Abstract
Selective capture of cells from bodily fluids in microchannels has broadly transformed medicine enabling circulating tumor cell isolation, rapid CD4+ cell counting for HIV monitoring, and diagnosis of infectious diseases. Although cell capture methods have been demonstrated in microfluidic systems, the release of captured cells remains a significant challenge. Viable retrieval of captured label-free cells in microchannels will enable a new era in biological sciences by allowing cultivation and post-processing. The significant challenge in release comes from the fact that the cells adhere strongly to the microchannel surface, especially when immuno-based immobilization methods are used. Even though fluid shear and enzymes have been used to detach captured cells in microchannels, these methods are known to harm cells and affect cellular characteristics. This paper describes a new technology to release the selectively captured label-free cells in microchannels without the use of fluid shear or enzymes. We have successfully released the captured CD4+ cells (3.6% of the mononuclear blood cells) from blood in microfluidic channels with high specificity (89% ± 8%), viability (94% ± 4%), and release efficiency (59% ± 4%). We have further validated our system by specifically capturing and controllably releasing the CD34+ stem cells from whole blood, which were quantified to be 19 cells per million blood cells in the blood samples used in this study. Our results also indicated that both CD4+ and CD34+ cells released from the microchannels were healthy and amenable for in vitro culture. Manual flow based microfluidic method utilizes inexpensive, easy to fabricate microchannels allowing selective label-free cell capture and release in less than 10 minutes, which can also be used at the point-of-care. The presented technology can be used to isolate and purify a broad spectrum of cells from mixed populations offering widespread applications in applied biological sciences, such as tissue engineering, regenerative medicine, rare cell and stem cell isolation, proteomic/genomic research, and clonal/population analyses.
Introduction
Capture, isolation and purification of specific cells from heterogeneous populations has enabled advancements in a broad variety of scientific fields including cell based diagnostics in microfluidic systems,1-5,45 cell specific genomic/proteomic analysis,6-8,46 clonal and population studies,9,10 stem cell purification for regenerative therapies,11,12 and circulating tumor cell capture for cancer research.6,13 For instance, isolation of CD4+ cells from blood has been widely used for HIV monitoring,1,2,14,15,47 for biological studies and pharmaceutical research.16-18 On the other hand, isolation of stem cells (e.g., CD34+ endothelial progenitor cells) from peripheral blood, cord blood, and bone marrow has found applications in regenerative medicine and tissue engineering.19-22 Fluorescence-activated and magnetic-activated cell sorting are commonly used methods for cell isolation, which require extensive preliminary sample processing and tagging of the cells with fluorophores or magnetic particles conjugated with antibodies. While these methods are powerful and sort cells from heterogeneous mixtures, the cost, complexity and the requirements for infrastructure (e.g., facility, technical personnel, and reagents) limit their use at the point-of-care (POC).1,16 There have been attempts to miniaturize these systems to simpler microfluidic platforms.23,24 However, the peripheral equipment required for detection and sorting using microfluidics remain bulky and costly. On the other hand, microfluidic systems have traditionally required thousands of cells for post-capture genomic and proteomic analysis (e.g., circulating tumor cell and neutrophil analysis).6,7,25 Genomic studies on captured cells in microfluidic systems are mainly hampered by the loss of genomic material in the channels due to the large surface to volume ratio. Retrieval and downstream processing of captured live cells can eliminate this limitation. Moreover, the release of selectively captured live cells is significantly needed for extensive post-processing capabilities and cultivation, and for clonal and population studies within the isolated cells.
Effectively releasing the selectively immuno-captured cells in microfluidic channels without compromising the viability and phenotypic characteristics of the cells remains a challenge.26-28 Chemical and physical means for cell detachment, such as enzymatic and fluid shear based methods are known to adversely affect cell viability and function.29,30 To minimize these effects, captured cells need to be released and recovered without using harsh chemical or physical mechanisms. Alginate based hydrogels have recently been used in combination with conjugated antibodies to isolate endothelial progenitor cells from blood in microchannels, in which the cells were released by chemically chelating the hydrogel that coats the channels.31 However, alginate based hydrogels allow a high level of non-specific binding28,32 and offer a limited number of sites for conjugating antibodies,31 which significantly reduce the capture specificity and efficiency. Furthermore, coating and chemical chelation of hydrogels in microfluidic channels add extra steps, lengthy processing time (e.g., 2+ days for gel preparation and channel coating), operational complexities (e.g. use of microfluidic pumps), and additional chemical reagents that render this approach costly and challenging to apply especially at the POC including bedside or the primary healthcare delivery settings.
Temperature responsive polymer (poly(N-isopropylacrylamide), PNIPAAm) interacts strongly with proteins (e.g., insulin chain A, serum albumin) above the lower critical solution temperature (LCST).33,34 Whereas, when the temperature of the polymer is reduced below the LCST, a complete desorption of the adsorbed proteins is possible.33,34 Even though responsive polymers have been used to release attached cells from surfaces without the use of fluid shear or enzymes,35-37 these studies relied on non-specific attachment of adherent cells (e.g., fibroblasts). There are three unaddressed challenges for temperature responsive polymers to be applicable in selective cell isolation. The first challenge is that the cells need to be selectively captured on the polymer, which requires immobilization of specific antibodies on the polymer. The second challenge is merging these polymers with microfluidic systems to process large volumes of heterogonous cell suspensions, such as bodily fluids and whole blood. The third challenge is releasing the captured cells from microfluidic channels. The challenge in release emerges from the fact that the cells tend to adhere strongly to the surface when they are immobilized via antibody–antigen interactions. Enzymatic and fluid shear based methods prove to be inefficient in detaching cells when immuno-based immobilization is used. Since the challenges listed have not been addressed so far, thermoresponsive microfluidic technology has not been available for selective cell isolation, purification, and diagnostics/monitoring applications with potential impact on clinical practice and outcome. Here, we introduce a new approach to rapidly release the selectively captured cells label-free in microchannels with high specificity and post-release viability by using simple manual pipettors and short processing times.
