Abstract
Vegetative fusion is essential for the development of an interconnected colony in many filamentous fungi. In the ascomycete fungus Neurospora crassa, vegetative fusion occurs between germinated conidia (germlings) via specialized structures termed “conidial anastomosis tubes” (CATs) and between hyphae within a mature colony. In N. crassa, both CAT and hyphal fusion are under the regulation of a conserved MAP kinase cascade (NRC1, MEK2, and MAK2). Here we show that the predicted downstream target of the MAK2 kinase pathway, a Ste12-like transcription factor known as PP1, regulates elements required for CAT and hyphal fusion. The PP1 regulatory network was revealed by expression profiling of wild type and the Δpp-1 mutant during conidial germination and colony establishment. To identify targets required for cell fusion more specifically, expression-profiling differences were assessed via inhibition of MAK2 kinase activity during chemotropic interactions and cell fusion. These approaches led to the identification of new targets of the cell fusion pathway that, when mutated, showed alterations in chemotropic signaling and cell fusion. In particular, conidial germlings carrying a deletion of NCU04732 (Δham-11) failed to show chemotropic interactions and cell fusion. However, signaling (as shown by oscillation of MAK2 and SO to CAT tips), chemotropism, and cell fusion were restored in Δham-11 germlings when matched with wild-type partner germlings. These data reveal novel insights into the complex process of self-signaling, germling fusion, and colony establishment in filamentous fungi.
Keywords: Neurospora crassa, signal transduction, cell fusion, STE12, chemotropism, transcriptional profiling
CELL fusion between genetically identical cells is important in development (for example, myoblast fusion during muscle formation) and occurs in many multicellular organisms from simple ascomycete fungi to mammals (Chen et al. 2007; Aguilar et al. 2013). Cell fusion between genetically identical cells can be mediated by cells that have differentiated, but in some cases, also between cells in an identical developmental state, for example, cell fusion between germinating asexual spores (conidia) of filamentous fungi (Pandey et al. 2004; Roca et al. 2005a; Read et al. 2010). In filamentous fungi, these fusions are integral to the formation of an interconnected hyphal network, which mediates genetic mixing and the sharing of resources (Simonin et al. 2012; Roper et al. 2013). How this process is initiated and maintained and what proteins are involved are still mostly unknown.
In filamentous ascomycete fungi, a conserved MAP kinase pathway that is involved in pheromone response and mating in Saccharomyces cerevisiae (Ste11, Ste7, and Fus3) (Bardwell 2005) is required for cell fusion and heterokaryon formation during vegetative growth (Hou et al. 2002; Wei et al. 2003; Pandey et al. 2004; Fu et al. 2011; Jun et al. 2011; Dettmann et al. 2012). This conserved pathway also plays a role in sexual development and secondary metabolism and is required for the virulence of both plant and animal fungal pathogens (Roman et al. 2007; Rispail and Di Pietro 2010; Bayram et al. 2012). In S. cerevisiae, reception of a mating-type-specific pheromone signal results in the activation of Ste11 (MEKK), Ste7 (MEK), and Fus3 (MAPK) (Bardwell 2005). After Fus3 is activated, it enters the nucleus and phosphorylates Ste12, a mating-type-specific transcription factor, as well as two regulators of Ste12, Dig1 and Dig2 (Blackwell et al. 2007). Activated Ste12 regulates the expression of many genes involved in the mating process, both indirectly and directly by binding pheromone response elements in target promoters (Zeitlinger et al. 2003).
Ste12-like proteins have also been studied in filamentous fungi, where they play essential roles in development and pathogenicity (Alspaugh et al. 1998; Vallim et al. 2000; Borneman et al. 2001; Park et al. 2002; Tsuji et al. 2003; Li et al. 2005; Nolting and Poggeler 2006; Ren et al. 2006; Tollot et al. 2009; Rispail and Di Pietro 2010; Wong Sak Hoi and Dumas 2010). Fus3-like proteins in filamentous fungi could directly or indirectly phosphorylate Ste12-like proteins, but this has not yet been shown in any system. However, strains carrying deletions of Fus3-like proteins or Ste12-like proteins often show a similar phenotype in many filamentous fungi, suggesting that Fus3-like kinases positively activate Ste12-like proteins. All Ste12-like proteins contain a divergent homeodomain (STE) near their N terminus, which is involved in DNA binding (Errede and Ammerer 1989). Ste12-like proteins in filamentous fungi also contain two C-terminal C2H2-Zn2+ motifs (Vallim et al. 2000; Chang et al. 2004) (Figure 1A) that are absent in S. cerevisiae and other related ascomycete yeast species. In the plant pathogen Magnaporthe grisea, both the STE domain and the C2H2-Zn2+ motifs of the STE12 homolog, mst12, are essential for the development of appressorial penetration pegs (Park et al. 2004), while in the human basidiomycete pathogen Cryptococcus neoformans, the STE domain and the C2H2-Zn2+ motifs were important for function, although mutagenesis of each domain resulted in strains with different phenotypes (Chang et al. 2004). In S. cerevisiae, Ste12-binding partners include Mcm1 and Tec1 (Primig et al. 1991; Madhani and Fink 1997). In Sordaria macrospora, a physical interaction between STE12 and MCM1 is required for sexual spore formation (Nolting and Poggeler 2006).
Figure 1.
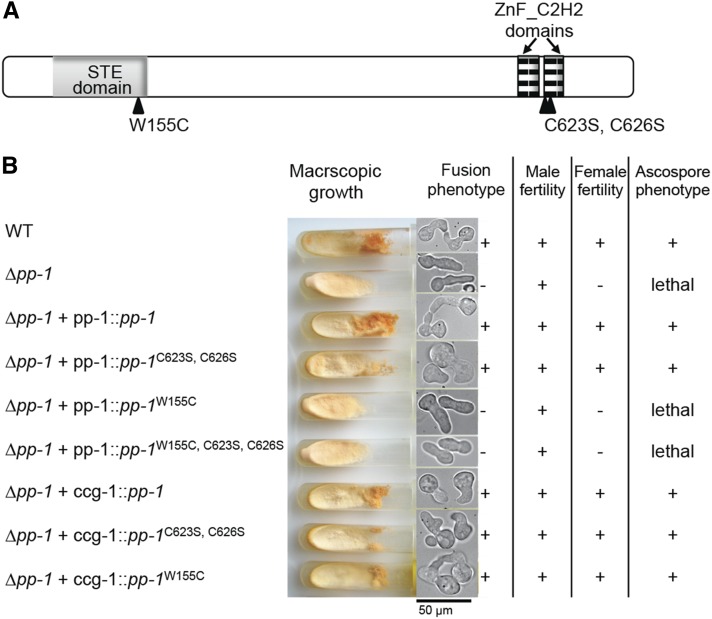
The homeodomain of PP1 is required for function. (A) Schematic representation of PP1 with protein domains depicted. Point mutations resulting in amino acid substitutions in the respective protein domains are indicated. (B) Phenotypic results of introduction of wild-type and mutated pp-1 alleles into a Δpp-1 mutant. Strains with alleles are listed on the left, and phenotypic analyses are depicted or listed on the right.
In Neurospora crassa, proteins orthologous to the Fus3 MAP kinase pathway (NRC1, MEK2, MAK2) regulate vegetative cell fusion (Pandey et al. 2004; Fleissner et al. 2008; Fu et al. 2011; Dettmann et al. 2012). Cell fusion events are particularly frequent in germinating asexual spores (conidia) and are associated with the production of specialized fusion structures termed “conidial anastomosis tubes” (CATs) (Roca et al. 2005b). Chemotropic interactions occur when two neighboring germlings sense each other and redirect the growth of CATs. During chemotropic interactions, NRC1, MEK2, and MAK2 oscillate together from CAT tip to cytoplasm. The oscillation of NRC1/MEK2/MAK2 occurs every 4 min and is perfectly out of phase with a second protein, SO, which also oscillates from CAT tip to cytoplasm in communicating germling pairs (Fleissner et al. 2009b; Dettmann et al. 2012). A STE12 homolog has also been identified in N. crassa (pp-1). A Δpp-1 mutant shows slow growth, is female sterile (they do not develop protoperithecia), and displays ascospore lethality (Li et al. 2005), phenotypes that are very similar to that displayed by Δmak-2 mutants. Previous analyses using a N. crassa partial-genome (13%) microarray identified genes whose expression is altered in a mature colony (7 days old) in the absence of mak-2 or pp-1. These analyses revealed that PP1 is involved in the regulation of sexual development, asexual development, and secondary metabolism (Li et al. 2005).
In this study, we evaluated the role of PP1 during cell fusion of asexual spores and expanded analyses using whole-genome arrays and RNA-seq to examine the function of MAK2 and PP1 during germling fusion and early colony establishment. These analyses revealed MAK2- and PP1-dependent gene networks involved in various aspects of colony development and identified novel genes with important roles in regulating communication and chemotropic attraction between N. crassa germlings.
Materials and Methods
Molecular techniques and strain construction
All strains constructed and used in this study are listed in Table 1 and Table 2. Strains were grown on Vogel’s minimal medium (VMM) (Vogel 1956) (with supplements as required) and were crossed on Westergaard’s medium (Westergaard and Mitchell 1947). Transformations and other N. crassa molecular techniques were performed as described (Colot et al. 2006) or using protocols available at the Neurospora home page at the Fungal Genetics Stock Center (FGSC) (http://www.fgsc.net/neurospora/neurosporaprotocolguide.htm).
Table 1. N. crassa strains used in this study.
| Name | Mutant genotype | Source |
|---|---|---|
| R13-5 | Δpp-1 A | Li et al. (2005) |
| Δpp-1 a | This study | |
| FGSC 9537 | Dip | FGSC |
| FGSC 6103 | his-3 A | FGSC |
| his-3; Δpp-1 A | This study | |
| his-3+::Pccg1 pp-1W155C a | This study | |
| his-3+::pp-1W155C a | This study | |
| his-3+::pp-1W155C, C623S, C626S a | This study | |
| his-3+::Pccg1 pp-1C623S, C626S a | This study | |
| his-3+::pp-1C623S, C626S a | This study | |
| his-3+::Pccg1 pp-1 a | This study | |
| his-3+::pp-1 a | This study | |
| mal1 | his-3+::Pccg1 mak-2Q100G; Δmak-2 A | Fleissner et al. (2009b) |
| FGSC 9718 | Δmus-51::bar a | FGSC |
| FGSC 988 | Oak Ridge WT a | FGSC |
| FGSC 2489 | Oak Ridge WT A | FGSC |
| R9-08 | hex-1 A | Jedd and Chua (2000) |
| AF-SoT8 | his-3+::Pccg1 so-gfp A | Fleissner et al. (2009b) |
| AF-M512 | his-3+::Pccg1 mak-2-gfp A | Fleissner et al. (2009b) |
| his-3; Δham-7 A | This study | |
| his-3+::Pccg1 so-gfp; Δham-7 A | This study | |
| his-3+::Pccg1 mak-2-gfp; Δham-7 A | This study | |
| his-3; Δham-11 A | This study | |
| his-3+::Pccg1 so-gfp; Δham-11 A | This study | |
| his-3+::Pccg1 mak-2-gfp; Δham-11 A | This study | |
| his-3+::Ptef1 ham-11-gfp; Δham-11 A | This study | |
| his-3+::Pccg-1 Prm-1-gfp; ΔPrm-1 A | Fleissner et al. (2009a) |
Table 2. Fusion phenotype of strains carrying deletions that showed significant differences in expression level in the Δpp-1 mutant relative to wild type.
