SUMMARY
Lysine63-linked ubiquitin (K63-Ub) chains represent a particular ubiquitin topology that mediates proteasome-independent signaling events. The deubiquitinating enzyme (DUB) BRCC36 segregates into distinct nuclear and cytoplasmic complexes that are specific for K63-Ub hydrolysis. RAP80 targets the five-member nuclear BRCC36 complex to K63-Ub chains at DNA double-strand breaks. The alternative four-member BRCC36 containing complex (BRISC) lacks a known targeting moiety. Here we identify Serine Hydroxymethyltransferase (SHMT) as a heretofore-unappreciated component that fulfills this function. SHMT directs BRISC activity at K63-Ub chains conjugated to the type 1 interferon (IFN) receptor chain 1 (IFNAR1). BRISC-SHMT2 complexes localize to and deubiquitinate actively engaged IFNAR1, thus limiting its K63-Ub mediated internalization and lysosomal degradation. BRISC deficient cells and mice exhibit attenuated responses to IFN and are protected from IFN-associated immunopathology. These studies reveal a novel mechanism of DUB regulation, and suggest a therapeutic use of BRISC inhibitors for treating pathophysiologic processes driven by elevated IFN responses.
INTRODUCTION
Ubiquitination is a critical modulator of protein activity and function (Ciechanover and Schwartz, 2004). The status of protein ubiquitination is determined by counteracting activities of E3 ubiquitin ligases that facilitate ubiquitin conjugation and de-ubiquitinating enzymes (DUBs) that remove the ubiquitin moiety from proteins (Kimura and Tanaka, 2010; Reyes-Turcu and Wilkinson, 2009). Accordingly, DUBs have emerged as important regulators of many physiologic processes including regulation of gene expression, cell division, DNA repair, signal transduction, receptor endocytosis and trafficking (Amerik and Hochstrasser, 2004; Clague and Urbe, 2006).
Whereas an ability to interact with a specific substrate represents an essential characteristic of numerous E3 ligases, it is poorly understood how DUBs are targeted to their substrates. As there are many fewer DUBs (compared to the number of E3 ligases), it is assumed that DUBs are likely to function within the context of multi-protein complexes and may rely on ancillary proteins for substrate recognition (Sowa et al., 2009; Ventii and Wilkinson, 2008).
We have previously reported the nuclear complex DUB containing MERIT40, BRCC45, Abraxas and BRCC36 that is capable of disassembling the K63-conjugated polyubiquitin chain (K63-Ub) upon targeting to DNA damage sites via its interaction with the tandem ubiquitin interaction motif containing protein RAP80 (Shao et al., 2009a; Shao et al., 2009b; Sobhian et al., 2007). A related complex appears to represent a major K63-Ub-directed DUB activity in the cytoplasm (Cooper et al., 2010; Cooper et al., 2009; Feng et al., 2010; Patterson-Fortin et al., 2010). This complex termed BRISC (the BRCC36 isopeptidase complex) utilizes KIAA0157/Abro instead of Abraxas and does not contain RAP80 (Figure 1A). Both complexes show exquisite specificity for K63-Ub hydrolysis, which relies on Zn2+ coordination within the active site of JAMM domain of BRCC36 (Cooper et al., 2010; Sobhian et al., 2007) as well as protein-protein interactions between BRCC36 and MPN- domain containing proteins Abraxas and KIAA0157, for the nuclear RAP80 and cytoplasmic BRISC complexes respectively (Cooper et al., 2010; Feng et al., 2010; Patterson-Fortin et al., 2010; Shao et al., 2009b). While both complexes rely on similar protein interactions for DUB activity, it is notable that the targeting component of the BRISC has remained elusive, as has a bona fide cytoplasmic substrate of its DUB activity (Figure 1A).
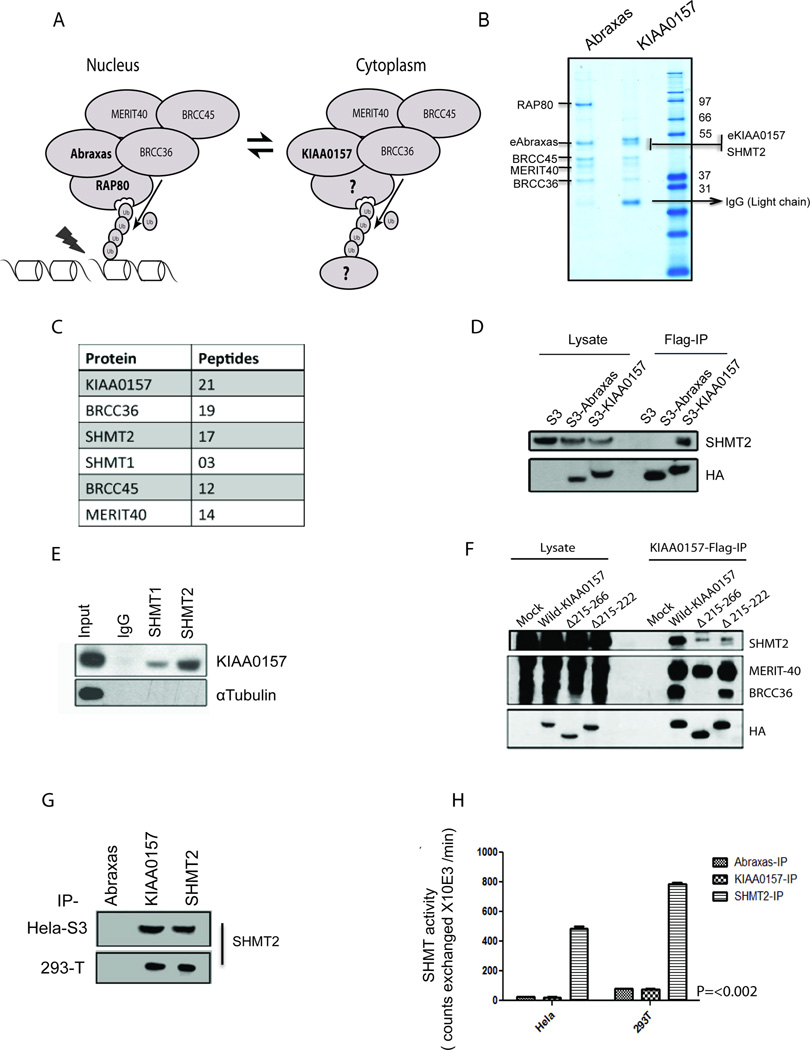
Figure 1. Identification of SHMT Species as a 5th Component of the BRISC Complex.
(A) Schematic comparison of the nuclear RAP80 complex that directs BRCC36-mediated DUB activity at K63-Ub chains formed at DNA damage sites with the cytoplasmic BRISC complex whose substrates and targeting components remain unknown.
(B) Proteins co-purified with either Flag-HA- Abraxas (eAbraxas) or KIAA0157 (eKIAA0157) were separated by SDS-PAGE and stained using Coomassie Brilliant Blue.
(C) Tabular display of the number of tryptic peptides from each of the indicated proteins that co-purified with eKIAA0157.
(D) Flag immunoprecipitation (IP) reveals association of SHMT2 with KIAA0157 but not Abraxas.
(E) KIAA0157 was detected by immunoblot following IP of endogenous SHMT1 or SHMT2.
(F) 293T cells transiently expressing wild and mutant Flag tagged versions of KIAA0157 were lysed and used for Flag-IP. Co- IPed proteins were detected using different antibodies as indicated.
(G) Immunoblot displaying similar amounts of SHMT2 from IP of either Flag-HA-KIAA0157 or endogenous SHMT2 from HeLa and 293T cells. SHMT catalytic activity was assayed from these immunoprecipitates (IPs).
(H) SHMT activity was observed in IPs of SHMT, but not KIAA0157. Similar results were observed in both cell lines.
Here we report identification of Serine Hydroxymethyltransferase (SHMT) as a specific BRISC component that targets the catalytic core of this complex to activated IFNAR1 chains of the type 1 interferon (IFN) receptor. BRISC-SHMT is capable of disassembling the K63-Ub chains from IFNAR1, thus limiting receptor endocytosis and lysosomal degradation. These activities are required for receptor-mediated cellular responses to IFN and its inducers in vitro and in vivo.
RESULTS
Identification of SHMT Species as KIAA0157 Interacting Partners and Regulators of BRISC Targeting to IFNAR1
To identify proteins that may target BRISC to its substrates, we utilized a proteomics screen approach in which interacting partners of Abraxas were compared with proteins co-purified with the BRISC-specific KIAA0157 expressed in HeLa S3 cells (Figure 1B). Examination of the Coomassie stained gel and tryptic peptides identified by mass spectrometry revealed common components of BRCC36, MERIT40 and BRCC45 for both complexes and the exclusive association of RAP80 with the nuclear Abraxas complex. In addition to the expected BRISC constituents MERIT40, BRCC45 and BRCC36, we identified Serine Hydroxymethyltransferase-2 (SHMT2) as a near stoichiometric binding partner and specific component of KIAA0157 purification (Figure 1B, C). Co-immunoprecipitation analysis further demonstrated that SHMT2 is associated with KIAA0157 but not with Abraxas (Figure 1D). A related metabolic enzyme SHMT1 was also co-purified with KIAA0157 albeit at a lesser peptide frequency (Figure 1C). Accordingly, KIAA0157 was detected in immunoprecipitates of endogenous SHMT1 and SHMT2 (Figure 1E). Consistent these observations, SHMT2 had been reported in a large-scale DUB proteomics screen to interact with BRCC36 (Sowa et al. 2009). However the functional significance of this association was not described nor was it appreciated that BRCC36 interacts with SHMT2 specifically in the context of the BRISC complex and not with the RAP80 complex. Our data indicate that SHMT associates with the BRISC complex via the interaction with KIAA0157. Mapping of the KIAA0157 that confer SHMT binding demonstrated that interaction with SHMT2 was less evident in the KIAA0157 mutant lacking the coil-coil domain. Importantly, the mutant lacking amino acids 215–222 within KIAA0157 (further termed Δ215–222) exhibited a decreased co-immunoprecipitation with SHMT2 while retaining the ability to interact with BRCC36 (Figures 1F and S1A). SHMT proteins localize to the mitochondria, cytoplasm, and nucleus. We were unable to detect any BRISC protein in the mitochondria, suggesting the interaction with SHMT occurs outside of the mitochondria (Figure S1B), however, purification of epitope tagged KIAA0157 from fractionated HeLa S3 cells revealed the presence of the BRISC-SHMT complex in both the cytoplasm and nucleus (Figure S1C and D).
