Abstract
S-glutathionylation is a redox-regulated modification that uncouples eNOS, switching its function from NO synthesis to •O2− generation, and serves to regulate vascular function. While in vitro or in vivo eNOS S-glutathionylation with modification of Cys689 and Cys908 of its reductase domain is triggered by high levels of GSSG or oxidative thiyl radical formation, it remains unclear how this process may be reversed. Glutaredoxin-1 (Grx1), a cytosolic/glutathione-dependent enzyme, can reverse protein S-glutathionylation; however, its role in regulating eNOS S-glutathionylation remains unknown. We demonstrate that Grx1 in the presence of GSH (1 mM) reverses GSSG-mediated eNOS S-glutathionylation with restoration of NO synthase activity. Since Grx1 also catalyzes protein S-glutathionylation with increased [GSSG]/[GSH], we measured its effect on eNOS-S-glutathionylation when [GSSG]/[GSH] was > 0.2, as can occur in cells and tissues under oxidative stress, and observed increased eNOS S-glutathionylation with a marked decrease in eNOS activity without uncoupling. This eNOS S-glutathionylation was reversed with decrease in [GSSG]/[GSH] to < 0.1. LC/MS/MS identified a new site of eNOS S-glutathionylation by Grx1 at Cys382, on the surface of the oxygenase domain, without modification of Cys689 or Cys908 that are buried within the reductase. Furthermore, Grx1 was demonstrated to be a protein partner of eNOS in vitro and in normal endothelial cells, supporting its role in eNOS redox-regulation. In endothelial cells, Grx1 inhibition or gene silencing increased eNOS S-glutathionylation and decreased cellular NO generation. Thus, Grx1 can exert an important role in the redox-regulation of eNOS in cells.
Endothelial nitric oxide synthase (eNOS) is responsible for the enzymatic production of nitric oxide (NO) at the expense of NADPH (1–3). NO is a critical small molecule generated within the endothelium, involved in regulating vascular tone, vascular growth, platelet aggregation, and modulation of inflammation (1, 2, 4). In many cardiovascular diseases, a decrease in bioavailable NO is a hallmark of endothelial dysfunction, and a common mechanism underlying this dysfunction is the over-production of reactive oxygen species (ROS) in the vasculature (5–7). Under conditions of oxidative stress, it has been demonstrated that eNOS can become uncoupled resulting in further ROS generation. Oxidation of the NOS cofactor, tetrahydrobiopterin (BH4), increase in cellular methylarginines, and eNOS S-glutathionylation all have been demonstrated to increase eNOS uncoupling with secondary eNOS-derived ROS generation contributing to many cardiovascular diseases (8–15).
eNOS S-glutathionylation was first observed to occur in endothelial cells under oxidative stress and in vessels of hypertensive rats (14). Cells treated with PABA/NO can also exhibit this post-translational modification of eNOS (15). We have previously shown that eNOS S-glutathionylation at two specific sites of the reductase domain (Cys689 and Cys908) triggers a marked increase in superoxide (•O2−) generation from the isolated enzyme, as well as from eNOS in endothelial cells and intact vessels. More recently, studies have further supported the importance of eNOS S-glutathionylation, as this modification was observed in endothelial cells treated with nitroglycerin (16), and streptozotocin-induced hyperglycemia in rats or with isosorbide-5-mononitrate treatment (17). Together, these results demonstrate that eNOS S-glutathionylation is a critical modification that can redox-modulate its function under cellular oxidative stress as occurs in a broad range of cardiovascular disease.
Similar to protein phosphorylation, protein S-glutathionylation is a reversible post-translational modification. It can redox-regulate protein function through the formation of a mixed-disulfide bond between active cysteine residues and glutathione. This modification has been shown to regulate protein function either through activation or inactivation under oxidative stress (18, 19). Moreover, the formation of this mixed-disulfide bond provides a protective effect that can prevent active protein thiols from further irreversible oxidation with an increase in ROS generation (20–22).
Because protein S-glutathionylation is a reversible modification, the reduction of this mixed-disulfide bond, a process termed deglutathionylation, is expected when the intracellular redox state is normalized after oxidative stress. Grx1, a cytosolic oxidoreductase, has been demonstrated to specifically and efficiently reduce protein mixed-disulfide bonds with deglutathionylation at the expense of reduced glutathione (GSH) (23, 24). This Grx1- catalyzed deglutathionylation process plays an important role in redox regulation and signaling under oxidative stress as occurs in cardiovascular disease, neurodegenerative disease, and cancer (23, 24). Several studies have demonstrated that Grx1 not only effectively reduces this thiol modification, but also can glutathionylate target proteins through Grx-catalyzed disulfide exchange or the formation of a stabilized Grx-glutathionyl disulfide anion radical (25–29). The dynamic steady-state of this Grx1-catalyzed thiol modification depends on the cellular ratio of GSSG to GSH (30). This dual role of Grx1 makes it an important player in redox modulation of proteins with active thiols and in signal transduction.
In this study, we demonstrate that eNOS S-glutathionylation of cysteine residues in the reductase domain can be reversed by Grx1 in the presence of GSH. However, when oxidant stress is increased with a corresponding increase in GSSG, the reaction by Grx1 switches from deglutathionylation to glutathionylation, and oxidatively modifies eNOS with S-glutathionylation and inactivation of NO production. Mass spectrometry identifies a new site of this modification at Cys382 on the surface of the eNOS oxygenase domain. We further demonstrate that inhibition of Grx1 or grx1 gene silencing leads to eNOS dysfunction in endothelial cells through oxidative modification and S-glutathionylation of eNOS. Thus, Grx1 has an important role in regulating eNOS function catalyzing deglutathionylation of eNOS under conditions of normal thiol-redox balance, while with a shift to oxidation of the glutathione pool, it induces glutathionylation at a unique site on the oxygenase down-regulating eNOS activity without uncoupling, providing a unique mechanism of eNOS redox regulation.
Materials and Methods
Materials
anti-NOS3 (C-20) HRP and anti-NOS3 (C-20) agarose conjugate (AC) antibodies were both obtained from Santa Cruz (Santa Cruz, CA), anti-Grx1 from Abcam (Cambridge, MA), anti-GSH from ViroGen (Watertown, MA), NADPH, L-Arg, calmodulin, hemoglobin, N-ethylmaleimide (NEM), GSH, GSSG, HEPES, and Tris from Sigma-Aldrich (St. Louis, MO). Secondary anti-rabbit and anti-mouse IgG-HRP antibodies were purchased from GE Life Sciences (Piscataway, NJ).
Protein and heme concentration determination
Protein concentration of purified eNOS was determined by the Bradford assay from Bio-Rad (Hercules, CA) using a bovine serum albumin standard. The heme content of the purified eNOS was determined by the pyridine hemochromogen assay as previously described (31, 32).
eNOS S-glutathionylation by GSSG
1 μg/μL of purified eNOS was incubated with 2 mM GSSG at room temperature (RT) for 20 min, within which time both Cys689 and Cys908 were S-glutathionylated (14). After incubation, the excess GSSG was removed by passage through a 5 mL HiTrap desalting column from GE Life Sciences (Piscataway, NJ) and concentrated, or through buffer exchange using a 50 KDa cutoff centricon from Millipore (Billerica, MA).
Deglutathionylation of S-glutathionylated eNOS
The deglutathionylation of eNOS was tested in a reaction in which S-glutathionylated eNOS was either incubated with 1 mM GSH or 1 mM GSH and 1 μM Grx1 at RT for 10 min. For immunoblotting analysis, the reaction was quenched by adding NEM to a final concentration of 10 mM at specified time points. For NO activity measurements, the reaction aliquot was directly incubated with all cofactors in a given volume for 1 min before starting the reaction (33).
Measurement of NOS activity
The NO production from purified eNOS was measured by the conversion of ferrous oxyhemoglobin to ferric methemoglobin at RT. The initial rate of NO production was determined from the slope of ΔA401-411 (Δε401-411 = 38,600 M−1 cm−1) using an Agilent (Santa Clara, CA) 8453 UV-Visible Spectrophotometer (33). The reaction mixture contained 10 μg of CaM, 100 μM BH4, 100 μM L-Arg, 200 μM NADPH, 200 μM CaCl2, and 10 μM oxyhemoglobin in 50 mM Tris-HCl, pH 7.4 in a total volume of 500 μL. Normally, 2–5 μg of eNOS was used and the reaction was initiated by addition of 10 μL of 10 mM NADPH. For FAD and FMN supplementation, 4 μM of FAD and FMN were incubated with glutathionylated or deglutathionylated eNOS for 5 min before NOS activity measurement. The activity of untreated eNOS is 85–100 nmol/mg/min at RT.
Immunoblotting for eNOS or eNOS S-glutathionylation
The procedure for immunoblotting for eNOS and eNOS S-glutathionylation was followed as previously described (14, 32).
Site-directed mutagenesis of eNOS at Cys689 to Ala
The single mutation of eNOS at C689 to Ala was generated to determine the effect of this specific cysteine residue on flavin binding. The wild-type eNOS gene was mutated at 689 to Ala using the QuikChange mutagenesis kit from Stratagene (Santa Clara, CA). The sequences of primers for C689 to Ala are “5′ GGCGACGAGCTGGCCGGCCAGGAGG 3′” and “5′ CCTCCTGGCCGGCCAGCTCGTCGCC 3′”.
FAD/FMN measurement using HPLC chromatography
The content of FAD/FMN of eNOS or S-glutathionylated eNOS was determined using HPLC chromatography (32, 34). The purified eNOS or S-glutathionylated eNOS (100 μg in 500 μL) was first boiled for 10 min to release FAD and FMN from the protein. Protein was then removed by filtration through a Microcon-3 (Millipore). 50 μL of the FAD/FMN-containing filtrates were separated on a Shimadzu (Columbia, MD) LC-2010AT HPLC system with a C18 reversed-phase column (Alltima Reversed-Phase C18 HPLC Columns, 4.6 mm × 150 mm). After injection, Buffer A (5 mM ammonium acetate, pH 6.0 and 7% methanol) with a flow rate of 0.5 mL/min was used for 2 min. Then, a linear gradient was developed to 70% methanol using Buffer B (5 mM ammonium acetate pH 6.0, 70% methanol) over 13 min. A Shimadzu RF-10A XL fluorescence detector with excitation wavelength set at 460 nm and emission wavelength set at 530 nm was used to detect FAD and FMN. FAD and FMN were completely separated with elution times of 13.0 min and 14.5 min, respectively. The peak area was used to calculate the content of FAD/FMN compared to FAD and FMN standards.