Materials and methods
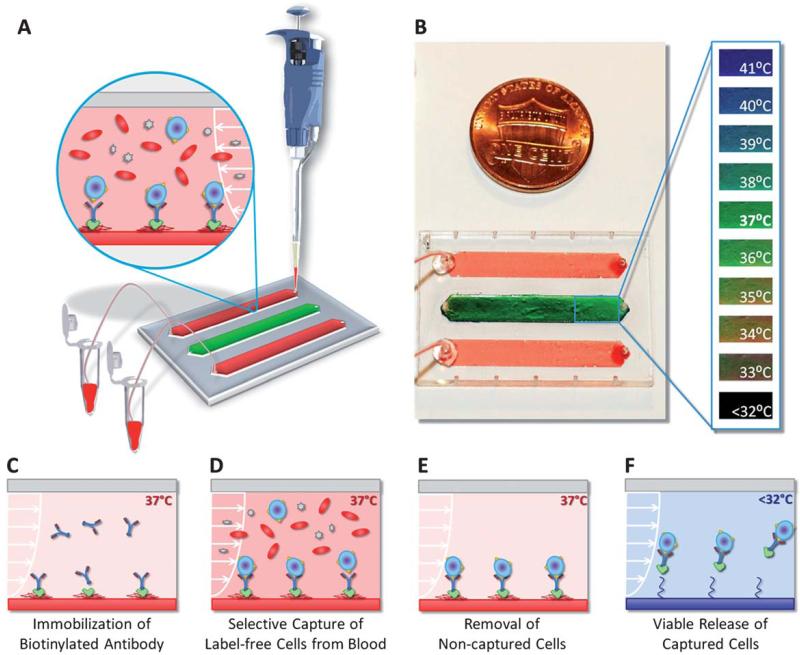
We developed a biotin binding protein (Neutravidin) and biotinylated antibody based surface chemistry on PNIPAAm microfluidic channels (Fig. 1). In this approach: first, Neutravidin followed by biotinylated antibody (human Anti-CD4 or human Anti-CD34) were immobilized in microfluidic channels at 37 °C (Fig. 1C). Next, the heterogeneous cell suspension (i.e., blood) was flown through the channels without any preliminary processing (Fig. 1D). During this step, the CD4+ cells or the CD34+ cells were captured on the channel surface with antibody–antigen interaction as we have shown before.38 Then, the non-captured cells in the channels were rinsed away with buffer solution at 37 °C (Fig. 1E). Rapid heating (37 °C) and cooling (<32 °C) of the microchannels was achieved by the small liquid volume (10 μL) in the channels. The Neutravidin–antibody complex together with the captured cells was then released from the surface by a reduction in temperature (Fig. 1F). The temperature of the microfluidic channels was controlled with a heating pad and monitored using a liquid crystal dye in a channel, working in the 32 °C to 40 °C range (sensitivity: 1 °C) (Fig. 1B). The temperature level and distribution in the thermoresponsive release channels were observed to be uniform throughout the experiments.
Fig. 1.
Thermoresponsive microfluidic chip developed for releasing selectively captured cells from blood. (A) The microfluidic chip was composed of three parallel channels (4 mm × 22 mm × 80 μm), one of which (middle channel) was used as the temperature indicator channel. Blood was introduced into the top and bottom release channels with a manual pipette. The tubing connected to the outlet ports allowed the collection of the released cells in microcentrifuge tubes. (B) The middle channel was coated with temperature sensitive liquid crystal dye, which was responsive between 35 °C (red-orange) and 40 °C (blue-purple). At the target temperature of 37 °C, the middle channel displayed green color. (C) Schematic drawing of the working principle of label-free selective capture from whole blood and controlled release of cells in thermoresponsive microfluidic channels. Biotin binding protein (Neutravidin) and biotinylated antibody (Anti-CD4 or Anti-CD34) were immobilized on the PNIPAAm channel surface at 37 °C. (D) Pre-warmed blood sample (at 37 °C) was injected into the microfluidic channel, and the CD4+ cells or the CD34+ cells in blood were captured on the channel surface. (E) The non-captured cells in the channels were rinsed off and the red blood cells were lysed. (F) The microchip was then cooled down below 32 °C (in less than 5 minutes). The released cells were rinsed out of the channels and collected at the channel outlet.
Quantification of CD4+ and CD34+ cells in blood samples
The CD4+ and CD34+ cells in the blood samples were quantified by using the standard flow cytometry methods.15 For flow cytometry analysis, 100 μL of buffy coat or whole blood was lysed with 1× fluorescence activated cell sorting (FACS) lysing solution diluted with deionized water (MilliQ Academic, Billerica, MA) from 10× BD FACS Lysing Solution (BD Biosciences, San Jose, CA). The sample was vortexed gently and incubated for 10 minutes at room temperature. Next, the sample was centrifuged at 2000 rpm for 5 minutes which was followed by discarding the supernatant. Cell debris were washed away with 2.5 mL of phosophate buffered saline (PBS), followed by a second centrifugation. Next, cells were suspended in 1 mL of PBS. 1938 μL of FACSFlow Sheath Fluid (BD Biosciences, Franklin Lakes, NJ) and 10 mL of cell solution prepared as a result of lysis step was mixed in a new FACS tube and eventually 12 μL of Alexa Fluor 488 anti-human CD34 antibody (Biolegend, San Diego, CA) or anti-human CD4 Alexa Fluor® 488 (eBioscience Inc., San Diego, CA) was added before 30 minutes of dark incubation at room temperature. With the addition of 50 μL of counting beads (BD Biosciences, San Jose, CA), cell solution was prepared for flow cytometry analysis, during which calibration beads (BD Biosciences, San Jose, CA) were used. The software used during FACS measurements was CellQuestPro (BD Biosciences). The results of these analyses are reported in Table 1.
Table 1.
Typical CD4+ and CD34+ cell counts in the blood samples used in this study quantified by flow cytometrya
| Cell count per microlitre (flow cytometry) |
Percentage of mononuclear blood cells |
Per million (106) blood cells (WBCs + RBCs) |
|
|---|---|---|---|
| CD4+ cells | 931 | 3.6% | 195 |
| CD34+ cells | 92 | 0.5% | 19 |
(Cytometry): the number of white blood cells (WBCs) in adults: 3.3 – 8.7 × 103 μL−1 (average: 6 × 103 μL−1); the number of red blood cells (RBCs) in adults: 3.93 – 5.6 × 106 μL−1 (average: 4.7 × 106 μL−1). (Source for the number of blood cells in adults: National Institutes of Health Clinical Center.)
Design and fabrication of microfluidic channels
We have optimized our microfluidic channel design to accommodate manual pipetting (Fig. 1A) and to result in optimum flow rates and shear stresses in the microchannels for the capture of cells from blood. Briefly, in each microfluidic chip, three channels (dimensions: 25 mm × 4 mm × 80 μm) were employed with either PNIPAAm layered polystyrene bottom surface (release channels) or polystyrene (control channels) bottom surface (Fig. S1 and S2†). To minimize the peripheral equipment needed to operate the microfluidic chips, we eliminated the need for syringes and pumps. We performed 40 manual pipetting experiments (0.5–10 μL manual pipettors with 0.1–10 μL pipette tips, VWR International, Batavia, IL) with microfluidic channels to estimate the resulting flow rates in channels for PBS and blood. Using the flow rates obtained from the experiment, the resulting shear stress on the channel surface was determined by employing the Navier–Stoke’s equation for incompressible fluids and infinite parallel flow. Eqn (1) was derived to evaluate the shear stress in the microchannels as described before.38
| (1) |
where the shear stress (τ) varies with the width (W) and height (H) of the channels, as well as the volumetric flow rate (Q), and the dynamic viscosity (μ) of the fluid. The channel dimensions (width and height) were optimized iteratively until the measured flow rates and the resulting estimated shear stress levels fell within the optimal range for the capture of CD4+ T-lymphocytes in microfluidic channels (Fig. 5B and C).15,38 The middle channel was stained with temperature sensitive liquid crystal dye to monitor the channel temperature throughout the experiment (Fig. 1B). To keep the fabrication costs low, microfluidic chips were made of inexpensive plastic components and adhesives with methods that would allow high fabrication yield (ESI†).
