Abstract
Automated diagonal capillary electrophoresis is a two-dimensional separation method that incorporates an immobilized enzyme reactor at the distal end of the first capillary and employs identical electrophoretic separation modes in both dimensions. Components undergo a preliminary separation in the first capillary. Fractions are parked in the reactor where some components undergo transformation. The fractions are then periodically transferred to the second capillary and replaced by the next components in the sample. Components that are not modified by the reactor will have identical mobility in both dimensions and fall on the diagonal of a reconstructed two-dimensional electropherogram, while analyte that undergo modification will fall off the diagonal. In this study, alkaline phosphatase was immobilized in a monolithic reactor. An LTQ-Orbitrap Velos mass spectrometer was used to monitor analyte as they migrated from the second capillary. The system was used to characterize the phosphorylation status of a tryptic digest of α-casein in a background prepared from a 22-fold excess of the tryptic digest of bovine serum albumin. 120 fractions underwent automated treatment in the alkaline phosphatase reactor and separation in the second dimension capillary over 40-minutes; nine phosphorylated α-casein peptides that produced 20 different phosphorylation states were detected with high confidence.
Keywords: Automated phosphopeptide analysis, diagonal capillary electrophoresis, alkaline phosphatase
INTRODUCTION
Protein phosphorylation is involved in many regulatory functions, such as cell cycle control, receptor-mediated signal transduction, differentiation, proliferation, transformation, and metabolism [1–4]. Mass spectrometry is used to characterize protein phosphorylation and has been extensively reviewed [1, 5–6]. In many experiments, a tryptic digest is subjected to an enrichment step based on the negative charged phosphate group or based on affinity of the phosphate group for selected metal ions. These procedures also enrich acidic peptides, which complicates subsequent analysis.
We recently reported a method that has the potential of unambiguously identifying phosphorylated peptides [7]. This technique, which we call diagonal electrophoresis, is a form of diagonal separation. We employed an integral microreactor located at the distal end of the first capillary of a two-dimensional capillary electrophoresis system. A mixture of peptides was introduced into the first capillary and separated based on zone electrophoresis. Fractions from this capillary were parked in the microreactor. After reaction, those fractions were transferred to the second capillary, where the components were again separated by zone electrophoresis. Components that passed through the reactor without change had identical mobility in the first and second separation dimensions, and those components form a diagonal in the reconstructed electropherogram. In contrast, components that underwent reaction formed spots off the diagonal.
We immobilized alkaline phosphatase in the microreactor. This enzyme is highly active, promiscuously removes phosphate groups from a wide range of molecules, but faithfully removes only phosphate groups. Since zone electrophoresis separates components based on their size-to-charge ratio, loss of the negatively charged phosphate group results in a significant mobility shift. In our first report, we analyzed fluorescently labeled standard peptides, and employed a laser-induced fluorescence detector. In this report, we describe the use of an Orbitrap-Velos mass spectrometer for the analysis of a mixture of the tryptic digest of α-casein and bovine serum albumin.
Diagonal separation techniques are fifty years old; the ancestral experiment used paper chromatography to screen disulfide bonds in peptides [8, 9]. Components were separated along one axis of the chromatography paper, treated to reduce disulfide bonds, and then developed in the orthogonal axis. Development revealed both on-axis and off-axis spots corresponding to components that were not bound by disulfide bonds (on-axis) and had been bound by disulfide bonds (off-axis). The original diagonal paper chromatography technique has been extended to other forms of chromatography, such as diagonal liquid chromatography [10] and diagonal gel electrophoresis [11]. Diagonal liquid chromatography requires fraction collection, reagent mixing to introduce a change in the properties of selected analyte, and a large number of second dimension chromatographic separations. The large number of pipetting and mixing steps, and the large number of second dimension chromatography cycles result in a tedious and slow procedure.
MATERIALS AND METHODS
Materials and Chemicals
Acrylamide (AA), ammonium persulfate (APS), tetramethylethylenediamine (TEMED), ammonium bicarbonate (NH4HCO3), N, N’-methylenebisacrylamide (MBA), acrylic acid N-hydroxyl-sccinimide ester (NAS), 3-(trimethroxysilyl)propyl methacrylate, alkaline phosphatase from bovine intestinal mucosa, α-casein, bovine serum albumin (BSA), TPCK-treated trypsin, and IMAC-Fe3+ beads were purchased from Sigma Aldrich (St. Louis, MO, USA). Poly(ethylene glycol) (PEG, Mw 10 000 Da) was purchased from Polyscience, Inc. (Warrington, PA, USA). Acetonitrile (ACN) and formic acid (FA) was ordered from Fisher Scientific (Pittsburgh, PA, USA). Water and methanol were purchased from Honeywell Burdick & Jackson (Muskegon, MI, USA). Fused capillaries were purchased from Polymicro Technologies (Phoenix, AZ, USA). ZipTipC18 was purchased from Millipore Co. (Gilbert, AZ, USA).
