Abstract
This study addresses the question of how purifying selection operates during recent rapid population growth such as has been experienced by human populations. This is not a straightforward problem because the human population is not at equilibrium: population genetics predicts that, on the one hand, the efficacy of natural selection increases as population size increases, eliminating ever more weakly deleterious variants; on the other hand, a larger number of deleterious mutations will be introduced into the population and will be more likely to increase in their number of copies as the population grows. To understand how patterns of human genetic variation have been shaped by the interaction of natural selection and population growth, we examined the trajectories of mutations with varying selection coefficients, using computer simulations. We observed that while population growth dramatically increases the number of deleterious segregating sites in the population, it only mildly increases the number carried by each individual. Our simulations also show an increased efficacy of natural selection, reflected in a higher fraction of deleterious mutations eliminated at each generation and a more efficient elimination of the most deleterious ones. As a consequence, while each individual carries a larger number of deleterious alleles than expected in the absence of growth, the average selection coefficient of each segregating allele is less deleterious. Combined, our results suggest that the genetic risk of complex diseases in growing populations might be distributed across a larger number of more weakly deleterious rare variants.
Keywords: purifying selection, exponential growth, deleterious mutations, demographic history, human
THE human population size has been growing rapidly, most notably since the advent of agriculture ∼10,000 years ago. The rate of population growth has increased over time to as much as 10–30% per generation during the last 500 years (Cohen 1996; Hawks et al. 2007; United Nations Department of Economic and Social Affairs Population Division 2011; Keinan and Clark 2012). This recent demographic growth is reflected in recent estimates from genetic data of the human current effective population size (Ne), with all estimates being much higher than the conventionally estimated historical variance effective population size of ∼10,000 (Coventry et al. 2010; Keinan and Clark 2012; Nelson et al. 2012; Tennessen et al. 2012). The growth in effective population size has resulted in an excess of rare alleles due to the very large number of recent mutations (Coventry et al. 2010; Fu et al. 2012; Keinan and Clark 2012; Nelson et al. 2012; Tennessen et al. 2012). Not only are the vast majority of protein-coding variants rare and recent, but also both the group of rarer and the group of more recent variants are enriched for deleterious mutations (Fu et al. 2012; Nelson et al. 2012; Tennessen et al. 2012). The extent to which recent population expansion has contributed to these empirical observations depends on how purifying selection has operated during the epoch of growth. Growth can potentially increase the number of deleterious alleles carried by each individual and the burden of disease-causing alleles in the human population.
It is crucial to understand the interaction of demographic expansion and natural selection, and in particular purifying (negative) selection, to assess whether the recent explosive growth has affected the genetic basis and architecture of complex disease. Several theoretical predictions of natural selection in a population of varying size have been formulated, but there is no adequate coverage of the situation where a population started growing only recently and is far from reaching a new mutation–selection–drift equilibrium. On one hand, it has been suggested that rapid growth of human populations may have led natural selection to be inefficient at removing deleterious mutations (Casals and Bertranpetit 2012), which could lead to a rapid accumulation of deleterious alleles (Kondrashov 1988; Lynch 2010). On the other hand, since an increase in Ne increases the efficacy of natural selection to raise the frequency of favorable mutations and reduce the frequency of deleterious mutations (Kimura 1955, 1957), it has also been suggested that with the current human Ne, even slightly deleterious mutations can be purged (Reed and Aquadro 2006). Indeed, for a population that has been exponentially growing throughout its history, it has been established that deleterious mutations have a lower probability of fixation compared to those in a nongrowing population, and most deleterious mutations will eventually be lost (Waxman 2011). However, for mutations that reach fixation, the expected time until fixation (conditional upon fixation) increases with population size (Kimura and Ohta 1969; Muruyama and Kimura 1974; Waxman 2012). Thus, weakly deleterious mutations can segregate longer, and increase in their number of copies in the population.
In this study, we examine the characteristics of deleterious mutations in a scenario of recent explosive growth of approximately the magnitude experienced by human populations, with the aim of answering three questions. First, is the population growth expected to lead to an increase in the number of deleterious mutations carried by each individual in the extant, large population? Second, how does the growth affect the purging of rare deleterious derived alleles? And third, has growth affected the average selection coefficient of segregating variants and, as a consequence, the average individual fitness?
Due to the limitations of theoretical results that assume equilibrium, we addressed these questions, using forward-in-time simulations that track the evolution of newly arisen mutations with known selective effects over time. Importantly, using simulations allowed the comparison of models with and without recent growth, as well as with and without purifying selection, thereby revealing how population growth alone has altered the efficacy of purifying selection.
