Abstract
We synthesized cationic random amphiphilic copolymers by radical copolymerization of methacrylate monomers with cationic or hydrophobic groups and evaluated their antimicrobial and hemolytic activities. The nature of the hydrophobic groups, and polymer composition and length were systematically varied to investigate how structural parameters affect polymer activity. This allowed us to obtain the optimal composition of polymers suitable to act as non-toxic antimicrobials as well as non-selective polymeric biocides. The antimicrobial activity depends sigmoidally on the mole fraction of hydrophobic groups (fHB). The hemolytic activity increases as fHB increases and levels off at high values of fHB, especially for the high-molecular-weight polymers. Plots of HC50 values versus the number of hydrophobic side chains in a polymer chain for each polymer series showed a good correlation and linear relationship in the log–log plots. We also developed a theoretical model to analyze the hemolytic activity of polymers and demonstrated that the hemolytic activity can be described as a balance of membrane binding of polymers through partitioning of hydrophobic side chains into lipid layers and the hydrophobic collapsing of polymer chains. The study on the membrane binding of dye-labeled polymers to large, unilamellar vesicles showed that the hydrophobicity of polymers enhances their binding to lipid bilayers and induces collapse of the polymer chain in solution, reducing the apparent affinity of polymers for the membranes.
Keywords: antibiotics, biocides, hemolysis, liposomes, polymers
Introduction
The development of synthetic antimicrobial agents is an emerging need due to antibiotic resistance of bacteria causing fatal infectious diseases, which significantly threaten public health in global regions. The resistance of bacteria was found initially in hospitals, however, the drug-resistant strains rapidly spread to other public places as well as communities across countries.[1]
New antimicrobial agents are urgently needed, which requires new design concepts and approaches to combat drug-resistant bacteria. Considering the wide use of agents as drugs and in consumer products, it would be ideal to create antimicrobials that can meet the following criteria: selective toxicity to bacteria versus human cells and low susceptibility to the development of resistance in bacteria. In this context, host-defense peptides have been extensively studied and their potential as alternatives for conventional antibiotics has been explored. These peptides consist of a relatively small number of residues (<40 aa). Upon binding to cell membranes, the peptides form facially amphiphilic helical structures with cationic and hydrophobic side chains regularly distributed on different sides of the helix surface. The amphiphilic features of peptides function by disrupting bacterial cell membranes, causing breakdown of the transmembrane potential, leakage of cytoplasmic components, and ultimately cell death. Natural antimicrobial peptides are believed to selectively target bacteria due to the preferable charge interaction between the cationic side chains of the peptides and the greater density of negative charges on the bacterial cell surface relative to eukaryotic cells. The application of peptide-based antimicrobials, however, is limited in practical terms by the high costs associated with their preparation and manufacturing, preventing their ubiquitous use in pharmaceutical and biomedical applications.
To this end, design and preparation of synthetic antimicrobial agents and the corresponding mechanistic studies have been conducted to gain a better understanding of how to create new agents including non-biological peptide mimics and synthetic polymers. Synthetic foldamers,[16] including β-peptides and peptoids,[21] have been utilized to mimic the amphiphilic structures and helical conformations of natural antimicrobial peptides. Facially amphiphilic welldefined oligomers of aryl amides and phenylene ethynylene also have been previously explored. These peptide mimics exhibited higher activity and selectivity than the natural antimicrobial sequences. Although these peptide mimics and oligomers can provide well-defined sequences and structures suitable for structure–activity studies, their preparation remains time- and cost-intensive.