SHMT1 and SHMT2 are tetrameric metabolic enzymes involved in one carbon metabolism. SHMT1/2 catalyze the reversible interconversion of Serine and tetrahydrofolate to Glycine and methylenetetrahydrofolate. This reaction generates single-carbon units for purine, thymidine, and methionine biosynthesis. We were unable to detect a SHMT catalytic activity associated with BRISC in KIAA0157 purifications, while similar amounts of the enzyme purified by SHMT2 immunoprecipitates demonstrated robust exchange of tritium from Glycine to H2O as measured by an isotopic assay with known specificity for SHMT activity (Scheer et al., 2005) (Figures 1 G, H). These observations are consistent with SHMT playing a structural function in the context of its association with BRISC, rather than a catalytic role in conjunction with BRISC DUB activity.
The preceding data suggest that SHMT proteins may function analogously to RAP80 in the BRISC DUB complex, potentially targeting BRISC to K63-linked polyubiquitin chains conjugated to substrate proteins. Intriguingly, SHMT2 proteins were also pulled down together with double co-immunoprecipitation with FLAG-tagged IFNAR1 chain and HA-tagged IFNAR2 chain of the type 1 interferon (IFN) receptor (Figure S2A, B). This receptor is rapidly downregulated in response to ligand and numerous non-ligand stimuli (Fuchs, 2013; Huangfu and Fuchs, 2010) that trigger robust phosphorylation-dependent ubiquitination of the IFNAR1 chain facilitated by the SCF-β-Trcp E3 ubiquitin ligase (Kumar et al., 2004; Kumar et al., 2003). Importantly, both the presence of K63-Ub on IFNAR1 and the role of these ubiquitin chain topologies in IFNAR1 endocytosis and degradation have been demonstrated (Fuchs, 2012; Kumar et al., 2007).
Co-immunoprecipitation experiments using either endogenous (Figure 2A) or FLAG-tagged recombinant (Figure S2C) IFNAR1 corroborated its ability to interact with SHMT2 in naïve cells and demonstrated that this interaction is further increased by treatment of cells with IFN. In addition, increased association between endogenous SHMT2 and FLAG-KIAA0157 was found in immunoprecipitates from the ligand-treated cells (Figure 2B), while the Δ215–222 protein failed to show such increases (Figure 2B). This result suggests that IFNAR1 engagement by ligand promotes a ternary complex with BRISC and SHMT2.
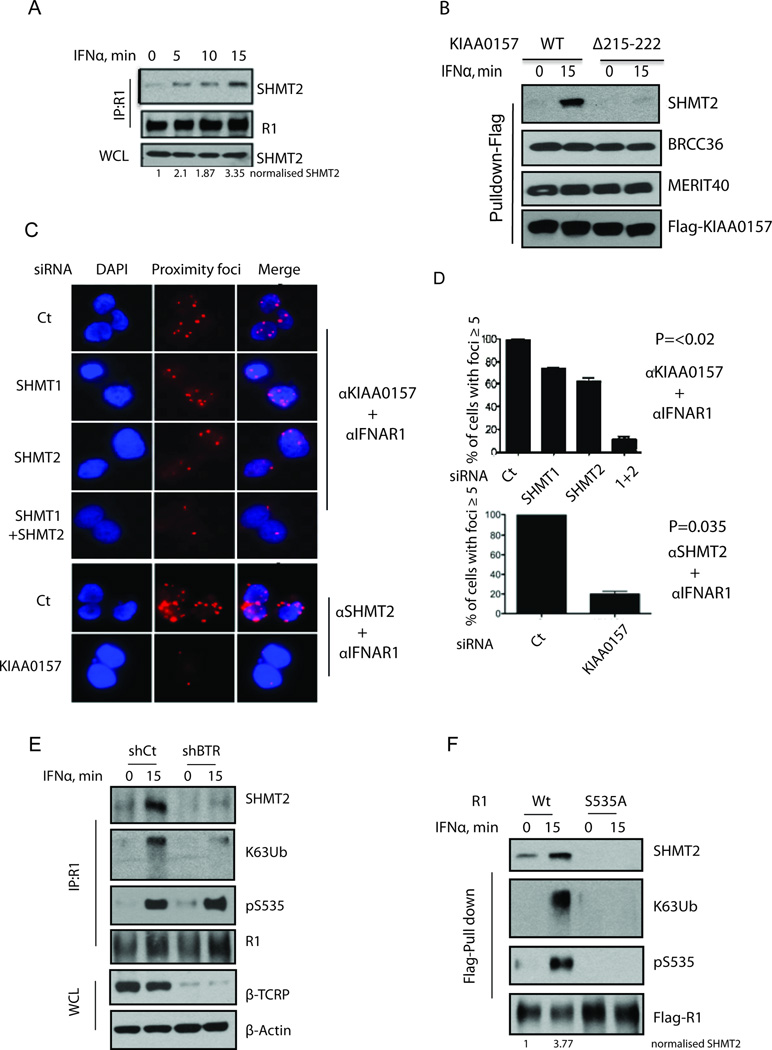
Figure 2. Interferon Stimulation Enhances BRISC-SHMT2 interaction and Association with IFNAR1.
(A) 293T cells were treated with IFNα for the indicated times. IFNAR1 was IPed and then SHMT2 and IFNAR1 were detected by immunoblotting using the indicated antibodies. WCL: Whole Cell Lysates.
(B) Cells expressing wildtype (WT) and Δ215–222 mutant KIAA0157 were treated with or without IFNα for 15 minutes. Flag-IP was performed and co-purified proteins were detected using the indicated antibodies.
(C) Proximity ligation assay showing interaction between KIAA0157 or SHMT2 and IFNAR1 (Proximity foci). U2OS cells with stable expression of Flag-HA-KIAA0157 were transfected with the indicated siRNAs and interaction between KIAA0157 or SHMT2 and IFNAR1 was studied using rabbit IFNAR1 and mouse anti-HA antibodies.
(D) Quantification of foci from A. Data and error bars here and thereafter represent Mean±S.E.M. from at least three independent experiments that included at least 100 cells per sample. P values were calculated using the Student’s t-test.
(E) 293T cells were transfected with shRNAs against GFP or β-Trcp (BTR). Cells were treated with IFNα for indicated times. IFNAR1 was IPed and SHMT2, K63-Ub, pS535 and IFNAR1 were detected by immunoblotting using the indicated antibodies. WCL was used for detection of β-Trcp and β-Actin by immunoblotting.
(F) 293T cells were transfected with Flag-IFNAR1 wild type (WT) or Flag-IFNAR1 S535A. Cells were treated with IFNα for indicated times. Flag-IFNAR1 was immunoprecipitated, and SHMT2, K63-Ub, pS535 and Flag-IFNAR1 were detected by immunoblotting using the indicated antibodies.
Proximity ligation assay analysis using the Duolink approach was employed to visualize colocalization of BRISC, SHMT2, and IFNAR1 in cells given the broad subcellular localization of SHMT and BRISC using standard immunofluorescence techniques. Duolink analysis demonstrated that IFNAR1 is capable of interacting with KIAA0157 and SHMT2 in intact cells. Importantly, knockdown of both SHMT1 and SHMT2 dramatically decreased the latter interaction, and similarly KIAA0157 knockdown decreased interaction between SHMT2 and IFNAR1 (Figure 2C–D) consistent with co-dependency of BRISC and SHMT for IFNAR1 targeting. Consistent with protein purification studies (Figure S1C and D), Duolink signal for interaction between all species was observed in both the cytoplasm and the nucleus (Figure S2D).
We next sought to delineate the mechanism underlying an increase in this recruitment in the IFN-treated cells. Treatment of cells with ligand was shown to stimulate the ubiquitination of endogenous IFNAR1 by increasing the Serine phosphorylation of its degron (including S535) that enables the recruitment of β-Trcp2 (also termed HOS or FBXW11, (Kumar et al., 2004; Kumar et al., 2003). In addition, overexpression of recombinant IFNAR1 and other inducers of unfolded protein responses can trigger ligand-independent IFNAR1 phosphorylation and ubiquitination (Fuchs, 2013; Liu et al., 2009). Importantly, the knockdown using shRNA targeted to both β-Trcp1 and β-Trcp2 noticeably decreased K63-Ub conjugation to the endogenous IFNAR1 as well as decreased the recruitment of SHMT2 to this receptor (Figure 2E). Moreover, mutant IFNAR1S535A that is incapable of binding β-Trcp and thus exhibits greater stability and signaling (Kumar et al., 2003) demonstrated negligible interaction with SHMT2 in either untreated or treated cells (Figure 2F). Collectively, the data suggest that ubiquitination of IFNAR1 stimulates the recruitment of SHMT2 by direct or/and indirect mechanisms.
The BRISC-SHMT Complex Regulates Ubiquitination, Endocytosis and Proteolytic Turnover of IFNAR1
Incubation of recombinant BRISC with IFNAR1 robustly de-conjugated the K63-linked chain from this receptor (Figure S3A) raising the possibility IFNAR1 is a BRISC substrate. Indeed, knockdown of either KIAA0157 or SHMT2 (Figure 3B) or of BRCC36 (Figure 3C) noticeably increased the extent of ligand-induced K63-linked ubiquitination of IFNAR1. This outcome might result from indirect regulatory effects of BRISC components on either protein kinases that phosphorylate IFNAR1 to enable the recruitment of β-Trcp or on the activity of the SCF-β-Trcp E3 ubiquitin ligase itself. However, we considered these possibilities unlikely because knockdown of BRISC components affected neither IFNAR1 phosphorylation on Ser535 (Figure 3B) nor the K48-linked chains on IFNAR1 (Figure 3C), revealing specificity for DUB activity on IFNAR1 conjugated to K63-Ub.
