich, Germany). PCR reaction protocols for mutagenic detection were 98°, 30 sec; 40 cycles of [98°, 5 sec; 62° (C9orf72)/66° (Fus), 5 sec; 72°, 5 sec]; 72°, 1 min; denaturation at 98°, 1 min; and rapid cool down to 25° for heteroduplex formation. Following the PCR, samples were analyzed with a LightScanner (BioFire Diagnostics, Salt Lake City) over a 65°–95° range. PCR products that contained mutant alleles were purified using the Qiaquick PCR purification kit (QIAGEN, Hilden, Germany), subcloned using the StrataClone Blunt PCR Cloning Kit (Agilent, Waldbronn, Germany), and sequenced (GATC Biotech, Konstanz, Germany). Sequences were compared to wild type, using the Vector NTI Advance 11.5 software suite (Life Technologies). To determine the detection limit of HRMA, cloned mutant PCR products from C9orf72 founders R5 and R12 were diluted with wild-type PCR product. The melting curves of three replicates of each test sample were analyzed and compared to wild-type controls, using the LightScanner software with Call-IT 2.0 (BioFire Diagnostics).
Off-target analysis
To assess potential TALEN off-target activity, the five highest-scored off-target sites (Table S2) were analyzed by HRMA in duplicate reactions, using locus-specific PCR primer pairs (see Table S3: Fus OS1–OS5 and Rik OS1–OS5). Four Fus and two C9orf72 founders were compared to a C57BL/6 wild-type control. Founder-derived PCR products amplified from the Fus off-sites 1 and 3 were subcloned and sequenced. These sequences were compared to the respective genomic sequences of the C57BL/6, DBA/2, and FVB/N mouse strains (Ensembl Resequencing database, http://www.ensembl.org, release 71, April 2013).
Results
Optimized expression of TALENs in one-cell mouse embryos
To target the Fus and the C9orf72 genes, we constructed TALEN pairs recognizing sequences within exon 15 and exon 2, respectively, using our TALENdesigner algorithm and modular construction protocol (Wefers et al. 2013). The TALEN coding regions were inserted into the mammalian expression vector pCAG-TALEN-pA, providing a CAG promoter and a polyadenylation signal sequence (Figure 1A). For the assessment of TALEN activity, expression vectors were cotransfected with customized nuclease reporter plasmids into HEK 293 cells as described in Wefers et al. (2013) and found to exhibit specific nuclease activity (Figure S1). For the expression of TALENs in one-cell embryos, the coding regions can be transcribed in vitro by T7 polymerase from linearized pCAG-TALEN-pA plasmids, followed by the polyadenylation of the coding RNA with poly(A) polymerase. Using this two-step protocol, we frequently noted an inconsistent production of single-species TALEN mRNAs, resulting in a smeared appearance of transcripts upon the polyadenylation step [Figure 1C, TALEN-poly(A)]. This effect occurred for TALEN but not for shorter, e.g., ZFN RNAs, possibly because the transcription of the 3-kb TALEN coding region leads to a larger fraction of truncated products, contaminating the polyadenylation reaction. To enable the reliable production of TALEN mRNAs, for optimal nuclease expression upon pronuclear injection, we inserted the TALEN coding regions into pT7-95A (Figure 1B). This vector provides a T7 promoter and a region of 95 adenine (A) residues located downstream of the TALEN coding region for the production of polyadenylated (TALEN-95A) mRNAs in a single step. Using pT7-TALEN-95A vectors for in vitro transcription, we reproducibly obtain full-length TALEN mRNAs of the expected size of 2948 nucleotides (Figure 1C, TALEN-95A). To determine the RNA concentration that supports efficient translation upon pronuclear microinjection, we used a 95A RNA encoding the Venus reporter. We found green fluorescence in all embryos microinjected with Venus-95A RNA at 90 ng/µl, upon culture to the two-cell stage (Figure 1D). To assess the potency of TALEN-95A RNAs for the mutagenesis of the C9orf72 and Fus genes we used concentrations of 90 ng/µl and 20 ng/µl, respectively.
Figure 1.
Production of TALEN mRNAs for embryo microinjection. (A) Plasmid pCAG-TALEN-Fus15-pA provides a CAG promoter (CAG) and a poly(A) signal sequence (pA) for expression of TALEN-Fus15 proteins in mammalian cells. TALEN mRNA can be produced in vitro from MluI-linearized plasmid DNA in a two-step procedure, using T7 polymerase for transcription and poly(A) polymerase for polyadenylation. (B) Plasmid pT7-TALEN-Fus15-95A provides a T7 promoter (T7) and a region of 95 adenine (95A) nucleotides, followed by an AleI site. TALEN RNA, including a plasmid-coded poly(A) sequence, can be produced in a single step, using T7 polymerase and AleI-linearized plasmid DNA. T7: T7 promoter region. (C) Agarose gel electrophoresis of reaction products transcribed with T7 polymerase from MluI-linearized pCAG-TALEN-Fus15-pA plasmids, followed by polyadenylation with poly(A) polymerase [left gel, TALEN-poly(A)], or produced in a single step with T7 polymerase from AleI-linearized pT7-TALEN-Fus15-95A plasmids (right gel, TALEN-95A). The size of full-length TALEN-95A transcripts is expected at 2948 nt. Marker: RNA size marker (×1000 nucleotides). (D) Microinjection of in vitro-produced Venus-95A RNA (90 ng/µl) into pronuclei of one-cell mouse embryos. The manipulated embryos were cultured to the two-cell stage and analyzed for Venus expression by fluorescence microscopy. Top, white light; bottom, green fluorescence.
