Abstract
This work is concerned with the historical progression, to fixation, of an allele in a finite population. This progression is characterized by the average frequency trajectory of alleles that achieve fixation before a given time, T. Under a diffusion analysis, the average trajectory, conditional on fixation by time T, is shown to be equivalent to the average trajectory in an unconditioned problem involving additional selection. We call this additional selection “fictitious selection”; it plays the role of a selective force in the unconditioned problem but does not exist in reality. It is a consequence of conditioning on fixation. The fictitious selection is frequency dependent and can be very large compared with any real selection that is acting. We derive an approximation for the characteristic trajectory of a fixing allele, when subject to real additive selection, from an unconditioned problem, where the total selection is a combination of real and fictitious selection. Trying to reproduce the characteristic trajectory from the action of additive selection, in an infinite population, can lead to estimates of the strength of the selection that deviate from the real selection by >1000% or have the opposite sign. Strong evolutionary forces may be invoked in problems where conditioning has been carried out, but these forces may largely be an outcome of the conditioning and hence may not have a real existence. The work presented here clarifies these issues and provides two useful tools for future analyses: the characteristic trajectory of a fixing allele and the force that primarily drives this, namely fictitious selection. These should prove useful in a number of areas of interest including coalescence with selection, experimental evolution, time series analyses of ancient DNA, game theory in finite populations, and the historical dynamics of selected alleles in wild populations.
Keywords: allele fixation, random genetic drift, diffusion analysis, genetic disease progression, population genetics theory, stochastic population dynamics
THE phenomenon of fixation, where an allele becomes fully established in a population, was first studied by Fisher (1922) and Haldane (1927). Building on results previously obtained by Fisher (1922), Haldane (1927) showed that the probability of fixation of a new beneficial allele in a large population is ∼2s, where s is the allele’s selective advantage (i.e., its selection coefficient). For typical values of s, the probability of fixation of a beneficial allele is very small. Hence even in a large population, the majority of positively selected alleles are unlikely to fix. The probability of fixation of neutral and deleterious mutations is nonzero but lies beyond the reach of Haldane’s result; they are even less likely to fix than beneficial mutations (Kimura 1962). However, despite the improbability of any new allele fixing, over time there is repeated production of new alleles in a population, so that eventually fixation will occur. Apart from the long-term changes in a population that fixation entails, we note that alleles “on the way” to fixation have intermediate and high frequencies for extended periods of time, and the associated variation has important implications for genetics and evolution. In the present work we are primarily concerned with the progression of alleles to fixation.
Since the early studies cited above, there have been various concepts of population genetics, including fixation, the probability of fixation, and the expected time to fixation, which have played a pivotal role in the development of the subject. These essential ingredients of the neutral theory of molecular evolution (Kimura 1983) appear in more recent developments, such as in methods to estimate the proportion of mutations under positive selection (see, e.g., Messer and Petrov 2013). The concept of fixation was somewhat eclipsed, in the 1990s, with the coming of age of retrospective population genetics and coalescent theory but today there are a number of good reasons to return to studying fixation.
First, despite important advances in modeling coalescence with selection, i.e., the selection ancestral graph (Krone and Neuhauser 1997), these new models have been hard to implement. The main approach used today to analyze the coalescent of a sample of DNA sequences, where both drift and selection occur, is to combine forward and backward simulations. Briefly, the first step is to perform simulations that go forward in time and incorporate selection, keeping track of the frequency of a beneficial allele. Then, via simulating backward in time, the coalescent is used to reconstruct the genealogy of the sampled individuals, conditional on the frequency of the beneficial allele—which was determined in the simulations of the first step (Teshima and Innan 2009; Ewing and Hermisson 2010).
The second reason comes from advances in experimental evolution. As has been stressed by Patwa and Wahl (2008) and exemplified by Gifford et al. (2012), models of fixation can now be tested. For example, Gifford et al. (2012) used the fungus Aspergillus nidulans to test a model of the probability of an allele surviving genetic drift—which is a major component of the probability of fixation.
Third, and in a similar vein, we note that in studies of ancient DNA (Skoglund et al. 2012), in studies of viruses (Rodrigo and Felsenstein 1999), and in experimental evolution (Illingworth et al. 2012), it is now possible to acquire the time series of an allele’s frequency. In experimental evolution, one major issue is that even strongly favored alleles take a long time to fix. In ancient DNA studies, there are generally only a limited number of time points available in the series. In both of these cases it would be very useful to be able to extrapolate the full trajectory of an allele’s frequency from limited information.
Fourth, a better understanding of the process of fixation will help to understand the dynamics at selected loci and may cast new light on well-known examples of allele trajectories in wild populations. Possibly the best known is that of the scarlet tiger moth, Callimorpha dominula, where a wing-color polymorphism has been tracked since 1939 in the Cothill reserve population in Oxfordshire, Britain. These longitudinal data do not trace the allele frequency from mutation to fixation, but from a time in 1939 where it had reached a frequency of ∼10%. The frequency then proceeded to decline to a recorded temporary loss during the 1960s (Ford and Sheppard 1969; O’Hara 2005). This study has been at the center of an ongoing debate, since the time Fisher and Ford (1947) dismissed genetic drift as the likely cause of this decline, principally on the basis of the estimated (large) population size. Instead, they argued for fluctuating selection (for recent work on fluctuating selection, see, e.g., Huerta-Sanchez et al. 2008; Waxman 2011a). Wright (1948) was the first to point out that there were probable scenarios consistent with an effective population size that is much smaller than the estimated census size. The most recent analysis is supportive of Wright’s notion that, although selection is operating, a greater proportion of variation in allele frequencies is explained by drift (O’Hara 2005). Less controversial examples include the fixation of SNPs associated with drug resistance in the malarial parasite Plasmodium falciparum (Taylor et al. 2012). Here, mutations found segregating at moderate frequencies (∼20%), in two genes, went to fixation in just 9 years after the widespread implementation of the drug. Strong directional trajectories, such as these, are atypical of drift and are often used as arguments for selection, simply by deduction. Another example is the peppered moth Biston betularia, whose melanic morph has been noted since the mid-19th century in the United Kingdom and recorded since 1959 at Caldy Common, England (Grant et al. 1996). Despite large fluctuations in the estimated population size at this site, the trajectory fits well with expectations that are based on a constant positive selection coefficient; explorations of multiple populations throughout England also found trajectories that are consistent with constant selection during fixation of a typical allele (Cook and Turner 2008). The above examples are valuable glimpses of the dynamics of mutant alleles in the wild, but provide limited scope for generalization.
Perhaps more useful are examples that present the known end point of a beneficial mutation’s path to fixation, such as human lactase persistence among European populations and fixation of the FY*O allele, which confers resistance to P. vivax in populations of Africa, where malaria is endemic (Sabeti et al. 2006). For cases such as these, the typical form of an allele’s frequency trajectory, combined with knowledge of additional historical events, can allow us to understand the path to fixation and the path from fixation to the allele’s origination. The typical trajectory can give us the opportunity to make a plausible reconstruction of events.
While there has been a lot of work on the probability of fixation (for recent work see, e.g., Otto and Whitlock 1997; Uecker and Hermisson 2011; Waxman 2011a), little or no theoretical work has been devoted to inferring the typical way that fixation actually occurs. The present work aims to remedy this omission. We present results that allow us to understand and calculate the typical route to fixation. We anticipate that the results presented here will be useful in future theoretical and experimental studies, for understanding and explaining the way factors, such as demographic and environmental changes, influence the progression of an allele to fixation.