Fig. 5.
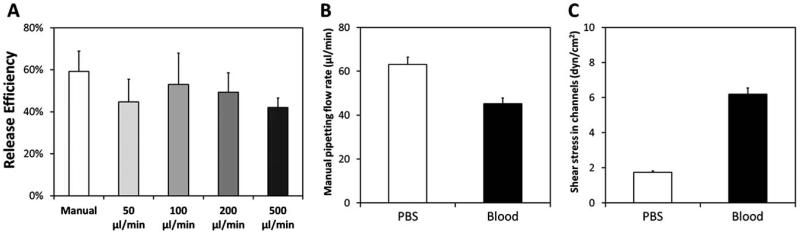
Effect of flow rate on release efficiency of captured CD4+ cells in microchannels. (A) The flow rates in the range of 50 μL min−1 to 500 μL min−1 were applied through a programmable syringe pump, whereas the manual flow rate was applied with standard pipettors as described in the methods section. A statistically significant effect on the release efficiency of captured CD4+ cells was not observed (P>0.05 for all comparisons). (B) Manual pipetting flow rate for phosphate buffered saline (PBS) and blood in microchannels. (C) The shear stress rate in channels when flowing PBS and blood. Error bars represent the standard error of the mean.
Functionalization of microchannels for selective label-free CD4+ and CD34+ cell capture
All components were kept at 37 °C throughout the experiment. The microchips were placed on a temperature controlled heating pad (Omega Engineering Inc, Stamford, CT) to maintain the hydrophobic state of the polymer. The channels were washed with Phosphate Buffered Saline (PBS, Mediatech, Herndon, VA). Next, biotin binding protein NeutrAvidin (Thermo Fisher Scientific Inc., Rockford, IL) was immobilized on the surface by incubating 100 μg mL−1 NeutrAvidin solution in PBS for 1 hour. The surface was then passivated with 1% Albumin from Bovine Serum (BSA) solution (Sigma-Aldrich Co., St Louis, MO) in PBS. Monoclonal biotinylated human anti-CD4 antibody (Beckman Coulter Inc., Brea, CA) was immobilized with two successive half hour incubations of 22 μg mL−1 solutions in PBS. Similarly, biotinylated anti-human CD34 antibody (BioLegend Inc., San Diego, CA) was immobilized in microchannels. After each incubation period, the channels were rinsed with PBS.
Selective CD4+ cell capture from human blood in microchannels
Following the completion of surface chemistry, blood was introduced into the channels. Discarded buffy coat samples from healthy patients were obtained from Blood Transfusion Services at Massachusetts General Hospital, Boston, MA under the approval of the Institutional Review Board. To minimize the effects of CD4+ cell number variations in the blood samples, we performed cell count on each buffy coat sample and normalized the number of white blood cells to the normal range observed in humans (Table 1). White blood cell concentrations were quantified using a hematology system (Drew-3, Drew Scientific Group, Dallas, TX) for each buffy coat sample obtained. Next, the serum was extracted from the buffy coat by centrifuging 5 mL of buffy coat at 2000 rpm for 12 minutes. Then, the buffy coat samples were diluted with the serum38 to obtain a white blood cell concentration within the normal range in humans (Table 1). It should be noted that these process steps are not required if whole blood samples from healthy patients are available, in which case a direct application of the whole blood into the microchannels would be possible as described below for our CD34+ capture experiments. For CD4+ capture, 20 μL of the blood sample (buffy coat) at 37 °C was introduced into the microchannels with a manual pipette (Fig. 1A). Immediately after sample injection, a 100 μL mL−1 FACS Lysing solution (BD Biosciences, San Jose, CA) in HyPure Cell Culture Grade Water (HyClone Laboratories Inc., Logan, UT) was introduced into the channels and incubated for 5 minutes to remove the remaining erythrocytes from the channels, followed by a PBS wash.
Selective CD34+ cell capture from human whole blood in microchannels
Discarded deidentified whole blood samples were obtained from Brigham and Women’s Hospital (Boston, MA), under the approval of the Institutional Review Board. Cell concentrations in whole blood were verified using a hematology system (Drew-3, Drew Scientific Group, Dallas, TX) to ensure that the levels are within the normal range per μL of blood (Table 1).39 For capturing the CD34+ cells, 100 μL of the whole blood was introduced into the microchannels, during which temperature was maintained at 37 °C. Immediately after sample injection, a 100 μg mL−1 FACS Lysing solution (BD Biosciences, San Jose, CA) in HyPure Cell Culture Grade Water (HyClone Laboratories Inc., Logan, UT) was introduced into the channels and incubated for 5 minutes to lyse and remove the erythrocytes from the channels, followed by a PBS wash.
Controlled release of captured CD4+ and CD34+ cells in channels
To release the captured cells, microchannels were removed from the heating pads and cooled down at room temperature for 5 minutes. Released cells were then rinsed out with PBS (or the culture medium of interest) and collected in 1.5 mL microcentrifuge tubes through tubing connected to the outlet port (Fig. 1A). After the release step, channels were assessed with a microscope to confirm and quantify the cell release. Release efficiency of captured cells was quantified by determining the ratio of the released cells to the remaining cells in channels. We have assessed the effect of flow rate (in the range of 50–500 μL min−1) in channels on release efficiency by using a programmable syringe pump.
Analysis of capture specificity in microchannels
The capture specificity of the channels can be defined as the total number of cells stained with anti-CD4 or anti-CD34 antibody-fluorophore, divided by the total number of cells counted in bright field images (Fig. 3A-D and 7A and B). Captured cells on a separate group of channels (n = 4 per group) were stained with anti-human CD4 antibody conjugated with Alexa Fluor® 488(eBioscience Inc., San Diego, CA) or anti-human CD34 antibody conjugated with Alexa Fluor 488(Biolegend, San Diego, CA) to assess capture specificity. The channels were imaged at 10 predetermined fixed locations to quantify captured cells in bright field and in fluorescent modes. Then, fluorescent cell counts were divided by the bright field cell counts to determine capture specificity.
Fig. 3.