| NCU ID no. | WT RPKM | Δpp-1 RPKM | Protein description | Deletion origin | Fusion % (WT = 86 ± 4)a |
|---|---|---|---|---|---|
| NCU00309b | 333 | 527 | Hypothetical | FGSC 19692 | WT |
| NCU00811 | 1302 | 630 | Hypothetical | This study | WT |
| NCU00881 | 145 | 67 | HAM7 | 13775 | 0 |
| NCU00995 | 274 | 0 | Hypothetical | This study | WT |
| NCU01380 | 413 | 1 | Hypothetical | This study | WT |
| NCU01697 | 130 | 2 | Hypothetical | FGSC 14575 | WT |
| NCU02361 | 42 | 117 | Formamidase | FGSC 21969 | WT |
| NCU02500 | 49 | 642 | CCG4 | FGSC 14944 | WT |
| NCU03013 | 368 | 25 | ACW10 | FGSC 11222 | WT |
| NCU03960 | 200 | 24 | HAM12 | FGSC 17233 | 66 ± 6 |
| NCU04122 | 269 | 19 | Malate dehydrogenase | FGSC 21282 | WT |
| NCU04192 | 70 | 42 | Vacuolar aspartyl aminopeptidase | FGSC 18884 | WT |
| NCU04732 | 179 | 65 | HAM11 | FGSC 17545 and this study | 0 |
| NCU05814 | 145 | 48 | Hypothetical | FGSC 17770 | WT |
| NCU06698 | 79 | 49 | Glycogenin | FGSC 12301 | WT |
| NCU07503 | 77 | 300 | Hypothetical | FGSC 14018 | WT |
| NCU07802 | 148 | 1 | Hypothetical | FGSC 20487 | WT |
| NCU08332 | 1422 | 448 | HEX1 | Jedd and Chua (2000) | WT |
| NCU08824 | 37 | 73 | Molybdopterin-binding domain | FGSC 15980 | WT |
| NCU09560 | 66 | 150 | Superoxide dismutase | FGSC 21068 | WT |
| NCU09562 | 501 | 5 | Hypothetical | FGSC 21874 | WT |
| NCU09693 | 116 | 5 | Hypothetical | This study | WT |
Mutants show fusion frequencies similar to their isogenic wild-type parent (FGSC 2489).
Genes in boldface type showed increased expression levels in the Δpp-1 mutant.
To construct the pp-1 site-directed mutant alleles, fusion PCR was performed with the mutation included in the overlapping primer region (Supporting Information, Table S1). Alleles were cloned into his-3 targeting plasmid pMF272 (AY598428) (Margolin et al. 1997; Freitag et al. 2004) and integrated into the his-3 locus of a his-3; Δpp-1 strain. Δpp-1 and Δpp-1 transformant strains showing an ascospore lethal phenotype were mated to the Diploid (Dip) mutant strain (FGSC 9537) as described in Hutchison et al. (2009) to construct homokaryotic strains or strains containing auxotrophic markers. In Dip crosses, ∼2/3 of the ascospores are large and diploid. Streaking Dip progeny onto sorbose plates results in restoration of haploid strains. The pp-1 sequence in all engineered strains was confirmed by DNA sequence analyses.
Deletion strains were obtained from the FGSC (McCluskey 2003) and were generated as part of the N. crassa functional genomics project (Colot et al. 2006; Dunlap et al. 2007). For each deletion strain, both the mating type A and mating type a strains were analyzed, if available. The phenotypes of deletion strains that were available only as a single mating type from the FGSC (McCluskey 2003) were confirmed by segregation analysis. Ascospore progeny were selected from crosses with FGSC 2489 (mating type A) or FGSC 988 (mating type a) and assessed for phenotype and hygromycin resistance. To pass this test, hygromycin resistance had to segregate with fusion phenotype. In cases where deletion strains were not publicly available (genes with locus identification numbers: NCU00811, NCU00995, NCU01380, and NCU09693), we constructed deletion mutants using a mus-51 background as described (Colot et al. 2006). Primers (Table S1) were designed to amplify the upstream and downstream flanks of these four genes, as well as the hygromycin cassette from plasmid pCSN44 (Colot et al. 2006). The two flanks, the hygromycin cassette and digested plasmid pRS426 (EcoRI and XhoI), were transformed into yeast, and recombined plasmids were obtained via plasmid rescue (Colot et al. 2006). Hygromycin transformants were crossed to wild type (WT) (FGSC 2489) to eliminate the mus-51 mutation and to obtain homokaryotic progeny. The genotype of all deletion strains was confirmed by PCR, using a primer for the hygromycin cassette (HygRupflank) paired with a gene-specific primer from the flank region, as well as a gene-specific primer outside the gene and a gene-specific primer within the open reading frame (Table S1).
To construct the internally GFP-tagged ham-11 construct, we amplified ham-11 using primers 04732F and 04732R (Table S1) and cloned it into a pCR-Blunt vector (Invitrogen). We sequenced and digested the amplicon from the vector with BglII and ApaI. The tef1 promoter was removed from plasmid ptef1-mek-2-gfp (Dettmann et al. 2012) with NotI and BamHI. Using a three-point ligation, we cloned the tef1 promoter and ham-11 amplicon into plasmid pmf272 (Freitag et al. 2004), using NotI and ApaI. GFP containing codons for eight glycine linkers and a BamHI restriction site on both ends was amplified from plasmid pMF272 using primers Fgfp+8xgly and Rgfp+8xgly (Table S1) and cloned into a pCR-Blunt vector (Invitrogen). We sequenced the amplicon, digested it with BamHI, and ligated it into the BamHI site in ham-11 situated between the two predicted transmembrane domains. This ham-11 construct with internally tagged gfp was transformed into the Δham-11 his3− strain with selection for His prototrophy. A homokaryotic strain was obtained via microconidia purification (Pandit and Maheshwari 1994).
Phenotypic analyses
Macroscopic growth differences were assessed by growing the different strains (WT, Δham-7, Δham-11, and Δham-12) in flasks for 1 week at 25° and on VMM plates for 2 days at 25°. Growth rate on VMM plates was measured at 25° by putting 1 × 106 conidia in the center of the plates and measuring the radial growth front after 24 and 48 hr. Aerial hyphae extension was determined by inoculating tubes containing 1 ml of liquid VMM with 1 × 106 conidia; aerial hyphae height was measured after 3 days of growth at 25° in constant light. Ten replicates were measured for each strain.
To assess the frequency of communication and fusion between conidia within a deletion strain as compared to wild type, slant tubes containing the strains were grown for 4–6 days or until significant conidiation occurred. Conidia were harvested by vortexing the slant tube with 2 ml ddH2O and subsequently filtered by pouring over cheesecloth to remove hyphal fragments. Conidia were diluted to a concentration of 3.3 × 107 conidia/ml. For each sample, 300 μl of spore suspension was spread on a 9-cm solid VMM plate. The plates were dried in open air for 20–30 min and incubated for 3–4 hr at 30°. Squares of 1 cm were excised and observed with a Zeiss Axioskop 2 using a ×40 Plan-Neofluor oil immersion objective. For each strain, 50–100 germling pairs were counted in triplicate. The ability to communicate is given as a percentage of pairs that display homing behavior when germinated conidia are within ∼15 μm of each other. To assess the frequency of communication between conidia of a deletion strain with wild-type germlings, mixtures of mak-2-gfp- or so-gfp-expressing strains and nonfluorescing strains were prepared and similarly treated as described above.
Fluorescence microscopy
Molecular techniques, including SO-GFP and MAK2-GFP strain construction, were previously described (Fleissner et al. 2009b). The strain used to cross so-gfp into deletion strains was AF-SoT8, and the strain used to cross mak-2-gfp into deletion strains was AF-M512 (Table 1) (Fleissner et al. 2009b).
Oscillation studies performed with MAK2-GFP and SO-GFP were conducted as described previously (Fleissner et al. 2009b); conidia (0.5 × 107 spores/9-cm plate) from MAK2-GFP or SO-GFP strains were mixed with equal amounts of conidia from the respective deletion mutants and plated on solid VMM, dried, and incubated at 30° for 3–4 hr before being used for microscopy. Images were taken using a Leica SD6000 microscope with a ×100 1.4 N.A. oil-immersion objective equipped with a Yokogawa CSU-X1 spinning disk head and a 488-nm laser controlled by Metamorph software. Multiple pairs of interacting germlings were analyzed per experiment, and representative pairs are shown for each strain.
Microarray analysis and quantitative RT-PCR
For the PP1 microarray experiment, RNA was isolated from wild-type or Δpp-1 conidia grown in constant light at 25° for 3–8 hr in VMM at 25° with constant light. Total RNA from frozen samples was isolated using Zirconia/Silica beads (0.5-mm diameter; Biospec) and a Mini- Beadbeater-96 (Biospec) with 1 ml TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The total RNA was further purified by digestion with TURBO DNA-free (Ambion) and an RNeasy kit (Qiagen). RNA concentration and integrity was checked by Nanodrop and agarose gel electrophoresis.
For the mak-2Q100G microarray experiment, strains were grown in liquid VMM for 5 hr at 25° with constant light; 20 min prior to harvesting, cells were treated either with 20 µM 1NM-PP1 in DMSO or with DMSO only.
Microarray hybridization and slide image analysis were as described previously (Tian et al. 2007). We chose to use a closed-circuit design for microarray comparisons, which is a statistically robust method of identifying differentially regulated genes over a time series, and used Bayesian Analysis of Gene Expression Levels (BAGEL) (Meiklejohn and Townsend 2005) to determine normalized gene expression values. Microarray profiling data are deposited at the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) as series record GSE46912 (mak-2 microarray) and GSE49679 (pp-1 microarray). Genes were clustered based on their differential expression in Δpp-1 over the time period using Hierarchical Clustering Explorer 3.0 (de Hoon et al. 2004). The MIPS functional catalog (FunCat) was used to assign functional annotation to genes (Ruepp et al. 2004). Enrichment of FunCat categories in gene sets vs. the whole genome was calculated using hypergeometrix distribution for P-value calculation.
Quantitative RT-PCR was used to confirm trends in the microarray and RNA-seq data, and was performed using the EX-PRESS One-Step SYBR GreenER Kit (Invitrogen) and the StepOnePlus Real-Time PCR System (Applied Biosystems). Reactions were performed in triplicate with a total reaction volume of 20 μl including 400 nM each of forward and reverse primers and 60 ng template RNA. Data analysis was performed by the StepOne Software (Applied Biosystems), using the Relative Quantitation/Comparative CT (ΔΔCT) setting. Data were normalized to the endogenous control actin. The primers used are listed in Table S1.
RNA-seq
RNA was isolated from conidia grown in constant light for 5 hr in VMM at 25°, originally inoculated at a concentration of 1 × 106 cells/ml. RNA-seq methods were as described in Ellison et al. (2011). The protocol used for library construction was modified from the standard Illumina method and has been described previously (Ellison et al. 2011). An Illumina Genome Analyzer (http://qb3.berkeley.edu/qb3/gsl/index.cfm) was used to sequence reads, and the analysis program TopHat (Langmead et al. 2009) was used to process the data. Reads per kilobase mapped (RPKM), a normalized measure of gene expression, were calculated using Cufflinks (Roberts et al. 2011). Fisher’s exact test was used to test the significance of differences between wild type and Δpp-1, with a significance cutoff of P < 0.001. Profiling data (pp-1 RNA-seq) are deposited at the GEO database (http://www.ncbi.nlm.nih.gov/geo/) series record (GSE50446).