Sample
A tryptic digest of α-casein was prepared as follows: 0.5 mg/mL α-casein in 100 mM NH4HCO3 (pH 8.0) was mixed with TPCK-treated trypsin in an enzyme-to-protein mass ratio of 1:30 for digestion at 37 °C overnight. The digests were acidified and desalted with a ZipTipC18 and lyophilized, followed by diagonal CE-ESI-MS/MS analysis. The BSA digest was prepared as previously reported [12]. 40 µg α-casein digest was added into 1 mg BSA digest. The peptide mixture was desalted with reverse-phase C18 SepPak cartridges from Waters (Milford, MA, USA), lyophilized and IMAC enriched, followed by analysis using diagonal capillary electrophoresis.
IMAC Enrichment
Phosphopeptides were enriched from the mixture of BSA and α-casein digests with IMAC as previously reported [13]. Briefly, 150 µL IMAC beads were washed with 1 mL of IMAC binding buffer (40% ACN, 25 mM FA in H2O) for three times, and then incubated with the tryptic digests for 60 min at room temperature with vigorous shaking. After 60-min incubation, IMAC beads were spun down and washed with 1 mL of IMAC binding buffer for three times. Phosphopeptides were then eluted from the IMAC beads for three times, and then by incubation with 40 µL of IMAC elution buffer (50 mM K2HPO4 pH 10) for 5 min at room temperature. The eluate was lyophilized and desalted for diagonal CE analysis.
Preparation of the immobilized alkaline phosphatase microreactor
A fused silica capillary (48 µm i.d./148 µm o.d.) was flushed with methanol (20 min), N2 gas (10 min), 1 M NaOH (20 min), H2O (10 min), 1 M HCl (20 min), H2O (10 min), methanol (20 min), and N2 gas (20 min). A portion of the treated capillary (~10 cm) was filled with a mixed solution of methanol and 3-(trimethroxysilyl) propyl methacrylate (1:1 (v/v)) by capillary action, and the reaction proceeded at room temperature for 24 h. The capillary was then washed with methanol to remove the unreacted components, and dried again with nitrogen. The treated capillary was stored at room temperature before use.
The general protocol of reference [14] was used for monolith polymerization and enzyme immobilization, with the following modifications. A mixture of 7.5 mg of AA, 11.25 mg of MBA, and 30 mg of PEG was dissolved in 1 mL of 0.2 M sodium bicarbonate/0.5 M sodium chloride (pH ~8.0) buffer. The mixture was vortexed for 30 s and then heated at ~50 °C for 15 min to completely dissolve the monomers. Then, 2 µL of 20% (v/v) TEMED was added into 0.5 mL of the prepared mixture, and the mixture was degassed for 15 min using nitrogen. Subsequently, 7 µL of NAS (140 mg/mL, dissolved in DMSO) was added on top of the solution, and degassed for 1 min. After that, 2 µL of 20% (w/v) APS was added into the middle part of the solution to initiate polymerization. After vortexing for several seconds, a 17.5 µL-aliquot of the solution was quickly mixed with 2.5 µL of alkaline phosphatase from bovine intestinal mucosa (13 mg/mL in a buffer containing Mg2+ and Zn2+). The activated portion of the treated capillary (~8 cm in length) was filled with the mixed solution, and reacted for 1 h at room temperature with the capillary ends sealed. After reaction, the capillary was successively washed with deionized water, glycine (10 mg/mL in phosphate buffer, pH 8.0), and 5 mM NH4HCO3 (pH ~8.0) for 30 min. The capillary was stored at 4 °C before use. The capillary was trimmed to the desired length before use. Typically, the overall capillary length was 28 cm, and the reactor occupied 0.5 cm at the distal end.
Diagonal CE-ESI-MS/MS Analys
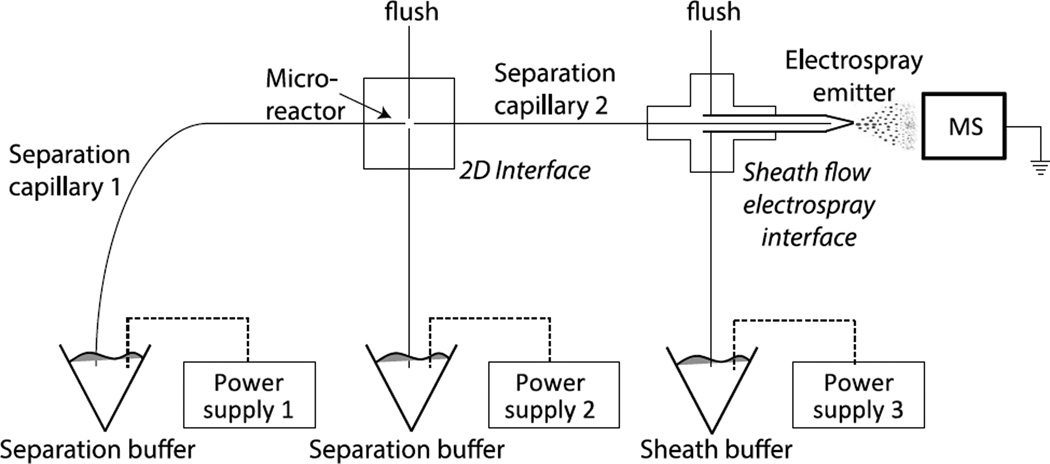
The diagonal capillary electrophoresis system was coupled to an LTQ-Orbitrap Velos mass spectrometer with an electrokinetically driven sheath-flow electrospray interface [15–17], Figure 1. Two capillaries (48 µm i.d./148 µm o.d., 23.0 cm length) were connected through a nicked-gap interface [18]. The capillary spacing was adjusted to ~50-µm. The interface was filled with the separation buffer. High voltage power supplies were connected to the injection and interface buffer vials and to the electrospray emitter.
