Abstract
Genome-wide association studies (GWASs) have unraveled a large number of cancer risk alleles. Understanding how these allelic variants predispose to disease is a major bottleneck confronting translational application. In this issue, Li and colleagues combine GWASs with The Cancer Genome Atlas (TCGA) to disambiguate the contributions of germline and somatic variants to tumorigenic gene expression programs. They find that close to half of the known risk alleles for estrogen receptor (ER)-positive breast cancer are expression quantitative trait loci (eQTLs) acting upon major determinants of gene expression in tumors.
Cancer is a complex trait affected by the interaction of numerous somatically acquired genetic and epigenetic lesions influenced by underlying germline genetic polymorphisms. Given that the most effective disease intervention is prevention, the detection of germline cancer susceptibility loci is a growing focus of several genome-wide association studies (GWASs). A consistent theme elaborating from these efforts is that common cancer risk alleles are, for the most part, located in intergenic and intronic chromosomal regions with unknown functions (Hindorff et al., 2011). Thus, the nature of the allele does not usually reveal much about the biology of the disease. Extensive public efforts supporting large-scale, high-resolution annotation of cancer genomes can be leveraged to identify genes that are directly modulated by cancer risk loci. For example, The Cancer Genome Atlas (TCGA) generates comprehensive profiles of gene expression, epigenetic modifications, copy-number variation, and somatic mutations in tumors together with matched constitutional DNA sequence information. Application of these kinds of orthogonal data sets to GWAS has lead to the identification of 3 of the 19 colorectal cancer risk alleles (Loo et al., 2012), 3 of the 12 chronic lymphocytic leukemia/small lymphocytic leukemia risk alleles (Sillé et al., 2012), and two completely linked risk loci in lung cancers of nonsmokers (Li et al., 2010) as cis-acting expression quantitative trait loci (eQTLs), genomic loci that regulate mRNA abundance.
In this issue of Cell, Li et al. (2013) pursue the germline determinants of gene expression in ER-positive and ER-negative breast tumors. The identification of such determinants may shed light on the mechanistic contribution of cancer risk loci on disease initiation and development. However, the detection of functionally relevant germline alleles, using tumor-derived gene expression data sets, is confounded by log differences in the sensitivity of gene regulatory programs to the somatic variation present in tumor tissue. The authors address this challenge by treating germline and somatic variation as independent variables in a multivariate linear regression model built upon the publicly available TCGA breast cancer data sets. Using copy-number variation and CpG island methylation to adjust for the contribution of tumor-acquired somatic abnormalities, the authors find that cis-acting eQTLs account for 1.2% of the total variance of gene expression in ER-positive tumors, as compared to somatic lesions, which account for 11%. Of note, three of the detected eQTLs directly mapped to 3 of 15 previously discovered breast cancer risk loci: 2q35 (IGFBP5), 5q11 (C5orf35), and 16q21 (TOX3). Fortified by this correlation, the authors next devised a novel and effective strategy to increase the sensitivity of detection of cancer-susceptibility loci that correspond to eQTLs. The basic premise was that GWAS-identified risk loci are enriched for cis-acting elements for transcription factors—a notion supported to some extent by the analysis of quantitative trait loci in model organisms (Gerke et al., 2009). If true, then detection of these relationships could be enhanced by leveraging the amplification of small changes in transcription factor abundance on the expression variation of the cohort of transcription factor client genes. To test this, the authors first asked whether any of the 12 unmapped breast cancer risk loci were located near transcription factors with DNA-binding motifs enriched in the promoters of genes with expression profiles that correlated with the presence of the susceptibility allele. They detected three new candidate eQTLs associated with the risk loci 6q25/ESR1, 9q31/KLF4, and 8q24/MYC. As expected for a bona fide eQTL target gene, RNA-seq data showed significant allelic imbalance for ESR1 and MYC in patients that were heterozygous for the associated risk locus. Importantly, chromosome conformation capture (3C) revealed physical interactions between the risk loci and their candidate target genes. The identification of 6 out of 15 breast cancer list loci as eQTLs, along with the annotation of the gene expression program they modify, is both technically and conceptually transformative for understanding the pathogenesis of breast cancer and holds potential for the identification of additional disease sites.
How can we improve our understanding of the 9 of 15 breast cancer risk alleles unexplained by the methods employed in this study? The answer lies in part with the challenge of annotating functional interactions between germline susceptibility loci and somatic variation. eQTLs that primarily act in concert with somatic mutations are likely to be difficult or impossible to detect when modeled as an independent variable. As mentioned by the authors, a more immediately addressable challenge would be the consideration of noncoding RNAs as eQTL target genes. MicroRNAs, which were missed in the present study due to the absence of a comprehensive data table, are under active investigation by many groups due to their pro- and antitumorigenic activities in a wide variety of tumor types. For example, four different miRNAs, miR-7, miR-128a, miR-210, and miR-516-3p, were shown to be associated with disease aggressiveness in a study of 38 ER-positive lymph-node-negative breast cancers (Foekens et al., 2008). Although cis-eQTL analysis requires miRNA expression information that is not yet available for the TCGA breast tumor data set, trans analysis may still be possible by employing miRNA/mRNA target predictions in much the same way as the authors employed transcription factor/mRNA target predictions here. Both cis and trans analyses that account for miRNAs as potential eQTL targets are immediately doable for the TCGA ovarian and glioblastoma multiform tumor data matrices.