Generation of C9orf72 knockout mice
To induce frameshift mutations within the mouse homolog of C9orf72 we designed a TALEN pair (TALEN-Rik2) targeting a sequence downstream of the start codon located within the second exon of the murine 3110043O21Rik gene (Figure 2A and Figure 3, A and C).
Figure 2.
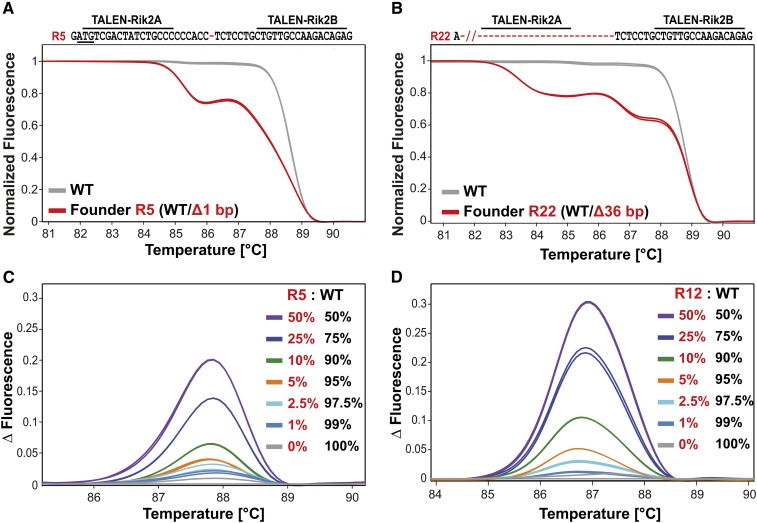
Identification of C9orf72 mutants by HRMA. (A and B) Melting analysis of PCR products amplified in duplicate from tail DNA of founder R5 (A) and of founder R22 (B) (red curves, overlaid) in comparison to wild-type controls (gray curves, overlaid). The TALEN-Rik2 target sequences within exon 2 are shown (start codon underlined); nucleotides deleted in mutant alleles are shown as red dashes and genotypes are given in parentheses. (C and D) The sensitivity of HRMA for mutant detection was determined by analyzing HRMA samples prepared with varying amounts of wild-type or mutant PCR products from founder R5 (1-bp deletion, C) and founder R12 (6-bp deletion, D). The limit to detect the R5 allele is at 5% for the mutant product (orange curve) and at 2.5% for the R12 allele (light blue curve).
Figure 3.
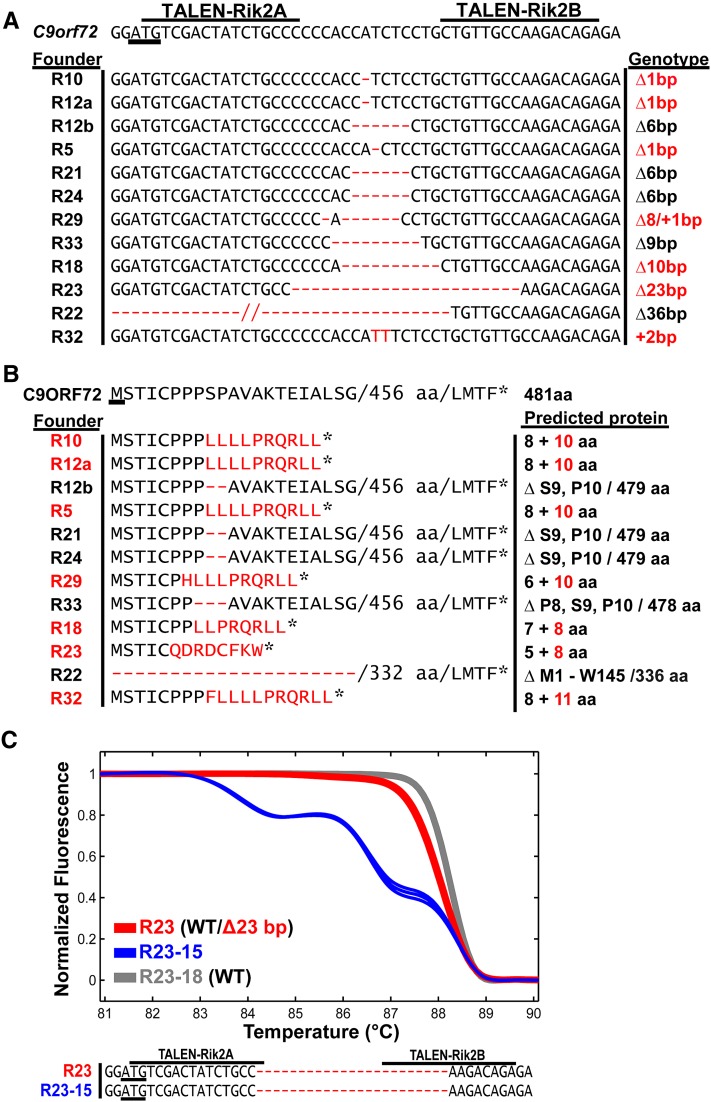
TALEN-induced C9orf72 alleles. (A) Sequence comparison of the TALEN-Rik2 target region within exon 2 of C9orf72 in comparison to mutant alleles amplified by PCR from tail DNA of the indicated founders. The start codon of C9orf72 is underlined and the recognition sequences of TALENs are indicated. Nucleotide deletions and insertions are shown as red dashes and red letters, respectively. The genotype classifies mutant alleles as products of NHEJ-associated deletion (Δ) or insertion (+); alleles exhibiting reading frameshifts are shown in red. Founder R12 contained two mutant alleles showing the deletion of 1 or 6 bp, respectively. (B) Predicted protein sequences of TALEN-induced C9orf72 alleles (start codon underlined). Upon the translation of the first five to eight wild-type codons, the mutant alleles R10, R12a, R5, R29, R18, R23, and R32 exhibit a reading frameshift, followed by a nonsense sequence of 8–11 residues (red letters) and a stop codon (asterisk). Allele R22 lost the start codon (Δ ATG) and may result in the complete loss of translation or in translational initiation at the downstream ATG codon 146 and the production of a truncated protein. The alleles R12b, R21, R24, and R33 show only deletions of codons 8–10 or 9–10 and preserve the downstream reading frame of C9orf72. (C) Melting analysis of triplicate PCR products from founder R23 (red curves, overlaid), its pup R23-15 (blue curves), and a wild-type C57BL/6 control (gray curves). The sequence analysis of cloned PCR products confirmed the germline transmission of the R23 allele.