Site-directed mutagenesis of eNOS at Cys689/C908 to Ala
A double mutant of eNOS at C689 and C908 to Ala was generated to test the effect and importance of these two specific cysteine residues in maintaining the structure and function of the eNOS reductase domain. The wild-type eNOS gene was first mutated at C908 to Ala, followed by another single mutant at Cys689 to Ala to form a double mutation of eNOS at Cys689 and C908 to Ala. The sequences of primers for C908 to Ala are “5′ GAAGTGGTTCCGCGCCCCCACGCTGCTG 3′” and “5′ CAGCAGCGTGGGGGCGCGGAACCACTTC 3′”. The sequences of primers for C689 to Ala are “5′ GGCGACGAGCTGGCCGGCCAGGAGG 3′” and “5′ CCTCCTGGCCGGCCAGCTCGTCGCC 3′”. The QuikChange mutagenesis kit from Stratagene (Santa Clara, CA) was used for this site-directed mutagenesis of eNOS(14, 32).
Flavin fluorescence measurement
Fluorescence spectra of eNOS flavins were measured using a SpexFluoroMax fluorescence spectrometer to determine the flavin content of eNOS. The total protein used for measurement was 50 μg for both wild-type and double mutant in 2 mL of PBS. The excitation spectrum of eNOS flavin was determined with an emission wavelength of 530 nm. The emission spectrum was determined with an excitation wavelength of 449 nm.
Overexpression and purification of Grx1
The Grx1 bacterial overexpression plasmid was a gift from Dr. John J. Mieyal (Case Western Reserve University, Cleveland, Ohio). The procedure for Grx1 overexpression and purification was followed as previously described. The activity of Grx1 was determined using a glutathione reductase coupled reaction with Cys-SG from Toronto Research Chemicals Inc. (Toronto, Ontario) as a substrate (35).
eNOS S-glutathionylation by Grx1
eNOS S-glutathionylation by Grx1 was carried out in a reaction containing 5 μg of eNOS in 50 mM Tris pH 7.4 with different ratios of GSSG/GSH in a final volume of 20 μL. The concentration of GSSG or GSH ranged from 0.1 mM to 1 mM. The reaction was initiated by addition of Grx1 to a final concentration of 1 μM at RT for 10 min. For mass spectrometric and immunoblotting analysis, NEM was added to a final concentration of 10 mM to quench the reaction. To measure eNOS activity, the oxyhemoglobin assay was carried out as previously described (33).
Measurement of •O2− generation by EPR spin-trapping
EPR spin-trapping measurements of oxygen radical production from eNOS (2 μg) or free FAD/FMN (1 μM) were performed as previously described (32).
Mass spectrometric analysis of eNOS S-glutathionylation by Grx1
The S-glutathionylated eNOS was subjected to SDS–PAGE on a 4–20% gradient polyacrylamide gel. Protein bands on the gel were then stained with Coomassie Blue. The band containing S-glutathionylated eNOS, which was confirmed by immunoblotting against anti-GSH antibody, was cut and digested in-gel with trypsin, chymotrypsin, or trypsin and chymotrypsin before mass spectrometric measurement. The S-glutathionylation of eNOS was determined with capillary-liquid chromatography tandem mass spectrometry (Nano-LC–MS/MS). The detailed parameters used in the MS measurements have been described in our previous study (14, 32). Sequence information from MS/MS data was processed with Mascot Distiller software, by using standard data processing parameters. Database searches were performed with the MASCOT (Matrix Science) program.
Site-directed mutagenesis of eNOS at Cys382 to Ala
The single mutation of eNOS at C382 to Ala was generated to test the effect of this specific cysteine residue in the resistance to the Grx1-directed eNOS S-glutathionylation. The wild-type eNOS gene was mutated at C382 to Ala using the QuikChange mutagenesis kit from Stratagene (Santa Clara, CA). The sequences of primers for C382 to Ala are “5′ GAGGATGTGGCTGTCGCCATGGACCTGGATAC 3′” and “5′ GTATCCAGGTCCATGGCGACAGCCACATCCTC 3′” (14, 32).
Cellular and in vitro analysis of eNOS and Grx1 interaction
To determine the eNOS and Grx1 interaction in endothelial cells, the co-immunoprecipitation of eNOS and Grx1 in bovine aortic endothelial cells (BAECs) was performed using anti-eNOS agarose conjugate (Santa Cruz, CA) or Grx1 antibody with protein A/G. The control experiment was performed using protein A/G beads only. The co-immunoprecipitation products were analyzed using immunoblotting against eNOS and Grx1. To determine the eNOS and Grx1 interaction in vitro, the purified eNOS with His-tag was first incubated with Ni-NTA magnetic beads from Invitrogen (Carlsbad, CA) followed by incubation with different concentrations of Grx1 at 4 °C for 1 hour before being washed three times with HEPES buffer (50 mM, pH 7.4, 150 mMNaCl) containing 5 mM imidazole to eliminate the non-specific binding. In the negative control, different concentrations of Grx1 were directly incubated with only Ni-NTA magnetic beads for 1 hour at 4 °C, and washed three times with HEPES buffer (50 mM, pH 7.4, 150 mMNaCl) containing 5 mM imidazole. Next, the co-precipitated product was eluted with HEPES buffer (50 mM, pH 7.4, 150 mMNaCl) containing 250 mM imidazole, followed by electrophoresis with 4–20% SDS-PAGE separation and immunostaining with anti-Grx1 and anti-eNOS.
Ex vivo eNOS S-glutathionylation by Grx1 and ex vivo analysis of eNOS and Grx1 interaction
For each experiment, one T-75 flask of BAECs was used to determine eNOS S-glutathionylation ex vivo. First, cells were lysed in 500 μL of 1xTBS buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl) with 1% NP-40, and 0.5% sodium deoxycholate. In the control experiment, the supernatant was treated with either 0.5 or 1 mM GSSG at RT for 10 min. In the reaction with Grx1, the supernatant was treated with either 0.5 mM or 1 mM GSSG and 1 μM of Grx1 at RT for 10 min. All reactions were quenched by addition of NEM to a final concentration of 10 mM. After the reaction, immunoprecipitation of eNOS was performed, followed by electrophoresis with 4–20% gradient SDS-PAGE and immunostaining with anti-GSH and anti-eNOS to determine the level of eNOS S-glutathionylation. The eNOS co-immunoprecipitated product was also separated on 4–20% SDS-PAGE and immunostained with anti-Grx1 to further analyze the eNOS and Grx1 interaction ex vivo.
Inhibition of Grx1 by Cd2+ in BAECs
The maintenance of BAECs cell culture was the same as previously described (14, 32). Cd2+ (200 μM) with 10% FBS complete medium was added to cells for 3 hours to inhibit Grx1 activity in endothelial cells(36). eNOS activity from BAECs was measured using EPR NO spin-trapping with Fe-N-methyl-D-glucamine dithiocarbamate (Fe2+-MGD) after treatment. To determine the level of eNOS S-glutathionylation and multimerization through inter-disulfide bond formation, BAECs were first lysed in RIPA buffer (50 mM Tris-HCl, pH 7.6, 150 mMNaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing 10 mM NEM followed by either immunostaining for eNOS or immunoprecipitation of eNOS, and then immunostained for protein S-glutathionylation with GSH antibody (14, 32).
Bovine aortic endothelial cells grx1 gene silencing
grx1 gene silencing from BAECs was used to confirm the effect of Grx1 on eNOS function. The sense siRNA strand is 5′-CUGUUGACACGGCUAAAGCUU-3′ and the antisense siRNA strand is 3′-UUGACAACUGUGCCGAUUUCG-5′. These siRNAs were custom synthesized by Thermo Scientific (Rockford, IL). DharmaFECT Transfection Reagent from Thermo Scientific (Rockford, IL) was used to deliver grx1 siRNAs to BAECs. After 48 h, Grx1 immunoblotting was used to determine the Grx1 knockdown efficiency. NOS activity was measured using EPR with Fe2+-MGD as a spin trap (14).
EPR spin-trapping measurements of NO production
Spin-trapping measurements of NO from BAECs were performed with a Bruker EMX spectrometer with Fe2+-MGD as the spin trap (14, 32). Spin-trapping experiments were performed on cells grown in six-well plates (106 cells per well). Before EPR spin-trapping measurements, control cells, cells treated with 200 μM Cd2+, and cells treated with grx1 siRNA were washed twice with PBS. Next, 0.8 ml of PBS containing glucose (1 g/L), CaCl2, MgCl2, the NO spin-trap Fe2+-MGD (0.5 mM Fe2+, 5.0 mM MGD) and calcium ionophore (1 μM) was added to each well, and the plates were incubated for 20 min at 37 °C in a humidified environment containing 5% CO2/95% O2. After incubation, the medium from each well was removed, and the trapped NO in the supernatants was quantified by EPR. Spectra recorded from these cellular preparations were obtained with the following parameters: microwave power 20 mW; modulation amplitude 4.0 G; modulation frequency 100 kHz.
Results
Deglutathionylation of S-glutathionylated eNOS by Grx1 and GSH
As reported previously, in the presence of elevated levels of GSSG, eNOS S-glutathionylation occurs with modification of Cys689 and Cys908. When eNOS is incubated with 2 mM GSSG for 20 minutes, ~ 50 – 75% modification occurs with loss of eNOS activity and uncoupling with loss of NO synthesis and gain of •O2− production (14). Consistent with our prior reports, following incubation of eNOS with 2 mM GSSG for 20 minutes and gel filtration to remove the GSSG, immunoblotting showed prominent eNOS S-glutathionylation with a ~55 % loss of eNOS activity, as measured by the rate of conversion of oxyhemoglobin to methemoglobin.