Trajectory of an Allele
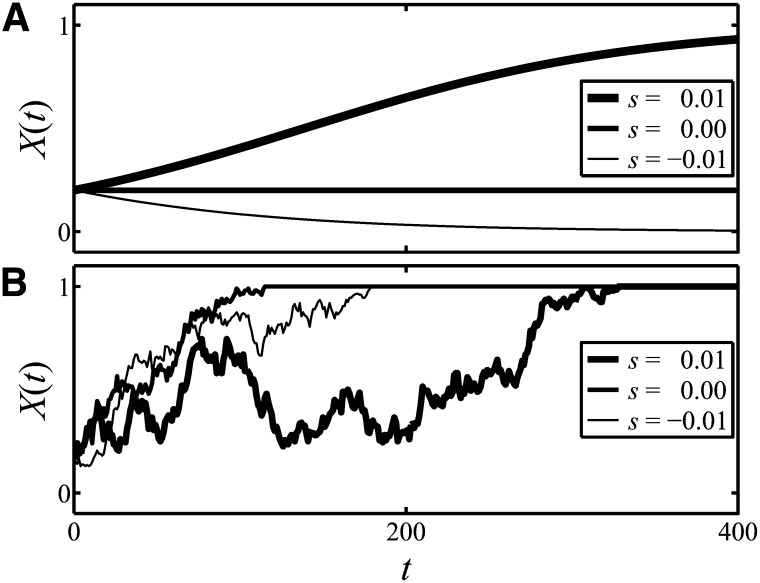
An important notion in this work is the trajectory of an allele. Knowing an allele’s trajectory means knowing its (relative) frequency for a range of relevant times. Examples of trajectories that result in fixation are given in Figure 1.
Figure 1.
(A) Three deterministic trajectories of the frequency of an allele in an infinite population. The trajectories are associated with three different values of the selection coefficient, s, in a model of additive selection (see Equation 1). The selection coefficients used are s = −0.01, 0.00, 0.01 and the initial frequency is y = 0.2. Given the infinite size of the population, random genetic drift does not occur and, hence strictly, neither does fixation. However, for positive selection coefficients, trajectories approach unity and can be reasonably described as ultimately fixing, while trajectories associated with negative or zero selection coefficients do not approach unity, and so do nothing resembling fixation. (B) Three particular trajectories of the allele frequency in a finite population of size N = 100. The trajectories are subject to random genetic drift and additive selection; the selection coefficients and initial frequency used are again s = −0.01, 0.00, 0.01 and y = 0.2. The trajectories displayed all undergo fixation. Trajectories associated with zero or negative selection coefficients have a nonzero probability of achieving fixation due to random genetic drift.
The present work is primarily concerned with understanding the characteristic trajectory of alleles that achieve fixation in a specific time. We adopt a natural definition of the characteristic trajectory, namely the average or expected trajectory. We can obtain the expected trajectory, which fixes by a specified time, by carrying out an average over many trajectories, all of which start with identical initial frequencies and all of which fix by the specified time. The expected trajectory is a key aspect of fixation. Apart from being a guide of the historical path, as noted above, it largely determines averages of quantities that depend on the allele’s frequency—when deviations from the expected value are either small or largely cancel out.
In this work we investigate (i) the shape of the characteristic trajectory, for alleles that achieve fixation; (ii) the important factors influencing the characteristic trajectory; and (iii) the extent to which observing fixation can bias our estimates of the level of selection that is acting.
We present an analysis that addresses (i), (ii) and (iii) and provides an approximation of the characteristic (i.e., expected) trajectory.
Given that we are asking questions about trajectories that actually achieve fixation means we are ignoring trajectories that lead to other outcomes. Focusing attention on only a subset of all trajectories is an act of conditioning in the language of probability theory. It may not be obvious at this point, but such conditioning leads to identical results to those derived from a related problem, where no conditioning is carried out (hence all trajectories are used in the analysis), but there is additional selection acting. This additional selection ensures that the trajectories in the unconditioned problem achieve the specified outcome (i.e., they achieve fixation by a given time). Since this additional selection does not exist in the biological system under consideration, but is a theoretical construct, we describe it as “fictitious”. The fictitious selection is a direct manifestation of the conditioning in the original problem.
There are advantages in viewing a conditioned problem as an unconditioned problem with what is generally an additional evolutionary force acting. First, an intuitive understanding can be gained of what factors are shaping the characteristic trajectory in such problems. Second, using the fictitious evolutionary force in a theoretical analysis suggests new ways of looking at the problem and new ways of proceeding.
Background
We present an analysis of an unlinked locus of interest in a randomly mating diploid sexual population. The locus has two alleles, denoted A and B, and generations are nonoverlapping. Censusing the population in adults, we reserve the term frequency for the proportion of all genes at the locus in adults that are the A allele. The frequency in generation t is written as X(t), where t = 0, 1, 2, … .
Infinite population
Consider first a population whose size is infinite, so there are no effects of random genetic drift and the frequency of the A allele at the locus of interest changes in a completely predictable (i.e., deterministic) manner over time. We assume the frequency of this allele, X(t), changes by only a small amount each generation so its dynamics are well described by the differential equation dX/dt = M(X, t). In this equation, it is useful to think of M(X, t) as the value of the “force” that acts when the frequency has the value X at time t. The function M(X, t) contains the effects of any migration, mutation, and selection that are acting. If M(X, t) is zero for all X, it signals the absence of all evolutionary forces and the frequency does not change; this describes a neutral locus. If there are evolutionary forces acting, then M(X, t) is generally nonzero, and it causes the frequency to change. For definiteness, we restrict the theoretical considerations of this work to the important case of additive selection (also called directional selection). In terms of a selection coefficient s of the A allele, additive selection is defined by the AA, AB, and BB genotypes having relative fitnesses of 1 + 2s, 1 + s, and 1, respectively. Throughout this work we assume, as is often the case, that natural values of s are very small (|s| ≪ 1). We thus neglect terms in s2 within M(X, t) and obtain M(X, t) = sX(1 − X). The resulting equation, which determines the trajectory of the frequency of the A allele, is
| (1) |
The solution of Equation 1 exhibits different behaviors, depending on the value of the selection coefficient s. When s is positive, the solution logistically grows toward unity, see Figure 1A, showing what we might call “fixation-like behavior”. Fixation cannot, strictly, occur in an infinite population: the allele frequency changes continuously and never reaches a value of unity. However, the approach of the frequency toward unity is so strongly reminiscent of fixation that, for practical purposes, its long-term behavior is not usefully distinguished from fixation. When s is zero or negative, no such approach of the frequency to unity occurs (Figure 1A).
Finite population
Consider now a finite population, where the locus of interest is again subject to additive selection, and neither mutation nor migration occur. In a finite population, the frequency no longer obeys Equation 1 because of random genetic drift, which amounts to a stochastic contribution to the dynamics. Such stochasticity is interesting and important. It can modify phenomena that occur in infinite populations and it can fuel phenomena that are impossible in infinite populations. For example, selection coefficients that are zero or negative do not lead to high allele frequencies or fixation in an infinite population, but in a finite population the stochastic contribution of genetic drift to the dynamics leads to a nonzero probability of high values or fixation of the allele frequency. Figure 1B contains two trajectories in a finite population that achieve fixation but are associated with selection coefficients that are not positive.
We consider a finite population where the locus of interest is subject to both additive selection and random genetic drift. Our aim is to describe some of the properties of an allele that achieves fixation before a given time has elapsed. Some results are also given for the case where the allele reaches a specific frequency at a specific time.
To frame our thoughts, it is helpful to think about the way we would learn from simulations about the properties of an allele that fixes before a specific time, say T. We would proceed by generating a large number of trajectories on the computer that are appropriate to a finite population and store only the subset of trajectories that fix by time T. This retention of only a subset of trajectories is equivalent to conditioning, in the sense of probability theory. From the stored trajectories, estimates can be made of properties of the focal allele. We may, however, be able to do better than simulations. Calculations, when we can perform them, give more accurate results than simulations. A calculation is equivalent to considering an infinite number of simulated trajectories; thus calculations do not suffer from the statistical errors associated with a simulation involving only a finite number of trajectories. Below, we give details of calculations, based on the diffusion approximation, which determine the implications of conditioning. The calculations presented have another benefit: they allow us to carry out transformations that relate apparently different scenarios and establish the following equivalence:
Either we carry out conditioning that corresponds, for example, to fixation occurring by a given time. This is equivalent to considering only a subset of the frequency trajectories. In this example these are only those trajectories that fix by a given time.