The main results of the simulations are that population growth largely increases the number of segregating sites in the population and slightly increases the average number of mutations carried by each individual, including that of deleterious mutations. Studying the relationship between the number of copies of a derived allele and its selection coefficient, our results indicate that the increased efficacy of natural selection in a growing population results in a faster elimination of newly introduced mutations that are strongly deleterious. As a consequence, while each individual in a growing population carries a larger number of deleterious variants, the average selection coefficient of each variant copy is less deleterious than in a population that has remained at constant size in recent history. Finally, we discuss the implications of these results on the overall individual fitness in extant human populations and on the genetic architecture of complex diseases.
Materials and Methods
Simulated loci
We performed forward-in-time population genetic simulations using the program SFS_CODE (Hernandez 2008), including a few minor revisions to its source code. We simulated two types of loci, one evolving neutrally and one evolving under purifying selection. For each type of locus, we considered two demographic scenarios, one that includes recent population growth and one without growth. For each of these four locus-by-demography combinations, we simulated genetic sequences of 5 kb with a mutation rate of 2 × 10−8 per nucleotide. The value chosen for the mutation rate is higher than recent estimates (Campbell et al. 2012; Keightley 2012), but does not affect our conclusions since these are based on comparing the different simulated combinations. For computational efficiency, each individual 5-kb locus is in complete linkage, with no recombination. We simulated 10,000 independent replicates of each of the four models, and considered summary statistics in each replicate and then averaged them across the 10,000 replicates. Relevant SFS_CODE command lines are provided in Supporting Information, File S1.
Models of population history
The two demographic scenarios (with growth and without growth) occur in two distinct populations that split from a common ancestral population. At the split time, SFS_CODE makes a copy of the ancestral population such that each new population conserves the full ancestral effective population size of Ne = 10,000 (throughout, quantities denote effective population sizes). This ensures that both populations start in identical states. Prior to the population split, the ancestral population follows a demographic model of European history with two population bottlenecks, as described in Keinan et al. (2007). Specifically, after the burn-in phase, the population undergoes a first bottleneck of intensity F = 0.264 at 4720 generations ago followed by a quick recovery to the ancestral population size of Ne = 10,000. The second bottleneck occurs 720 generations ago with an intensity of F = 0.09, also followed by a quick recovery to the ancestral population size (Keinan et al. 2007). Then, 420 generations before present the ancestral population splits into two populations. One population (referred to as the “with growth” population) starts growing exponentially 400 generations before present (∼10,000 years ago) at a rate of 1.74% per generation thereby reaching a final size of Ne = 10,000,000 at the end of the simulation (Keinan and Clark 2012). The other population (the “no growth” population) maintains a constant Ne = 10,000 from the split until the present. The with growth and no growth models are therefore identical over their entire history except for the last 400 generations (Figure S1). We followed mutations during the last 440 generations of the simulation, covering the entire exponential growth phase and the 40 preceding generations when the two models are still identical.
We considered additional models of population history, including varying recent growth and ancient history, as well as a baseline of a population that has been of constant size throughout history (File S1). These additional models show that all results presented throughout the study are robust to the details of ancestral model of European history (Keinan et al. 2007) and the recent explosive growth model (File S1). In all models we considered a generation time of 25 years.
Mutations and natural selection
For each model of population history, we simulated loci in which either all mutations are neutral (selection coefficient s = 0) or all mutations are deleterious (s < 0). For each deleterious mutation, s was obtained from the population-scaled selection coefficient, γ = 2Ns with γ following the opposite of a gamma distribution, (α, β). We chose the shape parameter α = 0.206 and rate parameter β = 1/2740 from Boyko et al. (2008), after rescaling to an ancestral Ne of 10,000. With these parameter values, the average s was −0.028. We simulated all loci as “noncoding” in SFS_CODE nomenclature since “coding” regions assume no fitness effect (s = 0) at approximately one-third of sites. Mutations with s < −1were set to s = −1.
Following the implementation in SFS_CODE (Hernandez 2008), the fitness of each individual is the product of the fitness effect of the mutations it carries, which for a mutation with selection coefficient s (s < 0 for deleterious mutations) is 1 + s in heterozygotes and (1 + s)2 in homozygotes. The selection model implemented in SFS_CODE is a model of shift in fitness, such that the selection coefficient of alleles that reach fixation is reset to 1. Throughout, we therefore ignored mutations that reach fixation. The use of a shift in fitness model has a minimal effect on the results since fixations are extremely rare (<0.3% of the mutations) during the 400 generations followed here. In addition, this model has recently been shown to be a realistic fit to human mutation load (Keightley et al. 2011; Lesecque et al. 2012). We refer to derived alleles (i.e., the new alleles introduced by the mutation process) as lost from the population when they reach 0 copies.