Figure 1.
Schematic diagram of the diagonal capillary electrophoresis-ESI-MS/MS system. A ~3-mm long monolithic microreactor was synthesized at the distal end of capillary 1.
Sample was injected by applying 24 kV to power supply 1, 12 kV to power supply 2, and 0 kV to power supply 3 for five seconds. After injection, the sample was replaced with running buffer, and the two-dimensional separation was performed by applying a potential of 19 kV to power supply 1,10.25 kV to power supply 2, and 1.5 kV to power supply 3. The mass spectrometer was held at ground potential. An initial separation was performed with these potentials for 130 s. A series of fraction transfers and second dimension separations was then started. Each cycle lasted for 33 seconds. The three-second duration transfers were driven by the same potentials as were used in the preliminary separation. The 30-s duration second-dimension separations were performed by applying a potential of 10.25 kV to power supplies 1 and 2, which resulted in a zero volt potential across capillary 1, and 1.5 kV to power supply 3.
An LTQ-Orbitrap Velos was used for detection. The ion transfer tube temperature was held at 300 °C. The mass spectrometer was programmed in data dependent mode. Full MS scans were acquired in the Orbitrap mass analyzer over m/z 350–1800 range with resolution 30,000 (m/z 400). The ten most intense peaks with charge state ≥ 2 were fragmented in the higher energy collisional dissociation (HCD) cell with normalized collision energy of 40%, isolation width 2 m/z and one microscan. The fragment ions were analyzed by the Orbitrap with resolution 7, 500. The maximum injection time for MS and MS/MS was 500 ms and 250 ms, respectively. Peaks selected for fragmentation more than once within 25 s were excluded from selection for 25 s.
Database searching of raw MS/MS files was performed in Proteome Discoverer 1.3 with MASCOT 2.2.4 against ipi.bovin.v3.68.fasta database. The database searching parameters included up to two missed cleavages allowed for tryptic digestion, precursor mass tolerance 10 ppm, fragment mass tolerance 0.05 Da. Cysteine carbamidomethylation was set as a fixed modification. Phosphorylation of serine, threonine, and tyrosine and oxidation of methionine were set as variable modifications. Database searching against the corresponding decoy database was also performed to evaluate the false discovery rate of peptide identification. Peptide confidence value as high was used to filter the peptide identification, and protein grouping was enabled. mzXML files were ported to Matlab for analysis. Selected ion electropherograms were treated with a lowess filter with one robust iteration, rastered to create a reconstructed electropherogram, and shifted to longer second-dimension separation times. The data were convoluted with a two-dimensional Gaussian with standard deviation equal to one in both dimensions. Finally, the data were normalized to unit height and summed.
RESULTS AND DISCUSSION
Diagonal capillary electrophoresis of α-casein phosphopeptides
The α-casein digest was mixed with the BSA digest at a molar ratio of 1:22. The peptide mixture was treated with IMAC-Fe3+ beads to mimic a real-world analysis, followed by diagonal CE-ESI-MS/MS analysis. Nine phosphorylated α-casein peptides with 20 different phosphorylation states were confidently identified by MS/MS (Table I). Phosphopeptide 1 and 2 share common sequence, as do phosphopeptides 3–5, and result from one or two missed cleavages.
Table I.
Phosphopeptides identified from α-Casein
| Phosphopeptide | Sequence | m/z | comments | Peptide number in figures 2–4, and 6. |
|---|---|---|---|---|
| 1* | KYKVPQLEIVPNSAEER | 1000.54480 | dephosphorylated | 1 |
| 2 | YKVPQLEIVPNSAEER | 936.49774 | dephosphorylated | 2 |
| YKVPQLEIVPNSAEER | 976.48145 | monophosphorylated | 2’ | |
| 3* | KTVDMESTEVFTK | 757.87488 | dephosphorylated | 3 |
| 4 | VPQLEIVPNSAEER | 790.91949 | dephosphorylated | 4 |
| VPQLEIVPNSAEER | 830.90198 | monophosphorylated | 4’ | |
| 5 | TVDMESTEVFTK | 693.82709 | dephosphorylated | 5 |
| TVDMESTEVFTK | 733.81036 | monophosphorylated | 5’ | |
| 6 | EQLSTSEENSKK | 690.33685 | dephosphorylated | 6 |
| EQLSTSEENSKK | 730.32050 | monophosphorylated | 6’ | |
| EQLSTSEENSKK | 770.30389 | diphosphorylated | 6’’ | |
| 7 | EKVNELSKDIGSESTEDQAMEDIK | 899.09314 | dephosphorylated | 7 |
| EKVNELSKDIGSESTEDQAMEDIK | 925.74847 | monophosphorylated | 7’ | |
| EKVNELSKDIGSESTEDQAMEDIK | 952.40387 | diphosphorylated | 7’’ | |
| EKVNELSKDIGSESTEDQAMEDIK | 979.06116 | triphosphorylated | 7’’’ | |
| 8** | VNELSKDIGSESTEDQAMEDIK | 813.38031 | dephosphorylated | 8 |
| VNELSKDIGSESTEDQAMEDIK | 840.03540 | monophosphorylated | 8’ | |
| VNELSKDIGSESTEDQAMEDIK | 866.69165 | diphosphorylated | 8’’ | |
| 9 | DIGSESTEDQAMEDIK | 884.38489 | dephosphorylated | 9 |
| DIGSESTEDQAMEDIK | 924.36877 | monophosphorylated | 9’ |
These peptides were completely dephosphorylated in the microreactor. Their mono-phosphate forms were not detected in MS/MS spectra
The tri-phosphate form of this peptide was not detected in the MS/MS spectra.