Or we do not condition in any way, but include a specific additional selective force. This is equivalent to considering all frequency trajectories; however, the additional selective force influences their behavior. In the above example of fixation, the additional selective force makes the trajectories fix by the given time.
The additional selection that is required to make the unconditioned problem, b, equivalent to a conditioned problem, a, does not actually exist. Rather, it encapsulates the way the data (the set of all frequency trajectories) have been sampled. That is, those trajectories we consider at any time, t, are included (sampled), because of properties they will have later, and so are, in fact, “conditioned on the future”. We call the additional selective force required in case b fictitious. Such a force is a conceptual construct that we believe provides a useful way of viewing and analyzing problems. Below we use this fictitious selective force to find an approximation for the trajectory that is characteristic of a fixing allele.
Fictitious Force
Origins
As already noted, the frequency of the A allele exhibits stochasticity in a finite population. Indeed, if we could inspect a number of copies of a population that are identical at time 0, then for positive times the frequency X(t) in different copies of the population would be likely to take different values—due to random genetic drift. Such variation requires a statistical description. Statistics of X(t) can be described by a Wright–Fisher model (Fisher 1930; Wright 1931). However, to make theoretical progress, we consider an analysis based on the diffusion approximation and use methods of Kimura (1955), along with some more recent results of McKane and Waxman (2007) and Waxman (2011b). Under the diffusion approximation, the frequency X(t) has a statistical description in terms of a distribution (a probability density). Assuming the frequency of the A allele has the definite value y, at an initial time of u, we write the distribution of X(t), when evaluated at a frequency of x, as K(x, t| y, u). This obeys the diffusion equation
| (2) |
(Kimura 1955, 1964), where Ne is the effective population size and
| (3) |
Note that generally, the diffusion equation in Equation 2 describes an unconditioned problem in which the evolutionary force that is acting is M(X, t).
To describe properties of trajectories that fix by time T, we need to construct their distribution, which is the analog of K(x, t|y, u), but is conditional on fixation occurring by time T. The construction involves two ingredients: (i) the probability of fixation by time T, given an initial frequency of x at time t, which we write as Pfix(T|x, t); and (ii) the unconditioned distribution, K(x, t|y, u).
The distribution, conditional on fixation by time T, can then be motivated from the following reasoning (formal arguments are given in Appendix A, where it is shown that the distribution is based on a mathematical transform that was introduced by Doob 1957). First, X(t) “propagates” from frequency y at time u to frequency x at time t; this propagation is characterized by K(x, t| y, u). Second, X(t) undergoes a transition from having a value x at time t to being fixed by time T. This occurs with probability Pfix(T|x, t). As a consequence, the distribution of X(t), conditional on fixation by time T, is proportional to Pfix(T|x, t)K(x, t|y, u). In other words, Pfix(T|x, t)K(x, t| y, u) is proportional to the distribution associated with trajectories that fix by time T. For the case where fixation ultimately occurs (T = ∞) the result is known in the literature (Ewens 1973, 2004). The result of the present work generalizes the result of Ewens to finite values of T.
It is natural to ask what equation the conditional distribution obeys. In Appendix A we show that the distribution obeys a diffusion equation that is of the same form as Equation 2; however, the force in the diffusion equation consists of the original force, sx(1 − x), plus an additional force Mfict(x, t), that we call fictitious. In the present case, Mfict(x, t) depends on the time-dependent probability of fixation, Pfix(T|x, t) (see Equation 5, below). The conditional distribution obeys a diffusion equation of the same form as Equation 2, but with a force of
| (4) |
Thus we can say that the conditional problem has the mathematical form of an unconditional problem, but with the force of Equation 4, which contains a fictitious component. The above reasoning gives the essence of the origin of the fictitious force. Generally, the specific form of the fictitious force Mfict(x, t) depends on the subset of all trajectories that we focus attention (condition) upon.
The fictitious force on trajectories that fix by a specific time
Let us return in more detail to the example just considered, namely the trajectories that achieve fixation by a specific time, T. For this case we find that the fictitious force, Mfict(x, t), is given by
| (5) |
(see Appendix A for details).
There are different ways we can view the fictitious force appearing in Equation 5. These different viewpoints do not affect any outcomes, but do determine the descriptive language used. We proceed by noting the presence of the factor x(1 − x) in Equation 5 and, on making the comparison with Equation 1, take the view that Mfict(x, t) is an additional contribution to the additive selection acting on the A allele. That is, we write
| (6) |
where sfict(x, t) is the selection coefficient associated with fictitious additive selection and
| (7) |
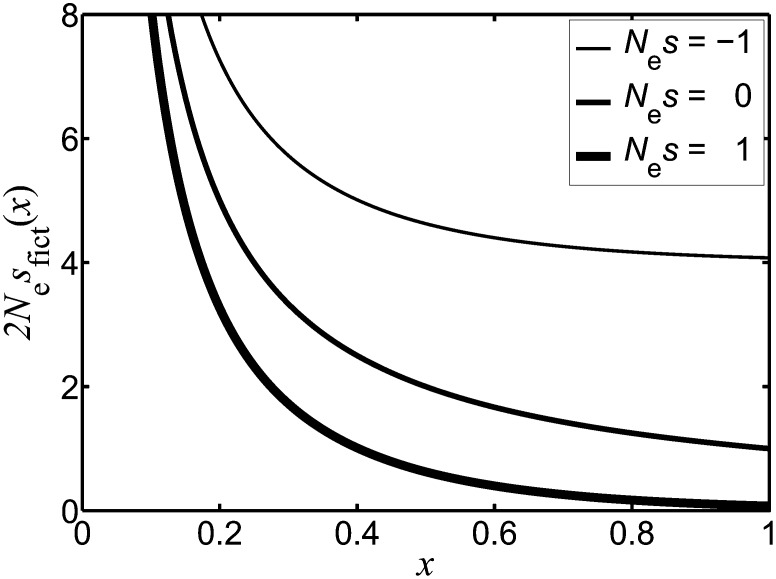
Having adopted the language of fictitious selection, we generally find that it is frequency and time dependent because sfict(x, t) depends on both x and t. Furthermore, the form of sfict(x, t) must be such that all trajectories fix by no later than time T. Thus no trajectory ever reaches zero frequency prior to time T and loss is prevented by sfict(x, t) being very large and positive at small frequencies and times (for an example of this, see Figure 2).
Figure 2.
For trajectories where fixation eventually occurs (T = ∞), the quantity sfict(x) represents the selection coefficient of the fictitious selection. We plot 2Nesfict(x) against the frequency, x. We give results when the parameter Nes (involving the real selection coefficient s) takes the values −1, 0, and +1. The result 2Nesfict(x) = 2Nes[coth(2Nesx) − 1] shows that sfict(x) explicitly depends upon x and therefore exhibits frequency dependence. The neutral limit of sfict(x) follows from taking s → 0 and is given by sfict(x) = 1/(2Nex).
Had we chosen to write the fictitious force in Equation 5 in the form Mfict(x, t) = mfict(x, t) × (1 − x), then mfict(x, t) would have the interpretation as either a rate of mutation from the B allele to the A allele or a rate of migration, from a population that is fixed for the A allele. In either interpretation, there is generally frequency and time dependence of mfict(x, t). We adopt only the selection viewpoint of the fictitious force in this work.
To gain insight into the properties of the fictitious selection, it is helpful to consider special cases of the associated selection coefficient, sfict(x, t) (Equation 7).