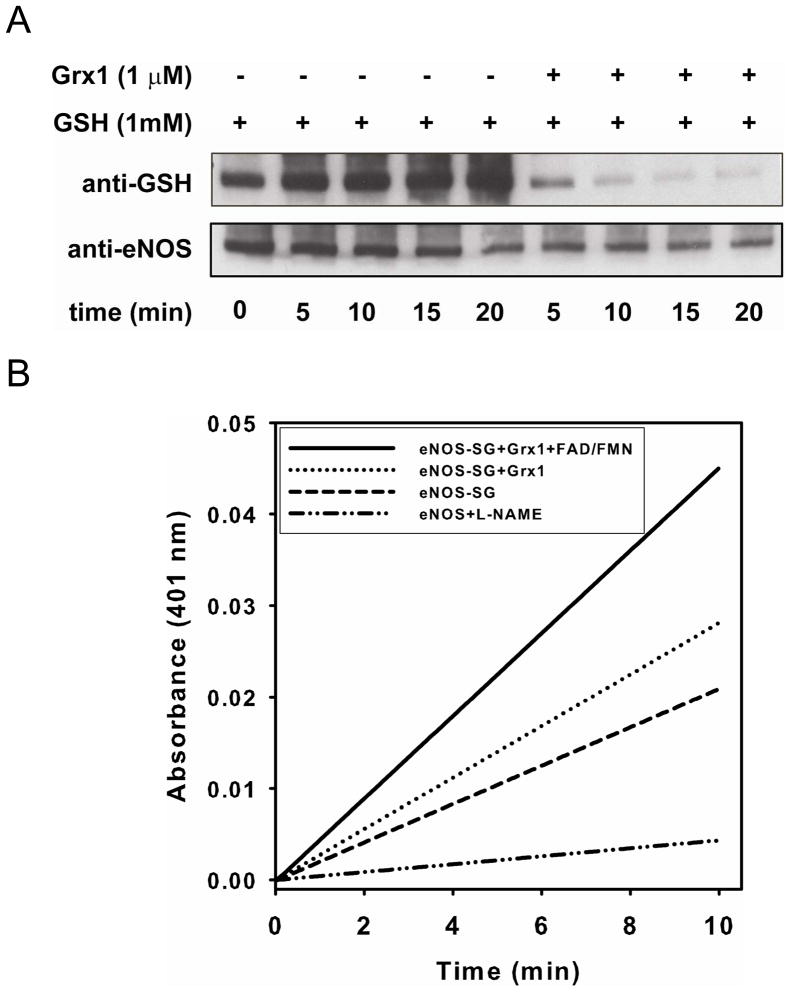
We next studied the effect of GSH and Grx1 on reversing eNOS S-glutathionylation. While in the presence of reduced GSH (1 mM) alone, no deglutathionylation of eNOS was seen even after 20 min of incubation (Fig 1A), with addition of both Grx1 (1 μM) and GSH (1 mM) a marked decrease in eNOS S-glutathionylation was seen after only 5 min of incubation. After 10 min of reaction, eNOS S-glutathionylation was almost totally reversed. In tandem with this, the oxyhemoglobin assay demonstrated a partial rescue of eNOS activity with rise from ~45% to 70% of basal activity (Fig 1B and C). Since S-glutathionylation of Cys689 and Cys908 within the eNOS reductase has been reported to increase the solvent accessibility of FAD and FMN, we hypothesized that it could weaken their binding resulting in partial loss and removal with the gel filtration that was performed. Therefore additional measurements were performed with addition of FAD and FMN (4 μM of each), and the activity was observed to fully recover to 100% of basal values, identical to unmodified enzyme. In contrast, supplementation of both FAD and FMN alone without GSH and Grx1 had no significant effect.
Figure 1. Deglutathionylation of S-glutathionylated eNOS requires Grx1.
A. Immunoblotting of eNOS deglutathionylation. Upper panel is immunoblotting against GSH antibody. Lower panel is immunoblotting against eNOS antibody. Grx1 is required for the reverse reaction. No eNOS deglutathionylation was seen in the absence of Grx1. B and C. NOS activity determined by the oxyhemoglobin assay. B. The kinetic traces of the absorbance change at 401 nm. The NOS activity was inhibited when 2 mM L-NAME was present. C. Reconstituted eNOS with 4 μM of FAD and FMN after deglutathionylation by Grx1 and GSH was required to fully recover activity to that without modification. The activity of untreated eNOS is 100 nmol/mg/min at RT. Data were expressed as mean ± SEM, n=3.
eNOS S-glutathionylation weakens eNOS flavin binding
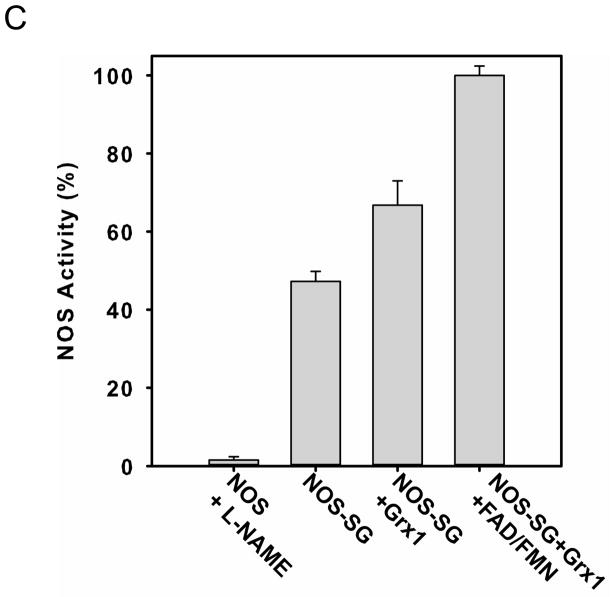
As described above, incubation of eNOS in the presence of GSSG, induces S-glutathionylation of two critical cysteine residues (Cys689 and Cys908) located in the eNOS reductase domain as previously identified by mass spectroscopy with confirmation by site-directed mutagenesis. Molecular modeling has shown that these cysteines are near the binding sites for both FAD and FMN cofactors and this led to the hypothesis that these modifications may weaken flavin binding. Therefore, HPLC measurements were performed to determine the level of eNOS-bound flavins before and after GSSG mediated S-glutathionylation. The excess GSSG and free or weakly bound FAD and FMN were removed through concentration and buffer exchange using a 50 KDa molecular weight cutoff Centricon from Millipore (Billerica, MA). The HPLC results (Fig 2A and B) indicated that the level of both eNOS-bound FAD and FMN after GSSG treatment and buffer exchange decreased to 45% ± 1% and 47% ± 1%, respectively, compared to that of untreated eNOS. Thus, S-glutathionylation weakened FAD and FMN binding to eNOS. When Cys908 was mutated to Ala in our previous study (32), the mutation of this residue slightly affected eNOS flavin binding (by ~ 20 %). To further determine the role of these residues on the effect of eNOS flavin binding, the level of eNOS-bound FAD and FMN of the C689A mutant measured by HPLC decreased to 20.5% ± 1.3% and 31.4 ± 4.4 %, respectively, compared to that of wild-type eNOS. These results suggested that C689 and C908 are both required for flavin binding. The results were expressed as mean ± s.e.m. (n = 3).
Figure 2. eNOS S-glutathionylation by GSSG or eNOS Cys double mutation to Ala induces flavin dissociation.
Purified eNOS was first incubated with 2 mM GSSG to induce protein S-glutathionylation followed by removal of GSSG with chromatography and sample concentration by ultrafiltration. The flavin content of the enzyme was determined using HPLC with a fluorescent detector with excitation wavelength at 460 nm and emission wavelength at 530 nm before and after treatment. A. The solid line is untreated eNOS, and the dashed line is GSSG-treated eNOS. The retention time of FAD is 13.0 min, and the retention time of FMN is 14.5 min. B. The content of eNOS bound FAD and FMN for the GSSG-treated enzyme decreased compared to that of untreated eNOS. All experiments were performed at least in triplicate. C. Fluorescence measurement of flavins. Wild-type eNOS (solid line) exhibited the intrinsic flavin fluorescence, but no fluorescent signal was seen from the eNOS Cys double mutant (dash line). The excitation spectrum of eNOS flavin was determined with an emission wavelength of 530 nm. The emission spectrum was determined with an excitation wavelength of 449 nm.
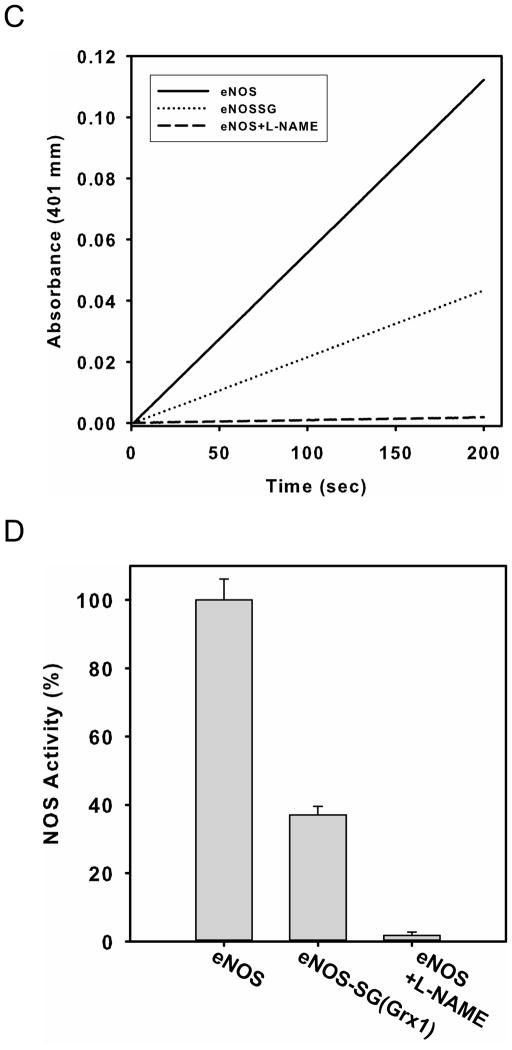
Consistent with the prior reports (14), we observe that while eNOS activity and NO generation is decreased by GSSG-mediated S-glutathionylation, •O2− generation is increased (Fig. 3E). The fact that this •O2− generation can not be inhibited by the oxygenase binding inhibitor L-NAME, indicates that this S-glutathionylation of eNOS at Cys689 and Cys908 opens the reductase providing access of O2 to the reduced flavins with resultant •O2− production (14).
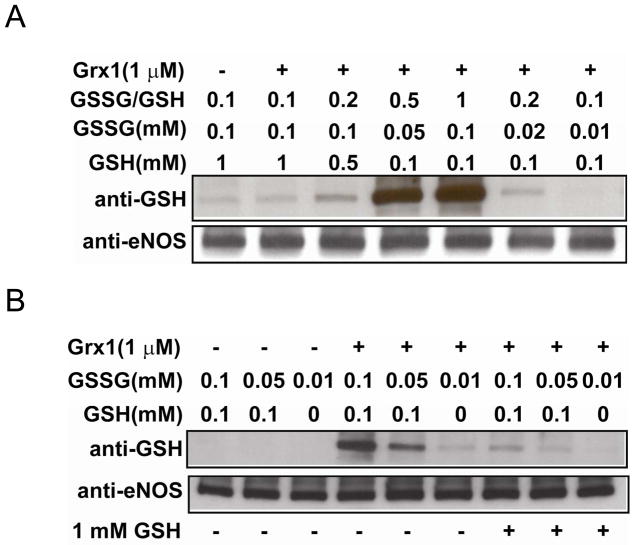
Figure 3. Grx1 enhances eNOS S-glutathionylation with increased ratio of GSSG/GSH, and inhibits enzyme activity.