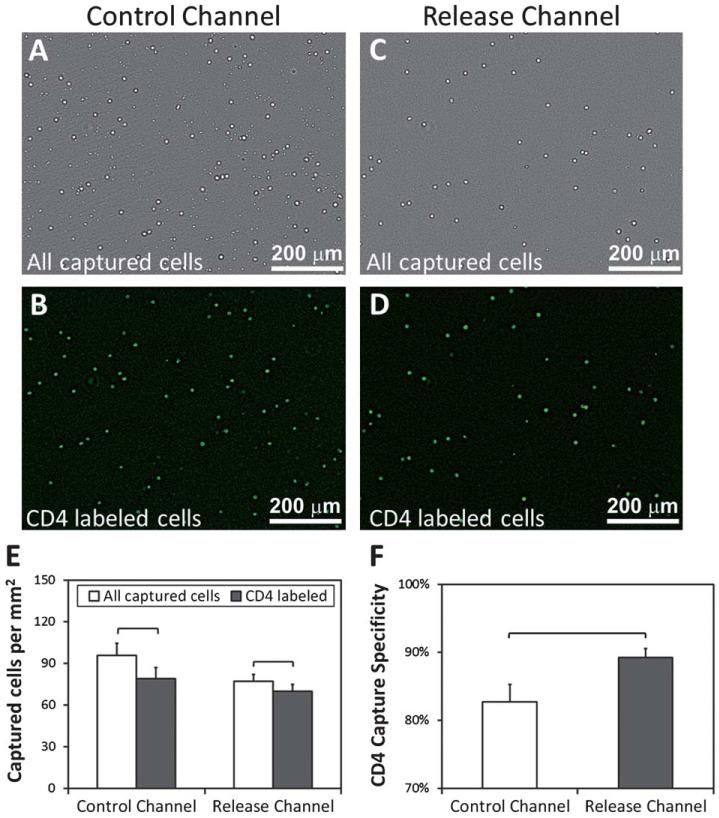
CD4+ cell capture specificity for control and release microchannels. (A) Bright field image of all the captured cells from blood (buffy coat) in control channels. (B) CD4 antibody stained cells in control channels. (C) All the captured cells in release channels. (D) CD4 antibody stained cells in release channels. (E) Specificity of captured cells at 37 °C was 83% (mean ± 2.5%) for control, and 91% (mean ± 1.3%) for thermoresponsive release channels. The brackets indicate statistically significant difference (n = 4 channels, 10 images per channel, t-test, p < 0.05). Error bars represent the standard error of the mean.
Fig. 7.
Validation of the microfluidic capture/release system with CD34+ stem cells from whole blood. (A) CD34+ stem cells were successfully captured from whole blood. (B) CD34 fluorescent labeling of captured cells in microchannels indicated capture specificity greater than 90%. (C–D) The released CD34+ cells displayed greater than 90% viability as indicated by live/dead assay. (E) Post-release cultivation of CD34+ cells was possible with healthy cellular morphology after a day of culture.
Post-release viability assessment and cultivation of CD4+ and CD34+ cells
To evaluate the post-release viability, CD4+ cells were collected and assessed by a dye exclusion viability method. The released cells were stained with trypan blue (Sigma Aldrich, St Louis, MO) and counted in a hemocytometer. The live and dead cell numbers were recorded and the counts were repeated 5 times to obtain an average. On the other hand, the released CD34+ cells were assessed for viability by using live/dead cell assay (Invitrogen, Carlsbad, CA). The released and collected CD4+ and CD34+ cells were cultured in RPMI 1640 media (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), and 100 UI mL−1 Penicillin-streptomycin. CD4+ cell medium was supplemented with phytohemagglutinin (PHA) (Thermo Fisher Scientific, Waltham, MA) to facilitate cellular mitosis.40 The CD4+ cells were cultured for 8 days at 37 °C and 5% CO2 incubator. During the cultivation period, cell morphologies and numbers were assessed with microscopy. Cell densities in culture per millimetre square were quantified to analyze the proliferation potential of CD4+ cells.
Microfluidic channel visualization and image processing
A Carl Zeiss Observer D1 Model Axio Inverted Microscope with the AxioVision LE (from Carl Zeiss) software was used to obtain microscopic images in this study. Carl Zeiss Plan-Apochromat (10×/0.45 ph1) objective lens was utilized for bright field and fluorescent imaging of microchannels. Ten images were taken per channel at predetermined fixed locations marked during microfluidic chipfabrication,which ensured consistency of the position of images before and after the cell release. ImageJ software (National Institutes of Health) was utilized to semi-automatically quantify the number of cells in each image using the Cell-Counter plug-in.
Statistical analysis
Data obtained in this study were reported as mean ± standard error of the mean. The collected data were tested for normal distribution with Anderson–Darling normality test. Parametric 2-sample t-test (release efficiency, n = 6 channels in each group with an average of 10 images per channel) and paired t-test (cell capture specificity, n = 4 channels in each group with an average of 10 images per channel) were used for statistical analysis. The effect of flow rate on release efficiency was assessed using analysis of variance (ANOVA) with Tukey’s post hoc comparison (n = 4 channels per flow rate). CD4+ cell proliferation was also analyzed using ANOVA with Tukey’s post hoc comparisons. Statistical significance was set at 95% confidence level for all tests (p < 0.05). The experiments were repeated at least four times with different blood samples to validate repeatability and reproducibility. Error bars in the figures represent the standard error of the mean.
Results
We showed that the protein adsorption/desorption mechanism on temperature-responsive polymer (PNIPAAm)34 can be adapted to develop a biotin-binding protein and biotinylated antibody based surface chemistry to release the selectively captured label-free CD4+ and CD34+ cells from blood with high viability and specificity. With this new approach, selective capture, controlled release and viable retrieval of cells can be achieved in less than 10 minutes with simple manual pipetting in a standard laboratory setting (Fig. 1).
CD4+ and CD34+ cell counts in blood samples
Quantification of cell numbers in the buffy coat samples indicated that 3.6% of the mononuclear cells were CD4+ cells, which accounted for 195 cells per million blood cells (including the red blood cells) (Table 1). CD34+ cell capture was performed using whole blood samples. The number of CD34+ cells in the whole blood samples was measured to be 0.5% of the mononuclear cells, which accounted for 19 cells per million blood cells (including the red blood cells) (Table 1).