(a) Fixation ultimately occurs T →∞:
When the time T becomes arbitrarily large (T → ∞), the quantity Pfix(T|x, t), which appears in Equation 7, becomes the probability that the A allele ultimately fixes. When the effective population size, Ne, and the strength of “real” selection, s (appearing in Equations 1 and 3) are independent of time, Pfix(T|x, t) becomes a function only of x, which we write as Pfix(x). Under the diffusion approximation, (Kimura 1962). Using this result and Equation 7 allows us to determine the selection coefficient of the fictitious selective force; in this case it is independent of time and given by
| (8) |
(cf. Ewens 1973, 2004; Lambert 2008). When the frequency x is small (x ≪ 1/(2Ne|s|)) or under neutrality (the limit of Equation 8 as s approaches zero), we obtain sfict(x) = 1/(2Nex). In this case, sfict(x) depends extremely strongly on frequency: it is positive and very large at low frequencies and this prevents loss of the A allele. Figure 2 illustrates the form of sfict(x) for several values of the real selection coefficient, s.
(b) Fixation occurs by finite time T:
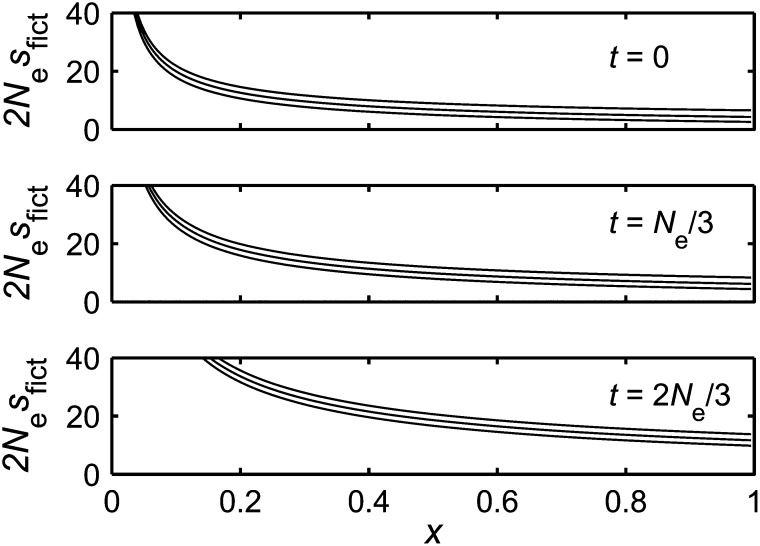
The result for sfict(x, t), when fixation occurs by the finite time T, is both frequency and time dependent. In Figure 3 we plot 2Nesfict(x, t) as a function of x, when the time T has the value T = Ne. Several different values of the real selection coefficient s and the time t have been chosen.
Figure 3.
We show how the selection coefficient of the fictitious selection, sfict(x, t), changes as a function of frequency and time. We have taken fixation to occur by time T = Ne = 99. Each panel covers a specific value of the time t; i.e., t = 0, Ne/3, or 2Ne/3. Each panel also contains three curves, where the top curve is for Nes = −1, the middle curve is for the neutral case Nes = 0, and the bottom curve is for Nes = 1. When the time t is small compared with the “final time” T, the quantity 2Nesfict(x, t) has a similar shape to the corresponding quantity in Figure 2, with one difference: its value is larger—to “compel” trajectories to fix by time T. When the value of the time t is increased, the quantity sfict(x, t) takes larger values. To illustrate this, at a frequency of x = 0.4, the ratios of the strength of fictitious selection sfict(x, t) at the times plotted, namely sfict(0.4, 0):sfict(0.4, Ne/3):sfict(0.4, 2Ne/3), are found to be ∼0.4:0.5:1. The quantity plotted, 2Nesfict(x, t), is defined by Equation 7. This involves a derivative, which we approximate by a discrete calculation, using a Wright–Fisher model with population size Ne. In particular, we use the approximation , where n is an integer, , and is the corresponding Wright–Fisher fixation probability (W is the Wright–Fisher transition matrix).
In Appendix B, we provide complementary material that gives details of trajectories that achieve an intermediate frequency at a specific time.
Characteristic Trajectory of a Fixing Allele
The above results, while interesting in their own right, also provide information that is relevant for understanding the behavior of alleles that fix by time T. As already stated, we take the frequency trajectory characterizing such fixations to be the expected trajectory. Determining the form of this involves averaging the frequency with respect to its distribution. Even from an approximate analysis, such as the diffusion approximation, this is not mathematically simple; the required distribution is the solution of the diffusion equation that is conditional on fixation occurring by the time T. However, the insight we have gained, from investigating the effects of conditioning on fixation, is that conditioning represents a very powerful force. Indeed, as we have shown, conditioning is equivalent to incorporating additional—fictitious—selection into the problem, and this selection is very strong for at least some frequencies. It is intuitively plausible that once the fictitious selection is taken into account, additional effects of random genetic drift are not so significant. This suggests that we can circumvent a great deal of the complexity of the computation of the expected trajectory by neglecting fluctuations around its expected value. This leads to the expected trajectory approximately obeying an equation of the same general form as that of the deterministic dynamics in an infinite population, Equation 1. When the real selection is additive, this leads to the expected trajectory, , approximately obeying
| (9) |
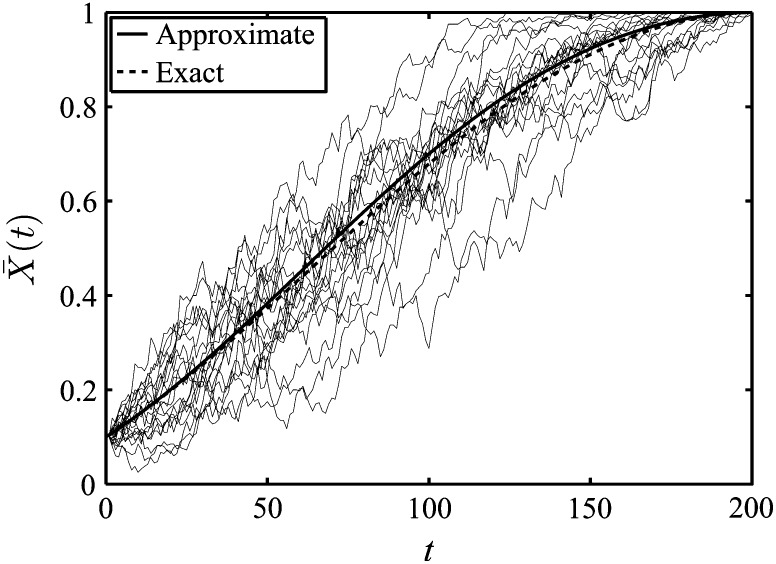
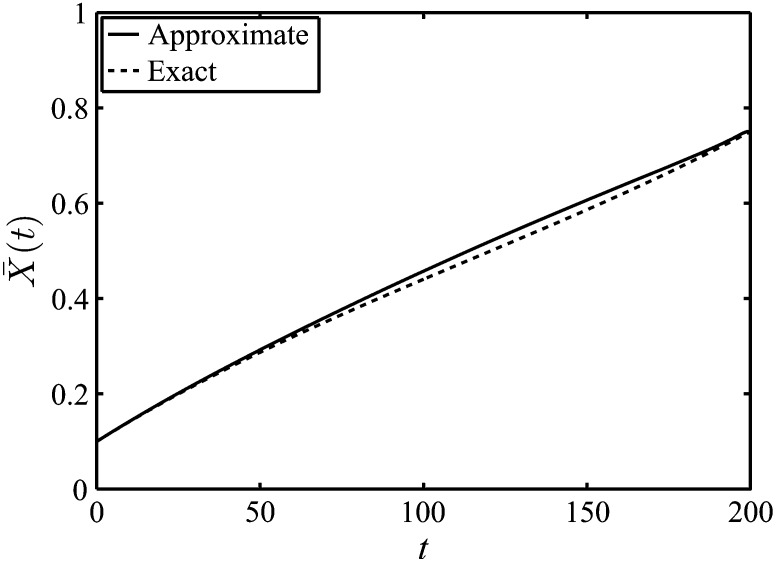
(see Appendix C for details). In Equation 9, sfict(x, t) is the selection coefficient associated with fictitious selection and is given in Equation 7. Approximate and exact forms of the expected trajectory, , are given in Figure 4.