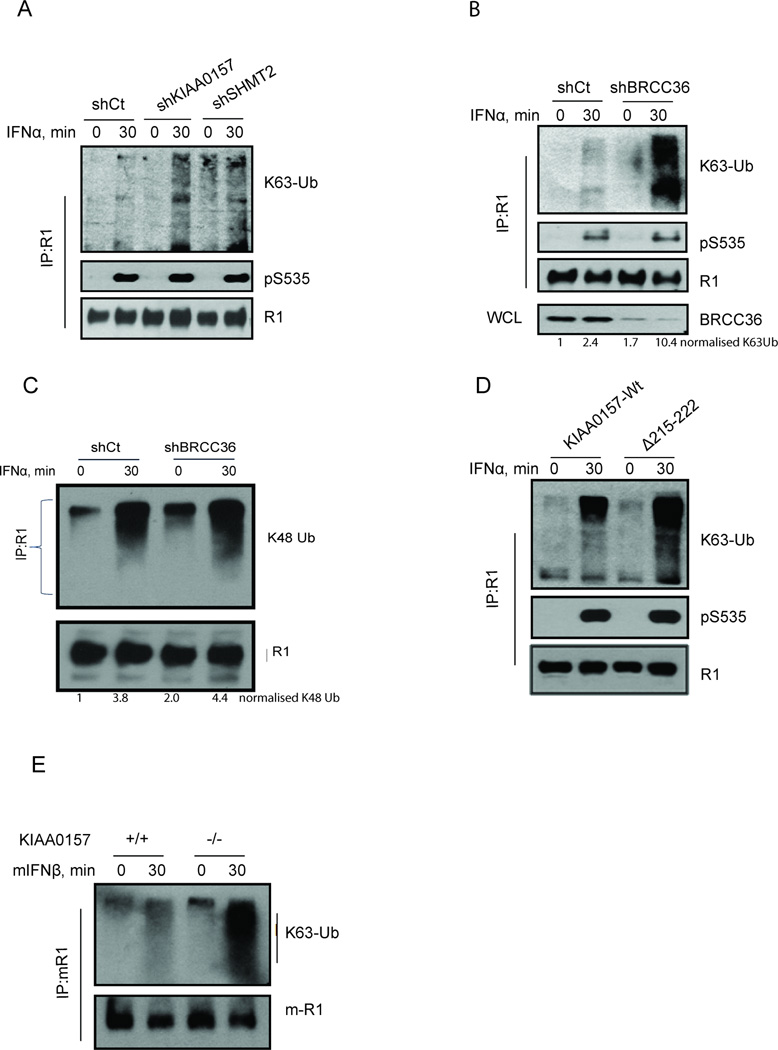
Figure 3. The BRISC-SHMT Complex Regulates K63-linked Ubiquitination of IFNAR1.
(A) 2fTGH cells with stable knockdown of KIAA0157 or SHMT2 were incubated with or without IFNα as indicated. IFNAR1 was IPed from cell lysates and K63-Ub, pS535 and IFNAR1 were detected using the indicated antibodies.
(B) 2fTGH cells with stable knockdown of BRCC36 were treated as in B. IFNAR1 was IPed from cell lysates and K63-Ub, pS535 and IFNAR1 were detected using the indicated antibodies.
(C) Flag-IFNAR1 was IPed from 293T cells and immunoblot was performed using antibodies specific to K48-Ub chains as indicated.
(D) K63-Ub modification and degron phosphorylation of endogenous IFNAR1 in cells expressing wild type KIAA0157 or its Δ215–222 mutant.
(E) MEFs from wild type or KIAA0157-null mice were treated with or without mIFNβ for 30 min. IFNAR1 was IPed from cell lysates and K63-Ub, pS526 (corresponding to human pS535) and IFNAR1 were detected using the indicated antibodies.
In line with this possibility, HeLa cells stably overexpressing wild type BRCC36 but not its enzymatically inactive BRCC36QSQ mutant (described in detail in (Shao et al., 2009b; Sobhian et al., 2007)) robustly decreased the K63-linked ubiquitination of IFNAR1 (Figure S3C). IFNAR1 ubiquitination was also noticeably increased in cells expressing KIAA0157 Δ215–222 protein suggesting SHMT2 dependent BRISC targeting is required to deubiquitinate IFNAR1 (Figure 3D). Finally, to obtain unequivocal genetic data on the role of BRISC in IFNAR1 ubiquitination we have generated KIAA0157 knockout mice by deleting exons 6 and 7 of the murine KIAA0157 locus (Figure S4). These mice were viable and fertile, and harbored an intact nuclear RAP80 complex (data not shown), but no detectable KIAA0157 protein. In embryonic fibroblasts (MEFs) obtained from KIAA0157−/− mice, we observed a much greater induction of K63-Ub on IFNAR1 compared to cells from wild type animals (Figure 3E). Together, these data strongly suggest that BRISC is recruited to IFNAR1 via interaction with SHMT and that this complex is a key regulator of receptor K63-ubiquitination.
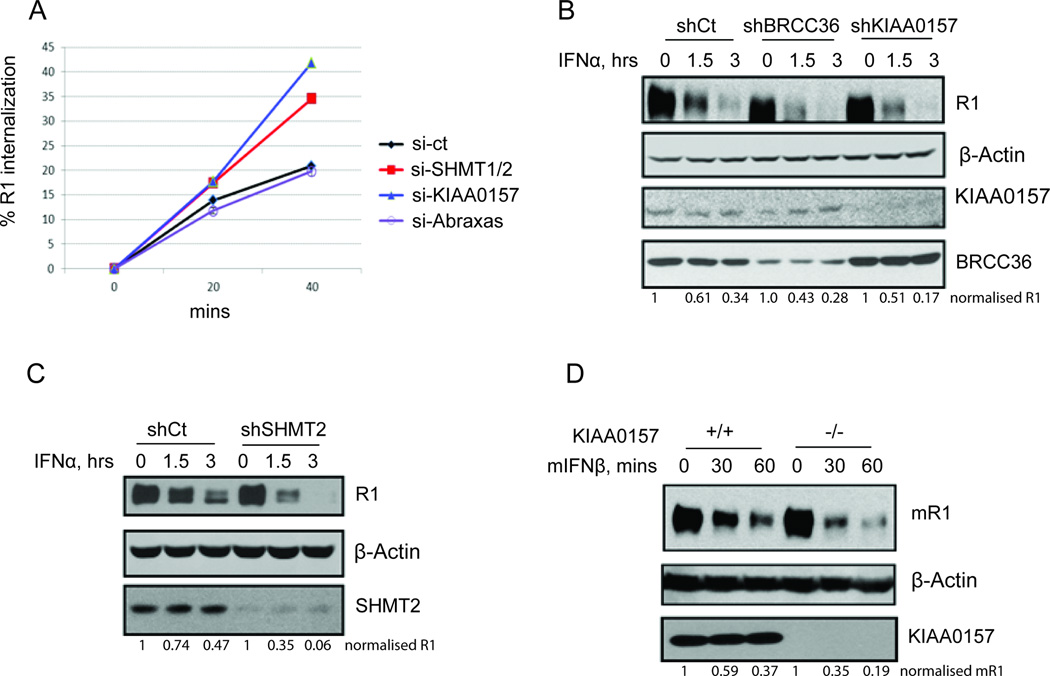
Ubiquitination of signaling receptors localized at the cell surface often regulates endocytosis of these proteins (Clague and Urbe, 2006; Haglund and Dikic, 2012). K63-Ub chains were shown to contribute to the stimulation of IFNAR1 endocytosis and acceleration of the proteolytic turnover of this receptor in the lysosomes (Kumar et al., 2007). Consistent with the proposed role of BRISC in regulating IFNAR1 ubiquitination (Figure 3) we observed that knockdown of either SHMT1/2 or KIAA0157 (but not of Abraxas) increased the internalization rate of IFNAR1 in the IFN-treated cells (Figure 4A). To determine whether BRISC regulates the turnover of IFNAR1 we conducted cycloheximide chase analyses on endogenous IFNAR1 in mouse and human cells treated with species-specific IFN (human IFNα or mouse IFNβ). Degradation of human IFNAR1 was accelerated upon knockdown of either BRCC36 or KIAA0157 (Figure 4B) or SHMT2 (Figure 4C). Furthermore, a greater rate of mouse IFNAR1 turnover was seen in KIAA0157 knockout MEFs (Figure 4D). These results suggest that BRISC-SHMT complex counteracts ubiquitination of IFNAR1 and, accordingly, functions to reduce the rate of receptor internalization and degradation.
Figure 4. BRISC-SHMT Regulates Endocytosis, Proteolytic Turnover and Abundance of IFNAR1.
(A) Internalization rate of endogenous IFNAR1 in IFN-stimulated 293T cells that had been transfected the indicated siRNAs.
(B) 2fTGH cells with stable knockdown of BRCC36 or KIAA0157 were treated with IFNα together with CHX (20 µg/ml) for the indicated times. IFNAR1 was immunoprecipitated and detected using specific antibodies. β-actin, KIAA0157, and BRCC36 were detected from WCL using the indicated antibodies.
(C) 2fTGH cells with stable knockdown of SHMT2 were treated with IFNα together with CHX (20 µg/ml) for the indicated times. IFNAR1 was immunoprecipitated and detected using specific antibodies. β-actin and SHMT2 were detected from WCL using the indicated antibodies.
(D) MEFs from wild type or KIAA0157 knockout mice were treated with mouse IFNβ together with CHX (20 µg/ml) for the indicated times. mIFNAR1 was immunoprecipitated and detected using specific antibodies. β-actin and MERIT40 were detected from WCL using the indicated antibodies.