A. Upper panel is the immunoblotting against GSH antibody and lower panel is the immunoblotting against eNOS antibody for loading control. Grx1 increased eNOS S-glutathionylation when the ratio of GSSG/GSH was greater than 0.2. B. Upper panel is the immunoblotting against GSH antibody and lower panel is the immunoblotting against eNOS antibody for loading control. When the concentrations of GSH and GSSG were both low, Grx1 was required for eNOS S-glutathionylation. This process was reversible when 1 mM GSH was added. C and D. eNOS was S-glutathionylated by Grx1 in the presence of 0.1 mM GSSG and 0.1 mM GSH. C. The kinetic traces of the absorbance change at 401 nm. D. The activity of control eNOS and S-glutathionylated eNOS by Grx1. (* P < 0.01 vs. respective controls). Data were expressed as mean ± SEM, n=3. The activity of untreated eNOS is 85 nmol/mg/min at RT. E. EPR spin-trapping measurements of oxygen radical generation. No increase in •O2− production was seen from this Grx1-modified eNOS compared to control eNOS. eNOS treated with 2 mM GSSG gave rise to a strong •O2− signal. No •O2− generation was seen from free FAD/FMN (1 μM) and NADPH (200 μM).
Mutation of eNOS at Cys689 and Cys908 to Ala decreases eNOS flavin binding
In order to further test the role of Cys698 and Cys908 for flavin binding, flavin fluorescence was used to investigate the eNOS Cys689A/C908A double mutant on the effect of eNOS flavin binding. Wild-type eNOS exhibited the intrinsic flavin fluorescence, but only very weak fluorescent signal was seen from the eNOS Cys double mutant (Fig 2C) indicating that these cysteines have an important role in facilitating or stabilizing FAD and FMN binding. Interestingly, as reported previously, with Cys689 and Cys908 mutation to serine, which has similar polarity and hydrogen bonding to cysteine, eNOS function and coupling is normal with no decline in NOS activity indicating no loss of FAD or FMN (14).
With increase in GSSG/GSH ratio Grx1 increases eNOS S-glutathionylation and inhibits eNOS activity
When the GSSG/GSH ratio was 0.1 in the absence or presence of Grx1, no or only low level eNOS S-glutathionylation was detected on immunoblotting. However, in the presence of Grx1 when the GSSG/GSH ratio was increased to 0.2, eNOS S-glutathionylation was increased and with a further increase of GSSG/GSH to 0.5 or 1, S-glutathionylation was dramatically enhanced (Fig 3A).
In order to determine if Grx1 was required for eNOS S-glutathionylation, studies were performed with low concentrations of GSSG and GSH (0.1 or 0.05 mM) in the absence of Grx1, and no eNOS S-glutathionylation was seen on immunoblotting (Fig. 3Aand B). However, when 1 μM of Grx1 was present, eNOS S-glutathionylation was greatly increased. With addition of 1 mM GSH, this oxidative modification was fully reversed within 10 minutes (Fig. 3B).
Further experiments were performed to measure the effects of Grx1-mediated S-glutathionylation on eNOS activity as assayed by the oxyhemoglobin to methemoglobin conversion assay. Following 10 min incubation of eNOS in the presence of Grx1 (1 μM) in the presence of 0.1 mM of GSSG and 0.1 mM GSH, NOS activity was ~65% decreased (Fig 3C and D). No increase in •O2− production was seen from this Grx1-modified eNOS using EPR DEPMPO •O2− spin-trapping (Fig. 3E), indicating that this decrease in activity was not associated with uncoupling.
Mass spectrometric analysis of eNOS S-glutathionylation by Grx1
Since the sites of Grx-1-mediated S-glutathionylation may be different than that caused by GSSG, mass spectroscopy analysis was performed to identify the specific sites of eNOS S-glutathionylation by Grx1. eNOS (~2 μg in 20 μl) was S-glutathionylated in the presence of 0.1 mM GSSG, 0.1 mM GSH, and 1 μM Grx1 at RT for 10 min. The reaction mixture was then subjected to 4–20% gradient SDS-PAGE separation under non-reducing conditions. The band corresponding to S-glutathionylated eNOS was cut and digested with trypsin, chymotrypsin or both, and then the digested peptide fragments were analyzed by LC/MS/MS. The percentage of sequence coverage determined by LC/MS/MS was 85%.
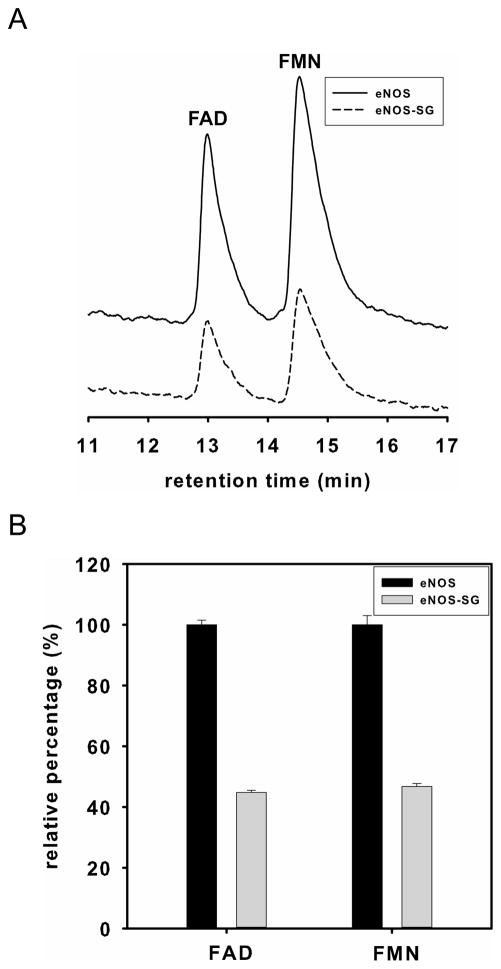
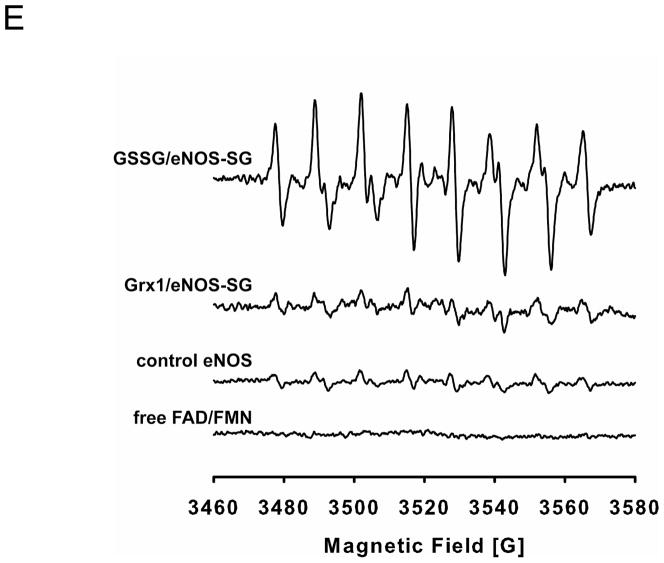
With the addition of one molecule of glutathione, the molecular weight of the glutathione-modified peptide fragment will increase by 305 Da compared to the unmodified peptide fragment. The peptides with the mass difference of 305 Da were identified by LC/MS, and the sequence of the modified peptides was further determined by MS/MS and analyzed by the Mascot software. From mass determination, Cys382 was identified as the major site for eNOS S-glutathionylation by Grx1 from both trypsin and chymotrypsin digestions. The percentage of this modification at Cys382 was >50%.
S-glutathionylated Cys382 was determined from fragment 373YNILEDVA VCMDLDTR388 (aa 373–388, Fig. 4A). To further validate the exact location of the glutathione-modified amino acid(s), the MS/MS spectrum of the tryptic and chymotryptic fragment ion at m/z 725.65973+ was obtained as shown in Fig. 4A. Under the conditions of the low energy CID, both y and b product ions were observed, corresponding to cleavages along the peptide backbone. The y series ions result from C-terminal peptide cleavages, while the b series ions result from cleavages at the N-terminus. In the spectrum of the tryptic and chymotryptic peptide ion, the molecular weight difference between fragment ions y6 and y7 was observed with a mass shift of 305 Da compared to the native fragment ions, allowing unequivocal assignment of the glutathionylated adduct to the Cys382 residue of the tryptic and chymotryptic peptides. Neither Cys689 nor Cys908 were S-glutathionylated by Grx1.
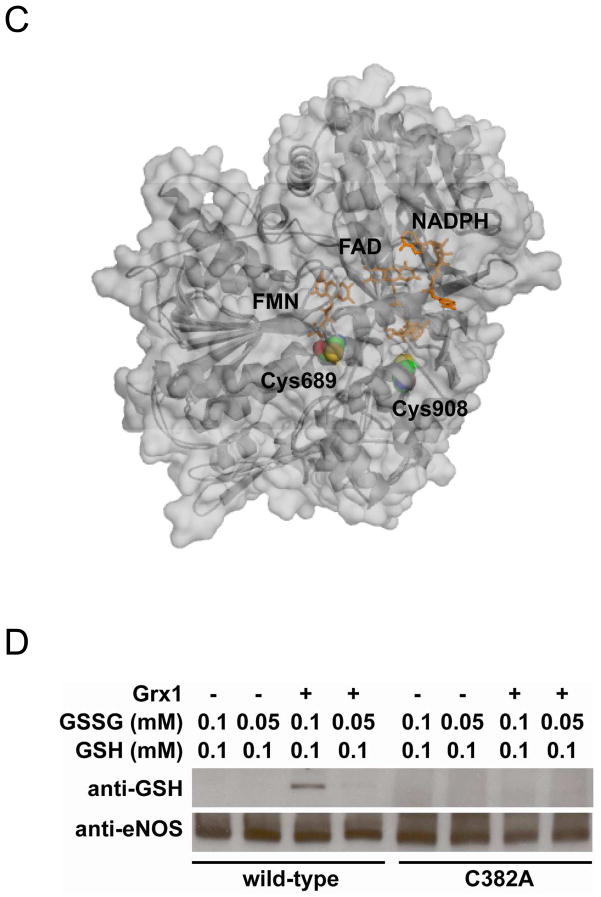
Figure 4. Mass spectrometry and molecular modeling reveal C382 is the site of eNOS S-glutathionylation by Grx1.