Figure 4.
Approximate and exact forms of the expected trajectory when fixation is achieved by a specific time T. We have used a final time of T = 200, an effective population size of Ne = 200, a value of the real selection coefficient of s = 0.01, and an initial frequency of y = 0.1. The approximate results follow from Equation 9, while exact results were determined from a Wright–Fisher model (Fisher 1930; Wright 1931). The expected trajectory from simulations is very close to the approximate and Wright–Fisher results and is not displayed; we have, however, plotted 20 simulated trajectories.
In Table 1 we give some numerical illustrations of the accuracy of the approximate expected trajectory of Equation 9 for some specific cases. The quantity D in Table 1 is a measure of the mean mismatch between the approximate and the exact trajectories and is small, suggesting very reasonable accuracy of the approximate trajectory for the parameter values considered.
Table 1. Comparing approximate and exact expected trajectories.
| Initial Frequency, y | Selection Coefficient, s | Effective Population Size, Ne | Time, T, by Which Fixation is Specified to Occur | D, Measure of Mean Mismatch Between Approximate and Exact Trajectories |
| 0.01 | 0.01 | 100 | 100 | 0.010 |
| 200 | 0.016 | |||
| 400 | 0.018 | |||
| 800 | 0.017 | |||
| 200 | 100 | 0.005 | ||
| 200 | 0.013 | |||
| 400 | 0.019 | |||
| 800 | 0.026 | |||
| −0.01 | 100 | 100 | 0.005 | |
| 200 | 0.011 | |||
| 400 | 0.013 | |||
| 800 | 0.014 | |||
| 200 | 100 | 0.002 | ||
| 200 | 0.007 | |||
| 400 | 0.013 | |||
| 800 | 0.022 | |||
| 0.1 | 0.01 | 100 | 100 | 0.009 |
| 200 | 0.016 | |||
| 400 | 0.018 | |||
| 800 | 0.017 | |||
| 200 | 100 | 0.005 | ||
| 200 | 0.011 | |||
| 400 | 0.019 | |||
| 800 | 0.027 | |||
| −0.01 | 100 | 100 | 0.005 | |
| 200 | 0.011 | |||
| 400 | 0.014 | |||
| 800 | 0.015 | |||
| 200 | 100 | 0.001 | ||
| 200 | 0.006 | |||
| 400 | 0.014 | |||
| 800 | 0.023 |
We compare the approximation of the expected trajectory, from Equation 9, with the exact numerical result, calculated from the Wright–Fisher model (Fisher 1930; Wright 1931). The quantity D, defined by , is a measure of the mean mismatch between the two trajectories.
Comparison of the Expected Trajectory with a Deterministic Trajectory
So far we have determined a very reasonable approximation to the expected trajectory, , which characterizes some of the properties of fixation of an allele by a given time. It is interesting to now consider an infinite population where the A allele is subject to additive selection, and ask what constant selection coefficient would be needed, to best reproduce the expected trajectory of a finite population. We thus look for a trajectory in an infinite population that obeys the deterministic equation dXinf/dt = sinfXinf(1 − Xinf) and then make the optimal choice of the constant selection coefficient sinf. A natural way of determining this optimal value is to minimize the mean mismatch between the two trajectories. That is, we minimize with respect to sinf.
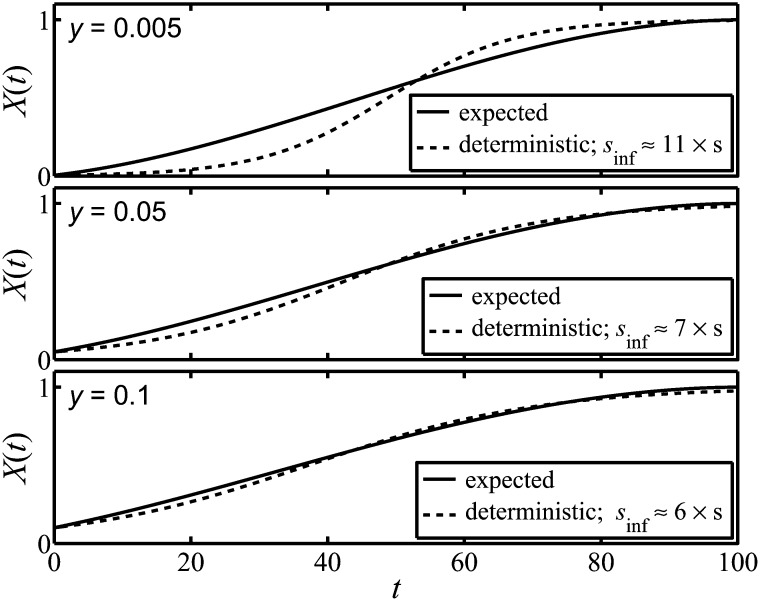
The parameters in the problem are the frequency y of the A allele at time t = 0 (y is common to both deterministic and expected trajectories), the real selection coefficient s (which appears in Equations 3 and 9), and the effective population size, Ne. The value of sinf that minimizes D generally depends on all of these parameters. Some results are illustrated in Figure 5. The value of the initial frequency can be seen to play an important role in the infinite population dynamics and can cause different shapes of the trajectory. For the deterministic trajectory to be as close as possible to the expected trajectory requires a value of sinf that can be substantially larger than s. For example, for N = 100, s = 0.01, T = 100, and y = 0.05, the value of sinf that minimizes D is sinf ≈ 0.0695, i.e., ∼700% of the value of s.
Figure 5.
We show the expected trajectory of a finite population with N = 100 and the most closely corresponding deterministic trajectory of an infinite population, Xinf(t), in which there is additive selection. The form of Xinf(t) follows from choosing the optimal value of the constant selection coefficient sinf of the additive selection. The different panels correspond to different values of the initial frequency, y, and the greatest difference between the expected and deterministic trajectories occurs at the lowest values of y. The behavior of Xinf(t) is determined by the initial frequency and the value of sinf. As a consequence it does not reach the value of unity at the time, T, by which the expected trajectory achieves fixation.
Discussion
In this work we have considered the phenomenon of allele fixation, which occurs only in finite populations, due to the occurrence of random genetic drift. We have focused on the characteristic trajectory of those alleles that reach fixation by a given time and the fictitious evolutionary force that appears to drive such alleles to fixation.
Characteristic trajectory
The typical or characteristic trajectory of an allele that fixes by a given time has been taken to be an average over all frequency trajectories that fix by the given time. Our presentation of the theory of this characteristic trajectory began by first describing the fictitious evolutionary force that can be thought of as pushing the trajectory to fixation (we say more about this force shortly). We then derived a simple differential equation that approximately determines the expected trajectory, assuming real additive selection, and showed that for a range of selection coefficients, population sizes, and values of the given time, the resulting approximation to the expected trajectory captures the exact result with good accuracy (Table 1). The differential equation determines the shape of the characteristic trajectory and in some cases explicitly displays parameters of the problem. This differential equation is straightforward to solve numerically and it demonstrates that for alleles segregating in a finite population, the typical path to fixation can be predicted across a range of scenarios. There are some implications of this finding.