BRISC-SHMT Controls Cellular IFNAR1-Mediated Signaling and Cellular Responses to IFN and Bacterial Lipopolysaccharide (LPS)
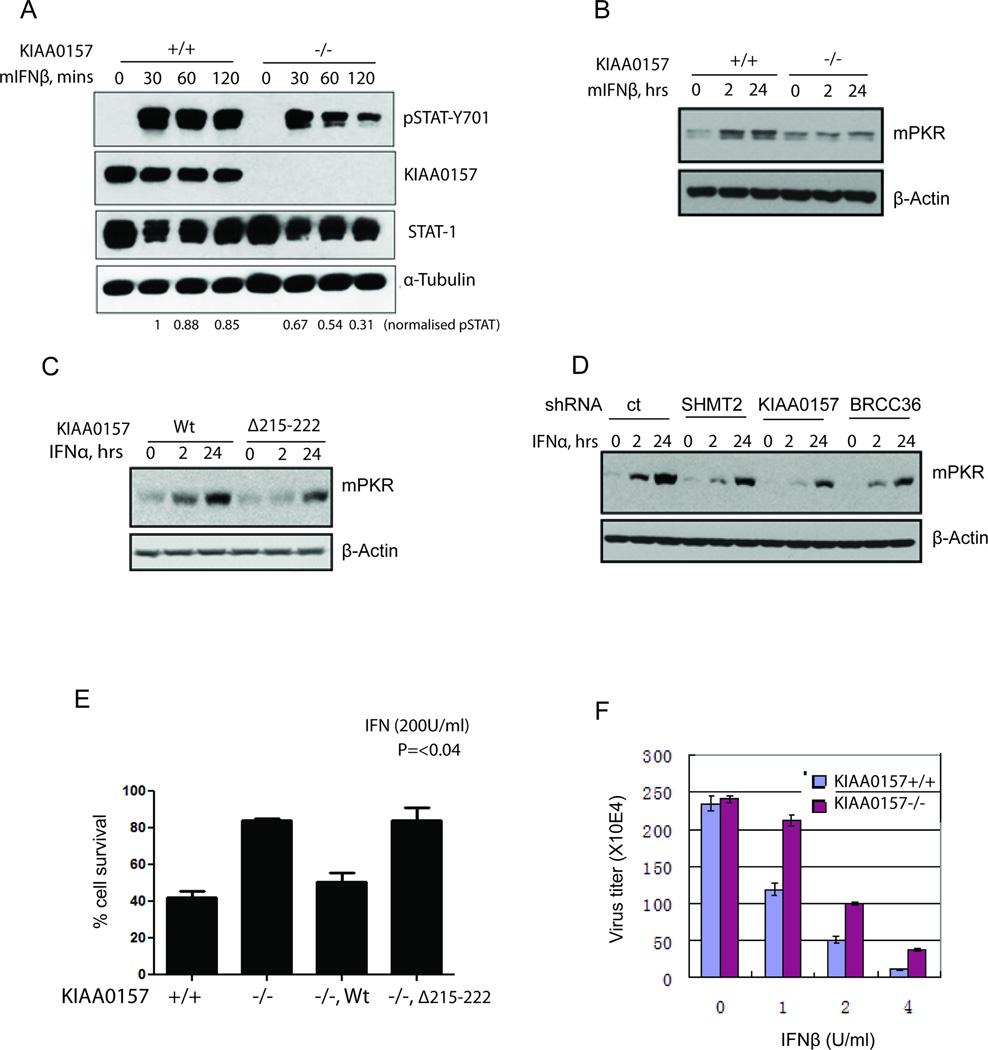
Given that the maintenance of IFNAR1 levels is critical for cellular responses to IFN (Fuchs, 2013), it is plausible that the magnitude and/or duration of cellular responses to IFN would be attenuated by impaired function of BRISC or its inadequate recruitment to IFNAR1. Consistent with this possibility, either activation of STAT1 or induction of PKR by IFN was noticeably weaker in KIAA0157 knockout MEFs compared to their wild type counterparts or where the KIAA0157-Δ215–222 mutant was overexpressed (Figures 5A–C). A similar attenuating effect on induction of STAT1 phosphorylation or induction of ISG15 and PKR was seen in primary human fibroblasts or in HeLa cells where either SHMT2 or BRCC36 or KIAA0157 were knocked down (Figure 5D). Furthermore, the anti-proliferative effects of IFN were less pronounced in KIAA0157−/− MEFs and derivatives of these cells reconstituted with KIAA0157-Δ215–222 mutant (compared to their wildtype counterparts or knockout MEFs where wild type KIAA0157 was re-expressed, Figure 5E). In agreement, IFN afforded a somewhat lesser protection from infection with vesicular stomatitis virus in KIAA0157 knockout MEFs (Figure 5F and S5), reinforcing the notion that BRISC-SHMT activities are required for the full extent of IFN signaling and its biological effects.
Figure 5. BRISC-SHMT Interaction Promotes Responses to IFN.
(A) KIAA0157 +/+ or −/− MEFs were treated with IFNβ for the indicated times. STAT1 Tyr701 phosphorylation and total STAT1 levels were detected from cell lysates using specific antibodies.
(B) MEFs of each genotype were either continuously exposed to mouse IFNβ for 24h or pulsed with IFN for 2h and then incubated without IFN for total of 24h. Levels of PKR and β-actin were detected from whole cell lysates using the indicated antibodies.
(C) 2fTGH cells stably expressing WT KIAA0157 or mutant KIAA0157-DM1 were treated with human FNα and analyzed as done previously in panel B.
(D) 2fTGH cells with stable knockdown of SHMT2 or KIAA0157 or BRCC36 were pulse-treated with IFNα for 2 hours and then IFNα was removed and cells harvested 24 hours after initial treatment. Alternatively, cells were treated with IFNα continuously for 24 hours. Whole cell lysates were used for detection of PKR and β-actin as indicated.
(E) KIAA0157 +/+, −/− and reconstituted null MEFs were treated with or without mIFNβ (200U/ml) for 48 hours and cells viability was analyzed by Trypan blue exclusion assay.
(F) Viral titers were measured in VSV infected KIAA0157+/+ and −/− MEFs from 2 independent experiments (each in triplicates).
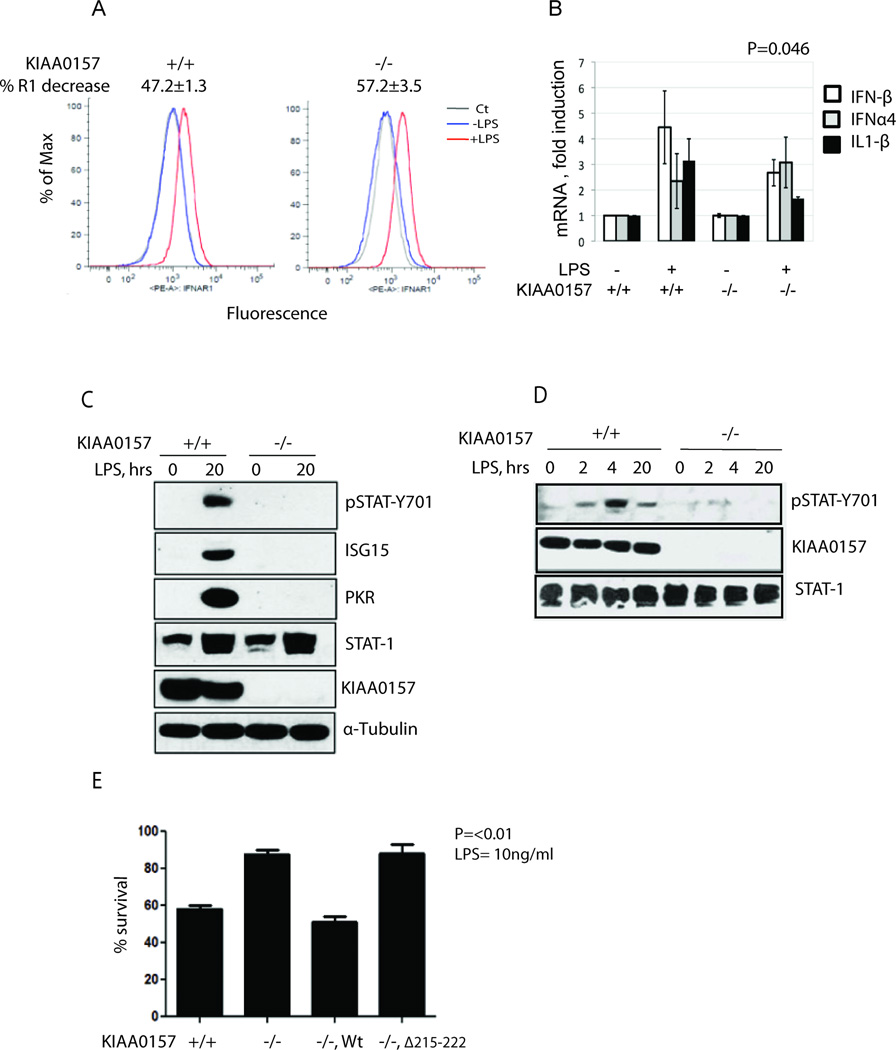
When cells respond to exogenously added IFN, both ubiquitination of IFNAR1 (counteracted by BRISCSHMT) and JAK-STAT signaling are activated concurrently. Accordingly, the role of BRISC-SHMT in regulating IFN signaling might be even more pronounced within the ligand-independent pathway of IFNAR1 ubiquitination (Fuchs, 2013; Liu et al., 2009; Liu et al., 2008). As bacterial lipopolysaccharide (LPS) induces IFNAR1 ubiquitination and downregulation even prior to stimulating the TLR4-mediated production of endogenous IFN (Qian et al., 2011), we examined the role of BRISC within the context of cellular responses to LPS. Consistent with the role of BRISC in regulating IFNAR1 endocytosis (Figure 4A) we observed a greater extent of IFNAR1 downregulation on the surface of peripheral blood leukocytes from LPS-treated KIAA0157-null mice compared to those from wild type animals (Figure 6A). Given a key role of weak IFNAR1-mediated tonic signaling in subsequent induction of type 1 IFN ligands (Gough et al., 2012; Taniguchi and Takaoka, 2001) it is anticipated that lesser induction of these genes would be observed in the BRISC-deficient mice. Indeed, LPS-triggered induction of mRNA for IFNβ (but not IFNα4) was significantly attenuated in leukocytes from KIAA0157−/− mice (Figure 6B). Similarly, an attenuated induction of IFNβ mRNA was seen in LPS-treated MEFs from these mice (data not shown). This combination of accelerated IFNAR1 downregulation and decreased IFN production would likely limit the extent of cellular responses to LPS in these animals.