Cell fractionation and Western blotting
Harvested conidia (1 × 106/ml) were inoculated in 100-ml VMM in flasks, incubated for 2.5 hr at 30° with shaking at 200 rpm and then an additional 2.5 hr at 30° without shaking. Germlings were harvested by vacuum filtration over a PVDF membrane and frozen in liquid nitrogen. Protein extraction from ground mycelium was performed using lysis buffer as described in Pandey et al. (2004) containing complete protease inhibitors but without phosphatase inhibitor and Triton X-100. Cell fractionation via centrifugation was performed as in Bowman and Bowman (1988). The P1 pellet (vacuoles, mitochondria, and endomembranes) and P2 pellet (plasma membrane) were dissolved in lysis buffer with 1% Triton X-100. The remaining supernatant from P2 was used for immunoprecipitation using Protein G Dynabeads (Invitrogen), according to manufacturer’s instructions, with the following exceptions: mouse anti-GFP antibody (Roche) was covalently bound to the beads using BS3 (Sulfo-DSS, Fisher Scientific) according to manufacturer’s instructions. Supernatant samples were incubated with the beads for 3 hr at room temperature. Protein was removed from the beads by heating at 70° for 10 min, and samples were run on a 4–12% Nu-Page Bis-Tris Gel (NOVEX, Life Technologies). Protein gels were subjected to Western blot analysis using standard methods. Samples for the MAK1 and MAK2 phosphorylation Western blot were treated similarly, except, after protein extraction with 1 ml lysis buffer, 25 µl of protein sample was directly loaded on a 7% NuPage Bis-Tris Gel (NOVEX, Life Technologies). Gels were subjected to Western blot analysis using standard methods, and detection of phosphorylated MAK1 and MAK2 was carried out using anti-phospho p44/42 MAP kinase antibodies (1:3000 dilution) (PhosphoPlus antibody kit; Cell Signaling Technology) as described (Pandey et al. 2004).
Results
The homeodomain is required for PP1 function
In filamentous fungi, Ste12-like proteins possess two C2H2-Zn2+ motifs, in addition to the well-conserved homeodomain-like STE domain (Park et al. 2002). The STE domain is essential for function and DNA binding in S. cerevisiae (Yuan and Fields 1991). The function of these two domains in N. crassa was investigated through the construction of alleles containing point mutations in the DNA-binding motifs. In the first pp-1 allele, a point mutation (W155C) was made in the STE domain; this residue is in a highly conserved region known to be crucial for homeodomain–DNA interactions (Yuan and Fields 1991). The cysteine residues within the C2H2-Zn2+ motifs are important for DNA-binding activity in zinc-finger transcription factors. We therefore constructed a second pp-1 allele, in which the two cysteine residues of one of the C2H2-Zn2+ motifs were mutated to serine (C623S and C626S); similar mutations in a Ste12-like gene in C. neoformans resulted in a strain showing an altered virulence phenotype (Chang et al. 2004). In the third pp-1 allele, the point mutations in both the STE and C2H2-Zn2+ motifs were combined to create a double-mutant pp-1 allele (Figure 1A). These alleles, plus a wild-type pp-1 allele, were fused to GFP at the C terminus and transformed into the his-3 locus of a his-3; Δpp-1 mutant; selection was for His prototrophy (Margolin et al. 1997). The wild-type pp-1 and the pp-1C623S;C626S alleles fully rescued the Δpp-1 phenotype; strains were indistinguishable from wild type when assessing vegetative growth, cell fusion, female fertility, and ascospore viability (Figure 1B). In contrast, strains bearing the pp-1W155C allele or the pp-1W155C;C623S;C626S allele showed a phenotype identical to the Δpp-1 phenotype. These strains showed no cell fusion and were sterile (Figure 1B). These data indicate that the STE domain is essential for all characterized functions of PP1, while the C2H2-Zn2+ motif is apparently dispensable.
To assess the phenotypic consequences of overexpression of the wild-type pp-1 and mutant alleles, the pp-1, pp-1W155C, and pp-1C623S;C626S alleles were cloned behind the ccg-1 promoter (McNally and Free 1988) and introduced into the Δpp-1 strain at the his-3 locus. Similar to the pp-1C623S;C626S strains regulated by the pp-1 promoter, strains carrying the pp-1C623S;C626S allele under the regulation of the ccg-1 promoter showed a wild-type vegetative, cell fusion, and protoperithecial development phenotype (Figure 1B). However, in contrast to strains bearing the pp-1W155C allele regulated by the pp-1 promoter, strains bearing the pp-1W155C allele under the regulation of the ccg-1 promoter also showed a wild-type vegetative growth phenotype, as well as normal cell fusion levels, protoperithecial development, and ascospore germination. These data suggest that the mutated STE domain can still bind DNA at very low efficiency, which is presumably enough to rescue the Δpp-1 phenotype when overexpressed.
PP1-GFP was undetectable when pp-1-gfp expression was driven by the native pp-1 promoter. However, PP1-GFP localized to nuclei during chemotropic interactions and cell fusion when pp-1-gfp was expressed from the ccg-1 promoter (Figure 2A). PP1-GFP driven by the ccg-1 promoter also showed nuclear localization during colony establishment, vegetative growth, and conidiation (data not shown); differential localization of PP1 during growth or reproduction was not apparent. Nuclear localization was also observed for strains bearing the pp-1W155C and pp-1C623S;C626S alleles, indicating that these point mutations did not interfere with nuclear localization of PP1.
Figure 2.
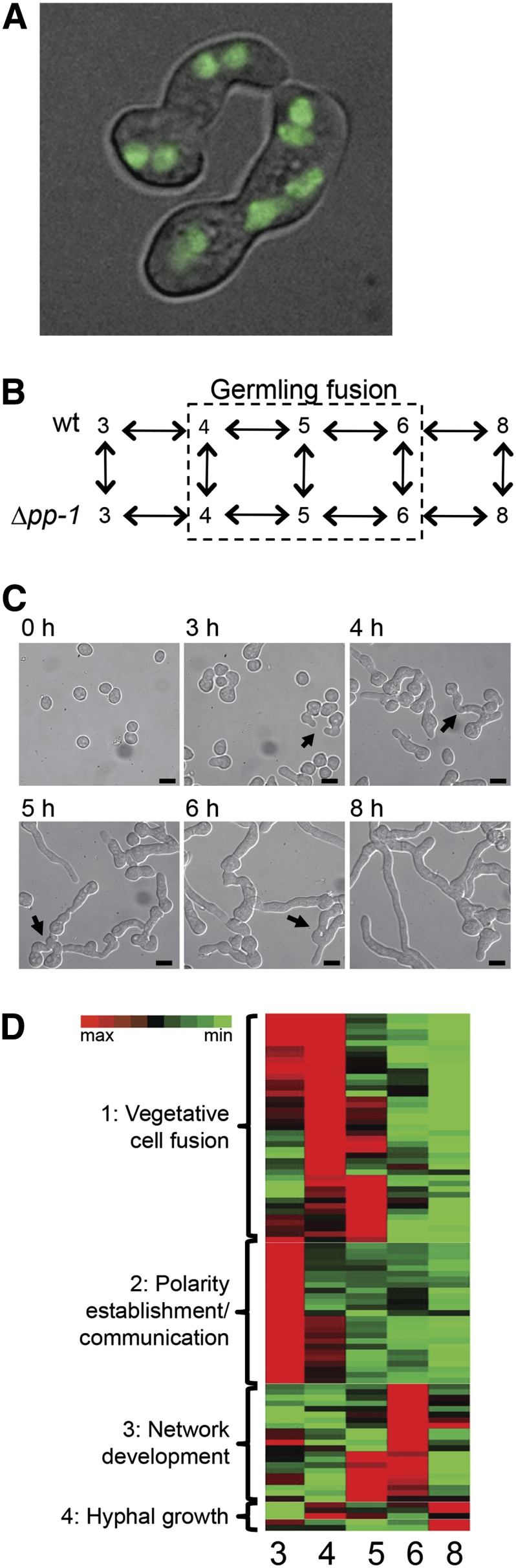
The role of pp-1 in hyphal network formation. (A) PP1-GFP shows nuclear localization during chemotropic growth in germlings. (B) The closed-circuit experimental design used for microarray analysis. The 4- to 6-hr subpopulation loop is marked in the dashed box. (C) Phenotypic analyses of conidial germination and hyphal network formation in wild type corresponding to identical time points as the 8-hr microarray time course. Arrows denote germling hyphal fusion events. Bar, 10 μm. (D) Visual representation of clustered gene expression profiles from Hierarchical Clustering Explorer analysis (de Hoon et al. 2004). Across each horizontal row (representing a single gene from the 3- to 8-hr germination time course), red indicates the highest dependence on PP1 for expression and green indicates the lowest dependence.
Transcriptional analysis of PP1 function during early colony development
We used full-genome microarrays (Tian et al. 2007) to monitor gene expression profiles in wild-type and Δpp-1 during germination, germling fusion, and early establishment of a colony. Gene expression differences between wild type and the Δpp-1 mutant were measured 3, 4, 5, 6, and 8 hr following inoculation of conidia into liquid media (Figure 2B). After 3 hr, newly germinated conidia and some chemotropic interactions were observed, which increased over time. Germling fusion events were prevalent after 5 hr, with ∼85% of conidia having undergone fusion events by this time point. Hyphal growth and branching started after 6 hr post-inoculation, and elongated and branched hyphae were abundant at 8 hr (Figure 2C).
A total of 5114 genes showed measurable expression levels throughout the time course, but only 92 genes showed expression at all time points and statistically significant expression differences of at least 1.5-fold between wild type and the Δpp-1 mutant at one or more time points (Table S2). Consistent with its role as a transcriptional activator, >50% of the genes showed decreased expression levels in the Δpp-1 mutant as compared to those with increased expression levels (60 down-regulated vs. 32 up-regulated).
Hierarchical clustering of the 92 genes during the 3- to 8-hr time course revealed four distinct clusters (Figure 2D; Table S2). The largest cluster of genes (cluster 1) showed differential expression levels during the 3- to 5-hr time course, when the process of chemotropic interactions and germling fusion events is most prevalent. A second cluster (cluster 2) showed differential expression specifically at the first time point (3 hr), suggesting that these genes may be important for the conidial germination process. The third and the fourth clusters showed differential expression during the 6- to 8-hr time course, suggesting that these genes are involved in the network formation and hyphal growth processes.
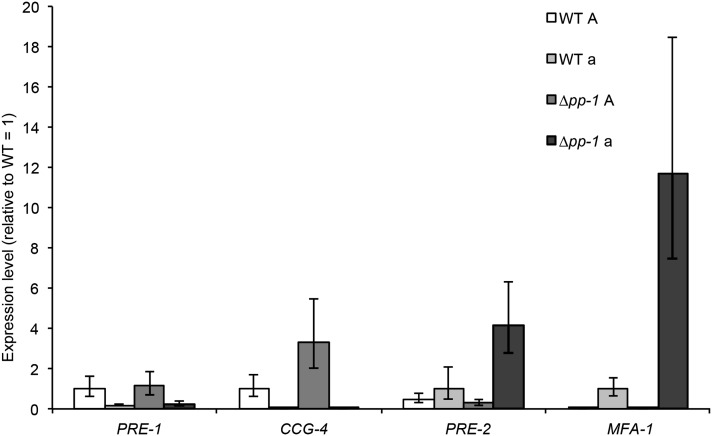
ccg-4, which encodes the mating type A pheromone, showed increased expression levels in a mating type A Δpp-1 mutant as compared to a wild-type mating type A strain (FGSC 2489) (Table S2). Increased expression levels of ccg-4 were also reported using partial genome microarrays and Δpp-1 A and Δmak-2 A strains (Li et al. 2005). We confirmed this observation by quantitative RT-PCR (qRT-PCR) from wild type A and Δpp-1 A strains (Figure 3). However, in mating type a strains as well as in Δpp-1 a strains, expression of ccg-4 was very low. We therefore also evaluated the expression of the mating type a pheromone gene, mfa-1, in wild type A and a strains, as well as in Δpp-1 A and a mating type strains; mfa-1 expression levels were significantly higher in the Δpp-1 a cells as compared to a wild-type a strain (Figure 3). To determine whether mutations in Δpp-1 also affect expression of the pheromone receptors, we assessed expression levels of pre-1 (a pheromone receptor) and pre-2 (A pheromone receptor) by performing qRT-PCR with mating type A and a Δpp-1 mutants as compared to wild-type mating type A and a strains. While expression levels for pre-1 were not significantly different between wild type and either A or a Δpp-1 cells, expression levels for pre-2 showed increased expression levels in the Δpp-1 a mating type strain (Figure 3).