Simulation scaling
For computational efficiency, and since the extant effective population size following growth is very large (10,000,000), we scaled down both the effective population size and the time by a factor of 10. This scaling approach has been shown to lead to little change in resulting patterns of variation (Hernandez 2008). While scaling does not broadly alter allele frequencies, it does affect the nominal number of copies of each allele. For example, the simulated ancestral population size is 1000 individuals, in which a singleton (allele appearing in a single copy) is of frequency 0.05%, while a singleton in a population of 10,000 individuals is of frequency 0.005%. For this reason, we do not make direct quantitative inference on the distribution of rare variants in the real-sized human population, but rather focus all analyses and conclusions on a comparison of the models with and without growth.
Analyses and summary statistics
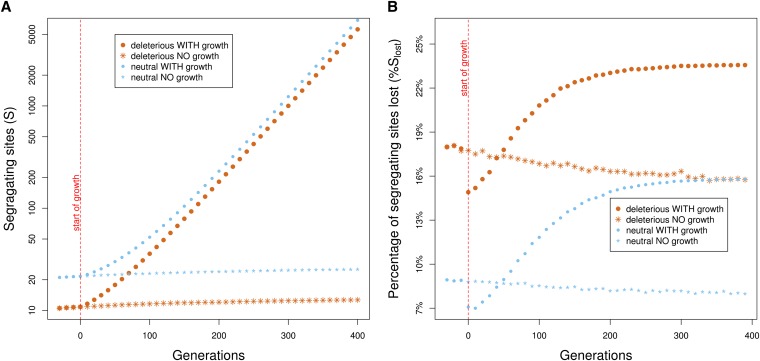
At each generation, we recorded the number of segregating sites, the number of copies of the derived allele at each segregating site, and their selection coefficients. We calculated and averaged the following quantities for each simulated scenario. S is the number of segregating sites, i.e., genomic positions that carry one or more copies of a derived allele (Figure 1A). The derived allele count (DAC) is the number of copies of the derived allele of a segregating site, which we denote by δi for site i. The proportion of segregating sites lost, , is given by , where R is the number of segregating sites lost (Figure 1B). The proportion of segregating sites lost is also computed within different categories of DAC, i.e., for different values of DAC k =1, . . . , 6 (Figure 2). The fraction of derived alleles at lost sites, , is given by . We partitioned deleterious segregating sites into three categories based on their selection coefficient s: “very deleterious” ([−1, −0.01]), “mildly deleterious” ((−0.01, −0.0001]), and “nearly neutral” ((−0.0001, 0]). The percentage of derived alleles in each of these categories is given by × 100, where Sk is the total number of segregating sites in category k (Figure 3). The average fitness effect across copies of derived alleles, wDA, is given by , where is the selection coefficient of the derived allele at site i (Figure 4). The average number of mutations per individual chromosome is calculated as L = , where Ne is the simulated (scaled) number of individuals (Figure 5).
Figure 1.
Population growth increases the number of segregating sites, but also the fraction of sites that are lost. (A) S, the number of segregating sites of the whole population (on a log scale); (B) %Slost, the percentage of segregating sites lost from the population in a single simulated generation (Materials and Methods). Both panels present the two simulated demographic scenarios (with growth and with no growth) for each selection model (neutral or deleterious). Results are presented every 10 generations (corresponding to a single simulated generation) during the last 440 generations. Population growth increases both S and %Slost. S is smaller for deleterious than for neutral mutations, while %Slost is higher. Trends with time in the models without growth are due to the preceding population bottlenecks (Figure S3).
Figure 2.
Rare variants are less likely to be lost during population growth, but deleterious ones are purged more efficiently. (A) Percentage of sites of a given derived allele count (DAC) that are lost (%Slost) in a single simulated generation. For example, at a neutral locus in the scenario without growth, just over 36% of all the singletons in the population are not observed in the next generation, with the other 64% being transmitted (with any number of copies) to the next generation. Note the different scaling of the y-axis in the different panels, which also explains the noisier trends as DAC increases. (B) For each demographic scenario, the same data as in A are presented as the ratio of %Slost at the deleterious over %Slost at the neutral loci. Population growth increases this ratio, which reflects the higher efficacy of natural selection.
Figure 3.
Higher efficiency of natural selection in a growing population decreases the percentage of the most deleterious allele copies. Segregating sites are classified into three discrete categories of fitness effect. For each category, the percentage of derived alleles (%DA) is the sum of the number of copies of derived alleles observed across all the segregating site in the category divided by the total number of derived alleles across all segregating sites in the population × 100 (%DA of the three categories sums up to 100). Data are shown for the last generation of the simulation, both with and without recent growth. Vertical bars denote ±SE based on 10,000 replicates. Population growth leads to a lower percentage of derived alleles in the most deleterious category.
Figure 4.
The average selection coefficient across alleles present in the population is increased by population growth. The average selection coefficient of a derived allele (wDA) is obtained by weighting the selection coefficient of each segregating site by its number of copies (Materials and Methods). The increase in average selection coefficient of derived alleles shows that alleles are on average less deleterious over time. The increase in the population model without growth is due to the preceding population bottlenecks (File S1). The increase is faster for the model with population growth.