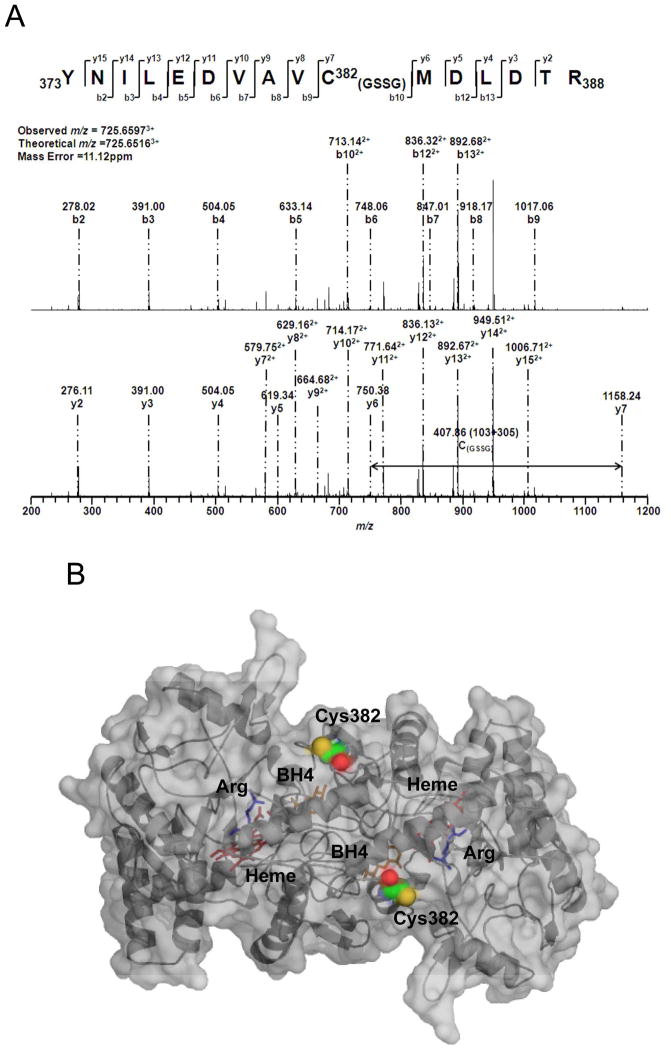
A. LC/MS/MS mass spectrometric analysis of eNOS S-glutathionylation by Grx1. Cys382 was identified as the specific site for S-glutathionylation by Grx1 in the presence of 0.1 mM GSSG and 0.1 mM GSH. The molecular weight difference between fragment ions y6 and y7 demonstrated a mass shift of 305 Da compared to the native fragment ions allowing unequivocal assignment of the GSH adduct to Cys382. B and C. Molecular modeling of eNOS oxygenase domain and reductase domain. The three dimensional structure of human eNOS oxygenase domain (PDB ID 3NOS) (B) and reductase domain (C) was visualized using PyMOL (Portland, OR). The eNOS oxygenase domain is shown in the dimeric form. Heme (red) shown in sticks, L-arginine (blue) shown in sticks, and BH4 (magenta) shown in sticks. The three-dimensional structure of human eNOS reductase domain was generated by homology modeling referencing the reductase domain of rat neuronal NOS (PDB ID 1F20) using Swiss molecular modeling as previous described (14). The eNOS reductase domain is shown in monomeric form. FMN, FAD and NADPH (orange) shown in sticks. Cys shown in spheres with the following atom colors: S in yellow, C in cyan, N in blue, and O in red. D. eNOS C382A mutant resists Grx1-induced S-glutathionylation. In the absence of Grx1 with GSSG/GSH ratio of 1 or 0.5, no eNOS S-glutathionylation was seen in both wild-type or C382A mutant. However, in the presence of Grx1 and GSSG/GSH, only wild-type eNOS exhibited Grx1-catalyzed S-glutathionylation, however, C382A mutant resisted Grx1-induced S-glutathionylation.
Molecular modeling of eNOS oxygenase and reductase domains
The published crystal structure of the human eNOS oxygenase domain (PDB ID 3NOS) was used to generate a three dimensional image. PyMOL (Portland, OR) was used to visualize the location of Cys382 in the oxygenase domain (Figure 4B). From the three dimensional structure of eNOS oxygenase domain, it is clear that Cys382 is solvent accessible and available for Grx1-catalyzed eNOS S-glutathionylation. In contrast from modeling of the eNOS reductase (Figure 4C), we can see that neither Cys689 nor Cys908 are fully solvent-exposed, with Cys 908 more deeply buried in the protein than Cys689. Thus, these sites would not appear to be susceptible to Grx1 binding and Grx1-mediated S-glutathionylation. However, following S-glutathionylation by GSSG these sites may become more solvent exposed and accessible to Grx1-mediated deglutathionylation. It is worth noting that Cys689 is very close to the C-terminal tail of the enzyme, which acts as one of the two CaM-modulated regulatory elements. Thus, S-glutathionylation of eNOS at Cys689 can potentially disrupt CaM modulation possibly affecting uncoupling.
The C382A eNOS mutant resists Grx1-catalyzed eNOS S-glutathionylation
When both wild-type eNOS and eNOS C382A mutant were incubated (10 min) with GSSG (0.1 or 0.05 mM) and GSH (0.1 mM) alone, no eNOS S-glutathionylation was seen (Fig 4C). However, when Grx1 was included in the reaction, the Grx1-catalyzed eNOS S-glutathionylation was only seen from wild-type eNOS with no Grx1-catalyzed eNOS S-glutathionylation of the eNOS C382A mutant (Fig 4D).
Grx1 plays an important role in redox regulation in endothelial cells and is associated with eNOS
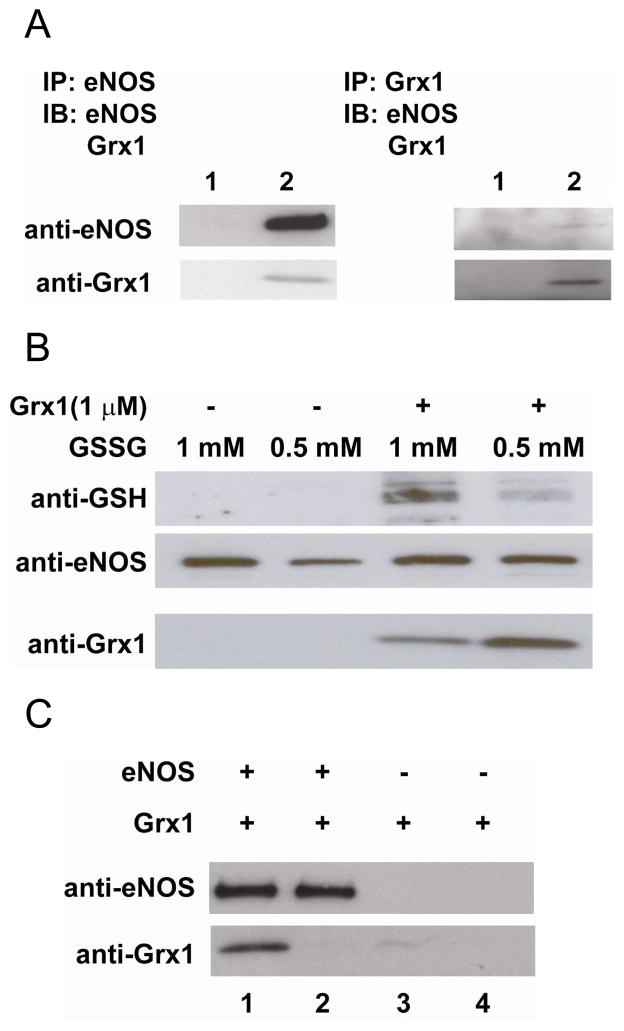
The co-immunoprecipitation of eNOS and Grx1 in BAECs demonstrated that eNOS and Grx1 can form a native complex in endothelial cells (Figure 5A). Grx1 co-immunoprecipitated with eNOS detected by immunoblotting against Grx1 antibody, and eNOS co-immunoprecipitated with Grx1 detected by immunoblotting against eNOS antibody. A post-lysate of endothelial cells was used to demonstrate that Grx1 can efficiently redox-regulate eNOS function. When a post-lysate was incubated with 1 or 0.5 mM GSSG alone for 10 min at RT, no eNOS S-glutathionylation was detected from GSH immunoblotting after immunoprecipitation with eNOS antibody. However, when GSSG and Grx1 were both incubated with the post-lysate, eNOS S-glutathionylation was detected, and the intensity of the modification was dependent on the concentration of GSSG (Fig 5B). Grx1 co-immunoprecipitated with eNOS detected by immunoblotting against Grx1 antibody
Figure 5. Grx1 is a protein partner of eNOS in endothelial cells and in vitro.
A. Co-immunoprecipitation of eNOS and Grx1 from BAECs. Left: Lane 1 is control with proteinA/G beads only. Lane 2 is IP with eNOS agaros-conjugated antibody. Upper panel is the immunoblotting against eNOS. Lower panel is the immunoblotting for Grx1. Right: Lane 1 is control with proteinA/G beads only. Lane 2 is IP with Grx1 antibody and proteinA/G. Upper panel is the immunoblotting against eNOS. Lower panel is the immunoblotting for Grx1. B. Co-immunoprecipitation of eNOS and Grx1 from ex vivo BAECs post-lysate. Upper panel is the immunoblotting against GSH. No eNOS S-glutathionylation was seen when post-lysate was incubated with 0.5 or 1 mM GSSG alone for only 10 min. When 1 μM Grx1 was included in the reaction, eNOS S-glutathionylation was enhanced. Middle panel is the immunoblotting for eNOS. Lower panel is the immunoblotting for Grx1. Grx1 co-immunoprecipitated with eNOS. C. Co-precipitation of eNOS and Grx1 using His-tagged magnetic beads. Upper panel is the immunoblotting for eNOS, and lower panel is the immunoblotting for Grx1. Lane 1 is co-precipitation of 0.2 μM eNOS and 0.4 μM Grx1. Lane 2 is co-precipitation of 0.2 μM eNOS and 0.04 μM Grx1. Lane 3 is 0.4 μM Grx1 only, and lane 4 is 0.04 μM Grx1 only. All experiments were performed at least in triplicate.