Figure 3.
Increased expression of genes encoding mating-type pheromones and receptors in the Δpp-1 mutant. qRT-PCR was performed on wild-type mating type A (FGSC 2489), wild-type mating type a (FGSC 988), and Δpp-1 A and Δpp-1 a strains, 5 hr post-inoculation.
To identify additional genes whose expression correlated with germling fusion events, we assessed expression differences using a subset of the time course, 4–6 hr, as some genes involved in colony development may not be expressed at either 3 or 8 hr, and our statistical analysis method requires data at all time points (see Materials and Methods). From analyzing data from the 4- to 6-hr loop, we identified an additional 26 genes (for a total of 86 genes) that showed decreased expression levels and 9 genes (for a total of 41 genes) that showed increased expression levels in the Δpp-1 mutant relative to wild type (Figure 4, A and B; Table S2). Together with genes identified from the 3- to 8-hr time course, FunCat analysis showed that most of the genes with lower expression in the Δpp-1 strain belonged to unclassified/hypothetical proteins (Table S3). Additional overrepresented categories included various signaling pathways and vacuolar degradation (P < 0.05; Table S3). Of the genes that showed increased expression in the Δpp-1 strain, enrichment for the functional category of transcription was identified (P < 0.05; Table S4).
Figure 4.
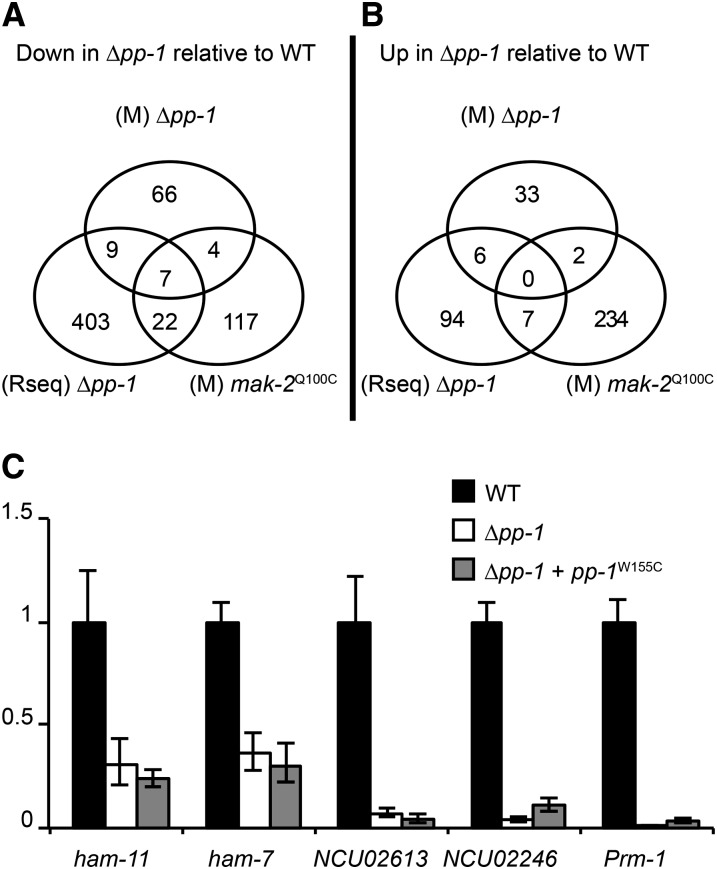
Summary of microarray and RNA-seq profiling data on Δpp-1 and mak-2Q100G mutants relative to wild type. (A) Venn diagram showing the overlap of genes that demonstrated a decrease in expression level identified by Δpp-1 (M Δpp-1) and mak-2Q100G (M mak-2Q100G) microarrays and RNA-seq analysis (Rseq Δpp-1) as compared to their wild-type isogenic parent (FGSC 2489). (B) Venn diagram showing the overlap of genes that demonstrated an increased expression level in the Δpp-1 (M Δpp-1) and mak-2Q100G (M mak-2Q100G) and RNA-seq analysis (Rseq Δpp-1) as compared to their isogenic wild-type parent (FGSC 2489). (C) qRT-PCR analysis showing relative expression levels of genes identified as being differentially expressed via microarrays/RNA-seq in the Δpp-1 and Δpp-1 + pp-1W155C mutants relative to wild type (FGSC 2489).
RNA-seq analyses were performed as a more sensitive method to identify transcriptional differences between wild-type and Δpp-1 cells at the time point with the highest number of germling fusion events, 5 hr post-inoculation. After statistical analysis (Materials and Methods), a total of 548 genes were identified as being significantly differentially expressed by at least 1.5-fold between wild type and Δpp-1; 441 genes showed reduced expression levels in the Δpp-1 mutant while 107 genes showed increased expression levels as compared to wild type (Figure 4, A and B; File S1).
Transcriptional analyses of MAK-2 function during early colony development
A Δmak-2 mutant in N. crassa shows a very similar phenotype to a Δpp-1 mutant, including lack of cell fusion, reduced vegetative growth, female sterility, and ascospore lethality (Pandey et al. 2004; Li et al. 2005). The question of whether MAK2 kinase activity is required for MAK2 oscillation, as well as for the oscillation of a second protein, SO, which is also required for cell fusion (Fleissner et al. 2009b), was tested by constructing a mak-2 allele (mak-2Q100G) containing a point mutation that allows specific inhibition of kinase activity with an ATP analog (1NM-PP1; Fleissner et al. 2009b). Addition of the inhibitor 1NM-PP1 to mak-2Q100G cells undergoing communication immediately blocks further oscillation of both MAK2 and SO and abruptly abolishes chemotropic interactions (Fleissner et al. 2009b). As a complement to our transcriptional profiling of the Δpp-1 mutant, and to identify targets more specifically related to the MAK2 kinase cascade, we performed transcriptional analysis of mak-2Q100G cells at the 5-hr time point, with and without exposure to 1NM-PP1, for the final 20 min of their growth (see Materials and Methods). We identified 392 genes with modified expression (≥1.5-fold) in the 1NM-PP1 mak-2Q100G-treated cells vs. mak-2Q100G (DMSO only). Of these, 149 genes showed a decreased expression level vs. 243 genes that showed increased expression levels (Figure 4, A and B; File S2). FunCat analysis of genes showing an increased expression level also showed an overrepresentation (P < 0.01) in categories of metabolism and in categories related to transport and defense (File S2), suggesting that treatment with 1NM-PP1 may elicit defense responses in N. crassa. Within the genes that showed decreased levels of expression was mak-2 itself, suggesting a feedback mechanism initiated as a result of the stalled signaling.
In total, 22 genes were identified as PP1 targets via both the microarray and the RNA-seq analyses, and an additional 6 genes were identified with the inclusion of the mak-2Q100G microarray data (Figure 4, A and B; Table S4). Altogether, 7 genes were identified in all three experiments, including ham-7 (NCU00881), a previously identified gene required for hyphal fusion (Fu et al. 2011; Maddi et al. 2012); NCU04122 (malate dehydrogenase); and 5 genes encoding the predicted hypothetical proteins NCU00995, NCU01697, NCU03960, NCU04732, and NCU09693. Genes encoding five additional hypothetical proteins were identified in the overlap between the Δpp-1 microarray and RNA-seq data (NCU00811, NCU01380, NCU05814, NCU07802, and NCU09562) as well as an anchored cell-wall protein (acw-10) (Maddi et al. 2009); hex-1, which encodes a protein involved in septal plugging (Jedd and Chua 2000); glycogenin (NCU06698); and vacuolar aspartyl aminopeptidase (NCU04192). In the comparison between the down-regulated genes in the Δpp-1 microarray and the mak-2Q100G microarray, only one additional gene encoding a hypothetical protein was identified (NCU07439). In the gene set that overlapped between the Δpp-1 RNA-seq data and the mak-2Q100G array data (decreased expression category), 5 additional genes were identified that, when mutated, resulted in strains blocked in germling/hyphal fusion. These include two components of the NADPH oxidase regulatory system, nox-1 (NCU02110) and nor-1 (NCU07850) (Fleissner et al. 2008; Fu et al. 2011); a transcription factor, adv-1 (NCU07392) (Fu et al. 2011); ham-6 (NCU02767), encoding a predicted transmembrane protein (Fu et al. 2011); and Prm-1. Prm-1 is involved in plasma membrane merger in N. crassa during both vegetative and sexual cell fusion (Fleissner et al. 2009a).
To confirm expression differences between Δpp-1 and wild type, we used qRT-PCR to analyze the expression of six genes, two from the set of seven overlapping genes from all three experiments, NCU04732 and ham-7 (NCU00881); two genes identified from the RNA-seq analysis, NCU02613 (hypothetical) and NCU02246 (hypothetical); and Prm-1 (NCU09337), which was identified in both the RNA-seq and mak-2Q100G microarray analyses. Our qRT-PCR recapitulated our RNA-seq/microarray analyses (Figure 4C), with the expression of all identified target genes being significantly reduced in both the Δpp-1 and pp-1W155C mutants relative to wild type.
PP1 and the regulation of genes required for hyphal fusion and network development
Approximately 40 genes have been identified with roles in colony establishment in N. crassa, specifically during germling or hyphal fusion (see review in Read et al. 2010, 2012). By evaluating our expression data, 16 of these genes were dependent on PP1 for their expression (>1.5-fold lower in Δpp-1 as compared to WT, P < 0.05, RNA-seq data) (Table 3). This list included ham-7, Prm-1, ham-6, nox-1, nor-1, and adv-1, as well as 2 genes encoding kinases in the cell-wall integrity pathway (mek-1 and mak-1) (Read et al. 2010); 2 genes encoding predicted components of the striatin complex (pp2A and mob-3) (Maerz et al. 2009; Fu et al. 2011); one transcription factor (rco-1) (Aldabbous et al. 2010); genes for two GPI-anchored cell-wall proteins (gpip-2 and gpip-1) (Bowman et al. 2006); ham-8, encoding a predicted transmembrane protein; ham-9, encoding a protein with pleckstrin homology domain and a sterile α-motif domain (Fu et al. 2011); and so (Fleissner et al. 2005). During germling fusion, SO also oscillates to CAT tips, with opposite dynamics to MAK2 (Fleissner et al. 2009b); correct oscillation of SO requires MAK2 kinase activity.