The reactor efficiency is related to the length of the reactor, the concentration of immobilized enzyme, temperature, and buffer composition. A relatively low activity reactor was used in this experiment. As a result, both the phosphorylated and dephosphorylated forms were observed for many peptides. Peptides containing more than one phosphate groups generated intermediates corresponding to the loss of one or more phosphates. Several digestion products (phosphopeptides 1, 3, and 8) generated tandem spectra from the dephosphorylated forms but not the fully phosphorylated form of the peptide.
Diagonal capillary electrophoresis of a set peptides generated from a single parent phosphopeptide
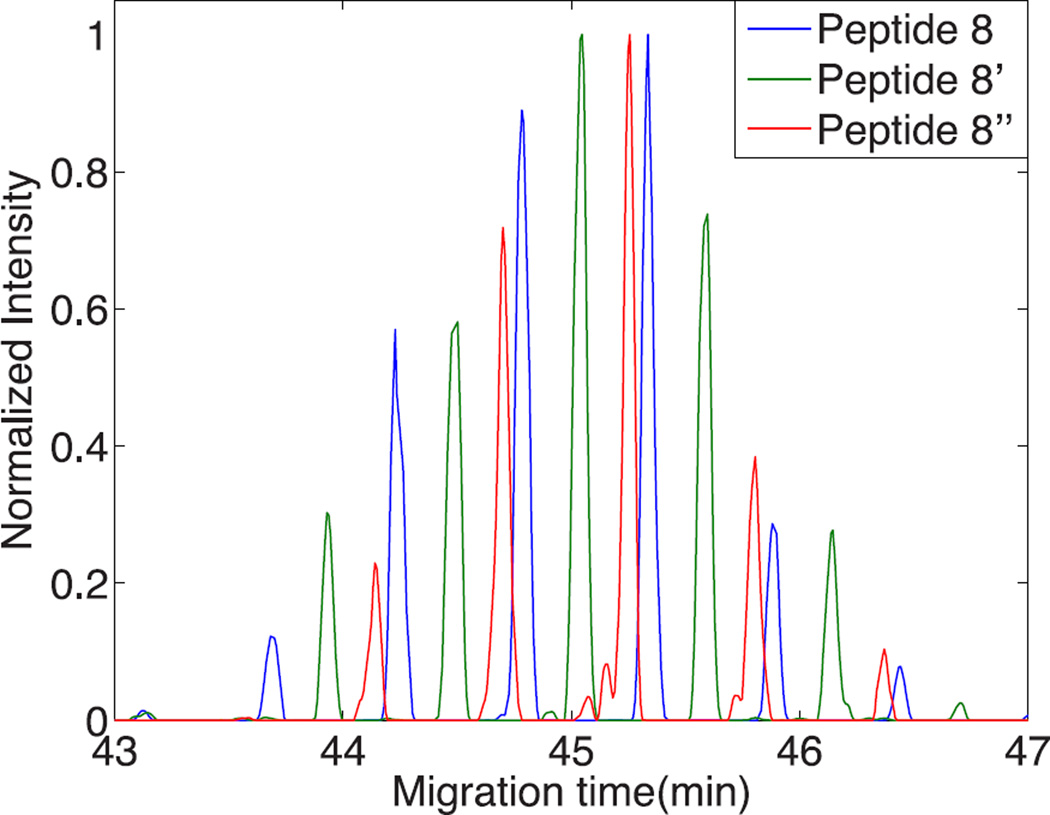
Figure 2 presents a close-up of the selected ion electropherograms generated from the reaction products produced from the tri-phosphate form of a peptide (VNELSKDIGSESTEDQAMEDIK) in the α-casein digest. The parent tri-phosphate form of the peptide did not survive passage through the reactor and was not detected by tandem mass spectrometry. It was instead converted to the di-phosphate (peptide 8’’, red trace), mono-phosphate (peptide 8’, green trace), and the dephosphorylated (peptide 8, blue trace) forms. The time trace consists of a set of 5 or 6 peaks for each peptide, and those peaks are separated by 33 seconds, which is the second dimension cycle time. The presence of five or six peaks from each peptide result from successive transfers of products from the microreactor to the second capillary. Each transfer requires three seconds. Based on the width of the envelop, the parent tri-phosphate form of the peptide had a ~6–9 second peak width in the first capillary, which is dominated by the original injection volume.