Figure 5.
The number of mutations carried by each individual chromosome is higher in a growing population. The number of mutations per chromosome L (Materials and Methods) is presented for neutral and deleterious loci. L increases slowly as the population grows: L increases by only 1.14% and 1.03% at the neutral and the deleterious locus, respectively, between the beginning and the end of the growth.
Results
Accumulation and loss of segregating sites
We first measured the accumulation of mutations in the population by tabulating S, the number of segregating sites. Since our simulations assume an infinite-sites model, each new mutation introduces a new segregating site. As expected (Watterson 1975; Tajima 1989), S increases rapidly over time as the population grows, culminating in over two orders of magnitude increase following 400 generations of growth, both with and without selection (Figure 1A). For both demographic scenarios, loci with deleterious mutations have on average fewer segregating sites than loci with solely neutral mutations (Figure 1A), as expected by purifying selection (e.g., Przeworski et al. 1999), but the relative difference between the two becomes smaller as the population grows (Figure 1A).
To further investigate the effect of genetic drift and natural selection on the number of segregating sites under population growth, we estimated at each generation the percentage of segregating sites that are not observed in the next generation (%Slost). After a few generations of mutation accumulation, %Slost becomes higher for the model with population growth, both for neutral and for deleterious loci (Figure 1B), implying that population growth increases not only the number of segregating sites, but also the rate at which they are lost. This phenomenon is explained by the larger fraction of singletons (Figure S2) and very rare variants in the growing population, which have a higher probability of loss (Figure 2).
Derived allele count of segregating sites
Each segregating site in the population can be categorized by the number of sequences that carry the derived allele. The average DAC per segregating site (see Materials and Methods) is a measure of the prevalence of those sites in the population. This measure is pertinent when comparing populations of different sizes since allele frequencies are difficult to interpret as the sample size (which is here the population size) increases in the population with growth. Furthermore, it is the allele count, rather than the allele frequency, that affects its probability of loss or transmission (File S1).
Our simulations reproduce a well-established effect of population growth (Slatkin and Hudson 1991; Wakeley 2008; Coventry et al. 2010; Keinan and Clark 2012; Nelson et al. 2012; Tennessen et al. 2012) by showing an increase in the proportion of singletons (sites with DAC = 1) (Figure S2). The proportion is further elevated at deleterious loci for both population models (Figure S2). To investigate the efficacy of purifying selection in a growing population free of the expected skew in the site frequency spectrum, we consider instead the DAC of lost segregating sites.
We computed the percentage of lost sites within each category of DAC, with %Slost for DAC = k being the percentage of segregating sites with k derived alleles that are lost within one generation (Materials and Methods). In contrast to the increase of %Slost when considered across all DACs (Figure 1B), we observe that within each DAC category, population growth decreases %Slost (Figure 2A), both for neutral (36.7% to ∼30.4%) and for deleterious loci (∼44.1% to ∼39.7% for singletons). This differential direction (Figure 1B vs. Figure 2A) is due to the greater percentage of variants with low DAC in the growing population. For example, singletons represent 46.8% of all segregating sites under growth with neutral mutations and only 17.9% in the same scenario without growth (Figure S2). As such, the percentage of variants that are singleton and lost (without conditioning on being a singleton) from all segregating sites is higher in the growing population than in the scenario without growth (30.4% × 46.8% = 14.2% vs. 36.7% × 17.9% = 6.6%, respectively).
In addition to the decrease in %Slost in each DAC, we also observe that %Slost is always higher at the loci under selection than at the neutral loci. Both these results follow expectations of the action of population growth and selection, respectively, but interestingly the increase in %Slost due to selection is proportionally higher in the growing population (Figure 2B). For singletons for instance, the proportion of segregating sites lost is 1.3 times higher under selection with growth, while it is only 1.2 times higher under selection without growth (Figure 2B). These results show that purifying selection further facilitates the purging of deleterious sites in a growing population.
Segregating sites that are lost are overwhelmingly sites with low DACs. Specifically, among all segregating sites that are lost in a given generation, singletons and doubletons make up >95% in both population models (Figure S4). Sites with DAC > 10 represent between 0% and 3.3 × 10−5% of the lost sites at deleterious loci for the model without and with growth, respectively. Hence, lost segregating sites represent only a small fraction of all the copies of derived alleles present in the population. While ∼16% of segregating sites are lost at the neutral locus with growth (Figure 1B), these sites compose only 0.1% of the total number of copies of derived alleles in the population (%DAlost, Materials and Methods; Figure S5). At a locus under purifying selection, %DAlost is an order of magnitude higher in both demographic models (Figure S5), thus showing the contribution of natural selection in the process of allele loss and removal.