Release efficiency of the captured cells in microchannels
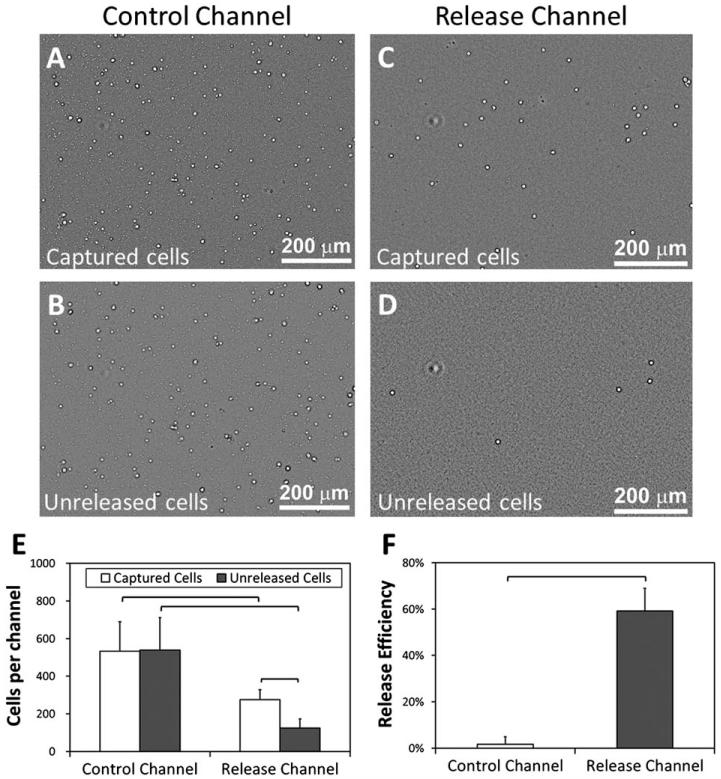
The microchannels were rendered chemically active by functionalization with anti-CD4 or anti-CD34 antibodies. We analyzed the release efficiency of captured CD4+ cells in thermoresponsive release channels in comparison to control channels (standard cell culture grade polystyrene) (Fig. 2A-D). A significant number of cells were released in release channels (paired t-test, p < 0.05) (Fig. 2E), while there was no statistically significant difference between the number of captured and unreleased cells in control channels (paired t-test, p>0.05). We observed the release efficiency to be 59% (±4%) in release channels, while the release efficiency was less than 2% in the control channels (Fig. 2F). The difference between the cell release efficiencies of control and release channels was statistically significant (t-test, p < 0.05).
Fig. 2.
Release efficiency for the captured CD4+ cells in control and release microchannels. (A) Bright field image of all captured cells from blood (buffy coat) in control channels at 37 °C. (B) Unreleased cells remaining in control channels after the temperature was reduced below 32 °C and the channels were rinsed. (C) All captured cells from blood in release channels at 37 °C. (D) Unreleased cells in the release channels after the temperature was reduced below 32 °C and the channels were rinsed. (E) When the temperature of control channels was decreased below 32 °C, a statistically significant release of captured cells was not observed after rinsing. On the other hand, when the temperature of the release channels was decreased below 32 °C, a significant (paired t-test, p < 0.05) release of captured cells was observed after rinsing. (F) Control microchannels displayed a release efficiency of less than 2%, whereas the thermoresponsive release channels allowed release efficiency in excess of 59%. The difference in release efficiencies of control and release channels was statistically significant (n = 6 channels, 10 images per channel, t-test, p < 0.05). Brackets connecting groups indicate statistically significant difference. Error bars represent the standard error of the mean.
Specificity of selective cell capture in microchannels
Selective cell capture specificity was analyzed by quantifying all the captured cells (Fig. 3A and C) and the number of CD4 fluorophore conjugated antibody labeled captured cells (Fig. 3B and D) in both control and release channels. For CD4+ cell capture, control channels displayed a capture specificity of 83% (±2.5%), while the thermoresponsive release channels displayed a significantly greater (t-test, p < 0.05), 91% (±1.3%) capture specificity (Fig. 3E and F).
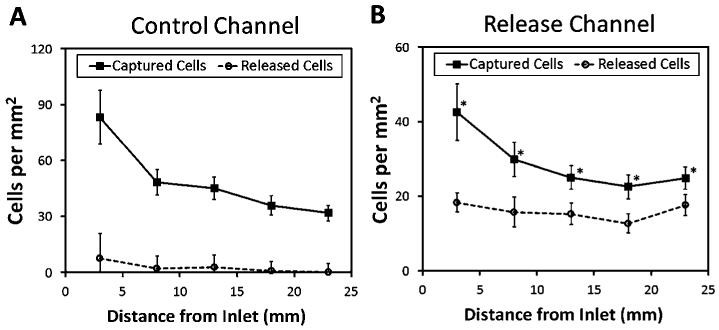
Cell capture and release distribution along the microchannels
Cell capture and release distribution along the channels were analyzed by imaging five predetermined locations in the channels. A higher density of captured cells was observed close to the channel inlets, which decreased towards the outlet (Fig. 4). When the channel temperature was reduced, less than 2% of the cells were released along the control channels (Fig. 4A). On the other hand, a significant release of captured cells was observed (59%) with a distribution along the release channels (Fig. 4B).
Fig. 4.
Distribution of captured and released cells along the microfluidic channels. (A) In control channels, captured cells were concentrated closer to the inlet with a decreasing distribution along the channel. After cooling down and rinsing, the number of released cells were significantly lower along the channels. (B) In release channels, more cells were captured closer to the inlet and the number gradually decreased along the channels. After release steps, a comparable number of cells were released along the channels. The difference between the number of captured and unreleased cells (i.e., the number of released cells subtracted from the number of captured cells) along the channel was statistically significant (n = 6 channels, 10 images per channel, * indicates p < 0.05, paired t-test). Error bars represent the standard error of the mean.
Effect of flow rate on release efficiency and optimized fluid shear rates in channels
Flow rates used in this study (50–500 μL min−1) did not have a major effect (p>0.05) on the release efficiency of captured cells in microchannels (Fig. 5A). To eliminate the dependence on peripheral pumping equipment, microchannel dimensions were optimized to result in manual pipetting flow rates of 63.1 μL min−1 (±3.3 μL min−1) for aqueous solutions (phosphate buffered saline, PBS) and 45.2 μL min−1 (±2.6 mL min−1) for blood (Fig. 5B). Based on these flow rates, channel dimensions, dynamic viscosity of water at 37 °C (7 × 10−4 Pa·s) and dynamic viscosity of blood at 37 °C (35 × 10−4 Pa·s),41 fluid shear stress induced on the channel surface was determined to be 1.7 dyn cm−2 (±0.1 dyn cm−2) for aqueous solutions, and 6.2 dyn cm−2 (±0.4 dyn cm−2) for blood (Fig. 5C). The estimated fluid shear stress levels were within the optimal range for the capture of CD4+ T-lymphocytes in microfluidic channels. We showed that manual pipetting allowed reproducible flow rates and shear stresses in the microchannels (Fig. 5B and C).