The approximation we have for the expected trajectory of an allele that achieves fixation constitutes, we believe, a workable and potentially useful description of the way fixation is achieved. A calculated trajectory can be viewed as establishing a relation—or a constraint—between parameters in the theory and trajectory observations. Indeed, with sufficient data, the trajectories obtained could shed light upon field observations. As one example of the sort of data where the current work could be applied, consider the human Duffy blood group antigens (FY). There are three common alleles of this gene (FY*A, FY*B, and FY*O) but in much of sub-Saharan Africa only the FY*O allele has come close to fixation. It has been hypothesized that the resistance to P. vivax malaria that this allele confers has resulted in positive selection (Miller et al. 1976). A plausible trajectory spanning the period of segregation and replacement of the non-FY*O alleles could, in principle, be determined, given estimates of the ancestral population size and the selection coefficient, when combined with other relevant information, such as the timing of the spread of agriculture and the accompanying spread of malaria (Seixas et al. 2002). Such an analysis would undoubtedly be complex. Perhaps simpler situations, where the results of the current work may be profitably applied, are where the effective population size is small and hence the times to fixation are correspondingly brief. This may well describe the situation of carcinogenic mutations in the minority of neoplastic cells that are capable of self-renewal (Pepper et al. 2009).
Fictitious force
When alleles have been observed to reach fixation in a relatively short period of time, it is possible that they were subject to very strong positive selection. The achievement of fixation might then occur in a near deterministic manner, according to something like the first equation in this work, Equation 1. But an alternative picture is possible.
It is reasonable to speculate that an ultimately fixing allele might be subject to selection at a typical level (i.e., selection that is not strong and not necessarily positive) along with random genetic drift. The trajectory of the allele, however, is one of the somewhat rare samples of all possible trajectories that achieve fixation due to a “fortunate” episode of random genetic drift. If so, it may appear that the allele is driven by a strong evolutionary force, but in this case the strong force does not exist. In the present work this force has been termed fictitious and we have found it most natural to interpret this force as selection, especially since it is likely to be identified as a major contribution to selection. As we pointed out, this is a matter of viewpoint and other viewpoints are possible. We note that the fictitious selective force is characterized by a selection coefficient that is generally both time and frequency dependent. The frequency dependence is enormously strong at low frequencies, ensuring that loss of the allele is completely prevented, and this originates in the conditioning on fixation, which should therefore not be viewed as being passive, but rather as being an agency with a major influence on the observed dynamics.
We can consider the fictitious selection to be the additional selection we would need to invoke to explain observations in a finite population, where we are presented only with fixing trajectories, but are not told that they are a subset of all trajectories (thus trajectories corresponding to loss of the allele are assumed to have not been presented to us). The fictitious selection could, from this viewpoint, be viewed as an artifact of a strong reporting bias. We can go farther and note that any attempt to identify the selection coefficient, in the assumed deterministic dynamics of an infinite population, could lead to substantial errors in the identified level of selection. In the examples given in Figure 5 in Comparison of the Expected Trajectory with a Deterministic Trajectory, we note that when the actual selection is positive, the selection coefficient that would be invoked in an infinite population would severely overestimate the real selection coefficient, in some cases by >1000%. If the actual selection were, in fact, negative (a fixing trajectory associated with a negative selection coefficient is given in Figure 1B), then the identified selection coefficient would not even have the correct sign. It seems possible to us that in experimental evolution, and related studies, where population sizes are modest, a misidentification of the strength of selection may be possible, but it seems less likely in laboratory bacterial populations, where population sizes can be extremely large. Furthermore, in such bacterial populations, competition assays against the wild-type strain can provide realistic estimates of the strength of selection.
We have shown in this work how the fictitious force arises from the diffusion approximation: the distribution of the frequency, conditional on fixation by a given time T, obeys a diffusion equation with an additional term, compared with the equation obeyed by the original, unconditional distribution. This additional term represents an evolutionary force and is directly identified with the apparent force that drives the allele to fixation—the fictitious force. The explicit presence of the fictitious force in the diffusion equation makes it a tangible mathematical object that can be subject to examination and analysis.
In a simple case, where the allele ultimately achieves fixation (T = ∞) and is subject to real additive selection (which would be the sole force acting in an infinite population), the selection coefficient describing the fictitious selection, sfict(x), where x is the frequency, is given by Equation 8. The form of the “fictitious selection coefficient”, sfict(x), depends on the effective size of the population, Ne, which confirms the obvious fact that at least some aspects of the fictitious force arise from random genetic drift. However sfict(x) also depends on the selection coefficient, s, of the real selection that is acting. The dependence of the fictitious force on drift is a key feature. In the absence of real selection (s → 0) the fictitious selection coefficient becomes sfict(x) = 1/(2Nex); this is frequency dependent and is extremely large at small frequencies—to prevent loss of the allele. However, the full s-dependent form of the fictitious selection coefficient in Equation 8 has a most interesting property in the limit of no drift.
When the effective population size becomes arbitrarily large (i.e., Ne → ∞), and the real selection coefficient, s, is positive, the fictitious selection coefficient vanishes. This is reasonable; the real selection, in large populations, is sufficient to drive the frequency to fixation. However, when the real selection coefficient, s, is negative, the fictitious selection coefficient sfict(x) becomes sfict(x) = −2s ≡ 2|s|. This feature is related to the known feature that all statistics of the fixation time, under additive or genic selection, are to high accuracy, the same for the selection coefficients s and −s (Nei and Roychoudhury 1973; Maruyama 1974, 1977; see also Taylor et al. 2006) and generally follows from time-reversal properties (Ewens 2004). This is an indication of the potentially strong effect of conditioning, which can effectively nullify the underlying force in the problem and replace it with something substantially different. Of course, the probability of actually observing fixation in a very large population, when selection is negative, is likely to be very small.
The present work has presented results for fixation by a given time. It can also be extended to some other situations, e.g., determining the approximate expected trajectory that achieves a given intermediate frequency at a given time (see Appendix B). The results of Appendix A can also be extended to determining the approximate trajectory that fixes during a specified time interval (results not given). Unlike the case where fixation occurs by a given time, the fictitious forces in these two cases may become negative for some fraction of the time, and this can be interpreted as a suppression of the frequency trajectories, so that the specified end condition is correctly achieved.
Other applications
We have studied Wright–Fisher dynamics, via the diffusion approximation. Closely related to this are stochastic population dynamics, such as those that occur in evolutionary game theory when the population size is finite (Traulsen and Reed 2012). Problems in this area typically lead to a diffusion equation that is closely equivalent to one derived from a Wright–Fisher model with frequency-dependent selection. Often, but not always, more than two types of alleles (or strategies/phenotypes) are considered in a game theory context. There are interesting and counterintuitive phenomena in this area. For example, sometimes an increase in the initial frequency of an allele can lead to an increased mean time of fixation of the allele (Altrock et al. 2010). This behavior is in contrast to that in the simple biological systems we have studied here, where an increased initial frequency would lead to a decreased mean time to fixation. Phenomena that appear to be related to this, in Wright–Fisher models with frequency-dependent selection, have been theoretically predicted to occur in the probability of fixation (Chalub and Souza 2013). It is plausible that in all such systems, the ideas and methods we have presented in this work, associated with the expected trajectory and fictitious selection, could shed new insights into the phenomena that these complex systems exhibit.
Overview
Fixation is generally an improbable event that is strongly influenced by random genetic drift. However, if we condition on fixation, then it appears that a lot of the effects of genetic drift are directly taken into account in the guise of a fictitious force. The effects of random genetic drift that remain, while not being negligible for small populations, appear to be relatively unbiased. Thus most of the dynamics of the mean trajectory result from the effects of the real selection (or other evolutionary forces) that would occur in an infinite population, combined with the effects of the fictitious force. It is easy to imagine that strong forces (or other processes) may be invoked in problems where conditioning, particularly conditioning on the future, has been carried out, but that the forces are largely an outcome of the conditioning and do not have a real existence. The work presented here clarifies these issues and provides two useful quantities for future analyses: the characteristic trajectory of a fixing allele and its primary driving force, namely fictitious selection. This work also provides a rationale for further interest in evolutionary trajectories that achieve high frequencies or fixation: they seem to rather directly encapsulate important information and insight about selective forces.