In contrast to peptide mimics, polymeric biocides/disinfectants are easy to make and are inexpensive. A number of polymeric antimicrobial agents have been prepared using scaffolds of conventional synthetic polymers including polystyrenes, polypryridines, and poly-(meth)acrylates. The polymer structures are often modified with cationic and hydrophobic side chains, which are randomly distributed in a polymer chain and thus display random amphiphilicity. Although the polymers display high antimicrobial activity, they often suffer from little or no selectivity to cell types or high levels of toxicity to human cells. However, a series of amphiphilic polynorbones have been prepared,[38] and their random copolymers displayed selective toxicity to bacteria over human red blood cells.[38] Recently, Mowery et. al hypothesized that the global amphiphilic conformation of random amphiphilic copolymers can be induced by a bacterial membrane surface.[39] Random copolymers of β-peptides with cationic and hydrophobic groups displayed cell-type selectivity in their antimicrobial and hemolytic activities. We previously reported antimicrobial and hemolytic activities of random copolymers based on polymethacrylates containing hydrophobic butyl and cationic side chains.[40] Although these polymers have a random sequence of hydrophobic and cationic groups and thus lack of secondary rigid structures, they displayed high antimicrobial activity and modest selectivity for bacteria over human cells.
Here, we systematically varied the nature of the hydrophobic side chains, the composition of the polymers, and the molecular weight to investigate the optimal balance of cationic side chains and hydrophobic groups, as well as the role of hydrophobic groups in the antimicrobial and hemolytic activities of random copolymers. We also examined the effect of the hydrophobic nature of polymers and lipid properties in membrane binding by using unilamellar vesicles with different lipid compositions.
Results and Discussion
Polymer design and synthesis
For the preparation of conventional antimicrobial polymers, quaternary amines are perhaps the most used cationic groups, and polymers are often of high molecular weights (MWs). In contrast, we chose primary amine groups as the cationic functionality and relatively low-MW polymethacrylate backbones (MW= 1000–10 000), mimicking structural features of antimicrobial peptides (MW=2500–3000 for magainin and melittin) (Scheme 1). Previously, we established that hydrophobic and charged side chains are required for antimicrobial activity, although the optimal balance had not been defined.[40] Here, we systematically varied the length and mole fraction of the hydrophobic groups (fHB) and degree of polymerization (DP). The polymers were synthesized by copolymerization of the appropriate monomers in the presence of methyl-3-mercaptopropionate as a chain-transfer agent, followed by deprotection of tert-butoxycarbonyl (Boc) groups in trifluoroacetic acid (TFA). The composition and molecular weight of the final polymers were determined by comparing the integrated intensities of the 1H NMR resonances from the terminal chain-transfer agent relative to the side chains (Table 1). The results were confirmed by size-exclusion chromatography using the Boc-protected precursors of selected polymers. In general, there was reasonable but not exact agreement between the mole ratio of the side chains used in the synthesis, and that obtained in the products. The lack of exact agreement reflects the selective enrichment of the polar products during the final ether-precipitation step. The polymers labeled with a dansyl group at the polymer end were prepared by the same procedure using a chain-transfer agent 2 modified with a dansyl group (Scheme 2 Table 2).
Scheme 1.
Synthesis of amphiphilic polymethacrylate derivatives.
Table 1.
Characterization of amphiphilic random copolymers
| Polymer series[a] |
Side chain | fHB [b] | DP range[c] |
Mnrange [g mol−1][d] |
Average Mn/103 [g mol−1] |
|---|---|---|---|---|---|
| C1-3.3 | methyl | 0.16–0.74 | 14–31 | 3000–3200 | 3.3 |
| C1-2.0 | 0.13–0.70 | 8–20 | 1700–2100 | 2.0 | |
| C1-1.6 | 0.12–0.64 | 7–11 | 1300–1900 | 1.6 | |
| C2-5.0 | ethyl | 0.10–0.55 | 20–32 | 3600–6100 | 5.0 |
| C2-2.9(1) | 0.11–0.70 | 11–22 | 2300–3500 | 2.9 | |
| C2-2.9(2) | 0–0.68 | 11–16 | 2300–3600 | 2.9 | |
| C2-1.8 | 0–0.50 | 7–9 | 1600–2200 | 1.8 | |
| C4-8.7[e] | butyl | 0–0.57 | 32–46 | 7900–10100 | 8.7 |
| C4-5.0[e] | 0–0.53 | 19–31 | 4500–6000 | 5.0 | |
| C4-1.6[e] | 0–0.47 | 5–9 | 1300–1900 | 1.6 | |
| C6-4.8 | hexyl | 0.09–0.51 | 20–24 | 4400–5600 | 4.8 |
| C6-3.3 | 0–0.36 | 11–19 | 2700–4100 | 3.3 | |
| C6-2.0 | 0–0.30 | 8–10 | 1900–2300 | 2.0 | |
| Bz-3.5 | benzyl | 0.09–0.80 | 13–17 | 2700–4100 | 3.5 |
| Bz-2.2 | 0.08–0.51 | 8–10 | 2000–2400 | 2.2 | |
| Bz-1.8 | 0.08–0.40 | 6–9 | 1600–2000 | 1.8 |
The series of polymers having alkyl groups were denoted by Cn-molecular weight averaged over the series, in which n is the number of carbons in the alkyl side chains.