Next, the purified His-tagged eNOS was first incubated with magnetic Ni-NTA beads followed by incubation with different concentrations of Grx1. The result from immunoblotting against Grx1 (Fig 5C) clearly indicated that Grx1 is a protein partner that binds to eNOS, consistent with the previous result from the co-immunoprecipitation of eNOS and Grx1 in BAECs experiments.
Inhibition of glutaredoxin leads to eNOS dysfunction with loss of NO generation
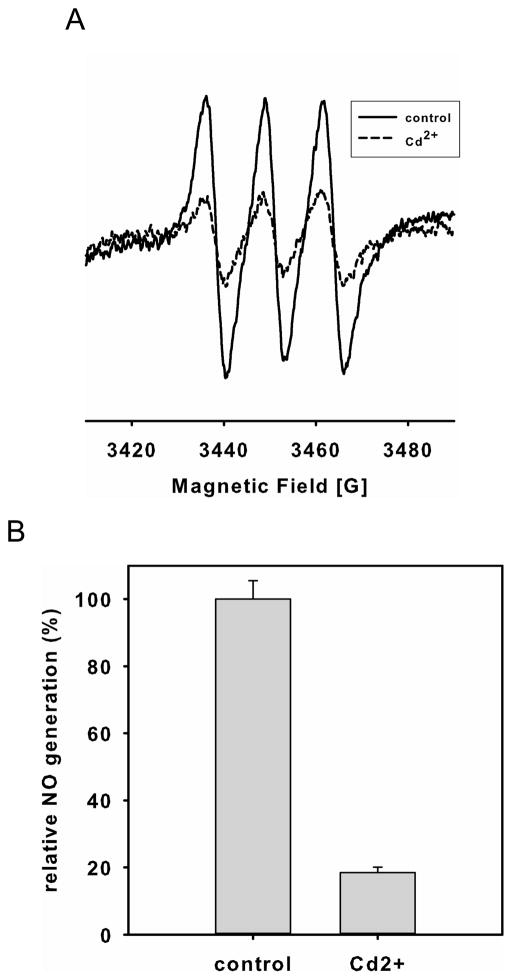
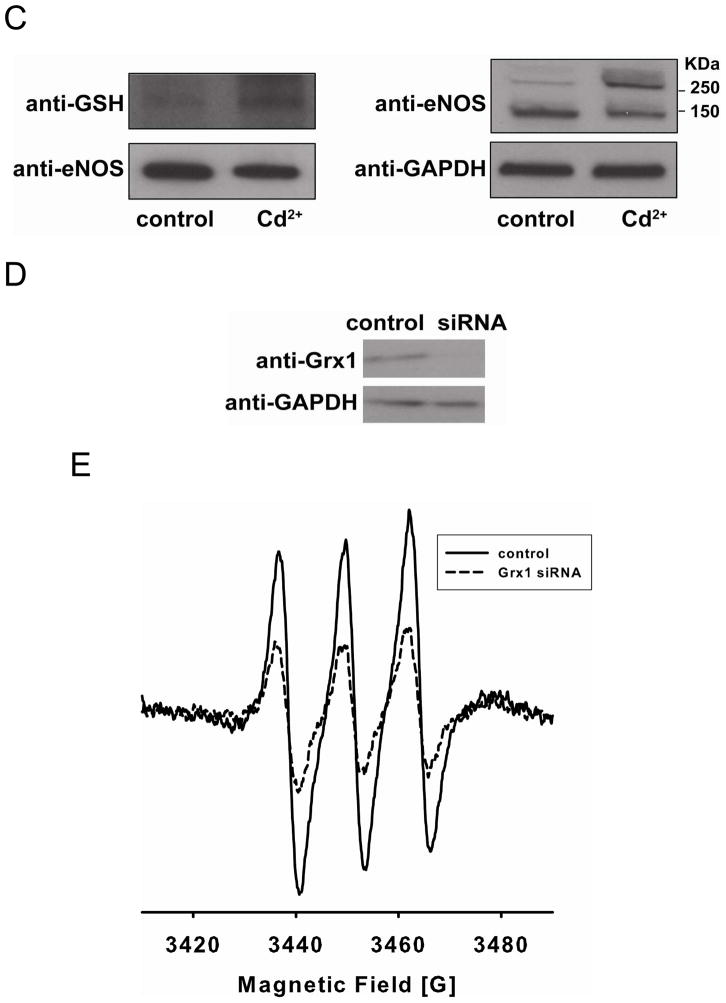
Cd2+ has been shown to inhibit Grx activity in endothelial cells (36, 37). Inhibition of Grx enzymes by Cd2+ (200 μM) in BAECs in complete media with 10% FBS, dramatically decreased NO production by 82 ± 2% as measured by EPR spin-trapping (Fig 6A and B). The decrease in NO production was correlated with an increase in eNOS S-glutathionylation determined from the immunoblotting of protein bound GSH from immunoprecipitated eNOS, as well as from the formation of inter-protein disulfide bonds determined by eNOS immunoblotting under non-reducing SDS-PAGE analysis (Fig 6C). While marked inhibition of eNOS activity was seen in the BAECs, only modest (<20%) inhibition of eNOS-mediated NO generation was seen with purified eNOS (10 μg/ml) in matched media. This confirms that Grx1 has an important role in regulating eNOS S-glutathionylation and preserving eNOS activity and NO generation.
Figure 6.
Inhibition of Grx enzymes by Cd2+ or grx1 gene silencing in endothelial cells decreases NO generation from endothelial cells with concomitant increase of eNOS oxidative modification. A and B. NO generation from BAECs measured by EPR spin-trapping using Fe2+-MGD. While a strong NO triplet signal was seen in untreated cells, Cd2+ treatment dramatically decreased this NO generation (* P < 0.01 vs. respective controls). Data were expressed as mean ± SEM, n=3. C. Immunoblotting of eNOS S-glutathionylation and dimerization. Left panel is the immunoprecipitation of eNOS followed by immunoblotting against GSH antibody (upper) and eNOS antibody (lower). Right panel is immunoblotting against eNOS antibody (upper) using non-reducing SDS-PAGE analysis, and GAPDH (lower). All experiments were performed at least in triplicate. D and E. grx1 gene silencing in endothelial cells decreases NO generation from endothelial cells. D. grx1 gene silencing. Lane 1 is control. Lane 2 is grx1 siRNA. Upper panel is the immunoblotting for Grx1, and lower panel is the immunoblotting for GAPDH as a loading control. After two days of post-transfection with grx1 siRNA, the efficiency of Grx1 knockdown was nearly complete. E. NOS activity from BAECs was measured using EPR with Fe2+-MGD as a spin trap. grx1 gene silencing deceases NO generation from BAECs compared to control.
grx1 gene silencing in BAECs leads to NOS dysfunction
grx1 gene silencing with siRNA was performed to further determine the effect of Grx1 on NOS function in endothelial cells. After two days of post-transfection with grx1 siRNA, immunoblotting of Grx1 indicated that the efficiency of Grx1 knockdown was nearly complete (Fig 6D). The NOS activity of control, or grx1 siRNA was measured using NO Fe2+-MGD EPR spin-trapping. The result (Fig 6E) demonstrated that grx1 gene silencing was associated with NOS dysfunction with a dramatic decrease in NO production in BAECs.
Discussion
Recently, we have demonstrated that eNOS can be S-glutathionylated in both hypertensive vessels and cells under oxidative stress, leading to uncoupling of the enzyme (14). S-glutathionylation of eNOS has been demonstrated through the direct reaction with GSSG (14) or through the formation of labile intermediate protein radicals (32). Several studies have also shown that eNOS S-glutathionylation is associated with many cardiovascular diseases and occurs in cells exposed to oxidative stress (15–17, 38). Protein S-glutathionylation, a reversible oxidative modification, plays an important role in redox signaling and can be protective against irreversible oxidation of protein thiols in many cardiovascular diseases (20–22). As such, it is important to identify the mechanism of the reverse process, deglutathionylation of eNOS protein thiols, and this can provide a potential therapeutic target to treat oxidant-induced diseases.
S-glutathionylation of eNOS has been shown to modulate its structure and function. Thus, it is important to understand the reversible nature of this oxidative modification, and whether this reversible thiol modification can redox-regulate eNOS function under oxidative stress as well as during disease progression. This reverse process is termed “deglutathionylation”, reduction of a mixed-disulfide bond between GSH and a protein thiol. Grx enzymes, GSH-dependent oxidoreductases, have been implicated in the reduction of mixed-disulfide bonds, especially S-glutathionylated proteins (23, 24).
In the current study, we demonstrated that no deglutathionylation occurred when GSH was the only reducing equivalent in the reaction, while there was a dramatic decrease in this oxidative modification when both Grx1 and GSH were present. The reduction of this mixed-disulfide bond (deglutathionylation) by GSH is via a bimolecular nucleophilic substitution (a SN2 reaction) (39). For GSH to be in the correct form for reducing this disulfide bond, the thiol of GSH must be deprotonated. The pKa of the GSH thiol is approximately 8.5; therefore, most of the GSH is expected to be protonated at physiological pH and not primed for this SN2 reaction or the charge repulsion between deprotonated GSH and S-glutathionylated eNOS (40). This explains why eNOS deglutathionylation was not seen with GSH as the only reducing equivalent in the reaction. This further demonstrates that Grx1 is required to overcome the activation energy barrier of this disulfide reduction for this deglutathionylation reaction. Thus, Grx1 is required to effectively reduce this oxidative modification.
We observed that following eNOS S-glutathionylation by GSSG, that FAD and FMN binding were weakened so that partial loss was seen. Therefore, addition of FAD and FMN was required to recovery the full activity of eNOS. The S-glutathionylated eNOS exhibits greatly increased •O2− generation, while comparable levels of free FAD, FMN, NADPH do not. Thus, S-glutathionylation of eNOS opens the FAD and FMN sites of the eNOS-reductase domain to react with O2 and weakens their binding so that dissociation with loss of binding to the enzyme can occur in the absence of free FAD and FMN.