Acknowledgments
We thank Jianfeng Feng, Joel Peck, Wenbo Sheng, and John Welch for helpful discussions. We are grateful to the handling editor, Lindi Wahl, and two anonymous reviewers for comments and suggestions that have helped us to improve this work. M.L. acknowledges the Centre for Computational Systems Biology and the School of Biological Sciences of Fudan University, particularly Ji Yang, for support during his stay at Fudan University. A.D.J.O. acknowledges the Centre for Computational Systems Biology of Fudan University for hospitality and the European Community Seventh Framework Programme (EU-FP7: Advanced Biomaterials for Regenerative Medicine) for support.
Appendices
Appendix A: Relating Conditioned and Unconditioned Problems
In this Appendix we determine the form of the fictitious evolutionary force that is equivalent to restricting frequency trajectories to a specific subset of all trajectories (i.e., to conditioning). We establish results involving (i) K(x, t|y, u), the (unconditioned) probability density of X(t), at frequency x, given that X(u) = y; (ii) Kseg(x, t|y, u), which represents the part of K(x, t|y, u) that includes only contributions from trajectories that are segregating; and (iii) Pfix(t|y, u), the probability of fixation by time t, given that X(u) = y.
The results we establish are based on the h-transform of the probability density by Doob (1957) and are as follows:
-
If we condition so that fixation occurs by time T (i.e., fixation can occur any time up to and including time T), and if the diffusion equation for K(x, t| y, u) is
(A1) - then the conditioned probability density obeys an equation of the same form as Equation A1 but with the replacement
(A2)
-
If we condition so that the frequency achieves the value z at time T, then the conditioned probability density obeys Equation A1 with
(A3) - In all of the cases considered in the present work,
(A4)
The term by which M(x, t) is augmented in Equations A2 and A3 represents the fictitious force, Mfict(x, t). Thus, in the fixation case, , and in the case where frequency z is achieved, . In each case we choose to write the expression for Mfict(x, t) as
| (A5) |
with sfict(x, t) defined to be the selection coefficient of the fictitious selection, and we can read off the form it takes in the “fixation” and “frequency achievement” cases from comparison with Equations A2 and A3.
Note that the replacement of M(x, t) via Equations A2 and A3 expresses the fact that under the diffusion approximation, a problem that is conditioned has a probability density that follows from an unconditioned problem, provided the force in the problem is modified from M(x, t) to M(x, t) + Mfict(x, t).
Calculation
To begin, we note that the unconditioned probability density we use, namely K(x, t| y, u), is complete in the sense that it describes populations where the A allele is segregating or has been lost or fixed (McKane and Waxman 2007; Waxman 2011b). As such, it possesses the property that for all t ≥ u, and K(x, t| y, u) generally contains spikes (Dirac delta functions) located at x = 0 and x = 1, which signify loss and fixation.
We first establish a general result for a probability density KC(x, t| y, u), which, later, we associate with problems involving conditioning. We take KC(x, t| y, u) to have the form
| (A6) |
where B(x, t) and F(x, t) obey backward and forward diffusion equations with respect to x and t. To maintain some generality, we write these equations as
| (A7) |
| (A8) |
where V = V(x, t) and M = M(x, t). We use these equations to determine the equation that KC = BF obeys. One way to proceed is to write F = KCB−1 and substitute this into Equation A8. Using Equation A7 leads, with some algebra, to the general result
| (A9) |
Thus KC(x, t| y, u) obeys an equation similar to that of F(x, t) except M(x, t) is replaced by :
| (A10) |
Next, we show that for the class of conditioned problems dealt with in this article, the distribution can be written in the form KC(x, t| y, u) = B(x, t)F(x, t), where F represents the unconditioned distribution.
In principle, the results we derive below hold when parameters such as the effective population size and the selection coefficient, etc., depend on time (cf. Waxman 2011a). We do not pursue any issues associated with such time dependence in this work.
Distribution when the frequency z is achieved at time T or fixation is achieved by time T
Let z denote a frequency that lies in the range 0 < z < 1 and corresponds to a segregating allele. We use Kz,T(x, t| y, u) to denote the solution of the diffusion equation that represents the probability density of X(t), at frequency x, given (i) that X(u) = y (with 0 < y < 1) and (ii) that the frequency z is achieved at time T. We have, by definition, Kz,T(x, t| y, u) = K(x, t| z, T; y, u). Using Bayes’ formula we obtain Kz,T(x, t| y, u) = K(z, T| x, t; y, u)K(x, t| y, u)/K(z, T| y, u). We use the Markov property to obtain Kz,T(x, t| y, u) = K(z, T| x, t)K(x, t| y, u)/K(z, T| y, u) and note that since z is a segregating frequency (0 < z < 1), all of the K’s on the right-hand side can be replaced by Kseg’s since none of the Dirac delta functions present can contribute. It follows that
| (A11) |
Consider now the frequency z = 1 that corresponds to a fixed allele. We use KT(x, t| y, u) to denote the probability density of X(t), at frequency x, given (i) that X(u) = y (with 0 < y < 1) and (ii) that fixation is achieved by time T. The analysis is similar to the segregating case just considered; we again start with Kz,T(x, t| y, u) but now allow z to take general values. We are, however, interested in only one part of Kz,T(x, t| y, u), namely the coefficient of the Dirac delta function δ(1 − z) that is located at z = 1. This coefficient is the contribution of frequency trajectories that fix by time T, and a term in δ(1 − z) is present in the solution whenever fixation can occur (McKane and Waxman 2007; Waxman 2011b). The required probability density, KT(x, t y, u), can be written as K(x, t| 1, T; y, u), where the “1, T” denotes formally conditioning on fixation by time T. We proceed as in the segregating case. With K(1, t| y, u) corresponding to the probability of fixation by time T, given that X(u) = y, i.e., K(1, T| y, u) = Pfix(T| y, u), we find that
| (A12) |
Appendix B: The Achievement of an Intermediate Frequency at a Specific Time
To complement the results for fixation that occurs by a given time, we now consider trajectories that achieve the intermediate frequency z at the specific time T. The fictitious force is now
| (B1) |
where Kseg(z, T| x, t) is the probability density of the allele frequency, evaluated at a segregating frequency of z at time T, given an original frequency of x at time t. Again we express this in terms of a selection of the fictitious selection, as in Equation 6. It follows from results in Appendix A that for t ≤ T
| (B2) |
In Figure A1 we illustrate the expected trajectory when a frequency of z = 3/4 is achieved at the time T = Ne.
Figure A1.
The expected trajectory when a frequency of 3/4 is achieved at a specific time, say T = 100. We adopt an effective population size of Ne = 100, a real selection coefficient of s = 0.01, and an initial frequency of y = 0.01. The approximate results follow from Equation 9 while the exact results come from a Wright–Fisher model (Fisher 1930; Wright 1931).
Appendix C: Determination of an Approximate Equation of the Expected Allele Trajectory
In this Appendix we determine an equation for the expected allele trajectory. We begin by writing the equation for the conditioned solution of the diffusion equation, Equation A9, as
| (C1) |
We assume that the probability current density of KC(x, t| y, u), namely
, vanishes at x = 0 and x = 1 (McKane and Waxman 2007), which automatically ensures conservation of probability .
Let us write the expected allele frequency at time t as . Then , where we have imposed the initial condition that . To fully determine we would need to solve the diffusion equation for KC(x, t| y, 0); this is generally hard. We aim for an approximation and proceed by multiplying Equation C1 by x and integrating from 0 to 1. The vanishing of the probability current density at x = 0 and x = 1 yields . Assuming V(0, t) = 0, V(1, t) = 0, yields . The most basic approximation is to replace x in M(x) + Mfict(x, t) by . This neglects fluctuations around the expected trajectory and yields
| (C2) |
This equation can be viewed as the beginning of an expansion around the expected value. We can include higher-order terms. If we terminated the expansion at second-order deviations from the mean, we would obtain coupled equations for the expected value and the variance; however, here we employ Equation C2.