Figure 2.
Time trace of the two-dimensional capillary electrophoresis analysis of peptides 8–8’’ in table 1 (VNELSKDIGSESTEDQAMEDIK and its mono- and di-phosphorylated forms).
The dephosphorylated peptide (peptide 8, m/z = 813.380) migrated first, followed ~14 seconds later by the mono-phosphorylated peptide (peptide 8’, m/z = 840.035), and followed ~14 seconds later by the unmodified di-phosphorylated peptide (peptide 8’’, m/z = 866.692). The separation was performed in an uncoated capillary at basic pH. At this pH, the electro-osmotic mobility was higher than the electrophoretic mobility of the phosphorylated peptides, which carry additional negative charges and migrate more slowly than unphosphorylated peptides.
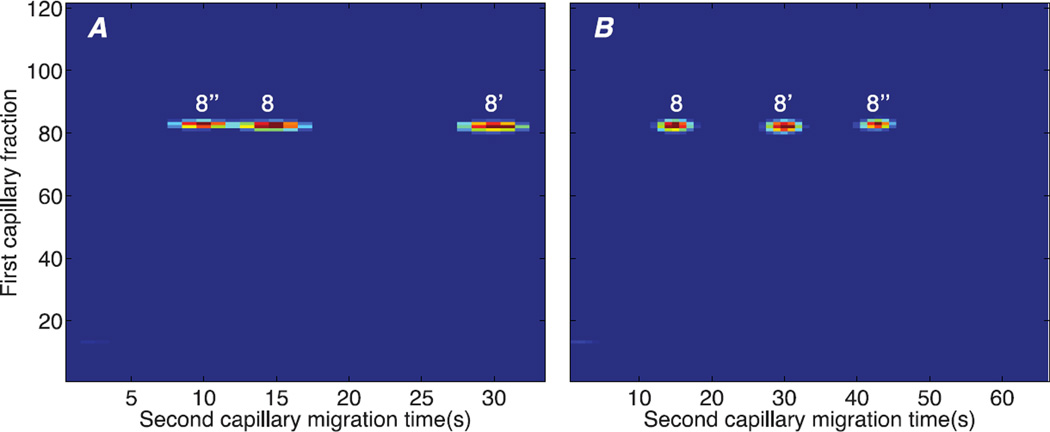
The data can be transformed into an image of the two-dimensional separation. This image is generated by rastering 33-s long slices from the data across the image, Figure 3a. The image consists of three horizontal spots. Peptide 8’’ generated a spot centered in the 83rd transfer cycle, while spots 8 and 8’ are centered in the 82nd transfer cycle. This peculiar ordering of the spots arises because the start of each transfer cycle is not synchronized with the arrival of the fastest moving component. This artifact can be addressed by pasting a second copy of the image to the right, and recording the position of the spots in the order in which they reached the detector, figure 3b.
Figure 3.
diagonal capillary electrophoresis of a di-phosphorylated (peptide 8’’, VNELSKDIGSESTEDQAMEDIK, Table 1), mono-phosphorylated (peptide 8’), and unphosphorylated (peptide 8) peptide. A – wrapped electropherogram where component 8’’ appears at a later first capillary time but earlier second capillary time than components 8 and 8’. B – unwrapped electropherogram where spot 8’’ is shifted one cycle (33 seconds) later in the second dimension by pasting its image to the right of the original data. The spots now appear in the correct temporal order in the second dimension, and the second-dimension spacing between the spots is uniform.
After unwrapping, the behavior of the diagonal capillary electrophoresis becomes clear. The parent form of the peptide arrived at the reactor at roughly the 75th fraction. It underwent partial dephosphorylation, generating the di-phosphate (peptide 8’’), the mono-phosphate (peptide 8’), and the unphosphorylated (peptide 8) forms. These peptides were separated in capillary 2, and produce a horizontal line because their arrival time in the first dimension is determined only by the parent peptide’s mobility. The dephosphorylated peptide has the shortest migration time and highest effective mobility, which is expected for this separation, which is performed under positive polarity and in an uncoated capillary that generates significant electro-osmotic flow.
Diagonal capillary electrophoresis of the tryptic digest of α-casein
Two terms need to be defined to describe the separation. The first is the separation window, which is the difference in migration time between the fastest and slowest migrating peptides during the capillary electrophoretic separation. The second term is the cycle time, which is the period between second dimension separations.
In this experiment, the α-casein peptides migrated in a separation window corresponding to the transfer of 120 fractions from capillary 1 to capillary 2. Each fraction was transferred by applying potential across capillary 1 for three seconds, which corresponds to 360 seconds of cumulative migration time in capillary 1. Since the same capillary length and voltage is used for capillary 2 separation, the set of peptides will also migrate over a 360 second separation window in capillary 2.