Figure 6. BRISC Deficiency Confers Severely Reduced Interferon Signaling in Response to LPS.
(A) FACS analysis of cell surface IFNAR1 levels in peripheral blood leukocytes (PBL) isolated from KIAA0157 +/+ and −/− mice (n=3 for each genotype) 3h after LPS treatment.
(B) Real time RT-PCR analysis of the indicated cytokine mRNA levels in PBL from KIAA0157 +/+ and −/− mice (n=4 for each genotype) in response to LPS treatment.
(C) KIAA0157 +/+ and −/− MEFs were treated with or without 1µg/ml of LPS for 20h and cell lysates were analyzed by immunoblotting as indicated.
(D) KIAA0157 +/+ and −/− MEFs were treated with or without 1µg/ml of LPS for indicated periods of time and levels activated STAT1 (pSTAT-Y701) and other proteins were analyzed in WCL using the indicated antibodies.
(E) KIAA0157 +/+, −/− and reconstituted −/− MEFs were treated with or without LPS (10ng/ml) and 10ug/ml CHX for 24 hours. Cells viability was analyzed by Trypan blue exclusion assay.
Consistent with this hypothesis, strongly reduced STAT1 phosphorylation and ISG15 protein levels were observed in KIAA0157−/− cells or in primary human fibroblasts following KIAA0157 knockdown in comparison to WT and control knockdown cells respectively (Figure 6C, D and S6A). In addition, KIAA0157−/− MEFs demonstrated resistance to the cytotoxic effect following treatment with LPS and cycloheximide, whereas reconstitution with wildtype, but not KIAA0157-Δ215–222 restored sensitivity to LPS (Figure 6E). These experiments implicate BRISC as an important regulator of cellular responses to LPS-induced endogenous IFN and suggest that this regulation specifically depends on loss of interaction with SHMT2.
Besides inducing production of IFN, LPS may elicit IFN-independent effects on cells including induction of inflammatory cytokines such as interleukin 1-beta (IL1β). Levels of functional secreted IL1β protein are determined by transcriptional regulation of mRNA of IL1β precursor as well as by its subsequent processing by caspase-1 (Andrei et al., 1999; Gabay et al., 2010; Watkins et al., 1999) that can be activated by multi-protein complexes termed inflammosomes (Schroder and Tschopp, 2010). Remarkably, a recent report demonstrated that pharmacologic administration of the nonspecific DUB inhibitor G5 suppressed the LPS-induced activation of Caspase 1 and ensuing activation of the inflammasome-dependent production of IL1β. Based on these findings and knockdown experiments, it was suggested that BRISC mediated deubiquitination of the NLRP3 inflammasome in response to LPS might be an event required for Caspase 1 activation (Py et al., 2013). To determine if reduced inflammasome activation could complement the effects elicited upon IFNAR1 within the context of LPS response, we monitored Caspase 1 cleavage products in primary bone marrow macrophages derived from KIAA0157−/− and wildtype mice (n=6 for each genotype). Remarkably, Caspase 1 activation was, at the very least, not impaired in LPS-treated KIAA0157−/− macrophages (Figure S6B), indicating that BRISC inhibition probably does not account for the pharmacologic effects of G5 on inflammasome activation. A decreased induction of IL1β mRNA observed in BRISC knockout blood leukocytes (Figure 6B) is likely to contribute to a somewhat lower level of IL1β (and other pro-inflammatory cytokines) detected in plasma from KIAA0157−/− mice (Figure S6C).
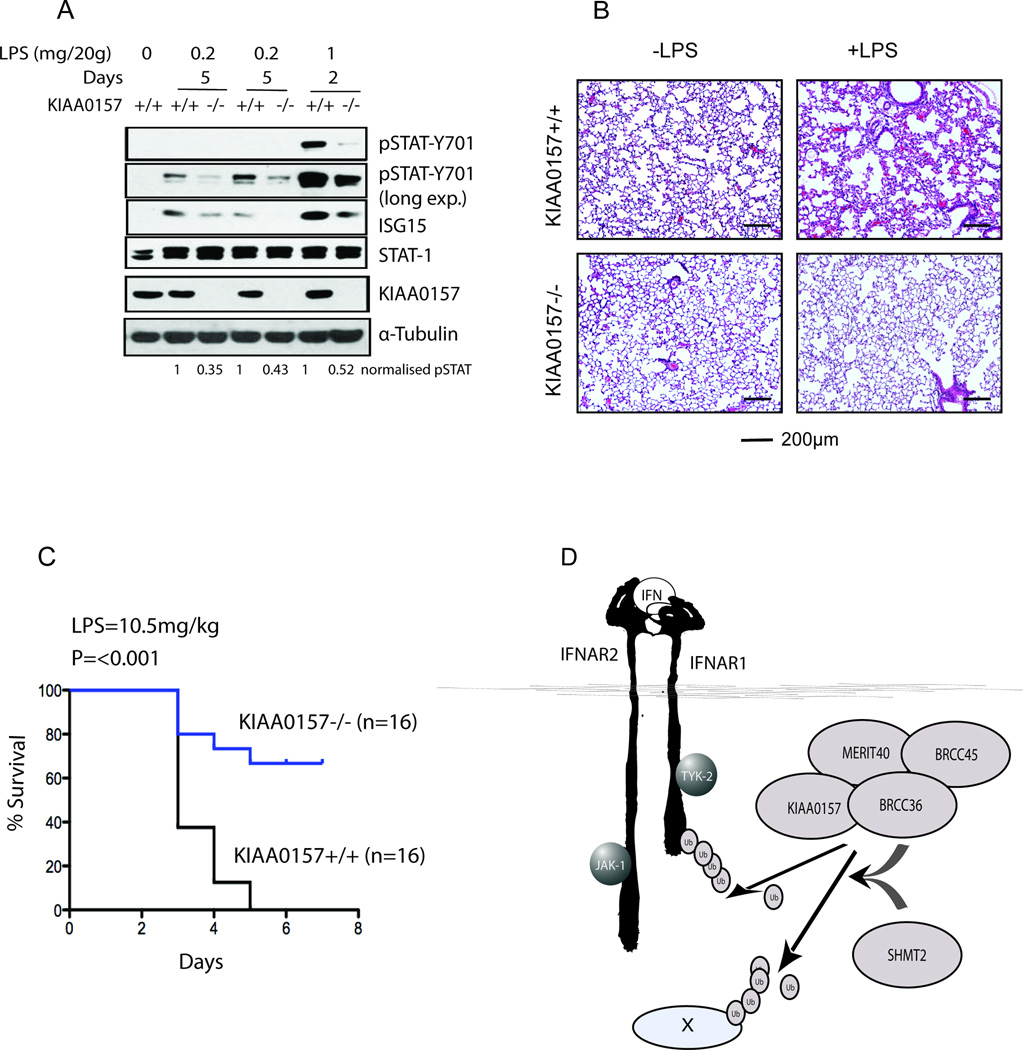
Importantly, while protecting from viral infections, IFN also plays a key role in mechanisms of immunopathology (Trinchieri, 2010). For example, in the model of septic shock induced by intraperitoneal LPS injections in vivo, IFNAR1 expression was required for mortality over a 5-day period (Karaghiosoff et al., 2003). To determine the in vivo role of BRISC in regulating these responses we injected cohorts of female littermate control KIAA0157+/+ or KIAA0157−/− mice with LPS and monitored IFNAR1 signaling, receptor levels, and mortality. As expected from a combination of lower ligand production and lesser receptor levels (Figure 6), BRISC deficiency resulted in strong attenuation of IFN responses in vivo as seen from reductions in LPS induced STAT1 phosphorylation and ISG15 induction (Figures 7A and S7). Moreover, KIAA0157−/− mice that received LPS injections exhibited a markedly decreased severity of LPS-induced lung injury, manifested in lesser erythrocyte extravasation, leukocytic infiltration of the interstitial and alveolar spaces, edema and alveolar distortion (Figure 7B). Accordingly, consistent with the importance of IFNAR1 levels for LPS toxicity (Karaghiosoff et al., 2003) and the requirement for KIAA0157 for IFNAR1 levels maintenance (Figures 4, 6A), BRISC-deficient animals displayed significantly reduced mortality (Figure 7C). Collectively, these results are consistent with SHMT-BRISC targeting to IFNAR1 as a contributing factor to mortality in response to LPS.
Figure 7. KIAA0157−/− Mice Display Reduced Interferon Signaling, Tissue Damage, and Mortality in Response to LPS in vivo.
(A) KIAA0157 +/+ and −/− mice were injected with the indicated amount of LPS for different time periods and lung and liver were harvested for protein analysis by western blot.
(B) Wild type and KIAA0157 null mice were injected with 10.5mg/kg LPS for 4–5 days and their lung were harvested, fixed and then stained with hematoxylin–eosin (H&E)
(C) Kaplan-Meier survival curves were generated after injecting 10.5mg/kg LPS in wild type and KIAA0157 null mice.
(D) Model depicting IFN inducible SHMT-BRISC assembly and de-ubiquitination activity on IFNAR1 and possibly other substrates (X).