Table 3. Previously identified cell fusion genes that showed dependence on pp-1 for wild-type expression levels in germlings.
| Gene | Locus ID no. | Gene description | Reference |
|---|---|---|---|
| Prm-1 | NCU09337 | Membrane protein | Fleissner et al. (2009a) |
| mek-1 | NCU06419 | MAP kinase kinase | Fu et al. (2011) |
| ham-6 | NCU02767 | Small hydrophobic protein | Fu et al. (2011) |
| so | NCU02794 | WW domain protein | Fleissner et al. (2005) |
| ham-8 | NCU02811 | Transmembrane protein | Fu et al. (2011) |
| nox-1 | NCU02110 | NADPH oxidase | Cano-Dominguez et al. (2008) |
| ham-9 | NCU07389 | Plekstrin domain protein | Fu et al. (2011) |
| ham-7 | NCU00881 | GPI-anchored protein | Fu et al. (2011); Maddi et al. (2012) |
| adv-1 | NCU07392 | Transcription factor | Fu et al. (2011) |
| gpip-2 | NCU07999 | GPI-anchored protein | Bowman et al. (2006) |
| nor-1 | NCU07850 | NADPH oxidase regulator | Cano-Dominguez et al. (2008) |
| gpip-1 | NCU06663 | GPI-anchored protein | Bowman et al. (2006) |
| mak-1 | NCU09842 | MAP kinase | Fu et al. (2011); Maddi et al. (2012) |
| pp2A | NCU06563 | Catalytic subunit of protein phosphatase 2A | Fu et al. (2011) |
| mob-3 | NCU07674 | Phocein | Maerz et al. (2009) |
| rco-1 | NCU06205 | Transcription factor | Aldabbous et al. (2010) |
Identification of novel proteins involved in hyphal fusion and network development
A comparison of the Δpp-1 microarray and RNA-seq analyses identified a 16-gene overlap that showed reduced expression levels, and a 6-gene overlap that showed increased expression levels, in the Δpp-1 mutant relative to wild type (Figure 4 and Table 2). We therefore evaluated the germling fusion/colony establishment phenotypes of strains containing deletions of these genes. Of these 22 genes, 18 deletion mutants were available from the FGSC (McCluskey 2003), and we constructed strains carrying deletions for the remaining 4 genes (NCU00811, NCU0995, NCU01380, and NCU09693) (Table 2). Germling fusion frequency of each deletion strain was determined and compared to the wild-type isogenic parent. Of the 22 deletion strains assayed, three mutants were affected in the ability to undergo germling fusion. As expected, ham-7 (NCU00881) mutants were completely unable to undergo germling fusion (Fu et al. 2011), and in addition so was a mutant carrying a deletion of NCU04732, which encodes a hypothetical protein (Figure 5A). Deletion of an additional gene, NCU03960, which also encodes a hypothetical protein, resulted in a strain that was capable of fusion, but with a significantly lower fusion frequency than wild type (Table 2 and Figure 5A).
Figure 5.
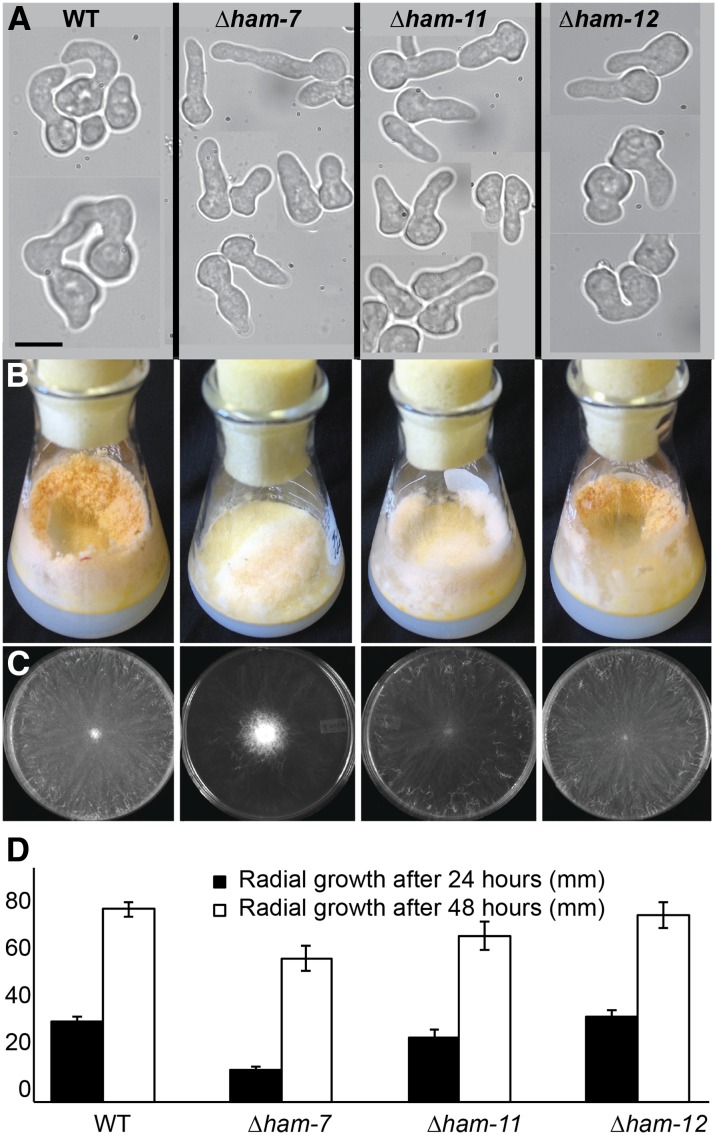
Fusion phenotypes of deletion mutants in PP1 target genes. (A) Chemotropic interactions and CAT fusion phenotypes of wild type (FGSC 2489), compared to the isogenic mutant strains Δham-7, Δham-11, and Δham-12. (B) Macroscopic growth phenotype of wild type, Δham-7, Δham-11, and Δham-12 mutants in flasks. Note the shortened aerial hyphae in the Δham-7 mutant. (C) Macroscopic growth phenotype on VMM plates of wild type, Δham-7, Δham-11, and Δham-12 2 days post-inoculation. (D) Radial growth (in mm) of wild type, Δham-7, Δham-11, and Δham-12 mutants over a 48-hr time period.
NCU04732 was named ham-11. HAM11 is a 633-amino-acid protein with two predicted transmembrane domains and is highly conserved among the Sordariomycetes. However, predicted proteins with strong homology in the N- and C-terminal regions to HAM11 were present in the genomes of most filamentous ascomycete species (no homologs were detected in Saccharomycetes or in Basidiomycetes). The Δham-11 mutants were fully fertile as a male or as a female, indicating that HAM11 is specifically required for germling fusion and not for sexual fusion. The Δham-11 growth phenotype differs from other fusion mutants (Δmak-2, Δso, and Δham-7) in that colonies were not “flat” due to reduced aerial hyphae extension (Figure 5, B and C). Aerial hyphae extension in Δham-11 mutants was similar to wild type (17 mm ± 1.7 as compared to 18.2 mm ± 2.6 for wild type), while aerial hyphae extension in Δham-7 mutants was only 1/2 of wild type (11 mm ± 1.3). The growth rate of Δham-11 over 48 hr was slightly slower than for wild type (Figure 5D).
A strain carrying a deletion of NCU03960, hereafter called ham-12, showed a significant (Student’s t-test: P < 0.01) reduction in fusion frequencies as compared to wild type (66 vs. 87%, respectively) (Table 2). ham-12 is predicted to encode a 157-amino-acid protein with one predicted transmembrane domain, but without any annotated function. ham-12 is more highly conserved than ham-11, with homologs identified across filamentous ascomycete fungi (Pezizomycotina). The growth, reproductive phenotype, and aerial hyphae extension of the Δham-12 mutant were identical to wild type (Figure 5, B–D).
Fusion defect of Δham-11 germlings is suppressed in the presence of a wild-type partner cell
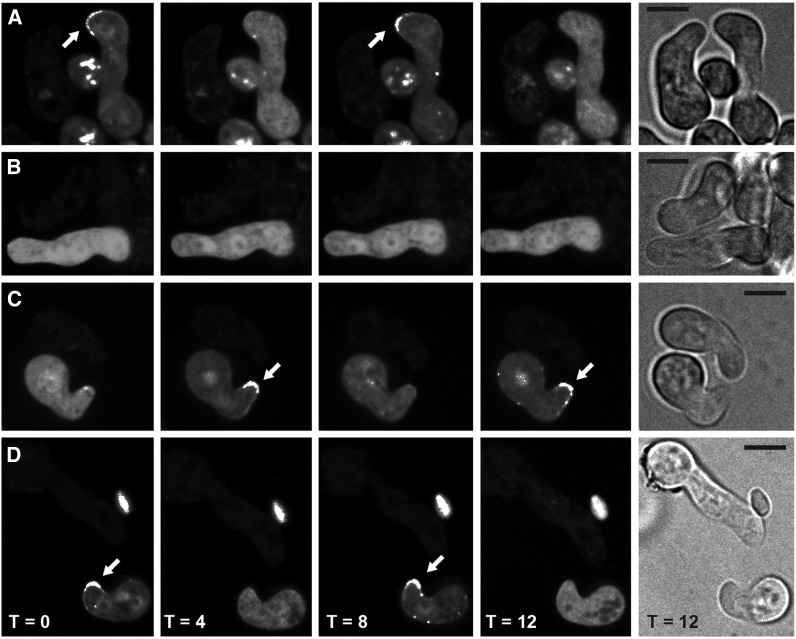
When wild-type germlings communicate, MAK2 oscillates from the cytoplasm to CAT tips every 4 min (8 min/cycle; Figure 6A). Similarly, when wild-type germlings communicate, SO oscillates from the CAT tips to the cytoplasm every 4 min (Figure 7A); MAK2 and SO oscillate in perfect opposition during chemotropic interactions (Fleissner et al. 2009b). To determine whether Δham-7 or Δham-11 germlings show an altered oscillation pattern of MAK2, we introduced mak-2-GFP constructs into wild type, Δham-7, and Δham-11 strains. For both Δham-7 and Δham-11 germlings, localization of MAK2-GFP was cytoplasmic, and localization to germling tips, chemotropic interactions, or cell fusion was not observed (Figure 6B and Figure 8). However, when Δham-11 germlings were confronted with wild-type cells, chemotropic interactions and cell fusion were observed. However, the percentage of Δham-11 germlings communicating with wild type was ∼25% lower as compared to wild-type–wild-type interactions, as wild-type–type fusions were overrepresented in mixtures of Δham-11 + wild-type germlings.
Figure 6.
Localization of MAK2-GFP in wild type (FGSC 2489; WT) and Δham-11 during germination and germling interactions. (A) MAK2-GFP oscillates to the tip every 8 min (arrows) in a wild-type cell bearing MAK2-GFP when communicating with an otherwise isogenic wild-type germling (WT mak-2-gfp + wild type). (B) MAK2-GFP shows no oscillation in Δham-11 mak-2-gfp germlings in proximity to otherwise isogenic Δham-11 cells (Δham-11 mak-2-gfp + Δham-11). (C) Chemotropic interactions and MAK2-GFP oscillation are apparent when wild-type germlings bearing MAK2-GFP are in proximity to Δham-11 germlings (WT mak-2-gfp + Δham-11). (D) Oscillation of MAK2-GFP is apparent in Δham-11 germlings in proximity to wild-type cells (Δham-11 mak-2-gfp + WT). Arrows indicate localization of MAK2-GFP to CAT tips when observed. Bright-field pictures on right show both germlings. Bar, 10 µm.
Figure 7.