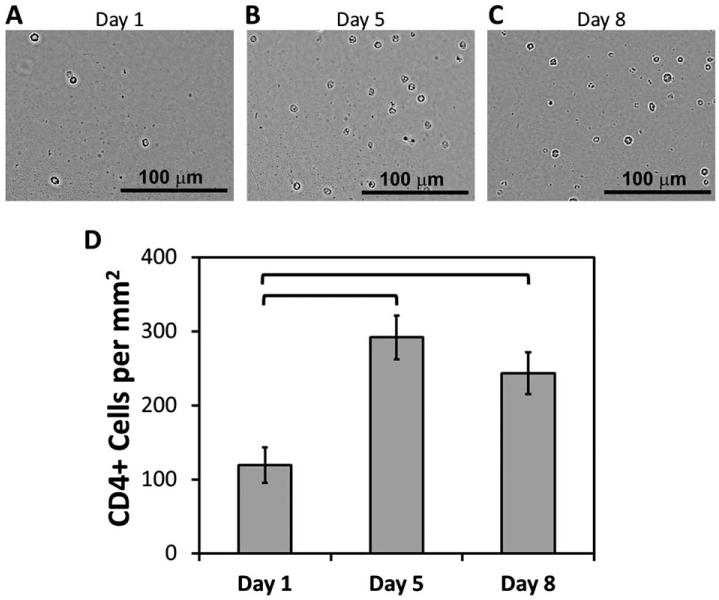
Post-release CD4+ cell viability and cultivation
The viability of released CD4+ cells from microchannels was analyzed by collecting the released cells at the outlet and by performing dye-exclusion viability assay. Post-release viability of CD4+ cells was observed to be 94% (±4%). The released CD4+ cells were cultured for 8 days and the viability was still observed to be greater than 90% at the end of the culture period. The morphological assessment of the cells over the 8 day culture period indicated intact and healthy cells (Fig. 6A-C) with a statistically significant proliferation at day 5 and at day 8 (Fig. 6D).
Fig. 6.
Post-release cultivation of CD4+ cells. (A–C) CD4+ cells were collected after release and cultured for up to 8 days. Microscopic imaging was performed at day 1, 5 and 8 to assess the cellular morphologies. (D) Cell densities in culture (per millimetre square) at day-5 and day-8 displayed a statistically significant increase compared to day-1 (p < 0.05). The brackets indicate statistically significant difference. Error bars represent the standard error of the mean.
Specific CD34+ capture, release and viability assessment
We validated our system by selectively capturing and releasing CD34+ stem cells, which are found to be at lower concentrations in peripheral blood compared to CD4+ cells (Table 1). CD34+ cells were selectively captured from whole blood samples, and released to result in isolated cells with high viability and cultivation potential. Capture specificity of the CD34+ cells was observed to be more than 90% (Fig. 7A and B), which was comparable to the specificity observed for CD4+ cells in the system (Fig. 3). The released CD34+ cells were viable (viability more than 90%, Fig. 7C and D) and displayed intact and healthy morphology after a day in culture (Fig. 7E).
Discussion
We have shown that specific antibodies can be immobilized in thermoresponsive microfluidic channels to selectively immobilize cells from blood with antibody–antigen interaction followed by controlled release and retrieval with high release efficiency, specificity and viability. To design thermoresponsive microfluidic channels that can effectively release selectively captured cells, it is important to understand the working principle of temperature responsive polymers and their interaction with proteins at different states. Temperature dependent interaction between the polymer and proteins is due to the changes in local environment around the hydrophobic isopropyl domains in the polymer. Below the LCST (<32 °C), the polymer becomes hydrophilic and swells in water42 at which point, the hydrophilic amide and carboxyl groups in the polymer form hydrogen bonds with water molecules, reducing the interaction with proteins. At and above the LCST (>32 °C), the polymer becomes hydrophobic and insoluble in water,34,42 where the hydrophobic isopropyl group in the polymer causes phase separation from the aqueous environment and therefore breakage of hydrogen bonds. In this case, the polymer becomes hydrophobic and associates less with the surrounding water and interacts more with the proteins in solution. Since the polymer has high affinity for adherence of proteins at 37 °C,34 Neutravidin (or other biotin binding proteins) can be immobilized on the polymer followed by a biotinylated antibody, and cells can be selectively captured at this temperature (Fig. 1D). By simply reducing the temperature of the surface below the LCST, protein–antibody–cell complex can be rapidly released from the surface (Fig. 1F). In this system, achieving rapid release of captured cells in a controllable manner with thermoresponsive polymers was possible by the small volumes employed in microchannels (Fig. 1C-F).
To ensure optimal performance of the release channels, a temperature indicator channel was included on-chip (stained with temperature responsive dye) to monitor the temperature distribution during the capture and release processes using a simple visual color change, sensitive down to 1 °C (Fig. 1B). All components were heated to 37 °C prior to injection into channels. Slight fluctuations in temperature were tolerated with the developed system since the surface behaves consistently in a wide temperature range (about 5 °C) above and below the LCST.
For the genomic studies on selectively isolated cells, it is critical that the genomic characteristics and gene expression of cells are not affected during the isolation process. It should be noted that in this study, the temperature change within the microchannels between the capture period (37 °C) and the release period (<32 °C) was about 5 °C. The release took place in less than 10 minutes facilitated by the small channel volume (~10 μL). It has been shown that the genomic and phenotypic expression of cultured embryonic stem cells (ESCs) were not affected from a temperature decrease of up to 27 °C (from 37 °C to 10 °C) over a period of 2 hours on thermoresponsive polymers.43
The number of captured cells per unit area of control channels was significantly more than the number of cells in release channels (Fig. 2E), which can be attributed to the higher extent of immobilization of biotin binding Neutravidin and biotinylated capture antibody on control channels. It is likely that the polymer layer does not fully cover the surface, or contains small gaps between the polymer chains above the LCST.34 This may have caused the adherence of the Neutravidin onto the polystyrene surface, which would not respond to a temperature change, and hence, not release the protein–antibody–cell complex. In addition, while the polymer was in a hydrated state (at 37 °C), proteins may have diffused through the polymer chains and got adsorbed directly onto the polystyrene surface via hydrophobic interactions as discussed before elsewhere.44 To enhance the release efficiency, polymer density on the surface can be increased by employing a chain transfer reaction on self-assembled monolayer chains terminated with thiol groups, as shown before.34 In this study, we used Neutravidin/biotin-antibody based surface chemistry, since it is a commonly used method in the literature,15,38 and the components are readily accessible through commercial vendors, which can be utilized by a wide group of researchers. Other protein–antibody immobilization methods (e.g., streptavidin/biotin-antibody) can potentially be adapted to thermoresponsive microfluidics to enhance capture and release efficiencies.