Discussion
It has become increasingly clear that many DUBs utilize protein-protein interactions to affect enzymatic activity, cellular localization, and access to substrates (Ventii and Wilkinson, 2008). For example, USP1 interaction with UAF1 is necessary for DUB activity in vitro and deubiquitination of cellular substrates PCNA and FANCD2 (Cohn et al., 2007; Huang et al., 2006; Nijman et al., 2005). Notably, several JAMM domain DUBs including Poh1 and BRCC36 fail to display proteolytic activity as a single polypeptide, instead necessitating additional protein-protein interactions (Cooper et al., 2010; Feng et al., 2010; Patterson-Fortin et al., 2010; Verma et al., 2002; Yao and Cohen, 2002). BRCC36 requires interaction with the KIAA0157 for minimal DUB activity in vitro while the nuclear RAP80 complex requires the combined association with Abraxas and BRCC45 (Cooper et al., 2010; Feng et al., 2010; Patterson-Fortin et al., 2010). RAP80 provides a recognition element for nuclear BRCC36 through its tandem ubiquitin interaction motifs, which show specificity for K63-Ub binding (Sobhian et al., 2007). This interaction is essential for BRCC36 DNA damage site targeting and deubiquitination of K63-Ub substrates γH2AX and RAP80 (Patterson-Fortin et al., 2010; Shao et al., 2009a).
As opposed to the nuclear complex, mechanisms of substrate targeting for the cytoplasmic BRISC had not been previously appreciated. This study provides the first insights into regulation of BRISC dependent K63-Ub deubiquitination (Figure 7D). A cytokine inducible interaction between BRISC and SHMT is necessary for efficient IFNAR1 deubiquitination and contributes to its subcellular localization. This interaction is important for restricting receptor degradation and, hence, for maximal responses to IFN at the cellular and organismal levels. The data suggest both similarities and differences to nuclear BRCC36 regulation. Both the BRISC and RAP80 complexes respond to signal induced ubiquitination at the plasma membrane and DNA double-strand breaks respectively. However, the nuclear complex relies on a constitutive interaction with a canonical ubiquitin binding protein, RAP80, while the BRISC demonstrated substrate inducible localization through IFNAR1 activated association between SHMT2 and BRISC. While phosphorylation dependent IFNAR1 ubiquitination by β-Trcp was required for this association, it is not clear if SHMT interaction with the BRISC increases affinity for K63-Ub or if another associated recognition event is responsible for localization. Notwithstanding, the utilization of ancillary recognition components may represent a general mechanism by which the DUB complexes are targeted to their substrates. Given that BRISC represents a major K63-Ub-specific DUB activity in the cytoplasm (Cooper et al., 2009), it is likely that it is also targeted to many other substrates (besides IFNAR1) through either SHMT or functionally similar moieties (Figure 7D). Furthermore, deubiquitination of these yet to be identified substrates could also contribute to in vivo responses to LPS and other stimuli. The identification of additional BRISC-SHMT substrates and the biochemical mechanisms of their recognition represent interesting topics for future investigation.
A second feature of this study is its potential medical significance. Elevated IFNAR1 signaling has been implicated in a variety of disease states, including gram negative endotoxic shock and autoimmune conditions such as rheumatoid arthritis and systemic lupus erythematosis. Mouse models of the latter demonstrate a remarkable attenuation of syndrome severity upon inactivation of IFNAR1 (Agrawal et al., 2009; Nacionales et al., 2007). Accordingly, means to limit this signaling (such as neutralizing antibodies) might benefit this group of patients (Stewart, 2003). Indeed human trials using antibodies against IFN to treat systemic lupus erythematosis have entered late stage clinical trials with evidence of efficacy (McBride et al., 2012).
Data presented here argue that inhibition of BRISC DUB activity may represent a novel and promising strategy to treat pathologic conditions that arise as a result of elevated IFNAR1 signals. KIAA0157−/− mice have ablated BRISC activity while sparing the nuclear RAP80 complex. These mice display no overt phenotype in the absence of stress, suggesting that BRISC inhibition may be well tolerated. However, we were able to document impaired IFNAR1 dependent signals in response to IFN and LPS in knockout cells and mice. More importantly, KIAA0157−/− mice could better tolerate LPS than their wild type counterparts. While rapid downregulation of IFNAR1 and ensuing weakening of tonic signaling towards expression and production of IFN ligands is likely to represent a major mechanism by which BRISC-deficient mice are de-sensitized to LPS, the contribution of other putative targets of BRISC cannot be excluded (Figure 7D).
Importantly, BRISC inhibition attenuates, but does not eliminate IFNAR1 levels and IFNAR1-mediated signaling (Figure 5). Given that the establishment of an anti-viral state requires much lower receptor density than the anti-proliferative/survival effects of IFN (reviewed in (Piehler et al., 2012), it is not surprising that the anti-viral phenotype of BRISC deficiency is fairly subtle (Figures 5F, S5). Importantly, the medical benefits of BRISC inhibition is that partial IFNAR1 destabilization should prevent immunopathology without substantially compromising anti-viral defenses. Accordingly, targeting BRISC may conceivably help to ameliorate diseases that have elevated IFNAR1 responses, while not producing unwanted susceptibility to viral pathogens. The data presented in this study suggest the need for developing potent and selective BRISC inhibitors to test this hypothesis in vivo.
EXPERIMENTAL PROCEDURES
Reagents
The following antibodies and other reagents were purchased from the following commercial sources: Sigma (M2 Flag, cycloheximide, anti-SHMT2, tetrahydrofolate), Biolegend (mouse IgG1-biotin), eBioscience (Streptavidin-PE, 12–4317, ELISA kit for detection of IL1β), Cell Signaling (anti-pSTAT1, STAT1, tubulin, ISG15), Leinco Technologies Inc (anti-mIFNAR1), PBL Laboratories (mouse and human IFN), Santa Cruz (PKR, caspase-1, STAT-1), Millipore (K63-Ub, K48-Ub), LS Biosciences (anti-KIAA0157). In-house produced purified antibodies were also used for detecting BRCC36, MERIT40 and Immunoprecipitating Abraxas, KIAA0157 and SHMT2 proteins. Antibodies against phosphoSer535-IFNAR1 were previously described (Kumar et al., 2004). 3H Glycine and Western lighting ECL substrate were procured from Perkin Elmer and Bradford reagent, AG-50 X8 ion exchange matrix were from BioRad. Duo-link assay kit was purchased from O-link Biosciences. 4–12% Bis-Tris gel, Trizol, Lipofactamine 2000, Lipofactamine RNAi, cell culture media and other materials for tissue culture were purchased from Invitrogen.
Cell and tissue Lysis, Immunoprecipitation and Immunoblotting
Cells were cultured in DMEM with 10% FBS. Cell lysis was performed in NETN-150 buffer (20mM Tris pH 7.4, 150mM NaCl, 0.5mM EDTA, 0.5% NP40 with 5mM 2-mercaptoethanol and 1mM PMSF). Tissue samples from mice were snap frozen in liquid nitrogen, crushed in mortar pestle and incubated with NETN-150 buffer on ice for 20 min, sonicated and spun down at maximum speed. Supernatant was used further for protein estimation and immunoblot. For immunoprecipitation, FLAG M2 agarose beads or antibody coated protein A/G (according to experiment) agarose were incubated for 1–2 hours in cell lysate and washed 3 times with NETN150 buffer. Immunoprecipitated material was eluted using either Flag peptide or Glycine and separated on 4–12% Bis-Tris gel.
For assessment of IFNAR1 ubiquitination in cells, endogenous or FLAG-tagged recombinant IFNAR1 species were immunoprecipitated with a corresponding antibody. The samples were washed three times with 50 mM Tris pH 7.4, 5% glycerol, 1M NaCl and one time with 50 mM Tris pH 7.4, 5% glycerol, 150mM NaCl and eluted with SDS-containing Laemmli buffer. Resulting samples were separated by SDS-PAGE and analyzed by immunoblotting using the chain-specific anti-K63-Ub or K48-Ub antibodies (Millipore).
Proximity ligation assay and immunofluorescence
A modified form of proximity ligation assay (Duo-link assay, Olink Biosceince) was performed according to vendor recommendations. U2OS cells with either stable or transient expression of FLAG-HA proteins were grown on cover slips and fixed using 3% paraformaldehyde/2% sucrose solution. Fixed cells were washed twice with PBS and permeabilized with 0.5% Triton X-100 in PBS for 5 minutes at 4°C. Cells were washed with PBST and incubated with a mouse anti-HA antibody and a rabbit primary antibody for another protein of interest together for 1 hour at 37°C in humidified chamber. Cells were washed with PBST and incubated with proximity ligation assay probes for another hour, and then subsequently incubated with ligation and amplification buffer according to the vendor’s recommendations. Coverslips were then mounted on glass slides using mounting media containing DAPI (Vectashield) and foci imaged and quantified using a florescence microscope.
Generation of KIAA0157 floxed mice and murine embryonic fibroblasts
Gene targeting of the murine KIAA0157 locus was performed by inGenious Targeting Laboratories. A targeting construct including a neomycin resistance cassette (neo) (Flanked by FLP site) and a 2.2-kb fragment containing exons 6 to 7, each flanked by loxP sites was generated. This construct was electroporated into C57BL/6N embryonic stem (ES) cells. C57BL/6N embryonic stem cells (ES) that had undergone homologous recombination were then microinjected into Balb/c blastocysta. The resulting chimeras that had high percentage black coat color were mated to wild-type C57BL/6N mice to generate F1 heterozygous mice. The F1 heterozygous mice were mated to ACTFLPe(B6.Cg) transgenic mice (The Jackson Laboratory) to generate F2 KIAA+/FL heterozygous mice. The F2 generated F2 KIAA+/FL heterozygous mice were mated to CMV-cre (B6.C) transgenic mice (The Jackson Laboratory) to generate F2 KIAA+/− heterozygous mice. The F2 KIAA+/− heterozygous mice were mated each other and viable KIAA−/− offspring were generated. Primers use for PCR genotyping: KIAA0157+/+ Forward primer: 5’-TTT CCA GCT GGT TCA TAG GG-3’ and Rev: 5’-GAA GGA CGG TCA GCC TGT AG-3’. Primers use to PCR genotype KIAA0157−/−: A 514bp band was generated using; forward primer: 5’-TGA TAG TGA GCC ATT ATG TGG GTG -3’ and Rev: 5’-GAA GGA CGG TCA GCC TGT AG-3’. Primary murine embryonic fibroblasts (MEFs) were generated from matings of KIAA+/− heterozygous mice and immortalized by infection with shRNA against p19ARF lentiviral. All experiments involving mice were approved by the University of Pennsylvania IACUC committee and performed in accordance with these standards.