From pronuclear microinjections of TALEN-Rik2 mRNAs (95A type, 90 ng/ul), we obtained 51 pups (Table 1) that were screened for the presence of mutant alleles by HRMA of PCR products covering the targeted exon. Twenty-one of these mice (41%) were identified as founders harboring mutant C9orf72 alleles, confirming that the pronuclear delivery of 95A-type mRNAs leads to a high mutagenesis rate. Representative HRMA results from 8 founders are shown in Figure 2 and Figure S2, exhibiting melting curves that deviate from the wild-type control. We selected 11 founders for the further characterization of modified C9orf72 alleles by subcloning and sequence analysis of PCR products. Among founders R5–R32 we identified 12 mutant alleles (Figure 3A) that exhibit deletions of 1–36 nucleotides or a 2-bp insertion within the TALEN target region. Seven of these deletions disrupt the C9orf72 reading frame in between codons 6–9 and are predicted for the translation of 8–11 additional amino acids (Figure 3B). For the establishment of a mutant breeding colony, founder R23 was mated to wild-type mice and its progeny genotyped by PCR and HRMA. Seven of 15 pups derived from R23 showed melting curves distinguished from the wild-type control and the sequencing of PCR products from pup R23-15 confirmed the germline transmission of the parental C9orf72 allele (Figure 3C).
Table 1. TALEN-mediated gene editing events.
| Microinjection experiment | TALEN mRNA concentration (ng/µl) | No. pups | No. mutant founders (%) | Founders’ NHEJ events (%) | Founders’ HR events (%) |
|---|---|---|---|---|---|
| 1. TALEN-Rik2 | 90 | 51 | 21 (41.2) | 21 (41.2) | — |
| 2. TALEN-Fus15 + ODNR513G | 20 | 83 | 8 (9.6) | 8 (9.6) | 7 (8.4) |
| 3. TALEN-Fus15 + ODNP517L | 20 | 50 | 2 (4) | 2 (4) | 2 (4) |
| TALEN-Fus15 ∑ = | 133 | 10 (7.5) | 10 (7.5) | 9 (6.8) |
Shown are mutant founder mice and mutant alleles obtained from the microinjection of Rik TALENs or Fus TALENs together with mutagenic ODNs into the pronuclei of one-cell embryos. The concentration of TALEN mRNAs and the number of pups obtained from embryo transfers are shown. Within these groups the overall frequency of TALEN-induced gene editing is indicated by the number of mice harboring mutant alleles (mutant founders), modified either by NHEJ or by HR events. Half of the founders derived from experiments 2 and 3 were mosaics, containing more than one modified allele. Therefore, the combined number of mice exhibiting alleles modified by NHEJ or HR exceeds the total number of mutant founders.