In summary, by carefully considering the DAC at lost sites, we have shown that the action of natural selection is not invalidated under population growth. In addition, although it decreases the proportion of segregating sites lost in each DAC category, the larger size of the growing population improves the efficacy of natural selection, and deleterious sites are more readily eliminated.
Fitness effect of deleterious alleles in a growing population
Average fitness effect of a deleterious mutation:
To go beyond the burden in the number of deleterious mutations and consider their effects, we compared the distribution of selection coefficients in the population models with and without growth. We computed the average fitness effect (selection coefficient) for derived alleles that are lost and derived alleles that are transmitted to the next generation by averaging the fitness effect of each allele weighed by its number of copies (Materials and Methods). As expected, in both demographic scenarios, lost sites are much more deleterious than sites that are transmitted (Figure S6). Interestingly, this phenomenon is more pronounced in a growing population, again pointing to the higher efficacy of selection in a larger population (Figure S6).
To obtain a snapshot of the fitness effect of all segregating variation (i.e., independently of whether sites are lost or transmitted to the following generation), we partitioned segregating sites into three categories corresponding to very deleterious, mildly deleterious, and nearly neutral (Materials and Methods). Considering the number of copies of each site, we measured the percentage of copies of derived alleles (%DA) that fall into each category. In the very deleterious category, %DA decreases progressively over time as the population grows (Figure S7). At the last generation of the simulation, %DA is significantly lower (by 8.4%) than in the model without growth (Figure 3). The effect of the population model on the other two categories is much smaller and nonsignificant (Figure S7 and Figure 3). The stronger effect of population growth on the most deleterious alleles is also visible in the site frequency spectrum (Figure S8). More generally, the selection coefficient s averaged across all derived allele copies (wDA) becomes less deleterious as the population grows (Figure 4). At the end of the simulation, an allele chosen randomly is 15.8% less deleterious in the population that has undergone growth (Figure 4). We note that the average selection coefficient also increases—although to a smaller extent—in the absence of growth (Figure 4). This is because the population model without growth is also not at equilibrium due to the preceding population bottlenecks. The role of the bottlenecks becomes evident in comparison to a model of a population that has been of constant size throughout history (File S1; Figure S9).
Despite the accumulation of deleterious segregating sites in the growing population, we show a stronger increase in the average fitness effect of derived alleles in the growing population (Figure 4). This effect is particularly evident when considering the relative amount of very deleterious alleles. In the scenario with growth, for one copy of a very deleterious allele there are 130 copies of nearly neutral ones; the respective is only 1 to 48 (Table S1A). The new mutations accumulated due to growth tend to be more deleterious due to their recency, but while less deleterious alleles increase in number of copies faster as the population grows, the very deleterious alleles are purged more effectively in this scenario.
Average number of mutations per chromosome:
We next considered the burden of deleterious mutations as the number of mutations present in each of the 2Ne chromosomes in the population. As expected, the average number of mutations per chromosome, L, is much larger at the neutral loci than at the deleterious loci (Figure 5). L is also larger—both with and without selection—in the growing population (Figure 5). This increase in L is steady over the generations of population growth, but in stark contrast to the several orders of magnitude increase of S (Figure 1A), L increases by only 0.9% relative to the model without growth at the neutral locus, and by only 6% at the deleterious locus (Figure 5; Figure S10). This can be understood by considering that the time to the most recent common ancestor (tMRCA) of a neutral locus, which underlies L, can increase only by as many generations as growth lasted, no matter how extreme that growth has been. For the demographic models considered here, tMRCA for a pair of chromosomes is on the order of 15,000 generations, and thus the very recent growth starting only 400 generations ago can lead only to a relatively small increase (File S1).
Average fitness of individuals:
We established that on the one hand, population growth slightly increases the average number of mutations carried by an individual and, on the other hand, each of these alleles is slightly less deleterious. The overall fitness of an individual is a function of the combined effect of all deleterious alleles it carries. In our simulations, the above two effects counteract each other such that individual fitness is similar between the two population models (Figure S11). We note, however, that this specific result might vary as a function of growth model parameters and dominance level.
Discussion
In this study, we address whether an extremely fast and recent population growth can hinder the action of purifying selection. We studied this independent of any effects that the transition to agriculture itself might have had on purifying selection (Eshed et al. 2004; Gage and DeWitte 2009). The joint effect of natural selection and population growth on deleterious alleles is not trivial to understand since these two forces have opposite effects: purifying selection purges deleterious alleles while population growth introduces an excess of deleterious mutations into the population. To disentangle the effects of population growth and purifying selection, we compared simulated loci with neutral mutations to those with deleterious mutations, each in population models with and without a recent epoch of exponential growth. The particular choice of the population growth model does not affect the results, as we showed by repeating the analyses using other published models of recent European history (File S1, Figure S12, Figure S13, and Figure S14).