How may these innovative approaches and novel findings be translated to the clinic? The discovery of cancer risk loci that are eQTLs may provide a path leading to early detection and prevention strategies (Figure 1). A productive journey down this path is predicated on firmly established causality. For example, the relevance of estrogen receptor expression in luminal breast cancer is undeniable; it is a master regulator of tumorigenic expression programs and is the target of first-line hormonal therapy in patients. However, it will be very important to determine whether the ESR1 eQTL controls gene expression in normal mammary epithelial cells in a similar manner to that in breast cancers. This could be greatly enabling for risk assessment, early detection, and prevention efforts. It remains to be assessed whether women who carry the ESR1 eQTL risk locus and ultimately develop breast cancer exhibit this differential expression in nonneoplastic mammary tissue as compared to women that do not develop the disease. The translation to the clinic would then be to identify women with the germline ESR1 eQTL, sample their breasts (e.g., with fine-needle aspirates) and screen for patients with the tumor-phenotype-associated gene expression pattern. These women could have both more early detection follow-up but could also be candidates for prevention therapy targeting ESR1 with the available antiestrogenic therapies currently used for treating clinically evident breast cancers.
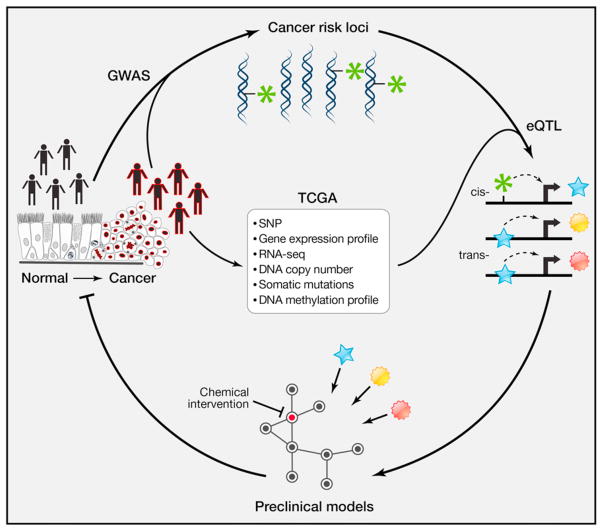
Figure 1. Bridging the Gap between Detection of Cancer Risk Loci and Development of Disease Prevention Strategies.
High-resolution annotation of molecular correlates in tumor samples can be dovetailed with GWAS to elaborate causal relationships between risk loci and target genes. These target genes serve as both functional and predictive biomarkers for personalized medicine. This, in turn, can lead to testable mechanistic hypotheses and nomination of early detection and prevention strategies.
Discovering how the modulation of eQTL target genes predisposes individuals to cancer, and whether reprogramming of the regulatory network mediated by the target genes can reverse the phenotype, also requires considerable investment in preclinical experimental models that would ideally be reflective of the genetic diversity found in patient populations. For example, excision of the equivalent of a human colon cancer risk locus upstream of MYC was sufficient to inhibit APCmin-driven intestinal tumorigenesis in the mouse (Sur et al., 2012). Validation of ESR1, the gene encoding ER, as the functional target of the 6q25 breast cancer risk eQTL will illuminate a solid path toward development of genome-tailored early detection and prevention tools.
Acknowledgments
H.K., J.M., and M.W. are supported by the Welch Foundation (I-1414), the National Institutes of Health (CA71443, CA129451, P50 CA70907), and the Cancer Prevention Research Institute of Texas (CPRIT).
This is a commentary on article Li Q, Seo JH, Stranger B, McKenna A, Pe'er I, Laframboise T, Brown M, Tyekucheva S, Freedman ML. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152(3):633-41.
References
- Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AWM, Klijn JGM, Wiemer EAC, Martens JWM. Proc Natl Acad Sci USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J, Lorenz K, Cohen B. Science. 2009;323:498–501. doi: 10.1126/science.1166426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Gillanders EM, Manolio TA. Carcinogenesis. 2011;32:945–954. doi: 10.1093/carcin/bgr056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sheu CC, Ye Y, de Andrade M, Wang L, Chang SC, Aubry MC, Aakre JA, Allen MS, Chen F, et al. Lancet Oncol. 2010;11:321–330. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Seo J-H, Stranger B, McKenna A, Pe’er I, LaFramboise T, Brown M, Tyekucheva S, Freedman ML. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo LWM, Cheng I, Tiirikainen M, Lum-Jones A, Seifried A, Dunklee LM, Church JM, Gryfe R, Weisenberger DJ, Haile RW, et al. PLoS ONE. 2012;7:e30477. doi: 10.1371/journal.pone.0030477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillé FCM, Thomas R, Smith MT, Conde L, Skibola CF. PLoS ONE. 2012;7:e29632. doi: 10.1371/journal.pone.0029632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur IK, Hallikas O, Vähärautio A, Yan J, Turunen M, Enge M, Taipale M, Karhu A, Aaltonen LA, Taipale J. Science. 2012;338:1360–1363. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]