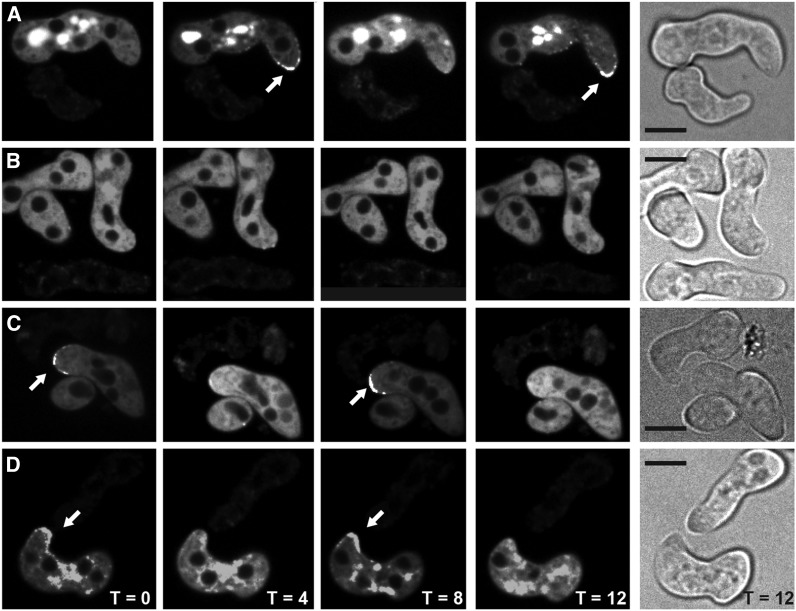
Localization of SO-GFP in wild-type and Δham-11 germlings. (A) A time course of SO-GFP oscillation during chemotropic interactions between wild-type germlings; oscillation of SO-GFP to CAT tips occurs every 8 min (WT so-gfp gfp + WT). (B) Δham-11 germlings bearing SO-GFP lack oscillation and localization of SO-GFP to CAT tips when in proximity to other Δham-11 germlings (Δham-11 so-gfp-gfp + Δham-11). (C) Wild-type cells bearing SO-GFP show oscillation when in proximity to Δham-11 cells and chemotropic interactions are restored (WT so-gfp + Δham-11). (D) Δham-11 germlings bearing SO-GFP show chemotropic interactions and oscillation of SO-GFP to CAT tips when in proximity to wild-type germlings (Δham-11 so-gfp + WT). Note that the oscillation dynamics of SO-GFP are identical to the WT + WT germling interactions shown in A. Arrows show localization of SO-GFP to CAT tips. Bright-field pictures show germlings. Bar, 10 µm.
Figure 8.
Lack of oscillation of MAK2-GFP and chemotropic interactions in Δham-7 germlings in proximity to wild type. (A) Cytoplasmic localization of MAK2-GFP in wild-type germlings when in proximity to Δham-7 germlings (WT mak-2-gfp + Δham-7). No oscillation of MAK2-GFP was observed during the time course. (B) Cytoplasmic localization of MAK2-GFP and lack of chemotropic interactions when mak-2-gfp; Δham-7 germlings are in proximity with wild-type cells (WT + Δham-7 mak-2-gfp). No localization of MAK2-GFP to CAT-like structures was observed during the time course. (C) Lack of chemotropic interactions between Δham-7 germlings and cytoplasmic localization of MAK2-GFP (Δham-7 + Δham-7 mak-2-gfp). Bright-field pictures show the germling proximity. Bar, 10 µm.
We hypothesized that in wild-type + Δham-11 germling pairs where chemotropic interactions were observed, MAK2 oscillation would be restored. As predicted, oscillation of MAK2-GFP in wild-type cells, when confronted with Δham-11 germlings, was identical to wild-type + wild-type pairings and showed identical MAK2 oscillation dynamics (8-min cycle from CAT tip to cytoplasm) (Figure 6C). Similarly, when Δham-11 germlings bearing MAK2-GFP were in proximity to wild-type cells, oscillation of MAK2 was coincident with chemotropic interactions and cell fusion (Figure 6D). In contrast, Δham-11 mak-2-gfp + Δham-11 germlings showed only cytoplasmic localization of MAK2 (Figure 6B). In contrast to the results obtained with Δham-11 mutants, no chemotropic interactions were observed between wild-type + Δham-7 germlings, and oscillation of MAK2 to CAT tips was not apparent in wild-type cells in proximity to Δham-7 germlings, nor in Δham-7 germlings bearing MAK2-GFP in proximity to wild-type cells (Figure 8).
We hypothesized that SO oscillation would also be restored in wild type + Δham-11 germlings when chemotropic interactions and cell fusion were observed. As predicted, SO oscillated in an 8-min cycle from CAT tip to cytoplasm in wild-type cells bearing SO-GFP communicating with Δham-11 cells (Figure 7C) and also when Δham-11 germlings bearing SO-GFP were undergoing chemotropic interactions with wild-type cells (Figure 7D). The dynamics of SO oscillation in wild type + Δham-11 pairings showed identical dynamics to wild-type + wild-type pairings. The phenotype of the Δham-11 mutant is unique among fusion mutants: communication and fusion are not restored when other fusion mutants are confronted with wild-type strains (mutants are unable to send or receive fusion signals) (Xiang et al. 2002; Pandey et al. 2004; Fleissner et al. 2005; Simonin et al. 2010; Fu et al. 2011).
Some fusion mutants are impaired in proper phosphorylation of the two MAPK kinases, MAK1 or MAK2, which are both required for germling fusion (Dettmann et al. 2012; Maddi et al. 2012). Since the Δham-11 mutant failed to initiate chemotropic interactions, we also evaluated phosphorylation of MAK1 and MAK2 in Δham-11 germlings with the Δham-7 mutant as a control. Conidia from wild type, Δham-7, or Δham-11 were germinated for 5 hr, and total protein was extracted and subjected to Western blot using phospho-specific P22/P24 antibodies, which recognize phosphorylated MAK1 and MAK2. As previously described (Maddi et al. 2012), Δham-7 shows a defect in phosphorylation of MAK1 and, to a modest extent, phosphorylation of MAK2. Surprisingly, Δham-11 germlings showed wild-type levels of phosphorylation of both MAK1 and MAK2 (Figure 9). These data indicate that, even though MAK2 oscillation to CAT tips is defective in Δham-11 germlings, phosphorylation of MAK2 by upstream factors is not.
Figure 9.
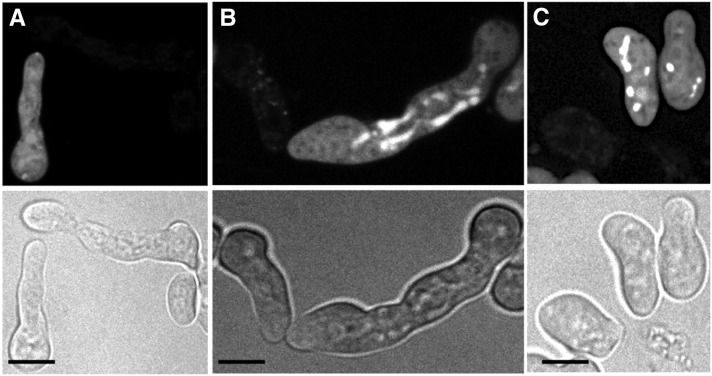
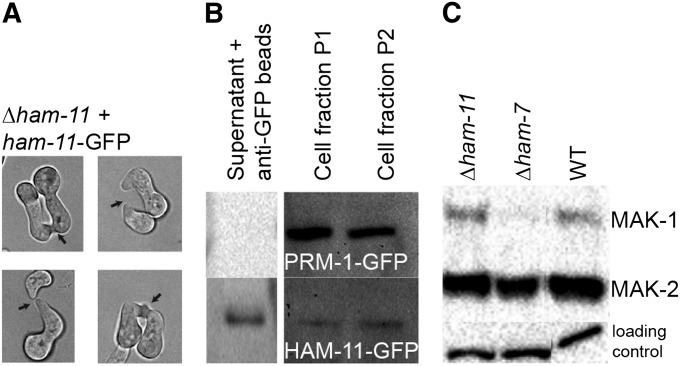
MAK1 and MAK2 phosphorylation, fusion phenotype, and Western blot of Δham-11 complemented with ham-11-gfp. (A) Four germling pairs of Δham-11 ham-11-GFP show chemotrophic growth and cell fusion (see arrows), showing full complementation of the Δham-11 phenotype by the ham-11-gfp construct. (B) Western blot of PRM1-GFP (which localizes to endomembranes and the plasma membrane of N. crassa by fluorescence microscopy) (Fleissner et al. 2009a). HAM11-GFP was detected in supernatant, endomembranes (cell fraction P1), and outer membranes (cell fraction P2). Cell fractionation purity was also assessed using antibodies to the plasma membrane ATPase (data not shown), which localized only to the membrane fractions. (C) Activation of MAK1 and MAK2 in wild type, Δham-11, and Δham-7 germlings. Protein samples from Δham-11 (lane 1), Δham-7 (lane 2), and wild-type (lane 3) 5-hr-old germlings from liquid VMM are shown. Phosphorylated MAK1 (51 kDa) and MAK2 (41 kDa) were detected using anti-phospho p44/42 MAP kinase antibodies (Cell Signaling Technology). (Bottom) Shows equal loading for each lane.
We constructed gfp-tagged alleles of ham-11 driven by the tef-1 promoter to determine localization of HAM11 in cells undergoing chemotropic interactions vs. those that are not communicating. The tef-1 gene is more highly expressed in germlings than ccg-1 and has been used to localize NCR1 and MEK2 to CAT tips (Dettmann et al. 2012). GFP was fused within the ham-11 ORF between amino acids 62 and 63, which lie between the two predicted transmembrane domains. This construct was transformed into a his-3; Δham-11 strain. Complementation was assessed in homokaryotic his-3::ham-11-gfp; Δham-11 progeny via evaluating germling fusion frequencies. In contrast to the Δham-11 strain that lacks germling fusion, germlings of the his-3::ham-11-gfp; Δham-11 strain showed fusion frequencies similar to wild-type + wild-type pairings (Figure 9A). However, although HAM11 could be detected via Western analyses, GFP fluorescence in live cells could not be detected above background.
Because we could detect HAM11 via Western blotting, but not by fluorescence microscopy, we resorted to cell fractionation to determine HAM11 localization. As a control, we utilized PRM1-GFP, which localizes to the plasma membrane and to internal membrane-bound structures (Fleissner et al. 2009a). We evaluated the presence of HAM11 and PRM1 during germling fusion (5-hr time point). From the cell lysate, three fractions were obtained: a resolubilized pellet containing endomembranes, a second resolubilized pellet containing plasma membranes, and a supernatant fraction from which cytoplasmic GFP-protein was immunoprecipitated. Antibodies to the N. crassa plasma membrane ATPase, which localizes to the plasma membrane (Bowman and Bowman 1988), were used to assure proper fractionation (data not shown). As expected, PRM1-GFP was observed in fractions containing membranes (endomembranes and plasma membrane), but not in the cytoplasmic fraction (Figure 9B), consistent with fluorescence mircroscopy results (Fleissner et al. 2009a). HAM11-GFP, however, was found in all three cellular fractions (Figure 9B). These data suggest that HAM11 is a protein that can shuttle between cytoplasm and vesicles/membranes in the cell.
Discussion
The MAK2 MAPK pathway in N. crassa is required for chemotropic interactions and cell fusion during vegetative growth. In this study, we show that one of the predicted targets of the MAK2 pathway, the transcription factor PP1, is also required for chemotropic interactions. This strongly supports the role of PP1 as a target of this MAPK pathway during cell fusion. We subsequently utilized a combination of transcriptional approaches during a time of intensive chemotropic interactions and germling fusion events to identify new components of the cell fusion pathway. When comparing the three data sets (Δpp-1 and Δmak-2Q100G microarrays and Δpp-1 RNA-seq), we identified an overlapping set of seven genes that contained three whose deletion affected germling fusion frequency. These included a previously identified fusion gene of unknown biochemical function (ham-7), which encodes a GPI-anchored protein. Recent data implicate HAM7 in functioning as a cell-wall sensor to regulate activation of the cell-wall integrity MAPK MAK1 (Maddi et al. 2012). Strains carrying a deletion of mak-1 show a pleiotropic phenotype and are also unable to undergo chemotropic interactions and germling fusion (Fu et al. 2011). Of the ∼40 genes that have been previously identified as being required for germling fusion, 15 were identified within our gene set that showed a reduction in expression level in Δpp-1 or mak-2Q100G strains, particularly in the Δpp-1 RNA-seq data set, reflecting the increased power of expression detection over traditional microarray data. In addition to genes that were coordinately regulated in the mak-2Q100G and the Δpp-1 data sets (genes up in both mutants or down in both mutants), genes that showed opposite regulation (increased expression level in one mutant was correlated with decreased expression in the second mutant) were also identified. In S. cerevisiae, Fus3p regulates negative effectors of Ste12p (Dig1p and Dig2p), in addition to direct regulation of Ste12p (Elion et al. 1993; Tedford et al. 1997; Blackwell et al. 2007), with activation of Fus3p being correlated with activation of Ste12p. In N. crassa, MAK2 also shows nuclear localization in germlings (Fleissner et al. 2009b), suggesting that MAK2 may influence activity of transcription factors in addition to PP1. Additional intricacies of PP1 regulation by MAK2 may occur in N. crassa as well as in other filamentous fungi, and gene sets that showed opposite expression patterns in the Δmak-2 mutant vs. the Δpp-1 mutant offer a opportunity to explore this issue.