Previously, we have identified two critical cysteinyl residues of eNOS involved in S-glutathionylation by GSSG that can uncouple eNOS leading to an increase in eNOS-derived •O2− generation from the reductase domain. These two highly conserved residues, Cys689 and Cys908 of eNOS, are located at the interface of the FAD and FMN binding domains. Introduction of a bulky and negatively-charged molecule (GSH) perturbs the eNOS structure in this region, increasing the solvent accessibility to the FAD and FMN cofactors (14). Our current HPLC analysis of the FAD and FMN content supports the hypothesis that S-glutathionylation of eNOS by GSSG perturbs this interface and weakens FAD and FMN binding to the S-glutathionylated eNOS. The importance of Cys689 and Cys908 for flavin binding was further supported by the loss of flavins seen in the eNOS C689A single mutant and C689A/C908A double mutant. Furthermore, the perturbation of the reductase domain induced by GSSG modification allows Grx1 access to these mixed-disulfide bonds (C689/C908) enabling deglutathionylation.
It has been asserted that, under normal physiological conditions, cellular GSSG concentrations may not reach a thermodynamically favorable level that can effectively S-glutathionylate protein thiols to redox-modulate protein function (41, 42). Previously, we have identified an efficient mechanism, in which the formation of S-glutathionylated protein thiols of eNOS can go through a labile intermediate protein thiyl radical generated by ROS followed by the reaction with GSH under oxidative stress (32). It is also important to identify whether there is an efficient enzyme-catalyzed regulation of eNOS S-glutathionylation sensitive to fluctuations in redox state, analogous to phosphorylation by the kinases and dephosphorylation by the phosphatases.
Grx enzymes are GSH-dependent enzymes that can specifically reduce a mixed disulfide bond from S-glutathionylated proteins generating GSSG as a product. When oxidant stress is increased, the cellular GSSG concentration is also increased. This increase will shift the cellular redox potential less negative in favor of S-glutathionylation. A recent study showed Grx can oxidatively modify its target protein when high GSSG concentrations are present (43).
In the current study, we demonstrate that indeed Grx1 can effectively S-glutathionylate eNOS. When Grx1 is included in the reaction, it can overcome the reaction energy barrier and accelerate the formation of eNOS S-glutathionylation. This eNOS S-glutathionylation by Grx1 depends on the ratio of GSSG/GSH and the total GSH pool (39, 44, 45). Thus, the increase in GSSG is thermodynamically driving this reaction towards S-glutathionylation, and the formation of S-glutathionylated eNOS is more efficient in the presence of Grx1. With the addition of 1 mM GSH, the reaction is favored in the reverse reaction, deglutathionylation, in which Grx1 can effectively reduce this mixed-disulfide bond of eNOS. The discovery of eNOS S-glutathionylation by Grx1 provides an additional pathway for redox modulation of eNOS function under oxidative stress that may be more efficient and sensitive than GSSG acting alone. During ischemia/reperfusion injury, the export of GSSG from cells was reported (42, 46). This can lead to an increase in the GSSG/GSH ratio and decrease in the total GSH pool. The local GSSG concentration is increased with the burst of •O2− generation during reperfusion (47, 48) which can dramatically increase the GSSG/GSH ratio locally. Thus, the increase in the GSSG/GSH ratio can provide a driving force shifting the reaction in favor of S-glutathionylation by Grx1.
eNOS S-glutathionylation by Grx1 decreases NO production by the enzyme, but stimulation of eNOS uncoupling is not seen. This is different from what is seen when eNOS is directly S-glutathionylated by GSSG, where S-glutathionylation at Cys689 and Cys908 of the reductase domain lead to •O2− generation from the enzyme. LC/MS/MS analysis of this Grx1-catalyzed eNOS S-glutathionylation reveals that the primary site of modification is located at Cys382, on the surface of the oxygenase domain (Fig. 4B), which is accessible for Grx1 modification. From the three-dimensional structure of the eNOS reductase domain (Fig. 4C), the two critical cysteine residues (Cys689 and C908) are buried in the interface of this domain, and are not accessible for Grx1-oxidative modification. Thus, neither Cys689 nor Cys908 are S-glutathionylated by Grx1 under these conditions.
The identification of eNOS S-glutathionylation at Cys382 provides evidence that this particular modification at the oxygenase domain of eNOS only affects NO production by shutting down its enzymatic function without a concomitant increase in the production of •O2−. The decrease in NO production from the Grx1-modified enzyme can prevent the formation of the more potent oxidant, peroxynitrite, which could cause further irreversible damage to the enzyme. Mutagenesis of eNOS at this specific residue resists Grx1-catalyzed eNOS S-glutathionylation indicating that Cys382 is the primary site for Grx1-induced S-glutathionylation. Thus, eNOS S-glutathionylation by Grx1 at Cys382 is sensitive to the fluctuation of [GSSG]/[GSH] ratio and provides a unique mechanism that may protect this eNOS protein thiol from further oxidation under oxidative stress.
The co-precipitation of Grx1 and eNOS in endothelial cells, ex vivo and in vitro experiments further supports the idea that Grx1 has a critical function in the redox-modulation of eNOS as a function of the cellular redox state. The Grx-1-mediated reduction of S-glutathionylated eNOS as well as its oxidation of eNOS to form protein S-glutathionylated adducts, suggests that Grx1 plays a dual role in redox regulation depending on the GSSG/GSH ratio, analogous to the function of kinases and phosphatases on protein phosphorylation and dephosphorylation.
Direct inhibition of Grx enzymes by Cd2+ or grx1 gene silencing in BAECs inactivated NOS through induction of eNOS S-glutathionylation and the formation of inter-protein disulfide bonds. It would be expected that S-glutathionylation of eNOS accumulates, possibly through disulfide exchange or thiyl radical intermediates, when Grx1 is knocked down or inhibited by Cd2+. This provides evidence that Grx1 is required to maintain eNOS protein thiols in the reduced state supporting proper eNOS coupling in cells. This finding supports our hypothesis that Grx1 is critical in redox modulation of eNOS function and maintenance of vascular tone.
In conclusion, Grx1 in the presence of GSH is required for efficient eNOS deglutathionylation and allows restoration of eNOS function. However, under mild to moderate oxidative stress with the increase in GSSG, Grx1 can also S-glutathionylate eNOS with inactivation of NO synthesis without uncoupling, thereby protecting eNOS from further oxidation. Cys382 of the eNOS oxygenase domain is identified as the major site modified by Grx1, in contrast to non-enzymatic S-glutathionylation of eNOS induced by GSSG or radical attack, in which the major modifications are within the reductase domain. Inhibition of Grx enzymes and grx1 gene silencing in cells lead to endothelial dysfunction with loss of NO production in part due to the increase in eNOS S-glutathionylation and inter-protein disulfide bond formation. The identification of the dual role of Grx1 in glutathionylation/deglutathionylation involved in reversible modulation of eNOS activity as a function of redox state provides a sensitive mechanism of redox regulation of vascular function and signaling during redox stress.
Acknowledgments
We thank Dr. Liwen Zhang at OSU CCIC proteomics center for support with mass spectrometric analysis. We thank Dr. John J. Mieyal (Case Western Reserve University, Cleveland, Ohio) for providing us glutaredoxin 1. This work was supported by R00 Grant HL103846 (C.-A. C.), and R01 Grants HL63744, HL65608, HL38324 (J. L. Z) from the National Institutes of Health.
Abbreviations
- BAECs
bovine aortic endothelial cells
- CaM
calmodulin
- eNOS
endothelial nitric oxide synthase
- EPR
electron paramagnetic resonance
- Fe2+-MGD
Fe-N-methyl-D-glucamine dithiocarbamate
- Grx1
glutaredoxin-1
- L-NAME
L-NG-nitroarginine methyl ester hydrochloride
- NO
nitric oxide
- eNOS
endothelial nitric oxide synthase
- RT
room temperature
- •O2−
superoxide
- BH4
tetrahydrobiopterin
Footnotes
This work was supported by R00 Grant HL103846 (C.-A. C.), and R01 Grants HL63744, HL65608, HL38324 (J. L. Z)
References
- 1.Rapoport RM, Draznin MB, Murad F. Endothelium-dependent relaxation in rat aorta may be mediated through cyclic GMP-dependent protein phosphorylation. Nature. 1983;306:174–176. doi: 10.1038/306174a0. [DOI] [PubMed] [Google Scholar]
- 2.Palmer RM, Ashton DS, Moncada S. Vascular endothelial cells synthesize nitric oxide from L-arginine. Nature. 1988;333:664–666. doi: 10.1038/333664a0. [DOI] [PubMed] [Google Scholar]
- 3.Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH. Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature. 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- 4.Hunley TE, Iwasaki S, Homma T, Kon V. Nitric oxide and endothelin in pathophysiological settings. Pediatr Nephrol. 1995;9:235–244. doi: 10.1007/BF00860758. [DOI] [PubMed] [Google Scholar]
- 5.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci U S A. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorescu D, Griendling KK. Reactive oxygen species, mitochondria, and NAD(P)H oxidases in the development and progression of heart failure. Congest Heart Fail. 2002;8:132–140. doi: 10.1111/j.1527-5299.2002.00717.x. [DOI] [PubMed] [Google Scholar]
- 7.Wolin MS. Reactive oxygen species and vascular signal transduction mechanisms. Microcirculation. 1996;3:1–17. doi: 10.3109/10739689609146778. [DOI] [PubMed] [Google Scholar]
- 8.Wever RM, van Dam T, van Rijn HJ, de Groot F, Rabelink TJ. Tetrahydrobiopterin regulates superoxide and nitric oxide generation by recombinant endothelial nitric oxide synthase. Biochemical and biophysical research communications. 1997;237:340–344. doi: 10.1006/bbrc.1997.7069. [DOI] [PubMed] [Google Scholar]
- 9.Xia Y, Tsai AL, Berka V, Zweier JL. Superoxide generation from endothelial nitric-oxide synthase. A Ca2+/calmodulin-dependent and tetrahydrobiopterin regulatory process. The Journal of biological chemistry. 1998;273:25804–25808. doi: 10.1074/jbc.273.40.25804. [DOI] [PubMed] [Google Scholar]