fHB=mole fraction of monomers with hydrophobic groups in a polymer chain.
DP=degree of polymerization (number of monomers in a polymer chain).
The number-average molecular weight of polymers (Mn) was calculated from fHB, DP, and the molecular weights of monomers and methyl 3-mercaptopropionate.
Data reported previously.[40]
Scheme 2.
General scheme for interactions of polymers with lipid bilayers.
Table 2.
Characterization of dansyl-labeled random copolymers.
| Polymer | Side chain | fHB | DP | Mn/103[g mol−1] |
|---|---|---|---|---|
| D0 | – | 0 | 13 | 3.4 |
| D27 | butyl | 0.27 | 16 | 3.7 |
| D49 | butyl | 0.49 | 12 | 2.6 |
Antimicrobial activity of homopolymers and random copolymers
We determined antimicrobial activity as the minimal inhibitory concentration (MIC) using E. coli as a test organism. Because we were interested in evaluating the activity of a single polymer chain to investigate the correlation between biological activity and the structural parameters of polymers including fHB and polymer length, the MIC data are given in molar concentration.
As a prelude to evaluating the antimicrobial copolymers, we examined the activity of the homopolymer with 100% aminoethyl side chains and 0% hydrophobic groups (fHB = 0) (Figure 1). The highest-molecular-weight polymer tested (DP=30) showed the greatest activity, and activity decreased as MW decreased.
Figure 1.
Antimicrobial activity of cationic homopolymers
The antimicrobial activity of the copolymers depended critically on their hydrophobic content and polymer length. In general, the antimicrobial activity for a given series of hydrophobic side chains depends sigmoidally on fHB (Figure 2). At low values of fHB, the activity of a series of polymers with a given molecular weight will level off at the value expected for the homopolymer. As the mole fraction of the hydrophobic substituents is increased, the activity increases, reaching approximately the same value for all polymers. The amount of data obtainable for polymers with high values of fHB was limited by the limited solubility of the polymers.
Figure 2.
Antimicrobial activity of cationic random copolymers of various molecular weights with A) methyl, B) ethyl, C) butyl, D) hexyls, and E) benzyl groups. The average molecular weights for a given polymer series are indicated in the legend. The Hill Equation [Eq. (1)] was used to fit smooth curves to the data in panels A–C. Fitting parameters are provided in the Supporting Information. [a] Data reported previously.[40]
The sigmoidal shapes of the fHB-versus-MIC curves is well described by the empirical Hill Equation (1):
| (1) |
in which fmid is the midpoint of the curve, n relates to the steepness of the curve, and c1 and c2 relate to the asymptotic limits (see Supporting Information for the fitting parameters). This treatment was used for the C1-, C2-, and C4-polymer series, although the hexyl and benzyl polymers were not soluble over a sufficient range to allow fitting of the parameters. From an examination of the C1 through C4-polymer series, we observed that as the hydrophobic group becomes smaller, the curves become steeper, with a “cooperativity value” of n ranging from approximately 28±10 to 3.5±2 (based on the quality of the fits). Furthermore, as the alkyl-chain length increased the curves shifted to the left, although there was also some dependence on MW; polymers with lower MW and smaller alkyl chains required a higher fHB to achieve maximal efficacy.