If we had performed an analysis that employed 120 separations in capillary 2, each with a 360 s cycle time, then the analysis would have required 12 hours of instrument time, which is unacceptably long. Instead, we employed a 33-second cycle time to reduce the overall analysis time to a much more manageable 40 minutes. However, the short cycle time results in the wrapping of the reconstructed diagonal capillary electrophoresis image and a superficially jumbled image. A simple example of this skewing was shown in figure 3, where the migration of the components was not in-phase with the transfer frequency.
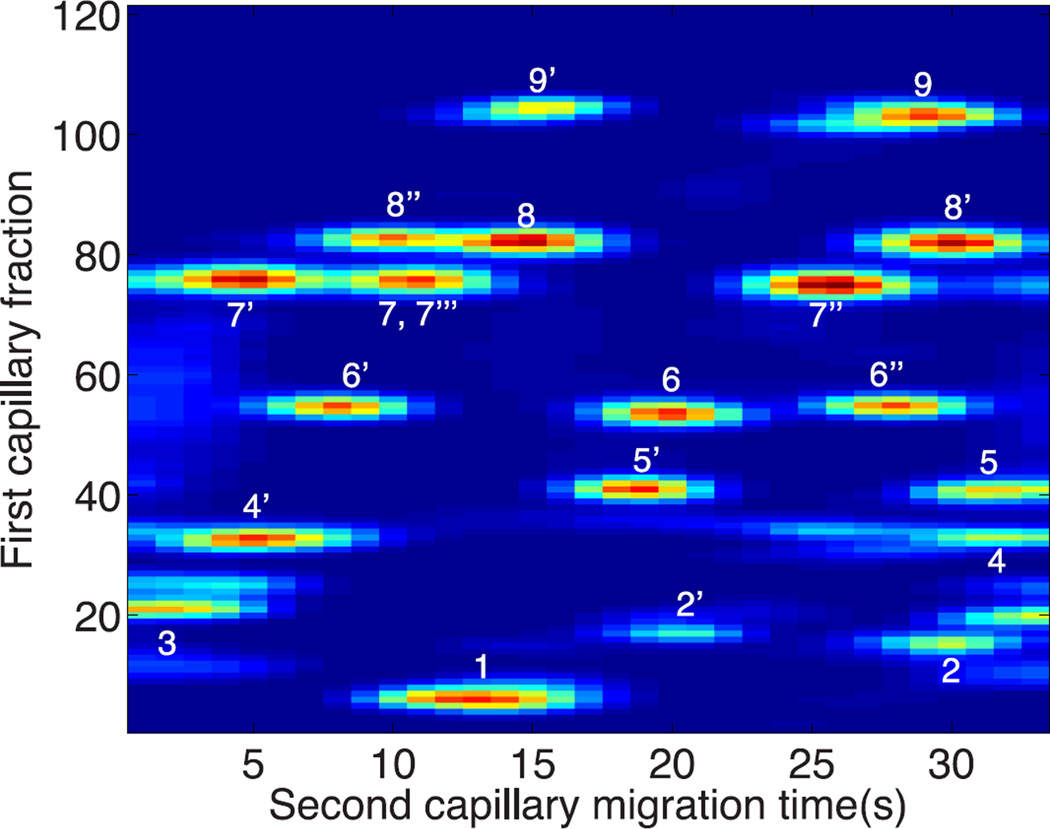
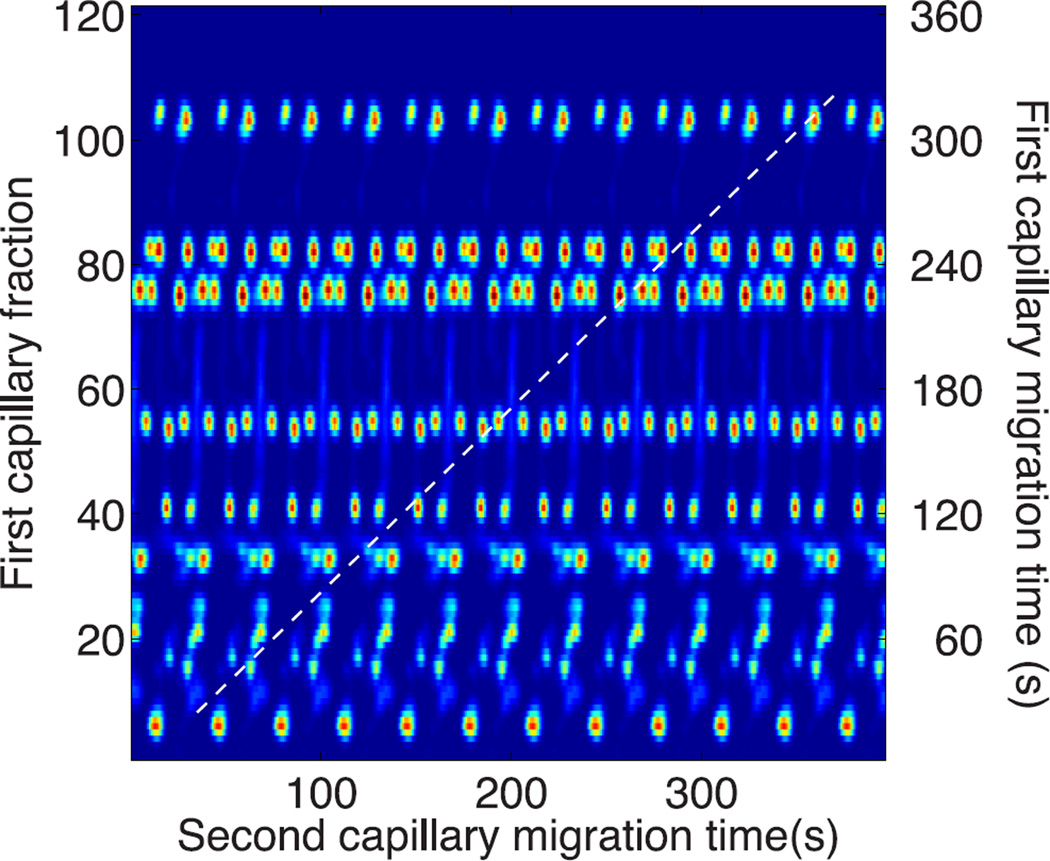
The complex mixture generated by the α-casein peptides results in a complex jumbling of peaks, figure 4, due to the short cycle time wrapping the data from successive fractions.