SHMT activity assay
SHMT Activity was analyzed by a radio-assay method (Scheer et al., 2005), where Tetrahydrofolate (THF) accelerates the exchange of the pro-2S proton from 2–3H Glycine to water. The counts exchanged from glycine to water are proportional to the enzymatic activity. In order to purify commercially available 2–3H Glycine, a fraction of crude 2–3H Glycine was acidified using 1M HCL and passed through a column containing AG-50 X8 ion exchange resin. After extensive washes, purified Glycine was eluted using 1M NH4OH. The eluted Glycine was dried under nitrogen spray and dissolved in 20mM phosphate buffer to ~1 million CPM/µl. For assaying SHMT activity, protein-A beads were incubated with purified SHMT2, Abraxas, KIAA0157 and IgG antibodies. These beads were used to pull respective proteins from HeLa or 293T cell lysates. Equal amount of SHMT2 (as analyzed by immunoblot) attached to the beads were incubated for 1 hour in 400µl of assay buffer containing 20 mM phosphate buffer pH 7.3, 1mM glycine, 10mM 2-mercaptoethanol and 4 µl of purified 2–3H at 37°C. Reactions were started by adding 1 mM THF and stopped by adding cold 50 mM HCl. The mixture was then loaded on the top of an AG-50 X8 matrix column and washed with 2 ml of 50mM HCl. The exchanged amount of 3H in collected washes was analyzed in scintillation counter and activity data was normalized by control beads activity.
Purification of DUB complexes and IFNAR1/2-interacting proteins
Hela S3 cells expressing Flag-HA-KIAA0157 and Flag-HA-Abraxas proteins were grown in suspension culture in spinner flasks. Purifications were performed according to Shao et al. (Shao et al., 2009b). Flag beads were washed, eluted by Flag-peptide elution and loaded on SDS-PAGE for visualizing protein bands upon Coomassie staining.
IFNAR1 purifications were performed in 293T cells that were transiently transfected with Flag-IFNAR1Y466F internalization deficient mutant of IFNAR1 (Kumar et al., 2007) and HA-IFNAR2 (a kind gift of J. Krolewski). Forty eight hours after transfection cells were washed, treated with IFNα (500U/mL for 2 min) and harvested. Cells were re-suspended in ice cold hypotonic buffer (10mM Tris-HCl pH 7.4, 7.5mM KCl, 1.5mM MgCl2, 5mM β-mercaptoethanol and 0.5mM PMSF). After 15 min incubation on ice cells were disrupted with 20 strokes in Dounce homogenizer the extracts were centrifuged to separate the nuclear and cytoplasmic fractions. The cytoplasmic fraction was subjected to overnight dialysis in NETENG-200 buffer (200mM NaCl, 0.5mM EDTA, 20mM Tris-HCl pH 7.4, 0.1% Nonidet P-40, 1.5mM MgCl2, 10% Glycerol, 5mM β-mercaptoethanol and 0.5mM PMSF) with gentle agitation. After dialysis the cytoplasmic fraction was clarified by centrifugation (~30000g for 30 min at 4°C) and used for immunoprecipitation with M2-agarose (Sigma; 10µL of packed agarose beads per 1mL of extract). After thorough washes bound proteins were eluted with 3xFlag peptide, re-precipitated using immobilized HA antibody, washed, eluted with Laemmli buffer and separated on 12–20% SDS-PAGE. Wide gel segments corresponding to different molecular weight range were cut and analyzed by mass spectrometry (Harvard Proteomics facility).
Viral infections
MEFs were treated with low doses of IFN (1–4 IU/ml) for 1 hour and then washed extensively prior to adding media lacking IFN. Following an 18 hour recovery period, VSV (0.1 MOI; Indiana serotype, propagated in HeLa cells) was used to infect MEFs for 1 h. After removing the virus inoculums, cells were then fed with fresh medium and incubated for 24 h. Culture supernatant was harvested and viral titer was determined in HeLa cells overlaid with methylcellulose as described elsewhere (Zheng et al., 2011). The expression of VSV-M protein in cell lysates was analyzed by immunoblot.
Cell viability
Immortalized MEFs were seeded at low confluency (~30%) and treated with or without 200U/ml of mouse-IFNβ for 48hs. Cells were harvested and cell viability was estimated using Trypan blue exclusion assay. For LPS treatment, cells at higher confluency (~80%) were treated with or without 10ng/ml LPS and 10µg/ml CHX. Cells treated only with CHX were used to normalize the cell viability after LPS treatment.
Caspase-1 Processing
A population of bone marrow cells was isolated from wildtype and KIAA0157 null mice and cells were plated in L929 conditioned media for macrophage differentiation. After one week, macrophages were treated with or without 200ng/ml of LPS for 12 hours. 30 minutes before cell harvesting, LPS stimulated cells were treated with or without 2mM ATP. Cells were lysed in NETN150 buffer and equal amount of protein was loaded on a 4–12% PAGE gel. A Caspase-1 antibody that recognizes pro-caspase-1 and processed caspase-1 (p20, p10) was used for immunoblot to analyze the extent of caspase processing.
In vitro DUB assay
BRISC complex ontaining active BRCC36-WT or catalytically inactive BRCC36-QSQ was purified from Sf9 cells as described before (Patterson-Fortin et al., 2010). 293T cells transiently expressing Flag-IFNAR1 were treated with αIFN for 2 hours and lysed. Recombinant R1 substrate was then immunopurified using Flag M2 agarose beads and eluted using Flag peptide. The R1 substrate was incubated with active or inactive BRISC protein in deubiqutination buffer (Fortin et al., 2010) for 2 hours. The reaction was stopped by adding 2X SDS loading buffer and loaded on 4–12% SDS PAGE gel. The proteins were blotted and K63-ubiqutination status of R1 was detected using specific antibodies.
Animals and LPS toxicity
The Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania approved all animal procedures of disease modeling, blood collection, euthanasia and tissue harvesting (protocols # 804095, 803995, 803170). Eight-ten week old female C57BL/6 littermate mice including wild type (Kiaa0157+/+) mice, and Kiaa0157−/− mice (generated during this study) were used in all experiments. For generalized inflammation model, mice were treated with LPS (10.5 mg/Kg, intraperitoneal injection) and sacrificed when they became moribund and displayed the loss of righting reflex, loss of >20% of body weight and non-responsiveness to footpad compression as previously described (Karaghiosoff et al., 2003). Lung tissues from LPS treated wild and KIAA0157-null mice were harvested at 4–5 days of injection. Lungs were inflated and fixed with 4% paraformaldehyde, dehydrated and embedded in paraffin. Tissue were further sectioned and stained with hematoxylin and eosin. Levels of IL1β protein in blood plasma were analyzed using ELISA kit (Ebioscience).
Flow cytometry
Blood was stained with biotinylated antibodies against mouse IFNAR1 or biotinylated isotype control antibodies. Red blood cells were lysed using the RBC lysis buffer (155mM NH4Cl, 12 mM KHCO3, and 0.1 mM EDTA) for 5 minutes and washed with PBS. Cells were then washed and stained with phycoerythrin conjugated streptavidin. All cells were analyzed using LSRFortessa flow cytometer (BD Biosciences).
For Flow cytometry of bone marrow derived macrophages (BMM) were harvested by enzyme free cell detachment solution (Invitrogen) and used for flow cytometry analysis of surface IFNAR1. BMM stained with viability dye (fixable eFluor 506 Ebioscience) were washed and stained with a cocktail containing APC labeled antibodies against F4/80, PerCP-Cy.5.5 labeled antibodies against CD11b (Biolegend), biotinylated antibodies against mouse IFNAR1 or biotinylated isotype control antibodies (MOPC-21, “Biolegend”). Cells were then washed and stained with phycoerythrin conjugated streptavidin. All cells were analyzed using LSRFortessa flow cytometer (BD Biosciences). Only viable non-aggregated CD11b/F480 double positive cells were analyzed for IFNAR1 surface expression (nearly 98% of cells were double positive and viable)
Real time PCR
Cells or tissues were flash-frozen and pulverized in liquid nitrogen, homogenized in Trizol reagent, and extracted with chloroform. Reverse transcription was carried out using Revertaid first strand cDNA synthesis kit (K1621, Thermo Scientific) and the cDNA was used for real time QPCR analysis of expression of cytokines and β-actin mRNA using the following primers: IFN-β (FW, 5’-AGCTCCAAGAAAGGACGAACAT-3’, REV, 5’-GCCCTGTAGGTGAGGTTGATCT-3’), IFN-α4 (FW, 5’-CCTGTGTGATGCAGGAACC-3’, REV, 5’-TCACCTCCCAGGCACAGA-3’, IL-1β (FW, 5’-CAACCAACAAGTGATATTCTCCATG-3’, REV, 5’-GATCCACACTCTCCAGCTGCA-3’), β-actin (FW, 5’-AGAGGGAAATCGTGCGTGAC-3’, REV, 5’-CAATAGTGATGACCTGGCCGT-3’) using Applied biosystems 7500 Fast real time PCR system. All analyses were carried out in triplicates.