On the other hand, the MIC values given in µgmL−1 displayed little or no dependence on polymer length (Supporting Information). The exception was the C4-polymer series that has a wide range of average molecular weights (1600– 8700 MWs) compared to those of other series, which is likely to create the evident dependence of maximum efficacy (lowest MIC values) on the MWs; the lowest-MW polymers in the C4 series displayed the lowest MIC values. This is probably because the low MW of the polymers increases the molar concentration (the number of polymer molecules per volume) for any given weight concentration, which decreases the MICs in µgmL−1 because the MICs in µm (an activity of each polymer molecule) are almost the same for all members of the C4-polymer series at high fHB values.
Hemolytic activity of random copolymers
The hemolytic activity is dependent on the hydrophobic properties of polymers and MW (Figure 3). The HC50 values are limited at low fHB because the hemolytic activity of polymers with low values of fHB was too low to give entire hemolysis curves for HC50 determination in the range of polymer concentrations used here. In general, the hemolytic activity increases as fHB increases and levels off at high values of fHB, especially for the high-MW polymers. The polymers with the largest MW in a given series of hydrophobic groups displayed the highest activity, except the C1 series, in which the activity of the smallest-MW polymers is highest. As the alkyl-chain length increased, the curves shifted to the smaller fHB region, indicating that the polymers with longer alkyl chains are more hemolytic at a given fHB. In contrast to the MICs, the HC50 values given in µgmL−1 displayed the same trends as those in µm (see the Supporting information).
Figure 3.
Hemolytic activity of cationic random copolymers with A) methyl, B) ethyl, C) butyl, D) hexyls, and E) benzyl groups. The average molecular weights for a given polymer series are indicated in the legend. [a] Data reported previously.[40]
The plots of HC50 values versus the number of hydrophobic side chains in a polymer chain (Nside chains=fHB ×DP) for each polymer series showed good correlation (Figure 4, panel A) and linear relationship in the log–log plots (Figure 4, panel B). The data of the C4- and C6-polymer series fit well to a simple power equation, and the data sets and curves of each series of polymers seem to be in parallel with each other. These results suggest that the hemolytic activity of polymers reflects the hydrophobic nature of polymer side chains.
Figure 4.
The HC50 of copolymers as a function of the number of side chains (Nside chains) (A and B) or ∑logP (C and D). Nside chains is defined as fHB × DP. The data for polymers having butyl and hexyl groups were fit to the equation HC50=c3(Nside chains)c4 in which c3 and c4 are constant. The HC50 data for C4- and C6-polymer series in panels C and D were fit to Equation (10). For the calculation of logP using Equation (7), the number of carbon atoms in benzyl groups was set as n=7. See Supporting Information for the fitting parameters.
Theoretical analysis of hemolytic activity
We further explored the hemolytic activity of polymers using our theoretical model to investigate the effect of the hydrophobic nature of polymers on their hemolytic activity. The polymers probably exist in multiple states during the binding and formation of polymer–lipid complexes. Because of its amphiphilic nature, the polymer chain undergoes a hydrophobic collapse or aggregation in an aqueous environment. The proposed models for the antimicrobial action of host-defense peptides suggest that the peptides bind to cell membrane surfaces by combination of electrostatic and hydrophobic interactions, following the insertion of peptides into the hydrophobic region of lipid bilayers. Based on this, the general scheme for the binding and insertion of polymers into lipid membranes and collapse of a polymer chain in solution is shown in Scheme 3.
Scheme 3.

Membrane binding and collapsing of polymers.
In this scheme, P and Pc are defined as free polymers and collapsed polymers inactive to the membranes, respectively. S, P·Sbound, and P·Sinserted relate to binding sites on the membranes, and polymers bound to the sites and inserted into the lipid bilayers, respectively. We assume that the hemolytic activity of polymers reflects the polymers bound to cell membranes, and the hydrophobic collapsing of polymer chains decreases the apparent affinity of polymers to the membranes or reduces the number of polymers interacting with the membranes, resulting in a decrease in hemolytic activity. It is, however, beyond the scope of this report to determine the active species in the mechanism of the hemolytic activity of polymers, and we assume all polymers bound to the membranes are involved in the pore formation causing lysis of red blood cells (RBCs). For this reason, we introduce here a more simplified scheme for creating a theoretical model (Scheme 4).