Figure 4.
Diagonal capillary electrophoresis of α-casein tryptic digest. Peptide numbers are listed in Table 1. This data has not been unwrapped to correct for the short cycle time used in the experiment. Spot numbering is described in Table 1.
The data can be unwrapped by pasting shifted copies of the reconstructed electropherogram, figure 5, and then drawing a diagonal at 45°, which connects the spots with identical mobility in the two dimensions.
Figure 5.
Twelve replicated copies of the electropherogram from figure 4. The dashed line is at 45° and connects components with identical migration time in the two dimensions.
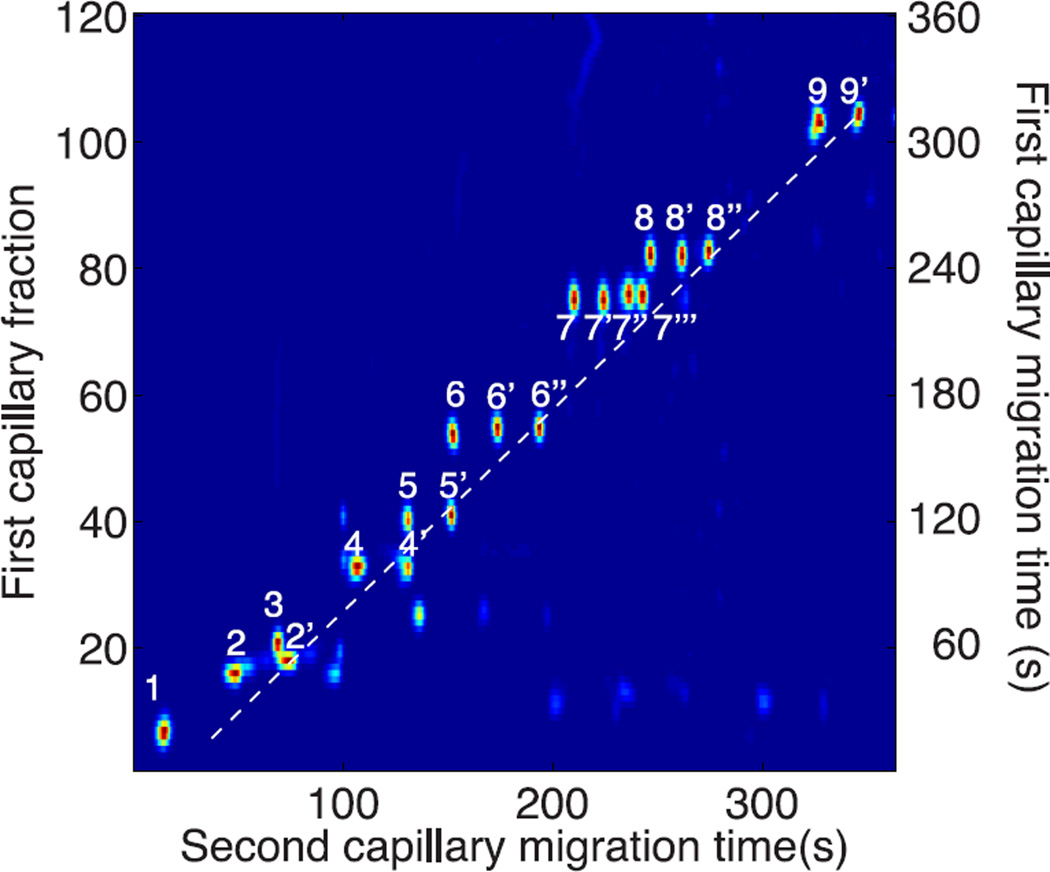
Finally, the spots corresponding to the unmodified components, which fall on the diagonal, and their dephosphorylated forms, which fall at immediately shorter times, are retained to generate the unwrapped diagonal capillary electropherogram, figure 6.