In addition to their role in germling fusion, it is clear that both PP1 and MAK2 are important in coordinating germination and hyphal growth. This aspect is also true for many of the previously identified fusion mutants, such as Δham-2, Δham-3, Δham-4, and Δham-7 (Xiang et al. 2002; Simonin et al. 2010; Fu et al. 2011), which also have pleiotropic phenotypes. Thus, our data may also be useful in the identification of components important for these processes in filamentous fungi, such as N. crassa. For example, in S. cerevisiae, Ste12 is important for invasive growth in haploid cells, while in diploid cells it plays a role in filamentation that occurs in response to nitrogen starvation (Gustin et al. 1998; Gancedo 2001). It is noteworthy that, in all our transcriptional profiling data sets during germination and colony establishment, the category showing the largest enrichment was that of unclassified proteins. This observation underscores our lack of understanding of developmental processes unique to filamentous fungi.
MAPK pathways orthologous to MAK2 have been implicated in sexual development in filamentous fungi; Δpp-1 and Δmak-2 mutants fail to form female reproductive structures but are fully fertile as males (although both display an ascospore germination defect). Surprisingly, we found a mating-type-specific effect on the expression of the mating pheromones, ccg-4 (A pheromone; up in Δpp-1 A) and mfa-1 (a pheromone; up in Δpp-1 a), and to a lesser extent the mating-type pheromone receptor, pre-2 (A pheromone receptor; up in Δpp-1 a). These data suggest interplay between the A and a mating-type-specific transcription factors believed to regulate pheromones and receptors in N. crassa (A-1 and a-1) and PP1. Pheromones (expressed by male cells) and receptors (expressed by trichogynes undergoing chemotropic interactions toward a male cell) are essential for mating in N. crassa (Kim and Borkovich 2004, 2006). In the closely related homothallic ascomycete fungus, S. macrospora, STE12 binds the mating-type protein SMTA-1, a small serine-threonine protein, SIP2, and the MADS box protein MCM1 during formation of reproductive structures (Nolting and Poggeler 2006). However, deletion of ste12 in S. macrospora does not affect vegetative growth or fruit body development. In S. cerevisiae, Ste12 via interaction with Mcm1 induces a-specific genes in response to pheromone activation, while Mcm1, α1, and Ste12 induce α-specific genes (Errede and Ammerer 1989; Yuan et al. 1993). Further experiments are needed to understand the interplay of PP1, the mating-type-specific transcription factors, pheromones, and receptors during both vegetative and sexual reproduction.
Ste12-like transcription factors are highly conserved among filamentous fungi. A comparison of our transcriptional data set with Ste12 chromatin-immunoprecipitation data from S. cerevisiae (Ren et al. 2000; Borneman et al. 2007; Zheng et al. 2010) identified nine conserved genes, including STE12/pp-1, FUS3/mak-2, PHD1/Asm-1 (NCU01414) (transcriptional activator), PRM1/Prm-1 (plasma membrane merger), ACO1/NCU02366 (aconitase), PGU1/NCU02369 (endopolygalacturonase), CHS1/chs-3 (chitin synthase), HAA1/NCU04830 (transcriptional activator), and PCL2/NCU04847 (cyclin) that were conserved. Of these genes, mutations in three affect germling fusion in N. crassa: pp-1, mak-2, and Prm-1. Interestingly, preliminary data show that a strain carrying a deletion of Asm-1 is also a hyphal fusion mutant (data not shown). An Asm-1 mutant shows a vegetative (“flat”) phenotype, female sterility, and an ascospore maturation defect (Aramayo et al. 1996), phenotypes that are reminiscent of Δpp-1. In S. cerevisiae, the Ste12-binding motif is TGAAACA, although not all target genes identified in S. cerevisiae show conservation of these sites (Ren et al. 2000; Borneman et al. 2007; Zheng et al. 2010). In N. crassa, only between 4–6.5% of the genes identified in the 1NM-PP1 MAK2Q100G and PP-1 RNA-seq experiments have a STE12-like motif present in their promoters at high confidence (data not shown). However, transcription-factor-binding sites are short and often degenerate, making computational detection in promoter regions of genes across an entire genome difficult (Tompa et al. 2005). Future experiments using ChIP-seq analyses would clarify direct target genes of PP1, especially if performed under different growth and developmental scenarios.
Of the seven genes that overlapped from all the expression profiling data sets, a deletion of one of them, ham-11, resulted in a phenotype that is unique among fusion mutants. All fusion mutants identified so far are blind and do not respond to the presence of a wild-type partner. The only reported exception is a N. crassa mutant in a gene identified by genome-wide association studies of germling communication (Palma-Guerrero et al. 2013). In this case, mutations in a predicted secreted protease (spr-7) actually increased fusion frequencies (from ∼86% to ∼97%). However, when the Δspr-7 mutant was mixed with wild-type germlings, fusion frequency was reduced to wild-type levels. In contrast, germlings of Δham-11 showed no fusion at all, but showed normal chemotropic interactions; oscillations of MAK2 and SO and near wild-type levels of cell fusion occur when in proximity to wild-type germlings. These data suggest that HAM11 function is essential for the initiation of chemotropic interactions. As ham-11 was identified as a target of PP1 and the MAK2 pathway, these results suggest a positive feedback mechanism associated with the initiation of signaling. Data showing normal levels of MAK2 phosphorylation in the Δham-11 mutant suggest that the defect is associated with the initiation of MAK2 oscillation to CAT tips and that HAM11 is not involved in the upstream cascade leading to MAK2 activation. Cells of N. crassa do not initiate chemotropic interactions, nor is localization of MAK2 or SO to CAT tips observed when cells are >15 μm apart. Thus, HAM11 might be involved in the regulation of a switch-like transition to signaling behavior when germlings are close enough to each other to initiate chemotropic interactions. HAM11 was not detected in the extracellular fraction (data not shown), but was identified in both the cytoplasmic and membrane fractions, suggesting that it may shuttle between compartments, perhaps depending upon MAK2-signaling status. Supporting this hypothesis is the finding that HAM11 was identified in a phosphoproteomic study to identify MAK2 substrates (A. Leeder and N. L. Glass, unpublished data).
Most of the fusion mutants previously identified affect chemotropic interactions and signaling of MAK2 and SO. However, a strain carrying a deletion of one of the transcriptional targets identified in this study, Prm-1, has normal chemotropic interactions, but is affected in plasma membrane merger (Fleissner et al. 2009a). In S. cerevisiae, PRM1 was identified by assessing selectively expressed genes in response to pheromone treatment (Heiman and Walter 2000) and is a direct target of Ste12 (Ren et al. 2000); prm1Δ mutants are also affected in plasma membrane merger. It is possible that genes encoding additional proteins associated with cell-wall breakdown, plasma membrane, and trafficking can be identified in our data sets. Such proteins that are involved in trafficking and late fusion events may not have been identified by our assays for chemotropic interactions and fusion frequency; ΔPrm-1 mutants show normal chemotropic interactions and oscillation of MAK2 and SO, but have a ∼50% reduction in plasma membrane merger (Fleissner et al. 2009a). In our data sets, we identified a number of genes dependent on PP1 for expression that encode proteins involved in vesicle trafficking, such as a homolog to S. cerevisiae SCN2 (NCU00566), which encodes a v-SNARE involved in the fusion of Golgi-derived secretory vesicles with the plasma membrane. Mutations in a gene encoding a homolog to calcium neuronal sensor-1 (NCU04379; cse-1)—which localizes to the Golgi and is involved in regulated secretion in neurons—reduce, but do not abolish, germling fusion frequency (Palma-Guerrero et al. 2013). Fusion frequencies are not restored when Δcse-1 germlings are paired with wild-type germlings. This phenotype is similar to that of Δham-12 mutants, where no differences in fusion frequencies were observed between Δham-12 + Δham-12 germlings and Δham-12 + wild-type germlings. In the future, epistasis and colocalization studies will provide additional clues for the function of these proteins during the processes of germling communication, chemotropic interactions, and cell fusion.
Supplementary Material
Acknowledgments
We thank Nick Salem for assistance with early screening of the mutants; David Kowbel for assistance with RNA-seq experiments; and Ken Allen (Yale Medical School) for kindly providing us with the PMA-1 antibody. We acknowledge use of material generated by a National Institutes of Health P01 GM068087 grant entitled “Functional Analysis of a Model Filamentous Fungus.” The work in this study was funded by a National Science Foundation grant to N.L.G. (MCB 1121311).