Founders obtained from microinjections of TALENs or ZFNs are frequently mosaics, harboring a mutation only in some of the somatic cells, if gene editing occurred after genome replication (Wefers et al. 2013). To assess whether mosaic mutant genotypes can be identified by HRMA, we performed control experiments to establish its detection limits. For this purpose we prepared HRMA test samples containing 1–50% of cloned, mutant C9orf72 PCR product (allele R5, 1-bp deletion; or R12, 6-bp deletion) and 99–50% of wild-type PCR product. In comparison to the wild-type controls, the presence of mutant alleles could be reliably detected in samples containing 5% (1-bp deletion) or 2.5% (6-bp deletion) of mutant DNA (Figure 2, C and D). These results indicate that mosaic founders harboring even a minor fraction of mutant cells can be recognized by melting analysis and validate HRMA as a simple and sensitive tool for the identification of mutants derived from embryo microinjection of TALENs.
Generation of FusR513G and FusP517L codon replacement mutants
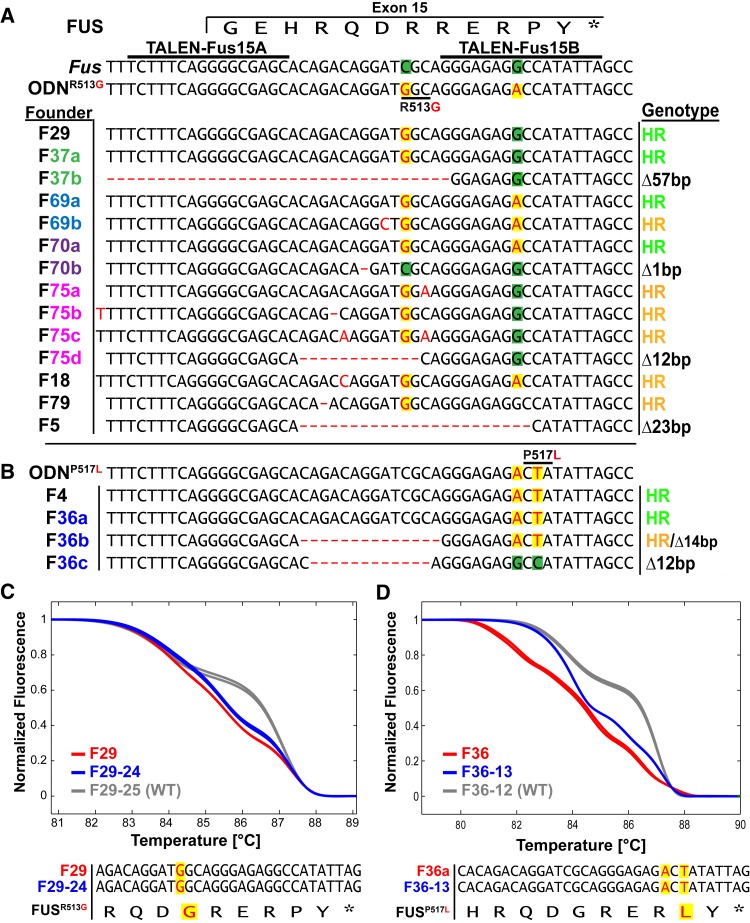
To recapitulate the patient-derived codon replacements R521G and P525L in the mouse Fus gene, we targeted the analogous positions R513 and P517, using synthetic oligodeoxynucleotides as template for TALEN-induced HR. We designed a TALEN pair (TALEN-Fus15) recognizing the C-terminal coding sequence located within exon 15 (Figure 4A). The oligonucleotides ODNR513G and ODNP517L cover 140 nt centered on exon 15 and include nucleotide replacements that redefine the codons 513 and 517 into glycine (R513G) or leucine (P517L), respectively. To exclude the potential reprocessing of recombined alleles by TALENs, each ODN included an additional, silent nucleotide replacement within the TALEN-Fus15B recognition sequence. TALEN-Fus15 mRNAs (95A type, 20 ng/µl) were co-injected with ODNR513G or ODNP517L into the pronuclei of one-cell embryos. We obtained 83 pups from the injections of ODNR513G and 50 pups from ODNP517L (Table 1), which were screened for mutations by HRMA of PCR products covering exon 15. The samples from 8 mice from ODNR513G (9.6%) and 2 mice from ODNP517L (4%) injections showed melting curves distinguished from the wild-type control (Figure S3) and were further analyzed by the subcloning and sequencing of each of the five PCR products. The founders derived from ODNR513G injections harbored 14 modified Fus alleles (Figure 4A). Four founders contained the desired R513G replacement together with (F69, F70) or without (F29, F37) the silent replacement within the TALEN-Fus15B binding region. Three founders (F18, F75, and F79) showed recombined R513G Fus alleles that included unintended single-nucleotide alterations (Figure 4A), likely resulting from ODN synthesis errors. Indeed, we noted that our ODNR513G synthesis used for microinjection contained a substantial fraction (∼1/3) of variant molecules, as determined by PCR, subcloning, and sequencing (data not shown). Furthermore, four of the founders harbored additional modified Fus alleles that underwent nucleotide deletions (F37b, F70b, F75d, and F5) or showed an unexpected nucleotide replacement within codon 512 (F69b). From the microinjection of ODNP517L both founders harbored recombined Fus alleles, including the P517L and adjacent silent replacements (F4, F36a, Figure 4B). Founder F36 was mosaic for another recombined allele, including a 14-bp deletion (F36b) and a deletion allele that lost 12 bp (F36c).