Figure 6.
Unwrapped diagonal capillary electrophoresis of a set of 20 peptides generated from the tryptic digest of α-casein. Peptide identities are listed in Table I. The dashed line has a slope of 1 and represents the diagonal on which peptides fall if they pass untransformed through the reactor. Spot identification is in Table 1.
Components 2’, 4’, 5’, 6’’, 7’’’, and 9’ were the fully phosphorylated peptides that passed through the reactor without loss of a phosphate group. Since these peptides have identical structure in the two capillaries, they have identical migration time in the two dimensions, defining the diagonal with unit slope.
The remaining peptides migrate more quickly in the second dimension capillary than their fully phosphorylated parent. The loss of a phosphate group results in an increase in net mobility under the positive polarity employed in this separation.
Peptides 1 and 3 are completely dephosphorylated; their corresponding phosphorylated forms were not observed. Instead, those fully phosphorylated forms were present in the original digest but then completely dephosphorylated in the reactor. The off-diagonal migration of these two peptides confirms that they had been phosphorylated in the original sample. As noted earlier, components 8–8’’ derived from a tri-phosphorylated peptide in the original sample. That parent peptide lost one or more phosphate groups and was not detected in the tandem spectra.
None of these dephosphorylated peptides were present in the original tryptic digest; if they had been present, they would have generated components on the diagonal at earlier first capillary fractions.
Highlights.
We coupled diagonal capillary electrophoresis to an Orbitrap mass spectrometer
We employed immobilized alkaline phosphatase for rapid and efficient removal of phosphate groups.
We developed an algorithm for the analysis of the resulting two-dimensional electropherograms.
ACKNOWLEDGEMENT
We thank Dr. William Boggess in the Notre Dame Mass Spectrometry and Proteomics Facility for his help with this project. This work was supported by grant R01GM096767 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Macek B, Mann M, Olsen JV. Annu. Rev. Pharmacol. Toxicol. 2009;49:199–211. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P. Nat. Cell Biol. 2002;4:127–130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 3.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 4.Wu R, Dephoure N, Haas W, Huttlin EL, Zhai B, Sowa ME, Gygi SP. Molecular & Cellular Proteomics. 2011;10 doi: 10.1074/mcp.M111.009654. M111.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thingholm TE, Jensen ON, Larsen MR. Proteomics. 2009;9:1451–1468. doi: 10.1002/pmic.200800454. [DOI] [PubMed] [Google Scholar]
- 6.Grimsrud PA, Swaney DL, Wenger CD, Beauchene NA, Coon JJ. Acs. Chem. Biol. 2010;5:105–119. doi: 10.1021/cb900277e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wojcik R, Vannatta M, Dovichi NJ. Anal. Chem. 2010;82:1564–1567. doi: 10.1021/ac100029u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JR, Hartley BS. Biochem. J. 1963;89:59–60. [Google Scholar]
- 9.Brown JR, Hartley BS. Biochem. J. 1966;101:214–228. doi: 10.1042/bj1010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gevaert K, Van Damme J, Goethals M, Thomas GR, Hoorelbeke B, Demol H, Martens L, Puype M, Staes A, Vandekerckhove J. Molecular & Cellular Proteomics. 2002;1:896–903. doi: 10.1074/mcp.m200061-mcp200. [DOI] [PubMed] [Google Scholar]
- 11.Chernov IP, Timchenko KA, Akopov SB, Nikolaev LG, Sverdlov ED. Anal. Biochem. 2007;364:60–66. doi: 10.1016/j.ab.2007.01.040. [DOI] [PubMed] [Google Scholar]
- 12.Sun L, Li Y, Yang P, Zhu G, Dovichi NJ. Journal of Chromatography. A. 2012;1220:68–74. doi: 10.1016/j.chroma.2011.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villen J, Gygi SP. Nat. Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palm AK, Novotny MV. Rapid Commun. Mass Spectrom. 2004;18:1374–1382. doi: 10.1002/rcm.1500. [DOI] [PubMed] [Google Scholar]
- 15.Wojcik R, Dada OO, Sadilek M, Dovichi NJ. Rapid Commun. Mass Spectrom. 2010;24:2554–2560. doi: 10.1002/rcm.4672. [DOI] [PubMed] [Google Scholar]
- 16.Sun L, Zhu G, Li Y, Wojcik R, Yang P, Dovichi NJ. Proteomics. 2012;12:3013–3019. doi: 10.1002/pmic.201200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Champion MM, Sun L, Champion PAD, Wojcik R, Dovichi NJ. Anal. Chem. 2012;84:1617–1622. doi: 10.1021/ac202899p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flaherty RJ, Huge BJ, Bruce SM, Dada OO, Dovichi NJ. Analyst. 2013;138:3621–3625. doi: 10.1039/c3an00284e. [DOI] [PMC free article] [PubMed] [Google Scholar]