Scheme 4.

Synthetic scheme of a chain-transfer agent with a dansyl group.
The dissociation constants Kc and Kd′ are defined by Equation (2):
| (2) |
HC50 can be presented as the total polymer concentration [Eq. (3)]:
| (3) |
Here, we define HC50 as the polymer concentration necessary for 50% lysis, at which 50% of the binding sites are occupied by polymers, and thus let [P·S] can be replaced by [S]total × 0.5=[S]1/2 [Eq. (4)]:
| (4) |
Because [S]=[P·S]=[S]1/2 at the polymer concentration= HC50, Kd′ is defined as [Eq. (5)]:
| (5) |
Here, we assume that the binding of polymers to the cell membranes can be described in terms of partitioning of hydrophobic side chains from the aqueous phase to the hydrophobic regions of lipid layers. The octanol/water partition coefficient of substances (P=[substance]octanol/[substance] water, or its decadic logarithm, logP) has been used to predict partitioning of substances or drugs to cells and organs by translocation through cell membranes.[42] Here, we similarly use logP for estimation of partitioning of alkyl side chains into hydrophobic regions of lipid layers. As the logP values for substances are experimentally obtained and previously reported,[43] those of methanol (−0.77), ethanol (−0.31), butanol (0.88), and hexanol (2.03) give the linear relationship defined in Equation (6):
| (6) |
in which n is the number of carbon atoms in the alkyl groups of alcohols. Based on this relationship, we have a relative partition coefficient [logP(n)] for alkyl side chains [Eq. (7)]:
| (7) |
The total partition coefficient of hydrophobic side groups in a polymer chain is given by the summation of logP of alkyl side chains in a polymer, defined as ∑logP=logP(n)× Nside chains. The coefficient Kd′ is further converted as shown [Eq. (8)]:
| (8) |
in which m is a variable to scale the relative logP values to the experimental data, and C0′ refers to the standard state. On the other hand, collapse of the polymer chain reflects the hydrophobicity of side chains, and we attribute the hydrophobicity of polymers to −∑logP [Eq. (9):
| (9) |
in which l is a variable for scaling the curves to the experimental data, and C0″ refers to the standard state. From Equations (5), (8), and (9), we obtain an equation describing the relationship between HC50 and total partitioning coefficient (hydrophobicity) of polymers [Eq. (10)]:
| (10) |
Equation (10) fits well to the HC50 data of the C4- and C6- polymer series, if [S]1/2 is fixed to 0.05 (Figure 4, panels C and D). Although we were unable to obtain good fitting to the data for C1 and C2 polymers, these data points were also found in the vicinity of the fitting curves for the C4-and C6- polymer series. These results indicate that lysis of RBCs reflects the membrane binding of polymers through partitioning of hydrophobic side chains of polymers into lipid layers. In addition, the hemolytic activity of polymers is described by the simple parameter of ∑logP, which is irrelevant to the identity of alkyl groups, suggesting that the hemolytic activity of polymers depends on the total hydrophobicity of polymers rather than the chemical structure of the side-chain groups. The fitting curves display a turnover at high ∑logP regions, at which the polymer chains are likely strongly collapsed or irreversibly aggregated, resulting in large Kc values or a decrease in the apparent affinity of polymers to the membranes. In addition, both C4-and C6-polymer series reached the same HC50 of about 0.07 µm after the turnover. These results indicate that this is the highest hemolytic activity that this polymer system can possibly access by increasing the hydrophobicity of polymers.