Our results show that population expansion is accompanied by an accumulation of segregating sites. At both locus types and in both demographic scenarios, mutations that are not transmitted to the next generation are typically singletons or doubletons, which constitute the majority of segregating sites but only a small fraction of all copies of derived alleles present in the population. Beyond known differences in the site frequency spectrum, we showed that mutations lost during population growth have on average more deleterious fitness effects than in a population that does not experience growth. This effect is attributable to the increased efficacy of purifying selection as the population size increases. As a result, derived alleles present in the growing population have on average a less deleterious effect when averaged per allelic copy. This result may seem at odds with recent sequencing studies that have shown that human populations carry a burden of recent mutations that tend to be more deleterious due to their recency (Fu et al. 2012; Nelson et al. 2012; Tennessen et al. 2012). However, recent mutations are expected to be more deleterious also in the absence of population growth. Thus, to understand how population growth has affected the selection coefficient of segregating mutations, the empirical comparison should be made to a human population that has not experienced recent growth. Here, we show that averaging all segregating sites, across frequency or age, the selection coefficient of an allele copy picked at random in the extant human population is less deleterious than it would have been had the population not gone through an epoch of extreme growth. We conclude that natural selection purges the most deleterious alleles more efficiently in the scenario with growth.
While the selection coefficient of derived alleles is on average less deleterious, our results also show that each individual carries a larger number of deleterious alleles in a growing population though only modestly so due to the recency of the growth in the human genealogical scale. Overall, the two effects balance out and the average individual fitness is not different in the growing population from that in the population without growth. Importantly, the simulations presented in this study considered only the case of mutations that are partially dominant, with fitness effect being 1 + s for heterozygotes and (1 + s)2 for homozygotes. For the vast majority (>94%) of mutations, the selection coefficient s is distributed between 0 and −0.01, for which s2 is negligible and this model is approximately additive (1 + s and 1 + 2s), which is commonly assumed in genome-wide association studies (GWAS). The actual distribution of dominance degree (h) is difficult to obtain in humans, except to note that most Mendelian disorders are largely recessive. In model organisms, estimates of dominance vary considerably and are biased in various ways (Fernandez et al. 2004; Agrawal and Whitlock 2011). In addition, it has been established that h covaries with s (Simmons and Crow 1977; Caballero and Keightley 1994; Phadnis and Fry 2005). While it remains to be tested how our results translate to alleles of varying dominance models, the effect of recent population growth is characterized by very rare alleles, which are seldom observed as homozygous.
Our results also show that deleterious derived alleles are expected to have fewer copies in the population than neutral ones and especially so in the growing population. This result is in agreement with empirical data showing that the odds ratio of rare variants being functional compared to variants with minor allele frequency >0.5% is 4.2 (Tennessen et al. 2012). Importantly, low frequency is not the sole predictor of functionality in a demographic expansion scenario, since we observed that growth also leads to accumulation of extremely rare neutral variants. Applying Murayama’s theory (Muruyama 1974) to population growth, Maher et al. (2012) and Kiezun et al. (2013) showed that—conditioned on allele frequency—allele age can be powerful in predicting selective effect.
The impact of changes in population size on deleterious variants has received considerable attention in recent years. Lohmueller et al. (2008), comparing 15 African American and 20 European American individuals, showed that human populations that went through a rapid and recent population bottleneck present a higher proportion of deleterious variation. Comparing the same two populations, Tennessen et al. (2012) found that this result was dependent on the criteria used to classify variants as putatively deleterious. In non-European samples that experienced population bottlenecks, Szpiech et al. (2013) showed that recent inbreeding increased the proportion of mildly deleterious homozygous mutations. The impact of ancient bottlenecks on the average selection coefficient of an allele is also visible in our simulations. While our simulations show that population growth does not have a downward impact on the average selection coefficient of a derived allele copy, population bottlenecks had more notably affected deleterious variation.
Population growth generated a strong increase in the number of segregating sites in the population that can potentially play a role in complex disease risk. Since the vast majority of these variants are extremely rare, recent growth leads only to a moderate increase in the number of derived alleles carried by each individual. This supports the claim that common diseases may frequently be subject to strong genetic heterogeneity (Lango Allen et al. 2010), with different patients that have a similar diagnosis carrying rare or private mutations at the same or different loci (Galvan et al. 2010; McClellan and King 2010; Ravanbod et al. 2012). At the same time, our results have no implications on the heterogeneity of de novo mutations since these are not affected by demographic history. Importantly, a larger population size by itself (and the consequent higher efficiency of selection), without population growth, would also result in increased genetic heterogeneity. In summary, we showed that the recent rapid growth experienced by many human populations can be partially responsible for increased levels of genetic heterogeneity in the architecture of complex disease and traits, via two effects: (1) the introduction of a much larger number of variants and (2) improved purging of the most deleterious alleles and maintenance of more mildly deleterious alleles, with the latter having smaller effect sizes on average for diseases that are under purifying selection.