The capture pattern observed along the microchannels (Fig. 4) can be explained by the fact that as the cell suspension (i.e., blood) flows along a channel, CD4+ cells are immobilized on the surface and subsequently a reduced number of cells are available for capture further along the channel. Even though a higher density of cells was captured closer to the inlet, a similar number of cells were released closer to the inlet compared to the middle and the outlet regions of the channels. Therefore, over-saturation of the channel surface with captured cells would not necessarily enhance the release efficiency, but may increase the total number of cells released.
In this study, our objective was to develop a time efficient and simple approach with an inexpensive microfluidic platform to achieve highly selective cell isolation from heterogeneous mixtures, which led to the use of manual pipetting as opposed to using a pump. The flow rates employed in this study did not have a major effect on the release efficiency (Fig. 5A), indicating that the cell release mechanism mainly depended on the temperature responsive behavior of channels. We have quantified the flow rates and fluid shear stresses achieved by manual pipetting for aqueous solutions and blood in microchannels (Fig. 5). Based on detailed characterization of flow rates in microchannels, we employed the optimal fluid shear range to capture cells from blood.
The number of CD4+ cells in whole blood samples available for this study was significantly low, as these whole blood samples were obtained from transplant patients under immunosuppressant treatment. Therefore, in this study, CD4+ cells were captured from fresh buffy coat samples obtained from healthy adults. Prior to utilization in experiments, white blood cell counts were determined in buffy coat samples, which were observed to be about 4 times higher than the normal range in humans (Table 1). Therefore, the buffy coat samples were diluted with their own serum to obtain normal cell concentrations observed in whole blood as described in Materials and Methods.
We have also performed preliminary analysis on the viability and post-release culture potential of selectively captured CD4+ cells (Fig. 6). The results indicated that release of CD4+ cells, which were captured in microchannels, did not significantly affect their viability and culture potential. Further, we have validated our system with CD34+ stem cell capture and release using whole blood (Fig. 7). The released CD34+ cells were viable and amenable to post-release cultivation (Fig. 7C-E), indicating the potential of the developed technology for stem cell isolation from sources such as peripheral blood, cord blood, and bone marrow.
Conclusion
Effective, easy to use, inexpensive, rapid, and selective label-free cell isolation technologies are significantly needed to enable a new era of down-stream processing of cells for broad applications in biological research and diagnosis of diseases. In this study, we have shown that cells can be rapidly and controllably released in microchannels with high viability and specificity after they were selectively and label-free captured from blood without any pre-processing steps. The release of live cells that are captured from heterogeneous cell suspensions, biological and bodily fluids by using inexpensive microfluidic systems can generate a great impact in areas such as genomic/proteomic analysis, stem cell isolation/purification for regenerative therapies, and circulating rare tumor cell isolation for cancer research.
Supplementary Material
Acknowledgements
This work was performed at the Demirci Bio-Acoustic-MEMS in Medicine (BAMM) Laboratories at Harvard-Massachusetts Institute of Technology Health Sciences and Technology, Brigham & Women’s Hospital Center for Bioengineering at Harvard Medical School. This work was partially supported by National institutes of Health (NIHR21AI087107, NIH R01AI081534) and the Center for Integration of Medicine and Innovative Technology (CIMIT) under the U.S. Army Medical Research Acquisition Activity Cooperative Agreements DAMD17-02-2-0006, W81XWH-07-2-0011, and W81XWH-09-2-0001. This research was made possible partially by a research grant that was awarded and administered by the U.S. Army Medical Research & Materiel Command (USAMRMC) and the Telemedicine & Advanced Technology Research Center (TATRC) at Fort Detrick, MD. The information contained herein does not necessarily reflect the position or policy of the government, and no official endorsement should be inferred.
Footnotes
Electronic supplementary information (ESI) available: See DOI: 10.1039/c1lc20487d
References
- 1.Gurkan UA, Moon S, Geckil H, Xu F, Wang S, Lu TJ, Demirci U. Biotechnol. J. 2011;6:138–149. doi: 10.1002/biot.201000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez WR, Christodoulides N, Floriano PN, Graham S, Mohanty S, Dixon M, Hsiang M, Peter T, Zavahir S, Thior I, Romanovicz D, Bernard B, Goodey AP, Walker BD, McDevitt JT. PLoS Med. 2005;2:663–672. doi: 10.1371/journal.pmed.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee WG, Kim Y-G, Chung BG, Demirci U, Khademhosseini A. Adv. Drug Delivery Rev. 2010;62:449–457. doi: 10.1016/j.addr.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee WG, Demirci U, Khademhosseini A. Integr. Biol. 2009;1:242–251. doi: 10.1039/b819201d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, Xu F, Demirci U. Biotechnol. Adv. 2010;28:770–781. doi: 10.1016/j.biotechadv.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotz KT, Xiao W, Miller-Graziano C, Qian W-J, Russom A, Warner EA, Moldawer LL, De A, Bankey PE, Petritis BO, Camp DG, Rosenbach AE, Goverman J, Fagan SP, Brownstein BH, Irimia D, Xu W, Wilhelmy J, Mindrinos MN, Smith RD, Davis RW, Tompkins RG, Toner M. Nat. Med. 2010;16:1042–1047. doi: 10.1038/nm.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salehi-Reyhani A, Kaplinsky J, Burgin E, Novakova M, Demello AJ, Templer RH, Parker P, Neil MAA, Ces O, French P, Willison KR, Klug D. Lab Chip. 2011;11:1256–1261. doi: 10.1039/c0lc00613k. [DOI] [PubMed] [Google Scholar]