Membrane binding of polymers
We also examined the binding behavior of the polymers to lipid bilayers using large unilamellar vesicles (LUVs). The main lipid component of the outer leaflet of human RBCs is neutral phosphatidylcholine (PC) lipids.[44, 45] In contrast, the cell membranes of E. coli consist mainly of neutral phospatidyethylamine (PE) (75–85%) with anionic phospatidylglycerol (PG) (10– 20%) and cardiolipin (CL) (≈5%), and liposaccharides (LPS) (in the outer membrane). The abundance of anionic PG, CL, and LPS in bacterial membranes results in a high density of negative charges on the bacterial cell surface.[46, 47] Based on this, we prepared LUVs from POPC as well as mixtures of POPE/POPG (8:2), POPC/POPG (8:2), or POPE/POPC (8:2) to investigate the effects of charge pairing of cationic polymers and anionic lipids and to examine the response to different chemical structures in the lipids, mimicking the surface properties of cell membranes. (Note that POPE cannot be used as a neutral control as it does not form LUVs in the absence of other lipids, and even in the presence of 20% POPC the vesicles are only stable for a short time.) The fluorescence properties of the dansyl group are sensitive to the surrounding environment: the emission spectrum undergoes a blue-shift, exhibits a narrowing in spectral width, and the quantum yield (overall emission intensity) increases upon transfer from a polar (aqueous) to a nonpolar (hydrophobic) environment.[48] The MIC of dansyl-labeled polymers was close to those of polymers with corresponding fHB and DP in the C4-polymer series in Figure 2, indicating the dye likely has little or no effect on the antimicrobial activity of the compounds.
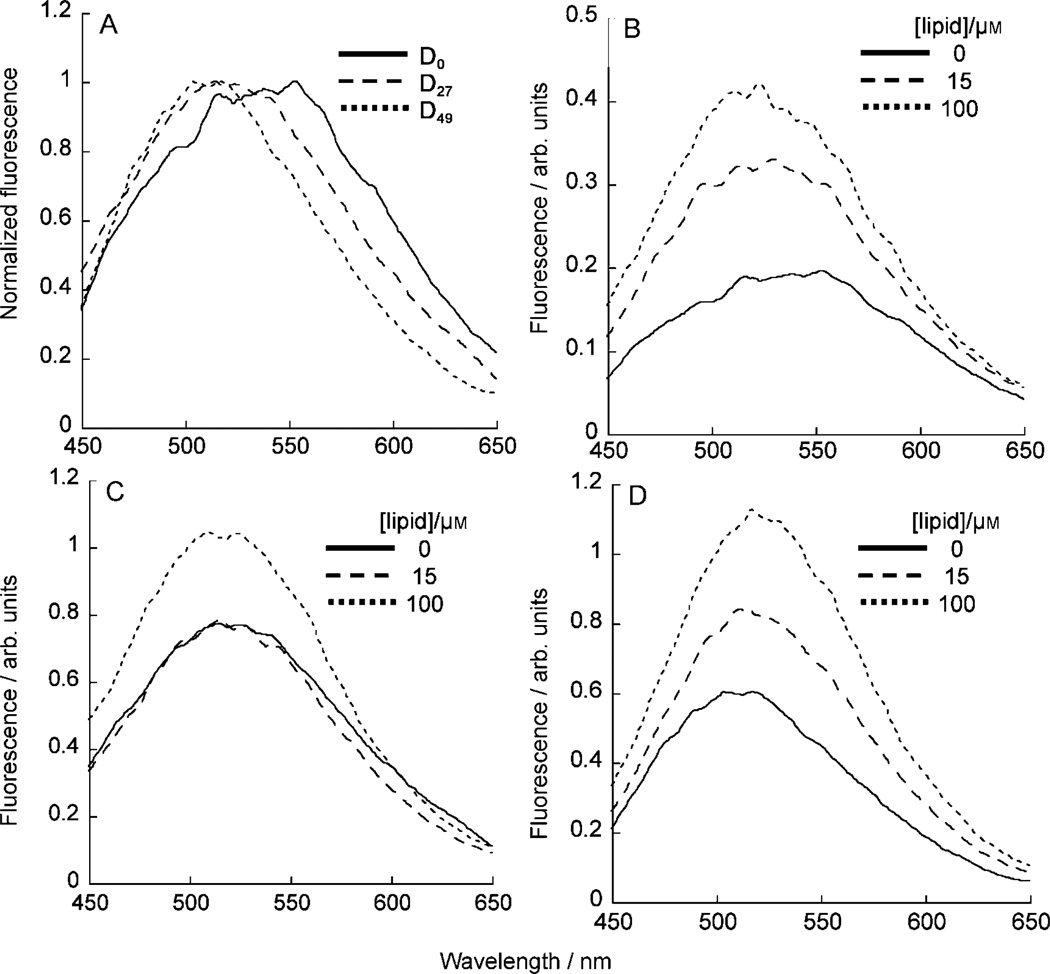
We first examined the inherent effect of polymer chains on the emission properties of the dansyl fluorophore. In the absence of any LUVs, as the fraction of butyl groups increased the fluorescence-emission intensity increased, and the spectra exhibited a blue-shift and narrowing (Figure 5, panel A). This is probably the result of the polymer chain adopting a collapsed conformation by association of butyl side chains in aqueous solution, which was denoted Pc in Schemes 3 and 4, and the dansyl group becoming sequestered in a more hydrophobic pocket of the collapsed polymer chain. We, however, observed a precipitate in the aqueous solution of the most hydrophobic polymer D49 over time (several weeks) and a significant decrease in its fluorescence intensity, whereas other hydrophilic polymers D0 and D27 appear to be stable in solution.