Using the form of in Equation 5 leads to Equation 9 of the main text. We have numerically solved this equation and find it leads to an that does not have large mean deviations from the exact solution—see Table 1 of the main text.
Footnotes
Communicating editor: L. M. Wahl
Literature Cited
- Altrock P. M., Gokhale C. S., Traulsen A., 2010. Stochastic slowdown in evolutionary processes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 82: 011925. [DOI] [PubMed] [Google Scholar]
- Chalub, F. A. C. C., and O. M. Souza, 2013 The frequency-dependent Wright–Fisher model: diffusive and non-diffusive approximations. J. Math. Biol. DOI: 10.1007/s00285-013-0657-7. [DOI] [PubMed] [Google Scholar]
- Cook L. M., Turner J. R. G., 2008. Decline of melanism in two British moths: spatial, temporal and inter-specific variation. Heredity 101: 483–489 [DOI] [PubMed] [Google Scholar]
- Doob J. L., 1957. Conditional Brownian motion and the boundary limits of harmonic functions. Bull. Soc. Math. France 85: 431–458 [Google Scholar]
- Ewens W. J., 1973. Conditional diffusion processes in population genetics. Theor. Popul. Biol. 4: 21–30 [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics: I. Theoretical Introduction. Springer-Verlag, New York [Google Scholar]
- Ewing G., Hermisson J., 2010. MSMS: a coalescent simulation program including recombination, demographic structure, and selection at a single locus. Bioinformatics 26: 2064–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1922. On the dominance ratio. Proc. R. Soc. Edinb. 42: 321–341 [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford [Google Scholar]
- Fisher R. A., Ford E. B., 1947. The spread of a gene in natural conditions in a colony of the moth Panaxia dominula L. Heredity 1: 143–174 [Google Scholar]
- Ford E. B., Sheppard P. M., 1969. The medionigra polymorphism of Panaxia dominula. Heredity 24: 112–134 [DOI] [PubMed] [Google Scholar]
- Gifford D. R., de Visser J. A. G. M., Wahl L. M., 2012. Model and test in a fungus of the probability that beneficial mutations survive drift. Biol. Lett. 9: 20120310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B. S., Owen D. F., Clarke C. A., 1996. Parallel rise and fall of melanic peppered moths in America and Britain. J. Hered. 87: 351–357 [Google Scholar]
- Haldane J. B. S., 1927. A mathematical theory of natural and artificial selection. V. Selection and mutation. Proc. Camb. Philos. Soc. 23: 838–844 [Google Scholar]
- Huerta-Sanchez M., Durrett R., Bustamante C. D., 2008. Population genetics of polymorphism and divergence under fluctuating selection. Genetics 178: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth C. J., Parts L., Schiffels S., Liti G., Mustonen V., 2012. Quantifying selection acting on a complex trait using allele frequency time-series data. Mol. Biol. Evol. 29: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1955. Stochastic processes and distribution of gene frequencies under natural selection. Cold Spring Harb. Symp. Quant. Biol. 20: 33–53 [DOI] [PubMed] [Google Scholar]
- Kimura M., 1962. On the probability of fixation of mutant genes in a population. Genetics 47: 713–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1964. Diffusion models in population genetics. J. Appl. Probab. 1: 177–232 [Google Scholar]
- Kimura M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK [Google Scholar]
- Krone S. M., Neuhauser C., 1997. Ancestral processes with selection. Theor. Popul. Biol. 51: 210–237 [DOI] [PubMed] [Google Scholar]
- Lambert A., 2008. Population dynamics and random genealogies. Stoch. Models 24: 145–163 [Google Scholar]
- Maruyama T., 1974. The age of an allele in a finite population. Genet. Res. 23: 137–143 [DOI] [PubMed] [Google Scholar]
- Maruyama, T., 1977 Stochastic problems in population genetics, pp. 1–245 in Lecture Notes in Biomathematics, Vol. 17, edited by S. Levin. Springer-Verlag, Berlin. [Google Scholar]
- McKane A. J., Waxman D., 2007. Singular solutions of the diffusion equation of population genetics. J. Theor. Biol. 247: 849–858 [DOI] [PubMed] [Google Scholar]
- Miller L. H., Mason S. J., Clyde D. F., McGuiness M. H., 1976. The resistance factor to Plasmodium vivax in Blacks: the Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295: 302–304 [DOI] [PubMed] [Google Scholar]
- Messer P. W., Petrov D. A., 2013. Frequent adaptation and the McDonald–Kreitman test. Proc. Natl. Acad. Sci. USA 110: 8615–8620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Roychoudhury A. A. K., 1973. Probability of fixation and mean fixation time of an overdominant mutation. Genetics 74: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara R. B., 2005. Comparing the effects of genetic drift and fluctuating selection on genotype frequency changes in the scarlet tiger moth. Proc. Biol. Sci. 272: 211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P., Whitlock M. C., 1997. The probability of fixation in populations of changing size. Genetics 146: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwa Z., Wahl L. M., 2008. The fixation probability of beneficial mutations. J. R. Soc. Interface 5: 1279–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper J. W., Findlay C. S., Kassen R., Spencer S. L., Maley C. C., 2009. Cancer research meets evolutionary biology. Evol. Appl. 2: 62–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P., Malmström H., Raghavan M., Stora J., Hall P., et al. , 2012. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science 336: 466–469 [DOI] [PubMed] [Google Scholar]
- Rodrigo A. G., Felsenstein J., 1999. Coalescent approaches to HIV population genetics, pp. 233–272 in The Evolution of HIV, edited by Crandall K. A. Johns Hopkins University Press, Baltimore [Google Scholar]
- Sabeti P. C., Schaffner S. F., Fry B., Lohmueller J., Varilly P., et al. , 2006. Positive natural selection in the human lineage. Science 312: 1614–1620 [DOI] [PubMed] [Google Scholar]
- Seixas S., Ferrand N., Rocha J., 2002. Microsatellite variation and evolution of the human duffy blood group polymorphism. Mol. Biol. Evol. 19: 1802–1806 [DOI] [PubMed] [Google Scholar]
- Taylor C., Iwasa Y., Nowak M. A., 2006. A symmetry of fixation time in evolutionary dynamics. J. Theor. Biol. 21: 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. M., Antonia A., Feng G., Mwapasa V., Chaluluka E., et al. , 2012. Adaptive evolution and fixation of drug-resistant Plasmodium falciparum genotypes in pregnancy-associated malaria: 9-year results from the QuEERPAM study. Infect. Genet. Evol. 12: 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima K. M., Innan H., 2009. mbs: modifying Hudson’s ms software to generate samples of DNA sequences with a biallelic site under selection. BMC Bioinformatics 10: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traulsen A., Reed F. A., 2012. From genes to games: cooperation and cyclic dominance in meiotic drive. J. Theor. Biol. 299: 120–125 [DOI] [PubMed] [Google Scholar]
- Uecker H., Hermisson J., 2011. On the fixation process of a beneficial mutation in a variable environment. Genetics 188: 915–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D., 2011a A unified treatment of the probability of fixation when population size and the strength of selection change over time. Genetics 188: 907–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D., 2011b Comparison and content of the Wright–Fisher model of random genetic drift, the diffusion approximation, and an intermediate model. J. Theor. Biol. 269: 79–87 [DOI] [PubMed] [Google Scholar]
- Wright S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1948. On the roles of directed and random changes in gene frequency in the genetics of populations. Evolution 2: 279–294 [DOI] [PubMed] [Google Scholar]