- 9.McKenna BK, Selim AA, Bringhurst FR, Ehrlich DJ. Lab Chip. 2009;9:305–310. doi: 10.1039/b811889b. [DOI] [PubMed] [Google Scholar]
- 10.Ramadan Q, Samper V, Poenar D, Liang Z, Yu C, Lim TM. Sens. Actuators, B. 2006;113:944–955. [Google Scholar]
- 11.Hosokawa M, Arakaki A, Takahashi M, Mori T, Takeyama H, Matsunaga T. Anal. Chem. 2009;81:5308–5313. doi: 10.1021/ac900535h. [DOI] [PubMed] [Google Scholar]
- 12.Wu HW, Lin CC, Lee GB. Biomicrofluidics. 2011;5:26. doi: 10.1063/1.3528299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Phillips JA, Yan JL, Li QG, Fan ZH, Tan WH. Anal. Chem. 2009;81:7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandstrom E, Urassa W, Bakari M, Swai A, Mhalu F, Biberfeld G, Pallangyo K. Int. J. STD AIDS. 2003;14:547–551. doi: 10.1258/095646203767869165. [DOI] [PubMed] [Google Scholar]
- 15.Moon S, Gurkan UA, Blander J, Fawzi WW, Aboud S, Mugusi F, Kuritzkes DR, Demirci U. PLoS One. 2011;6:8. doi: 10.1371/journal.pone.0021409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Cometti B, Pappas D. Anal. Chim. Acta. 2007;601:1–9. doi: 10.1016/j.aca.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 17.Busch R, Cesar D, Higuera-Alhino D, Gee T, Hellerstein MK, McCune JM. J. Immunol. Methods. 2004;286:97–109. doi: 10.1016/j.jim.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Sethu P, Moldawer LL, Mindrinos MN, Scumpia PO, Tannahill CL, Wilhelmy J, Efron PA, Brownstein BH, Tompkins RG, Toner M. Anal. Chem. 2006;78:5453–5461. doi: 10.1021/ac060140c. [DOI] [PubMed] [Google Scholar]
- 19.Hristov M, Erl W, Weber PC. Trends Cardiovasc. Med. 2003;13:201–206. doi: 10.1016/s1050-1738(03)00077-x. [DOI] [PubMed] [Google Scholar]
- 20.Boyer M, Townsend LE, Vogel LM, Falk J, Reitz-Vick D, Trevor KT, Villalba M, Bendick PJ, Glover JL. J. Cardiovasc. Surg. 2000;31:181–189. doi: 10.1016/s0741-5214(00)70080-2. [DOI] [PubMed] [Google Scholar]
- 21.Yang JJ, Ii M, Kamei N, Alev C, Kwon SM, Kawamoto A, Akimaru H, Masuda H, Sawa Y, Asahara T. PLoS One. 2011;6:14. doi: 10.1371/journal.pone.0020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winslow JM, Liesveld JL, Ryan DH, Dipersio JF, Abboud CN. Bone Marrow Transplant. 1994;14:265–271. [PubMed] [Google Scholar]
- 23.Sung-Yi Y, et al. Meas. Sci. Technol. 2006;17:2001. [Google Scholar]
- 24.Fu AY, Chou H-P, Spence C, Arnold FH, Quake SR. Anal. Chem. 2002;74:2451–2457. doi: 10.1021/ac0255330. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Cui DF, Liu CC, Li H, Chen J. Anal. Chim. Acta. 2007;584:237–243. doi: 10.1016/j.aca.2006.11.057. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Jones P, Haswell SJ. Chem. Eng. J. 2008;135:S82–S88. [Google Scholar]
- 27.Wankhede SP, Du Z, Berg JM, Vaughn MW, Dallas T, Cheng KH, Gollahon L. Biotechnol. Prog. 2006;22:1426–1433. doi: 10.1021/bp060127d. [DOI] [PubMed] [Google Scholar]
- 28.Plouffe BD, Brown MA, Iyer RK, Radisic M, Murthy SK. Lab Chip. 2009;9:1507–1510. doi: 10.1039/b823523f. [DOI] [PubMed] [Google Scholar]
- 29.Jung K, Hampel G, Scholz M, Henke W. Cell. Physiol. Biochem. 1995;5:353–360. [Google Scholar]
- 30.Fujioka N, Morimoto Y, Takeuchi K, Yoshioka M, Kikuchi M. Appl. Spectrosc. 2003;57:241–243. doi: 10.1366/000370203321535187. [DOI] [PubMed] [Google Scholar]
- 31.Hatch A, Hansmann G, Murthy SK. Langmuir. 2011;27:4257–4264. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suárez-González D, Barnhart K, Saito E, Vanderby R, Hollister SJ, Murphy WL. J. Biomed. Mater. Res. 2010;95:222–234. doi: 10.1002/jbm.a.32833. Part A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanazawa H, Yamamoto K, Kashiwase Y, Matsushima Y, Takai N, Kikuchi A, Sakurai Y, Okano T. J. Pharm. Biomed. Anal. 1997;15:1545–1550. doi: 10.1016/s0731-7085(96)02004-3. [DOI] [PubMed] [Google Scholar]
- 34.Huber DL, Manginell RP, Samara MA, Kim BI, Bunker BC. Science. 2003;301:352–354. doi: 10.1126/science.1080759. [DOI] [PubMed] [Google Scholar]
- 35.Okano T, Yamada N, Okuhara M, Sakai H, Sakurai Y. Biomaterials. 1995;16:297–303. doi: 10.1016/0142-9612(95)93257-e. [DOI] [PubMed] [Google Scholar]
- 36.von Recum HA, Kim SW, Kikuchi A, Okuhara M, Sakurai Y, Okano T. J. Biomed. Mater. Res. 1998;40:631–639. doi: 10.1002/(sici)1097-4636(19980615)40:4<631::aid-jbm15>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 37.Yamato M, Utsumi M, Kushida A, Konno C, Kikuchi A, Okano T. Tissue Eng. 2001;7:473–480. doi: 10.1089/10763270152436517. [DOI] [PubMed] [Google Scholar]
- 38.Moon S, Keles HO, Ozcan A, Khademhosseini A, Haeggstrom E, Kuritzkes D, Demirci U. Biosens. Bioelectron. 2009;24:3208–3214. doi: 10.1016/j.bios.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. Garland Science; New York: 2002. [Google Scholar]
- 40.Lefort CT, Kim M. J. Vis. Exp. 2010;40:e2017. doi: 10.3791/2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boutsianis E, Dave H, Frauenfelder T, Poulikakos D, Wildermuth S, Turina M, Ventikos Y, Zund G. Eur. J. Cardio Thorac. Surg. 2004;26:248–256. doi: 10.1016/j.ejcts.2004.02.041. [DOI] [PubMed] [Google Scholar]
- 42.Takei YG, Aoki T, Sanui K, Ogata N, Sakurai Y, Okano T. Macromolecules. 1994;27:6163–6166. [Google Scholar]
- 43.Dey S, Kellam B, Alexander MR, Alexander C, Rose FRAJ. J. Mater. Chem. 2011;21:6883–6890. [Google Scholar]
- 44.Tekin H, Ozaydin-Ince G, Tsinman T, Gleason KK, Langer R, Khademhosseini A, Demirel MC. Langmuir. 2011;27:5671–5679. doi: 10.1021/la200183x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim YG, Moon S, Kuritzkes DR, Demirci U. Quantum dot-based HIV capture and imaging in a microfluidic channel, Biosensors & Bioelectronics. 2009;25(1):253–258. doi: 10.1016/j.bios.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moon S, Ceyhan E, Gurkan UA, Demirci U. Statistical modeling of single target cell encapsulation. PLoS One. 2001;6(7) doi: 10.1371/journal.pone.0021580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alyassin MA, Moon S, Keles HO, Manzur F, Lin RL, Haeggstrom E, Kuritzkes DR, Demirci U. Rapid automated cell quantification on HIV microfluidic devices. Lab on a Chip. 2009;9(23):3364–3369. doi: 10.1039/b911882a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.