IFNAR1 internalization assays
Internalization of endogenous IFNAR1 was determined using a high throughput fluorescence-based method described in detail elsewhere (Kumar et al., 2007; Kumar et al., 2008). Briefly, cells in 60-mm dishes were transfected with the indicated constructs or induced with tetracycline and plated equally on 24 well poly-D-lysine plates (Becton, Dickinson and Company). Cells were starved in Dulbecco’s Modified Eagle’s medium lacking FBS and were either treated with starvation media supplemented with IFNα2a for the indicated time periods, or were kept on ice and not exposed to the IFNα2a supplemented media (time point 0). The cells were washed, blocked, and incubated with the anti-IFNAR1 AA3 antibody (Goldman et al., 1999) for human IFNAR1 or anti-mIFNAR1 antibody for mouse receptor. The primary antibodies were then removed and the cells were washed extensively before adding a goat anti mouse IgG H + L HRP conjugated antibody. Following another series of washes, the cells were incubated with AmplexRed Ultra Reagent 10-acetyl-3,7-dihydroxyphenoxazine (Molecular Probes). Aliquots were transferred to black 96 well plate (NUNC) and fluorescence was measured by reading with Beckman Coulter DTX880 Mutimode Detector plate reader using a 530 nm filter for excitation and a 590 nm filter for emission. Results were calculated using the following formula: % Internalized= 100−[(Vs−Vb) × tn/(Vs−Vb) × t0] × 100, where: Vs=Value of samples, Vb=Value of background (mock-transfected or probed with irrelevant antibody), tn= time point n, t0=time point 0.
Highlights.
The K63-Ub specific DUB complex BRISC forms an interferon inducible interaction with Serine Hydroxymethyltransferase (SHMT) enzymes.
BRISC-SHMT deubiquitinates IFNAR1 and is required for responses to type 1 interferons.
KIAA0157−/− mice display impaired responses to interferon and are resistant to bacterial lipopolysaccharide.
Acknowledgments
We thank J. Krolewski for the gift of reagents and the members of JA Diehl, C Koumenis, W Tong, E. Witze, SY Fuchs and RA Greenberg laboratories (all from the University of Pennsylvania) for critical suggestions. We also thank JF Gregory (University of Florida) and P. Stover (Cornell University) for advice on SHMT activity assays. This work was supported by NIH/NCI grants CA092900 and CA142425 (to SYF) and CA138835, CA17494, and GM101149 (to RAG who is also supported by a Research Scholar Grant from the American Cancer Society, a DOD Breast Cancer Idea Award, a Harrington Discovery Institute Scholar-Innovator Award, a Pennsylvania Breast Cancer Coalition grant, and funds from the Abramson Family Cancer Research Institute and Basser Research Center for BRCA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interests.
REFERENCES
- Agrawal H, Jacob N, Carreras E, Bajana S, Putterman C, Turner S, Neas B, Mathian A, Koss MN, Stohl W, et al. Deficiency of type I IFN receptor in lupus-prone New Zealand mixed 2328 mice decreases dendritic cell numbers and activation and protects from disease. J Immunol. 2009;183:6021–6029. doi: 10.4049/jimmunol.0803872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Andrei C, Dazzi C, Lotti L, Torrisi MR, Chimini G, Rubartelli A. The secretory route of the leaderless protein interleukin 1beta involves exocytosis of endolysosome-related vesicles. Mol Biol Cell. 1999;10:1463–1475. doi: 10.1091/mbc.10.5.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Schwartz AL. The ubiquitin system: pathogenesis of human diseases and drug targeting. Biochim Biophys Acta. 2004;1695:3–17. doi: 10.1016/j.bbamcr.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Clague MJ, Urbe S. Endocytosis: the DUB version. Trends Cell Biol. 2006;16:551–559. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Cooper EM, Boeke JD, Cohen RE. Specificity of the BRISC deubiquitinating enzyme is not due to selective binding to Lys63-linked polyubiquitin. J Biol Chem. 2010;285:10344–10352. doi: 10.1074/jbc.M109.059667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, Cohen RE. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 2009;28:621–631. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Wang J, Chen J. The Lys63-specific deubiquitinating enzyme BRCC36 is regulated by two scaffold proteins localizing in different subcellular compartments. J Biol Chem. 2010;285:30982–30988. doi: 10.1074/jbc.M110.135392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SY. Ubiquitination-mediated regulation of interferon responses. Growth Factors. 2012;30:141–148. doi: 10.3109/08977194.2012.669382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SY. Hope and fear for interferon: the receptor-centric outlook on the future of interferon therapy. J Interferon Cytokine Res. 2013;33:211–225. doi: 10.1089/jir.2012.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay C, Lamacchia C, Palmer G. IL-1 pathways in inflammation and human diseases. Nat Rev Rheumatol. 2010;6:232–241. doi: 10.1038/nrrheum.2010.4. [DOI] [PubMed] [Google Scholar]
- Goldman LA, Zafari M, Cutrone EC, Dang A, Brickelmeier M, Runkel L, Benjamin CD, Ling LE, Langer JA. Characterization of antihuman IFNAR-1 monoclonal antibodies: epitope localization and functional analysis. J Interferon Cytokine Res. 1999;19:15–26. doi: 10.1089/107999099314379. [DOI] [PubMed] [Google Scholar]
- Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36:166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Dikic I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci. 2012;125:265–275. doi: 10.1242/jcs.091280. [DOI] [PubMed] [Google Scholar]
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Huangfu WC, Fuchs SY. Ubiquitination-dependent regulation of signaling receptors in cancer. Genes Cancer. 2010;1:725–734. doi: 10.1177/1947601910382901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaghiosoff M, Steinborn R, Kovarik P, Kriegshauser G, Baccarini M, Donabauer B, Reichart U, Kolbe T, Bogdan C, Leanderson T, et al. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Tanaka K. Regulatory mechanisms involved in the control of ubiquitin homeostasis. J Biochem. 2010;147:793–798. doi: 10.1093/jb/mvq044. [DOI] [PubMed] [Google Scholar]
- Kumar KG, Barriere H, Carbone CJ, Liu J, Swaminathan G, Xu P, Li Y, Baker DP, Peng J, Lukacs GL, Fuchs SY. Site-specific ubiquitination exposes a linear motif to promote interferon-alpha receptor endocytosis. J Cell Biol. 2007;179:935–950. doi: 10.1083/jcb.200706034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KG, Krolewski JJ, Fuchs SY. Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination and degradation of the IFNAR1 subunit of type I interferon receptor. J Biol Chem. 2004;279:46614–46620. doi: 10.1074/jbc.M407082200. [DOI] [PubMed] [Google Scholar]
- Kumar KG, Tang W, Ravindranath AK, Clark WA, Croze E, Fuchs SY. SCF(HOS) ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-alpha receptor. EMBO J. 2003;22:5480–5490. doi: 10.1093/emboj/cdg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KG, Varghese B, Banerjee A, Baker DP, Constantinescu SN, Pellegrini S, Fuchs SY. Basal ubiquitin-independent internalization of interferon alpha receptor is prevented by Tyk2-mediated masking of a linear endocytic motif. J Biol Chem. 2008;283:18566–18572. doi: 10.1074/jbc.M800991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, HuangFu WC, Kumar KG, Qian J, Casey JP, Hamanaka RB, Grigoriadou C, Aldabe R, Diehl JA, Fuchs SY. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe. 2009;5:72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Plotnikov A, Banerjee A, Suresh Kumar KG, Ragimbeau J, Marijanovic Z, Baker DP, Pellegrini S, Fuchs SY. Ligand-independent pathway that controls stability of interferon alpha receptor. Biochem Biophys Res Commun. 2008;367:388–393. doi: 10.1016/j.bbrc.2007.12.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JM, Jiang J, Abbas AR, Morimoto A, Li J, Maciuca R, Townsend M, Wallace DJ, Kennedy WP, Drappa J. Safety and pharmacodynamics of rontalizumab in patients with systemic lupus erythematosus: results of a phase I, placebo-controlled, double-blind, dose-escalation study. Arthritis Rheum. 2012;64:3666–3676. doi: 10.1002/art.34632. [DOI] [PubMed] [Google Scholar]
- Nacionales DC, Kelly-Scumpia KM, Lee PY, Weinstein JS, Lyons R, Sobel E, Satoh M, Reeves WH. Deficiency of the type I interferon receptor protects mice from experimental lupus. Arthritis Rheum. 2007;56:3770–3783. doi: 10.1002/art.23023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Patterson-Fortin J, Shao G, Bretscher H, Messick TE, Greenberg RA. Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex. J Biol Chem. 2010;285:30971–30981. doi: 10.1074/jbc.M110.135319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehler J, Thomas C, Garcia KC, Schreiber G. Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol Rev. 2012;250:317–334. doi: 10.1111/imr.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell. 2013;49:331–338. doi: 10.1016/j.molcel.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Qian J, Zheng H, Huangfu WC, Liu J, Carbone CJ, Leu NA, Baker DP, Fuchs SY. Pathogen recognition receptor signaling accelerates phosphorylation-dependent degradation of IFNAR1. PLoS Pathog. 2011;7:e1002065. doi: 10.1371/journal.ppat.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Wilkinson KD. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem Rev. 2009;109:1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer JB, Mackey AD, Gregory JF., 3rd Activities of hepatic cytosolic and mitochondrial forms of serine hydroxymethyltransferase and hepatic glycine concentration are affected by vitamin B-6 intake in rats. J Nutr. 2005;135:233–238. doi: 10.1093/jn/135.2.233. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci U S A. 2009a;106:3166–3171. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, Greenberg RA. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009b;23:740–754. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart TA. Neutralizing interferon alpha as a therapeutic approach to autoimmune diseases. Cytokine Growth Factor Rev. 2003;14:139–154. doi: 10.1016/s1359-6101(02)00088-6. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Takaoka A. A weak signal for strong responses: interferon-alpha/beta revisited. Nat Rev Mol Cell Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventii KH, Wilkinson KD. Protein partners of deubiquitinating enzymes. Biochem J. 2008;414:161–175. doi: 10.1042/BJ20080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Watkins LR, Hansen MK, Nguyen KT, Lee JE, Maier SF. Dynamic regulation of the proinflammatory cytokine, interleukin-1beta: molecular biology for non-molecular biologists. Life Sci. 1999;65:449–481. doi: 10.1016/s0024-3205(99)00095-8. [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Zheng H, Qian J, Varghese B, Baker DP, Fuchs S. Ligand-stimulated downregulation of the alpha interferon receptor: role of protein kinase D2. Mol Cell Biol. 2011;31:710–720. doi: 10.1128/MCB.01154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]