Figure 4.
Generation of FusR513G and FusP517L mutants. (A and B) Sequence comparison of the TALEN target region covering the Fus exon 15, of ODNR513G, ODNP517L, and cloned PCR products amplified from tail DNA of ODNR513G (A) and ODNP517L (B) founder mutants, identified by HRMA. The exon 15 coded FUS sequence, the TALEN binding sites, and codons 513 and 517 are indicated; nucleotides deviating from wild type (green background) are shown in red on a yellow background. Nucleotide deletions or insertions are shown as red dashes or red letters. The genotype describes the mutant alleles as a product of homologous recombination (HR) or NHEJ-associated deletion (Δ) or insertion (+). (C and D) Melting analysis of duplicate PCR products from founder F29 (ODNR513G) (C) (red curves) its pup F29-24 (blue curves) and founder F36 (ODNP517L) (D) (red curves) and its pup F36-13 in comparison to wild-type controls (gray curves). The sequence analysis of cloned PCR products from pups F29-24 and F36-13 confirmed the germline transmission of the FusR513G and FusP517L alleles.
For the establishment of Fus mutant lines we mated founders F29 (FusR513G, Figure 4C) and F36 (FusP517L, Figure 4D) to wild-type mice and genotyped their offspring by PCR and HRMA. Eight of 15 pups derived from F29 and 8 of 12 pups from F36 showed melting curves differing from the wild-type control and the cloning and sequencing of PCR products confirmed the germline transmission of the parental FusR513G (pup F29-24, Figure 4C) and FusP517L alleles (pup F36-13, Figure 4D). Five additional founders were mated to wild-type mice and transmitted modified Fus loci to their offspring, as confirmed by HRMA (Table S1).
These results show that TALENs and ODNs created recombined Fus loci in 6.8% of mice derived from microinjections (1 recombined founder per 15 pups, Table 1) and that mutations identified in the founders’ tail DNA were faithfully transmitted through the germline. To further confirm the integrity of the targeted FusR513G locus we amplified genomic sequences covering 3.4 kb upstream of codon 513 and 3.2 kb of the downstream region, using tail DNA of the heterozygous FusR513G pup F29-24. The direct sequencing of both PCR products, representing molecules derived from the wild type and the FusR513G allele, revealed a uniform reading pattern of the Fus wild-type sequence, except for the C to G replacement within codon 513 that showed a mixed G/C peak (Figure S5). This result proves the genomic integrity of the FusR513G allele within a region of 6.6 kb centered on codon 513. To further analyze the functionality and transcription of the FusR513G allele we isolated mRNA from the tail of pup F29-24, prepared Fus cDNA, and PCR amplified a 341-bp region of the Fus transcript covering exons 14 and 15. The sequence analysis of cloned PCR products revealed spliced cDNA sequences including the R513G replacement, confirming the functionality of the FusR513G allele (Figure S4).
Analysis of TALEN off-target activity
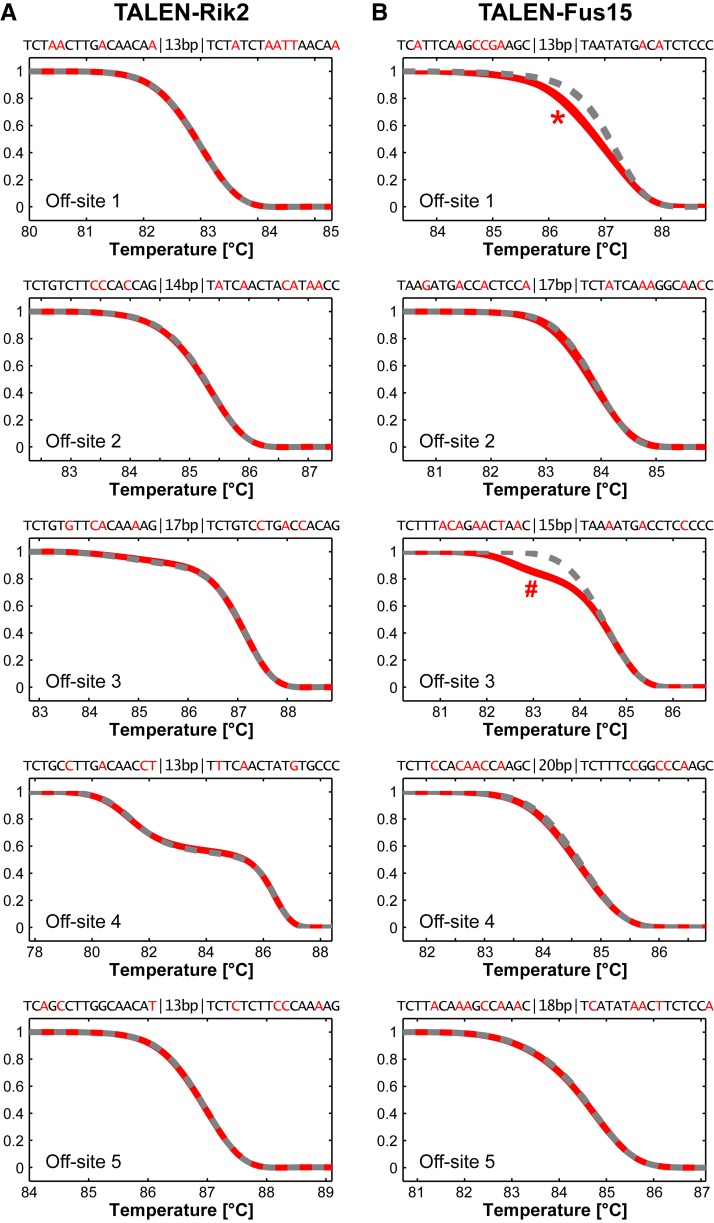
TALENs may recognize genomic (off-target) sites, which are similar to the intended target sequence and cause undesired genetic modifications. To assess the frequency of such off-target events in mutants derived from TALEN microinjections, we analyzed for each pair of our TALENs five off-target sites (Table S2) (Doyle et al. 2012), using tail DNA from four Fus and two C9orf72 founder mutants. PCR products covering these regions were analyzed by HRMA in comparison to a C57BL/6 wild-type control (Figure 5). Whereas the melting curves of all TALEN-Rik2 and of three TALEN-Fus15 off-target sites were identical to the controls, the PCR products from the Fus off-sites 1 and 3 were distinguished from the control. These PCR products were subcloned and the analysis of five sequences per sample revealed single-nucleotide substitutions that represent known polymorphisms in the genome of the inbred strains (Figure S6) we used for embryo production. Hence, we found no evidence for the presence of off-target mutations in founders derived from microinjections of our TALENs.