Our results indicate that the recent rapid expansion of human populations has perturbed different population genetic attributes of our species. These can be used to suggest directions of exploration, define strategies in medical genetics, refine association methods, and tests of positive natural selection optimized for the genetic diversity segregating in human populations (Yu et al. 2009). Our results have clear relevance to the issue of missing heritability in genome-wide association studies (Maher 2008; Manolio et al. 2009; Eichler et al. 2010). The impact of population growth on individual mutation load and the genetic architecture of complex diseases deserve further study, both theoretically and empirically. For example, a careful study of recombination and linkage patterns would benefit the association methods based on identity-by-descent (Gusev et al. 2011; Browning and Thompson 2012; Zhuang et al. 2012). Comparing large studies of deep sequencing between functional and nonfunctional regions will provide further empirical insight into ways that rapid growth affects the balance of mutation, drift, and selection. Finally, theoretical models need to be extended to accommodate additional factors, including variation in the degree of dominance, variation in family size (Wakeley et al. 2012), and changes in variance in reproductive success over time.
Supplementary Material
Acknowledgments
The authors thank Leonardo Arbiza for help with optimization of analysis tools, and to Joshua Akey, Leonardo Arbiza and Kevin Mitchell for comments on earlier versions of this paper. A.G.C. and A.K. were supported in part by the National Institutes of Health (U01-HG005715). E.G. was supported in part by a Cornell Center for Comparative and Population Genomics fellowship. A.K. was also supported by The Ellison Medical Foundation, an Alfred P. Sloan Research Fellowship, and the Edward Mallinckrodt, Jr. Foundation.
Footnotes
Communicating editor: N. A. Rosenberg
Literature Cited
- Agrawal A. F., Whitlock M. C., 2011. Inferences about the distribution of dominance drawn from yeast gene knockout data. Genetics 187: 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A. R., Williamson S. H., Indap A. R., Degenhardt J. D., Hernandez R. D., et al. , 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., Thompson E. A., 2012. Detecting rare variant associations by identity-by-descent mapping in case-control studies. Genetics 190: 1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A., Keightley P. D., 1994. A pleiotropic nonadditive model of variation in quantitative traits. Genetics 138: 883–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. D., Chong J. X., Malig M., Ko A., Dumont B. L., et al. , 2012. Estimating the human mutation rate using autozygosity in a founder population. Nat. Genet. 44: 1277–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals F., Bertranpetit J., 2012. Genetics. Human genetic variation, shared and private. Science 337: 39–40 [DOI] [PubMed] [Google Scholar]
- Cohen J. E., 1996. How Many People Can the Earth Support? Ed. 1. W. W. Norton, New York [Google Scholar]
- Coventry A., Bull-Otterson L. M., Liu X., Clark A. G., Maxwell T. J., et al. , 2010. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat. Commun. 1: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler E. E., Flint J., Gibson G., Kong A., Leal S. M., et al. , 2010. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11: 446–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed V., Gopher A., Gage T. B., Hershkovitz I., 2004. Has the transition to agriculture reshaped the demographic structure of prehistoric populations? New evidence from the Levant. Am. J. Phys. Anthropol. 124: 315–329 [DOI] [PubMed] [Google Scholar]
- Fernandez B., Garcia-Dorado A., Caballero A., 2004. Analysis of the estimators of the average coefficient of dominance of deleterious mutations. Genetics 168: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W., O’Connor T. D., Jun G., Kang H. M., Abecasis G., et al. , 2012. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493: 216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage T. B., DeWitte S., 2009. What do we know about the agricultural demographic transition? Curr. Anthropol. 50: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan A., Ioannidis J. P., Dragani T. A., 2010. Beyond genome-wide association studies: genetic heterogeneity and individual predisposition to cancer. Trends Genet. 26: 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Kenny E. E., Lowe J. K., Salit J., Saxena R., et al. , 2011. DASH: a method for identical-by-descent haplotype mapping uncovers association with recent variation. Am. J. Hum. Genet. 88: 706–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawks J., Wang E. T., Cochran G. M., Harpending H. C., Moyzis R. K., 2007. Recent acceleration of human adaptive evolution. Proc. Natl. Acad. Sci. USA 104: 20753–20758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez R. D., 2008. A flexible forward simulator for populations subject to selection and demography. Bioinformatics 24: 2786–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., 2012. Rates and fitness consequences of new mutations in humans. Genetics 190: 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Eory L., Halligan D. L., Kirkpatrick M., 2011. Inference of mutation parameters and selective constraint in mammalian coding sequences by approximate Bayesian computation. Genetics 187: 1153–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A., Clark A. G., 2012. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science 336: 740–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A., Mullikin J. C., Patterson N., Reich D., 2007. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat. Genet. 39: 1251–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A., Pulit S. L., Francioli L. C., van Dijk F., Swertz M., et al. , 2013. Deleterious alleles in the human genome are on average younger than neutral alleles of the same frequency. PLoS Genet. 9: e1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1955. Stochastic process and distribution of genes frequencies under natural selection. Cold Spring Harb. Symp. Quant. Biol. 20: 33–53 [DOI] [PubMed] [Google Scholar]