- 10.Vasquez-Vivar J, Kalyanaraman B, Martasek P, Hogg N, Masters BS, Karoui H, Tordo P, Pritchard KA., Jr Superoxide generation by endothelial nitric oxide synthase: the influence of cofactors. Proc Natl Acad Sci U S A. 1998;95:9220–9225. doi: 10.1073/pnas.95.16.9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardounel AJ, Xia Y, Zweier JL. Endogenous methylarginines modulate superoxide as well as nitric oxide generation from neuronal nitric-oxide synthase: differences in the effects of monomethyl- and dimethylarginines in the presence and absence of tetrahydrobiopterin. The Journal of biological chemistry. 2005;280:7540–7549. doi: 10.1074/jbc.M410241200. [DOI] [PubMed] [Google Scholar]
- 12.Druhan LJ, Forbes SP, Pope AJ, Chen CA, Zweier JL, Cardounel AJ. Regulation of eNOS-derived superoxide by endogenous methylarginines. Biochemistry. 2008;47:7256–7263. doi: 10.1021/bi702377a. [DOI] [PubMed] [Google Scholar]
- 13.Harrison DG. Cellular and molecular mechanisms of endothelial cell dysfunction. The Journal of clinical investigation. 1997;100:2153–2157. doi: 10.1172/JCI119751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CA, Wang TY, Varadharaj S, Reyes LA, Hemann C, Talukder MA, Chen YR, Druhan LJ, Zweier JL. S-glutathionylation uncouples eNOS and regulates its cellular and vascular function. Nature. 2010;468:1115–1118. doi: 10.1038/nature09599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manevich Y, Townsend DM, Hutchens S, Tew KD. Diazeniumdiolate mediated nitrosative stress alters nitric oxide homeostasis through intracellular calcium and S-glutathionylation of nitric oxide synthetase. PloS one. 2010;5:e14151. doi: 10.1371/journal.pone.0014151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knorr M, Hausding M, Kroller-Schuhmacher S, Steven S, Oelze M, Heeren T, Scholz A, Gori T, Wenzel P, Schulz E, Daiber A, Munzel T. Nitroglycerin-induced endothelial dysfunction and tolerance involve adverse phosphorylation and S-Glutathionylation of endothelial nitric oxide synthase: beneficial effects of therapy with the AT1 receptor blocker telmisartan. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:2223–2231. doi: 10.1161/ATVBAHA.111.232058. [DOI] [PubMed] [Google Scholar]
- 17.Oelze M, Knorr M, Kroller-Schon S, Kossmann S, Gottschlich A, Rummler R, Schuff A, Daub S, Doppler C, Kleinert H, Gori T, Daiber A, Munzel T. Chronic therapy with isosorbide-5-mononitrate causes endothelial dysfunction, oxidative stress, and a marked increase in vascular endothelin-1 expression. European heart journal. 2012 doi: 10.1093/eurheartj/ehs100. [DOI] [PubMed] [Google Scholar]
- 18.Okamoto T, Akaike T, Sawa T, Miyamoto Y, van der Vliet A, Maeda H. Activation of matrix metalloproteinases by peroxynitrite-induced protein S-glutathiolation via disulfide S-oxide formation. The Journal of biological chemistry. 2001;276:29596–29602. doi: 10.1074/jbc.M102417200. [DOI] [PubMed] [Google Scholar]
- 19.Reddy S, Jones AD, Cross CE, Wong PS, Van Der Vliet A. Inactivation of creatine kinase by S-glutathionylation of the active-site cysteine residue. The Biochemical journal. 2000;347(Pt 3):821–827. [PMC free article] [PubMed] [Google Scholar]
- 20.Biswas S, Chida AS, Rahman I. Redox modifications of protein-thiols: emerging roles in cell signaling. Biochem Pharmacol. 2006;71:551–564. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 21.Zweier JL, Chen CA, Druhan LJ. S-glutathionylation reshapes our understanding of endothelial nitric oxide synthase uncoupling and nitric oxide/reactive oxygen species-mediated signaling. Antioxidants & redox signaling. 2011;14:1769–1775. doi: 10.1089/ars.2011.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghezzi P. Regulation of protein function by glutathionylation. Free Radic Res. 2005;39:573–580. doi: 10.1080/10715760500072172. [DOI] [PubMed] [Google Scholar]
- 23.Fernandes AP, Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxidants & redox signaling. 2004;6:63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- 24.Shelton MD, Chock PB, Mieyal JJ. Glutaredoxin: role in reversible protein s-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxidants & redox signaling. 2005;7:348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 25.Starke DW, Chock PB, Mieyal JJ. Glutathione-thiyl radical scavenging and transferase properties of human glutaredoxin (thioltransferase). Potential role in redox signal transduction. The Journal of biological chemistry. 2003;278:14607–14613. doi: 10.1074/jbc.M210434200. [DOI] [PubMed] [Google Scholar]
- 26.Qanungo S, Starke DW, Pai HV, Mieyal JJ, Nieminen AL. Glutathione supplementation potentiates hypoxic apoptosis by S-glutathionylation of p65-NFkappaB. The Journal of biological chemistry. 2007;282:18427–18436. doi: 10.1074/jbc.M610934200. [DOI] [PubMed] [Google Scholar]
- 27.Gallogly MM, Starke DW, Leonberg AK, Ospina SM, Mieyal JJ. Kinetic and mechanistic characterization and versatile catalytic properties of mammalian glutaredoxin 2: implications for intracellular roles. Biochemistry. 2008;47:11144–11157. doi: 10.1021/bi800966v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallogly MM, Starke DW, Mieyal JJ. Mechanistic and kinetic details of catalysis of thiol-disulfide exchange by glutaredoxins and potential mechanisms of regulation. Antioxidants & redox signaling. 2009;11:1059–1081. doi: 10.1089/ars.2008.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy MP. Mitochondrial thiols in antioxidant protection and redox signaling: distinct roles for glutathionylation and other thiol modifications. Antioxidants & redox signaling. 2012;16:476–495. doi: 10.1089/ars.2011.4289. [DOI] [PubMed] [Google Scholar]
- 30.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Current opinion in pharmacology. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Chen CA, Druhan LJ, Varadharaj S, Chen YR, Zweier JL. Phosphorylation of endothelial nitric-oxide synthase regulates superoxide generation from the enzyme. The Journal of biological chemistry. 2008;283:27038–27047. doi: 10.1074/jbc.M802269200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CA, Lin CH, Druhan LJ, Wang TY, Chen YR, Zweier JL. Superoxide induces endothelial nitric-oxide synthase protein thiyl radical formation, a novel mechanism regulating eNOS function and coupling. The Journal of biological chemistry. 2011;286:29098–29107. doi: 10.1074/jbc.M111.240127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen W, Druhan LJ, Chen CA, Hemann C, Chen YR, Berka V, Tsai AL, Zweier JL. Peroxynitrite induces destruction of the tetrahydrobiopterin and heme in endothelial nitric oxide synthase: transition from reversible to irreversible enzyme inhibition. Biochemistry. 2010;49:3129–3137. doi: 10.1021/bi9016632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stuehr DJ, Cho HJ, Kwon NS, Weise MF, Nathan CF. Purification and characterization of the cytokine-induced macrophage nitric oxide synthase: an FAD- and FMN-containing flavoprotein. Proc Natl Acad Sci U S A. 1991;88:7773–7777. doi: 10.1073/pnas.88.17.7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chrestensen CA, Eckman CB, Starke DW, Mieyal JJ. Cloning, expression and characterization of human thioltransferase (glutaredoxin) in E coli. FEBS letters. 1995;374:25–28. doi: 10.1016/0014-5793(95)01066-n. [DOI] [PubMed] [Google Scholar]
- 36.Chrestensen CA, Starke DW, Mieyal JJ. Acute cadmium exposure inactivates thioltransferase (Glutaredoxin), inhibits intracellular reduction of protein-glutathionyl-mixed disulfides, and initiates apoptosis. The Journal of biological chemistry. 2000;275:26556–26565. doi: 10.1074/jbc.M004097200. [DOI] [PubMed] [Google Scholar]
- 37.Murata H, Ihara Y, Nakamura H, Yodoi J, Sumikawa K, Kondo T. Glutaredoxin exerts an antiapoptotic effect by regulating the redox state of Akt. The Journal of biological chemistry. 2003;278:50226–50233. doi: 10.1074/jbc.M310171200. [DOI] [PubMed] [Google Scholar]
- 38.Schuhmacher S, Oelze M, Bollmann F, Kleinert H, Otto C, Heeren T, Steven S, Hausding M, Knorr M, Pautz A, Reifenberg K, Schulz E, Gori T, Wenzel P, Munzel T, Daiber A. Vascular dysfunction in experimental diabetes is improved by pentaerithrityl tetranitrate but not isosorbide-5-mononitrate therapy. Diabetes. 2011;60:2608–2616. doi: 10.2337/db10-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagen.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 40.Srinivasan U, Mieyal PA, Mieyal JJ. pH profiles indicative of rate-limiting nucleophilic displacement in thioltransferase catalysis. Biochemistry. 1997;36:3199–3206. doi: 10.1021/bi962017t. [DOI] [PubMed] [Google Scholar]
- 41.Dalle-Donne I, Milzani A, Gagliano N, Colombo R, Giustarini D, Rossi R. Molecular mechanisms and potential clinical significance of S-glutathionylation. Antioxidants & redox signaling. 2008;10:445–473. doi: 10.1089/ars.2007.1716. [DOI] [PubMed] [Google Scholar]
- 42.Ambrosio G, Santoro G, Tritto I, Elia PP, Duilio C, Basso A, Scognamiglio A, Chiariello M. Effects of ischemia and reperfusion on cardiac tolerance to oxidative stress. The American journal of physiology. 1992;262:H23–30. doi: 10.1152/ajpheart.1992.262.1.H23. [DOI] [PubMed] [Google Scholar]
- 43.Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, Murphy MP. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant DEFENSE. The Journal of biological chemistry. 2004;279:47939–47951. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 44.Flohe L. The fairytale of the GSSG/GSH redox potential. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagen.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 45.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free radical biology & medicine. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 46.Ko YE, Lee IH, So HM, Kim HW, Kim YH. Mechanism of glutathione depletion during simulated ischemia-reperfusion of H9c2 cardiac myocytes. Free Radic Res. 2011;45:1074–1082. doi: 10.3109/10715762.2011.595407. [DOI] [PubMed] [Google Scholar]
- 47.Zweier JL, Flaherty JT, Weisfeldt ML. Direct measurement of free radical generation following reperfusion of ischemic myocardium. Proc Natl Acad Sci U S A. 1987;84:1404–1407. doi: 10.1073/pnas.84.5.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramires PR, Ji LL. Glutathione supplementation and training increases myocardial resistance to ischemia-reperfusion in vivo. American journal of physiology Heart and circulatory physiology. 2001;281:H679–688. doi: 10.1152/ajpheart.2001.281.2.H679. [DOI] [PubMed] [Google Scholar]