Figure 5.
Genome-wide off-target analysis of TALEN-Rik2 and TALEN-Fus15. (A and B) Melting analysis of predicted off-target sites of TALEN-Rik2 (A) and TALEN-Fus15 (B) in wild-type and mutant founder mice (R5, R23 and F5, F29, F4, F36). The potential TALEN target sequences, spacer length, and mismatches (red letters) to the Fus and C9orf72 target sites are indicated. HRMA revealed no differences from the C57BL/6 wild-type control (dashed gray curves) and founder-derived PCR products (red curves), except for the Fus15 off sites 1 (*) and 3 (#), which were identified as polymorphisms present in the different genetic backgrounds. Details on these polymorphisms are shown in Figure S6.
Discussion
Single-nucleotide polymorphisms leading to codon replacements within human genes are increasingly identified as disease-related mutations by high-throughput genomic analysis such as exome sequencing. Genetic mouse models that recapitulate such mutations will be instrumental to studying the underlying disease mechanisms and to developing therapeutic interventions. We recently provided proof-of-principle that nucleotide and codon replacements can be directly introduced into the mouse genome by microinjection of TALEN mRNAs (15 ng/µl) and ODNs into the pronuclei of one-cell embryos (Wefers et al. 2013). For the Rab38 locus we achieved a rate of 1.8% for targeted replacements and of 4.8% for NHEJ-mediated deletions, using as yet nonoptimized conditions. To enable the routine production of targeted mouse models at high efficiency, we aimed to enhance gene editing by increasing the incidence of TALEN-induced DSBs upon pronuclear delivery. High rates (>40%) of NHEJ-mediated deletions have been achieved by the microinjection of TALEN mRNAs into the embryonic cytoplasm (Sung et al. 2013), tolerating large volumes, but this route provides no option for the codelivery of DNA templates into the nucleus. Since pronuclei tolerate only minimal volumes and narrow injection capillaries, restricted for the delivery of a few picoliters, we sought to optimize the activity and concentration of the co-injected TALEN mRNAs. To this end we used mRNAs with template-coded poly(A) regions (95A) and found that pronuclear delivery at 90 ng/µl leads to the effective translation of a fluorescent reporter. As a validation of this protocol we found that deletions in C9orf72 were induced in 41% of pups derived from injections of TALEN mRNAs at 90 ng/µl. For the targeting of Fus we used TALEN-95A mRNAs at a lower concentration (20 ng/µl) together with ODNs, comparable to the earlier targeting of Rab38, and found targeted replacements to occur at a rate of 6.8%. These rates of gene editing are four- to eightfold higher compared to our previous results from Rab38, suggesting that TALEN-95A mRNAs lead to enhanced mutagenesis. At these rates one or more targeted alleles can be obtained from a single day of microinjection, typically resulting in ∼25 pups if FVB-derived embryos are used for microinjection. TALENs have been further used to induce knockout mutations in embryos of the C57BL/6 inbred strains upon cytoplasmic delivery (Davies et al. 2013; Sung et al. 2013; Qiu et al. 2013). Therefore we are confident that it will be also possible to generate targeted mutations by the delivery of TALEN-95A mRNAs and targeting DNA molecules into the pronuclei of C57BL/6 embryos. Nevertheless, the number of live births from C57BL/6 embryos is about half compared to that from the FVB strain such that 2 days of microinjection will be required to recover 25 pups.