Footnotes
Communicating editor: E. U. Selker
Literature Cited
- Aguilar P. S., Baylies M. K., Fleissner A., Helming L., Inoue N., et al. , 2013. Genetic basis of cell-cell fusion mechanisms. Trends Genet. 29: 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldabbous M. S., Roca M. G., Stout A., Huang I. C., Read N. D., et al. , 2010. The ham-5, rcm-1 and rco-1 genes regulate hyphal fusion in Neurospora crassa. Microbiology 156: 2621–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alspaugh J. A., Perfect J. R., Heitman J., 1998. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet. Biol. 25: 1–14 [DOI] [PubMed] [Google Scholar]
- Aramayo R., Peleg Y., Addison R., Metzenberg R., 1996. Asm-1(+), a Neurospora crassa gene related to transcriptional regulators of fungal development. Genetics 144: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L., 2005. A walk-through of the yeast mating pheromone response pathway. Peptides 26: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram O., Bayram O. S., Ahmed Y. L., Maruyama J., Valerius O., et al. , 2012. The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism. PLoS Genet. 8: e1002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell E., Kim H. J., Stone D. E., 2007. The pheromone-induced nuclear accumulation of the Fus3 MAPK in yeast depends on its phosphorylation state and on Dig1 and Dig2. BMC Cell Biol. 8: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Hynes M. J., Andrianopoulos A., 2001. An STE12 homolog from the asexual, dimorphic fungus Penicillium marneffei complements the defect in sexual development of an Aspergillus nidulans steA mutant. Genetics 157: 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Gianoulis T. A., Zhang Z. D., Yu H., Rozowsky J., et al. , 2007. Divergence of transcription factor binding sites across related yeast species. Science 317: 815–819 [DOI] [PubMed] [Google Scholar]
- Bowman E. J., Bowman B. J., 1988. Purification of vacuolar membranes, mitochondria, and plasma membranes from Neurospora crassa and modes of discriminating among the different H+-ATPases. Methods Enzymol. 157: 562–573 [DOI] [PubMed] [Google Scholar]
- Bowman S. M., Piwowar A., Al Dabbous M. e., Vierula J., Free S. J., 2006. Mutational analysis of the glycosylphosphatidylinositol (GPI) anchor pathway demonstrates that GPI-anchored proteins are required for cell wall biogenesis and normal hyphal growth in Neurospora crassa. Eukaryot. Cell 5: 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Dominguez N., Alvarez-Delfin K., Hansberg W., Aguirre J., 2008. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot. Cell 7: 1352–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Wright L. C., Tscharke R. L., Sorrell T. C., Wilson C. F., et al. , 2004. Regulatory roles for the homeodomain and C2H2 zinc finger regions of Cryptococcus neoformans Ste12alphap. Mol. Microbiol. 53: 1385–1396 [DOI] [PubMed] [Google Scholar]
- Chen E. H., Grote E., Mohler W., Vignery A., 2007. Cell-cell fusion. FEBS Lett. 581: 2181–2193 [DOI] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon M. J., Imoto S., Nolan J., Miyano S., 2004. Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Dettmann A., Illgen J., Marz S., Schurg T., Fleissner A., et al. , 2012. The NDR kinase scaffold HYM1/MO25 is essential for MAK2 map kinase signaling in Neurospora crassa. PLoS Genet. 8: e1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J. C., Borkovich K. A., Henn M. R., Turner G. E., Sachs M. S., et al. , 2007. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv. Genet. 57: 49–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Satterberg B., Kranz J. E., 1993. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol. Biol. Cell 4: 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison C., Hall C., Kowbel D., Welch J., Brem R., et al. , 2011. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc. Natl. Acad. Sci. USA 7: 2831–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Ammerer G., 1989. STE12, a protein involved in cell-type-specific transcription and signal transduction is yeast, is part of protein-DNA complexes. Genes Dev. 3: 1349–1361 [DOI] [PubMed] [Google Scholar]
- Fleissner A., Sarkar S., Jacobson D. J., Roca M. G., Read N. D., et al. , 2005. The so locus is required for vegetative cell fusion and postfertilization events in Neurospora crassa. Eukaryot. Cell 4: 920–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner A., Simonin A. R., Glass N. L., 2008. Cell fusion in the filamentous fungus, Neurospora crassa. Methods Mol. Biol. 475: 21–38 [DOI] [PubMed] [Google Scholar]
- Fleissner A., Diamond S., Glass N. L., 2009a The Saccharomyces cerevisiae PRM1 homolog in Neurospora crassa is involved in vegetative and sexual cell fusion events but also has postfertilization functions. Genetics 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner A., Leeder A. C., Roca M. G., Read N. D., Glass N. L., 2009b Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc. Natl. Acad. Sci. USA 106: 19387–19392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M., Hickey P. C., Raju N. B., Selker E. U., Read N. D., 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41: 897–910 [DOI] [PubMed] [Google Scholar]
- Fu C., Iyer P., Herkal A., Abdullah J., Stout A., et al. , 2011. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot. Cell 10: 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J. M., 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25: 107–123 [DOI] [PubMed] [Google Scholar]
- Gustin M. C., Albertyn J., Alexander M., Davenport K., 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62: 1264–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M. G., Walter P., 2000. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 151: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Xue C., Peng Y., Katan T., Kistler H. C., et al. , 2002. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 15: 1119–1127 [DOI] [PubMed] [Google Scholar]
- Hutchison E., Brown S., Tian C., Glass N. L., 2009. Transcriptional profiling and functional analysis of heterokaryon incompatibility in Neurospora crassa reveals that reactive oxygen species, but not metacaspases, are associated with programmed cell death. Microbiology 155: 3957–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G., Chua N. H., 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2: 226–231 [DOI] [PubMed] [Google Scholar]
- Jun S. C., Lee S. J., Park H. J., Kang J. Y., Leem Y. E., et al. , 2011. The MpkB MAP kinase plays a role in post-karyogamy processes as well as in hyphal anastomosis during sexual development in Aspergillus nidulans. J. Microbiol. 49: 418–430 [DOI] [PubMed] [Google Scholar]
- Kim H., Borkovich K., 2004. A pheromone receptor gene, pre-1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol. Microbiol. 52: 1781–1798 [DOI] [PubMed] [Google Scholar]
- Kim H., Borkovich K. A., 2006. Pheromones are essential for male fertility and sufficient to direct chemotropic polarized growth of trichogynes during mating in Neurospora crassa. Eukaryot. Cell 5: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Bobrowicz P., Wilkinson H. H., Ebbole D. J., 2005. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics 170: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddi A., Bowman S. M., Free S. J., 2009. Trifluoromethanesulfonic acid-based proteomic analysis of cell wall and secreted proteins of the ascomycetous fungi Neurospora crassa and Candida albicans. Fungal Genet. Biol. 46: 768–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddi A., Dettman A., Fu C., Seiler S., Free S. J., 2012. WSC-1 and HAM-7 are MAK-1 MAP kinase pathway sensors required for cell wall integrity and hyphal fusion in Neurospora crassa. PLoS ONE 7: e42374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H. D., Fink G. R., 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275: 1314–1317 [DOI] [PubMed] [Google Scholar]
- Maerz S., Dettmann A., Ziv C., Liu Y., Valerius O., et al. , 2009. Two NDR kinase-MOB complexes function as distinct modules during septum formation and tip extension in Neurospora crassa. Mol. Microbiol. 74: 707–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin B. S., Freitag M., Selker E. U., 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44: 34–36 [Google Scholar]
- McCluskey K., 2003. The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52: 245–262 [DOI] [PubMed] [Google Scholar]
- McNally M. T., Free S. J., 1988. Isolation and characterization of a Neurospora glucose-repressible gene. Curr. Genet. 14: 545–551 [DOI] [PubMed] [Google Scholar]
- Meiklejohn C. D., Townsend J. P., 2005. A Bayesian method for analysing spotted microarray data. Brief. Bioinform. 6: 318–330 [DOI] [PubMed] [Google Scholar]
- Nolting N., Poggeler S., 2006. A STE12 homologue of the homothallic ascomycete Sordaria macrospora interacts with the MADS box protein MCM1 and is required for ascosporogenesis. Mol. Microbiol. 62: 853–868 [DOI] [PubMed] [Google Scholar]
- Palma-Guerrero J., Hall C. R., Kowbel D., Welch J., Taylor J. W., et al. , 2013. Genome wide association identifies novel loci involved in fungal communication. PLoS Genet. 9: e1003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Roca M. G., Read N. D., Glass N. L., 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3: 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit A., Maheshwari R., 1994. A simple method of obtaining pure microconidia in Neurospora crassa. Fungal Genet. Newsl. 41: 64–65 [Google Scholar]
- Park G., Xue G. Y., Zheng L., Lam S., Xu J. R., 2002. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 15: 183–192 [DOI] [PubMed] [Google Scholar]
- Park G., Bruno K. S., Staiger C. J., Talbot N. J., Xu J. R., 2004. Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol. Microbiol. 53: 1695–1707 [DOI] [PubMed] [Google Scholar]
- Primig M., Winkler H., Ammerer G., 1991. The DNA binding and oligomerization domain of MCM1 is sufficient for its interaction with other regulatory proteins. EMBO J. 10: 4209–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read N. D., Fleissner A., Roca M. G., Glass N. L., 2010. Hyphal fusion, pp. 260–273 in Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. ASM Press, Washington, DC [Google Scholar]
- Read N. D., Goryachev A. B., Lichius A., 2012. The mechanistic basis of self-fusion between conidial anastomosis tubes during fungal colony initiation. Fungal Biol. Rev. 26: 1–11 [Google Scholar]
- Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., et al. , 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306–2309 [DOI] [PubMed] [Google Scholar]
- Ren P., Springer D. J., Behr M. J., Samsonoff W. A., Chaturvedi S., et al. , 2006. Transcription factor STE12alpha has distinct roles in morphogenesis, virulence, and ecological fitness of the primary pathogenic yeast Cryptococcus gattii. Eukaryot. Cell 5: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispail N., Di Pietro A., 2010. The homeodomain transcription factor Ste12: connecting fungal MAPK signalling to plant pathogenicity. Commun. Integr. Biol. 3: 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Trapnell C., Donaghey J., Rinn J. L., Pachter L., 2011. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca G. M., Read N. D., Wheals A. E., 2005a Conidial anastomosis tubes in filamentous fungi. FEMS Microbiol. Lett. 249: 191–198 [DOI] [PubMed] [Google Scholar]
- Roca M., Arlt J., Jeffree C., Read N., 2005b Cell biology of conidial anastomosis tubes in Neurospora crassa. Eukaryot. Cell 4: 911–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman E., Arana D. M., Nombela C., Alonso-Monge R., Pla J., 2007. MAP kinase pathways as regulators of fungal virulence. Trends Microbiol. 15: 181–190 [DOI] [PubMed] [Google Scholar]
- Roper M., Simonin A., Hickey P. C., Leeder A., Glass N. L., 2013. Nuclear dynamics in a fungal chimera. Proc. Natl. Acad. Sci. USA 110: 12875–12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A., Zollner A., Maier D., Albermann K., Hani J., et al. , 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32: 5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin A. R., Rasmussen C. G., Yang M., Glass N. L., 2010. Genes encoding a striatin-like protein (ham-3) and a forkhead associated protein (ham-4) are required for hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 47: 855–868 [DOI] [PubMed] [Google Scholar]
- Simonin A., Palma-Guerrero J., Fricker M., Glass N. L., 2012. Physiological significance of network organization in fungi. Eukaryot. Cell 11: 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedford K., Kim S., Sa D., Stevens K., Tyers M., 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7: 228–238 [DOI] [PubMed] [Google Scholar]
- Tian C., Kasuga T., Sachs M. S., Glass N. L., 2007. Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot. Cell 6: 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollot M., Wong Sak Hoi J., van Tuinen D., Arnould C., Chatagnier O., et al. , 2009. An STE12 gene identified in the mycorrhizal fungus Glomus intraradices restores infectivity of a hemibiotrophic plant pathogen. New Phytol. 181: 693–707 [DOI] [PubMed] [Google Scholar]
- Tompa M., Li N., Bailey T. L., Church G. M., De Moor B., et al. , 2005. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 23: 137–144 [DOI] [PubMed] [Google Scholar]
- Tsuji G., Fujii S., Tsuge S., Shiraishi T., Kubo Y., 2003. The Colletotrichum lagenarium Ste12-like gene CST1 is essential for appressorium penetration. Mol. Plant Microbe Interact. 16: 315–325 [DOI] [PubMed] [Google Scholar]
- Vallim M. A., Miller K. Y., Miller B. L., 2000. Aspergillus SteA (sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol. Microbiol. 36: 290–301 [DOI] [PubMed] [Google Scholar]
- Vogel H. J., 1956. A convenient growth medium for Neurospora. Microbiol. Genet. Bull. 13: 42–46 [Google Scholar]
- Wei H., Requena N., Fischer R., 2003. The MAPKK kinase SteC regulates conidiophore morphology and is essential for heterokaryon formation and sexual development in the homothallic fungus Aspergillus nidulans. Mol. Microbiol. 47: 1577–1588 [DOI] [PubMed] [Google Scholar]
- Westergaard M., Mitchell H. K., 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573–577 [Google Scholar]
- Wong Sak Hoi J., Dumas B., 2010. Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot. Cell 9: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q., Rasmussen C., Glass N. L., 2002. The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160: 169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y. L., Fields S., 1991. Properties of the DNA-binding domain of the Saccharomyces cerevisiae STE12 protein. Mol. Cell. Biol. 11: 5910–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y. O., Stroke I. L., Fields S., 1993. Coupling of cell identity to signal response in yeast: interaction between the alpha 1 and STE12 proteins. Genes Dev. 7: 1584–1597 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Simon I., Harbison C. T., Hannett N. M., Volkert T. L., et al. , 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113: 395–404 [DOI] [PubMed] [Google Scholar]
- Zheng W., Zhao H., Mancera E., Steinmetz L. M., Snyder M., 2010. Genetic analysis of variation in transcription factor binding in yeast. Nature 464: 1187–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.