- Kimura M., 1957. Some problems of stochastic processes in genetics. Ann. Math. Stat. 28: 882–901 [Google Scholar]
- Kimura M., Ohta T., 1969. The average number of generations until fixation of a mutant gene in a finite population. Genetics 61: 763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A. S., 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336: 435–440 [DOI] [PubMed] [Google Scholar]
- Lango Allen H., Estrada K., Lettre G., Berndt S. I., Weedon M. N., et al. , 2010. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467: 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesecque Y., Keightley P. D., Eyre-Walker A., 2012. A resolution of the mutation load paradox in humans. Genetics 191: 1321–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller K. E., Indap A. R., Schmidt S., Boyko A. R., Hernandez R. D., et al. , 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature 451: 994–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2010. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. USA 107: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B., 2008. Personal genomes: the case of the missing heritability. Nature 456: 18–21 [DOI] [PubMed] [Google Scholar]
- Maher M. C., Uricchio L. H., Torgerson D. G., Hernandez R. D., 2012. Population genetics of rare variants and complex diseases. Hum. Hered. 74: 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J., King M. C., 2010. Genetic heterogeneity in human disease. Cell 141: 210–217 [DOI] [PubMed] [Google Scholar]
- Muruyama T., 1974. The age of a rare mutant in a large population. Am. J. Hum. Genet. 26: 669–673 [PMC free article] [PubMed] [Google Scholar]
- Muruyama T., Kimura M., 1974. A note on the speed of gene frequency changes in reverse directions on a finite population. Evolution 28: 161–163 [DOI] [PubMed] [Google Scholar]
- Nelson M. R., Wegmann D., Ehm M. G., Kessner D., St Jean P., et al. , 2012. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337: 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N., Fry J. D., 2005. Widespread correlations between dominance and homozygous effects of mutations: implications for theories of dominance. Genetics 171: 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M., Charlesworth B., Wall J. D., 1999. Genealogies and weak purifying selection. Mol. Biol. Evol. 16: 246–252 [DOI] [PubMed] [Google Scholar]
- Ravanbod S., Rassoulzadegan M., Rastegar-Lari G., Jazebi M., Enayat S., et al. , 2012. Identification of 123 previously unreported mutations in the F8 gene of Iranian patients with haemophilia A. Haemophilia 18: e340–e346 [DOI] [PubMed] [Google Scholar]
- Reed F. A., Aquadro C. F., 2006. Mutation, selection and the future of human evolution. Trends Genet. 22: 479–484 [DOI] [PubMed] [Google Scholar]
- Simmons M. J., Crow J. F., 1977. Mutations affecting fitness in Drosophila populations. Annu. Rev. Genet. 11: 49–78 [DOI] [PubMed] [Google Scholar]
- Slatkin M., Hudson R. R., 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129: 555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech Z. A., Xu J., Pemberton T. J., Peng W., Zollner S., et al. , 2013. Long runs of homozygosity are enriched for deleterious variation. Am. J. Hum. Genet. 93: 90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. The effect of change in population size on DNA polymorphism. Genetics 123: 597–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. A., Bigham A. W., O’Connor T. D., Fu W., Kenny E. E., et al. , 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337: 64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations, Department of Economic and Social Affairs, Population Division , 2011. World Population Prospects: The 2010 Revision, Volume I: Comprehensive Tables. ST/ESA/SER.A/313. United Nations, New York.
- Wakeley J., King L., Low B. S., Ramachandran S., 2012. Gene genealogies within a fixed pedigree, and the robustness of Kingman’s coalescent. Genetics 190: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276 [DOI] [PubMed] [Google Scholar]
- Waxman D., 2011. A unified treatment of the probability of fixation when population size and the strength of selection change over time. Genetics 188: 907–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D., 2012. Population growth enhances the mean fixation time of neutral mutations and the persistence of neutral variation. Genetics 191: 561–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Keinan A., Chen H., Ferland R. J., Hill R. S., et al. , 2009. Detecting natural selection by empirical comparison to random regions of the genome. Hum Mol Genet 18: 4853–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z., Gusev A., Cho J., Pe’er I., 2012. Detecting identity by descent and homozygosity mapping in whole-exome sequencing data. PLoS ONE 7: e47618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.