Several Fus founders recombined with the 140-nt ODNR513G showed unintended nucleotide insertions, deletions, or replacements. These alterations likely result from the error-prone synthesis of oligonucleotides and identify ODN quality and length as important factors to optimize the net rate of correctly modified alleles. Since the synthesis of shorter ODNs correlates with an increased fraction of correct molecules, it will be of future interest to determine the in vivo recombination rate of ODNs in relation to the molecules’ length. In mammalian cell lines it has been shown that a minimum of 50 bp of homology is sufficient to achieve a high recombination rate (Chen et al. 2011).
As previously observed for ZFN- and TALEN-induced mutagenesis, a part of our Fus and C9orf72 founders were mosaic for one or more modified alleles, resulting from multiple, independent editing events that may occur before or after the first or second cycle of genome replication. Since modified loci are first identified in the founder’s tail DNA, it is essential that the same alleles are present in the germ cell population to establish breeding colonies. Each of eight mated founders transmitted mutant alleles to 10–67% of its progeny, thereby confirming the contribution of mutant cells to the germline. Furthermore, the resequencing of Fus and C9orf72 alleles from heterozygous pups confirmed the identity to the parental loci, indicating that the analysis of tail DNA is predictive of the mutational spectrum in the germline.
At present little is known about the potential processing of sites that are similar to the intended TALEN target sequence and which degree of sequence divergence is necessary to exclude off-target recognition. Using tail DNA from Fus and C9orf72 founders we analyzed five potential off-target sites by HRMA and found no indication for processing at these sites. Since for TALEN-Fus15 and Rik2 the closest genomic off sites are distinguished by seven or more nucleotide substitutions, our results suggest that under this condition TALENs do not cause modifications at sites that are predictable with the known binding code. Whether TALENs also recognize other, presently unpredictable target sites requires further clarification by whole-genome sequencing.
Besides the one-time generation of mutant alleles by embryo manipulation, the genotyping of mutant offspring adds a constant workload to the maintenance of breeding colonies. Mutant alleles harboring nucleotide replacements cannot be identified through the mere size of PCR products. Therefore, present PCR genotyping protocols require differentiating between wild-type and mutant alleles by digestion with restriction enzymes and gel electrophoresis. This rationale often requires the incorporation of additional, undesired nucleotide substitutions to create or delete enzyme recognition sites. All of these drawbacks are relieved by automated HRMA that requires just the melting analysis of PCR reactions. Our characterization of mutant Fus and C9orf72 founders and their offspring validated HRMA as a universal and sensitive tool for the identification and genotyping of TALEN-induced nucleotide replacements and deletions. By the inclusion of wild-type control DNA it will be further possible to differentiate heterozygous and homozygous mutant genotypes by HRMA.
Taken together, our advanced TALEN mutagenesis and analysis procedure enables the accelerated, routine production of new genetic mouse models. Since TALENs combined with ODNs allow genome-wide targeting at high precision, this technology supports expedited in vivo analysis of newly discovered disease-associated mutations.
Supplementary Material
Acknowledgments
We thank R. Kneuttinger, P. Kunath, A. Krause, A. Tasdemir, S. Weidemann and O. Yefremova, for excellent technical assistance. This work was supported by the European Union within the European Conditional Mouse Mutagenesis (EUCOMM) project (LSHG-CT-2005-018931, to W.W.), by the German Ministry of Education and Research within the Disease Genes to Proteins (DIGTOP) project (01GS0858, to W.W. and R.K.) of the National Genome Research Network (NGFN)-Plus program, by the excellence cluster for systems neurology (SyNergy) 1010 (to. C.H., B.S., W.W.), by the Kompetenznetzwerk Degenerative Demenzen (KNDD2) (FKZ01GI1005D to T.F), and by the Indian Council of Agricultural Research (No.29-1/2009-EQR/Edn, to S.K.P.). The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013)/European Research Council grant agreement n°321366-Amyloid, to C.H.).
Footnotes
Communicating editor: D. Voytas
Literature Cited
- Ash P. E. A., Bieniek K. F., Gendron T. F., Caulfield T., Lin W.-L., et al. , 2013. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77: 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., et al. , 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Bosco D. A., Lemay N., Ko H. K., Zhou H., Burke C., et al. , 2010. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum. Mol. Genet. 19: 4160–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi M. R., 2005. Gene targeting in mice: Functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6: 507–512 [DOI] [PubMed] [Google Scholar]
- Carbery I. D., Ji D., Harrington A., Brown V., Weinstein E. J., et al. , 2010. Targeted genome modification in mice using zinc-finger nucleases. Genetics 186: 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., et al. , 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Pruett-Miller S. M., Huang Y., Gjoka M., Duda K., et al. , 2011. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8: 753–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court D. L., Sawitzke J. A., Thomason L. C., 2002. Genetic engineering using homologous recombination. Annu. Rev. Genet. 36: 361–388 [DOI] [PubMed] [Google Scholar]
- Davies B., Davies G., Preece C., Puliyadi R., Szumska D., et al. , 2013. Site specific mutation of the Zic2 locus by microinjection of TALEN mRNA in mouse CD1, C3H and C57BL/6J oocytes. PLoS ONE 8: e60216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M., Mackenzie I. R., Boeve B. F., Boxer A. L., Baker M., et al. , 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D., Haass C., 2013. Fused in sarcoma (FUS): an oncogene goes awry in neurodegeneration. Mol. Cell. Neurosci. DOI: 10.1016/j.mcn.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Doyle E. L., Booher N. J., Standage D. S., Voytas D. F., Brendel V. P., et al. , 2012. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40: W117–W122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flisikowska T., Thorey I. S., Offner S., Ros F., Lifke V., et al. , 2011. Efficient immunoglobulin gene disruption and targeted replacement in rabbit using zinc finger nucleases. PLoS ONE 6: e21045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal J., Zhang J., Kwinter D. M., Zhai J., Jia H., et al. , 2011. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol. Aging 32: 2323.e27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts A. M., Cost G. J., Freyvert Y., Zeitler B., Miller J. C., et al. , 2009. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P., Abuin A., Bradley A., 2000. Gene targeting, principles, and practice in mammalian cells, pp. 1–35 in Gene Targeting: A Practical Approach, The Practical Approach Series, edited by A. Joyner. Oxford University Press, Oxford [Google Scholar]
- Hicks G. G., Singh N., Nashabi A., Mai S., Bozek G., et al. , 2000. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat. Genet. 24: 175–179 [DOI] [PubMed] [Google Scholar]
- Ito D., Seki M., Tsunoda Y., Uchiyama H., Suzuki N., 2011. Nuclear transport impairment of amyotrophic lateral sclerosis-linked mutations in FUS/TLS. Ann. Neurol. 69: 152–162 [DOI] [PubMed] [Google Scholar]
- Kabashi E., Bercier V., Lissouba A., Liao M., Brustein E., et al. , 2011. FUS and TARDBP but not SOD1 interact in genetic models of amyotrophic lateral sclerosis. PLoS Genet. 7: e1002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino Y., Washizu C., Aquilanti E., Okuno M., Kurosawa M., et al. , 2011. Intracellular localization and splicing regulation of FUS/TLS are variably affected by amyotrophic lateral sclerosis-linked mutations. Nucleic Acids Res. 39: 2781–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski T. J., Jr, Bosco D. A., Leclerc A. L., Tamrazian E., Vanderburg C. R., et al. , 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208 [DOI] [PubMed] [Google Scholar]
- Lieber M. R., 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79: 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew M., Pryor R., Palais R., Meadows C., Erali M., et al. , 2004. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 50: 1156–1164 [DOI] [PubMed] [Google Scholar]
- Meyer M., de Angelis M. H., Wurst W., Kuhn R., 2010. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 107: 15022–15026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C., Tan S., Qiao G., Barlow K. A., Wang J., et al. , 2011. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Mori K., Weng S.-M., Arzberger T., May S., Rentzsch K., et al. , 2013. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339: 1335–1338 [DOI] [PubMed] [Google Scholar]
- Moscou M. J., Bogdanove A. J., 2009. A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Porteus M. H., Carroll D., 2005. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 23: 967–973 [DOI] [PubMed] [Google Scholar]
- Qiu Z., Liu M., Chen Z., Shao Y., Pan H., et al. , 2013. High-efficiency and heritable gene targeting in mouse by transcription activator-like effector nucleases. Nucleic Acids Res. 41: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton A. E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., et al. , 2011. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72: 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D., Tsai S. Q., Khayter C., Foden J. A., Sander J. D., et al. , 2012. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30: 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P., Smih F., Jasin M., 1994. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. USA 91: 6064–6068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J., Sung P., Klein H., 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77: 229–257 [DOI] [PubMed] [Google Scholar]
- Sasayama H., Shimamura M., Tokuda T., Azuma Y., Yoshida T., et al. , 2012. Knockdown of the Drosophila fused in sarcoma (FUS) homologue causes deficient locomotive behavior and shortening of motoneuron terminal branches. PLoS ONE 7: e39483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y. H., Baek I.-J., Kim D. H., Jeon J., Lee J., et al. , 2013. Knockout mice created by TALEN-mediated gene targeting. Nat. Biotechnol. 31: 23–24 [DOI] [PubMed] [Google Scholar]
- Taylor J. P., 2013. Neuroscience. RNA that gets RAN in neurodegeneration. Science 339: 1282–1283 [DOI] [PubMed] [Google Scholar]
- Wefers B., Meyer M., Hensler S., Panda S., Ortiz O., et al. , 2012. Gene editing in one-cell embryos by zinc-finger and TAL nucleases. Curr. Protoc. Mouse Biol. 2: 347–364 [DOI] [PubMed] [Google Scholar]
- Wefers B., Meyer M., Ortiz O., Hrabé de Angelis M., Hansen J., et al. , 2013. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc. Natl. Acad. Sci. USA 110: 3782–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.