Figure 5.
Emission spectra of dansyl-labeled polymers in the absence or presence of POPC LUVs. A) Normalized emission spectra of D0, D27, and D49 in the absence of LUVs. Emission spectra of B) D0, C) D27, and D) D49 in the presence of 0, 15, and 100-µm POPC
In the presence of lipid (POPC), both the overall fluorescence intensity increased and the spectra narrowed as the lipid concentration increased for all polymers tested. Both observations are consistent with the dansyl group being more shielded from the aqueous environment (Figure 5, panels B–D). The spectra of D0 and D27 blue-shifted in higher lipid concentrations as the dansyl groups were transferred to the hydrophobic region of lipid layers from aqueous solutions or the hydrophobic environment provided by the polymers. In contrast, D49 exhibited a small red-shift in the spectra as lipid concentration increased, indicating that the environment around the dansyl group in the polymer is slightly less polar (more hydrophobic) than that in the polymer–lipid complex. The spectra of all polymers exhibited λmax of similar wavelength (≈530 nm) at 100-µm lipid, suggesting that the dansyl groups are eventually located in a similar hydrophobic environment of polymer–lipid complex in high lipid concentrations. The spectra of polymers for LUVs consisting of POPC/POPG and POPE/POPC displayed a trend similar to that of POPC and exhibited λmax values within the same wavelength region (≈530 nm) at high lipid concentrations (see Supporting Information).
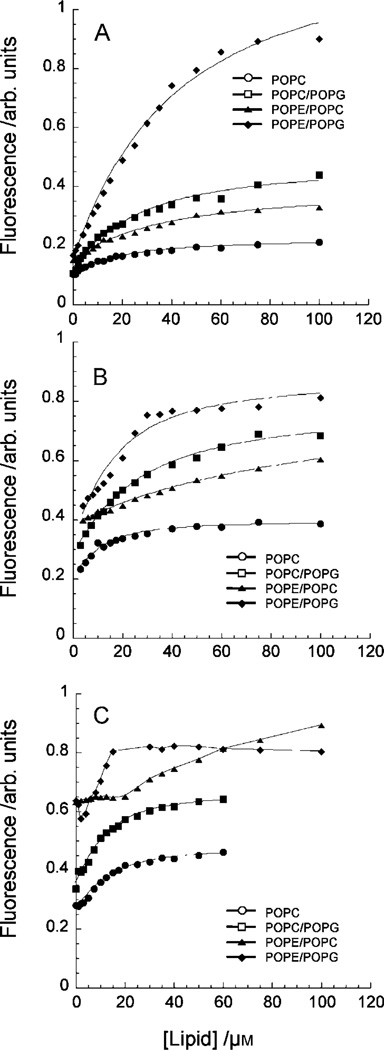
We obtained binding isotherms of polymers by monitoring an increase in emission intensity at 500 nm from the dansyl group by titrating LUVs into the sample (Figure 6). The fluorescence-emission data were fit to a single site-binding isotherm [Eq. (13)], Experimental Section) to give the overall dissociation constant, Kd″, in the equilibrium between the polymers in solution and the polymer–lipid complex, assuming equilibrium between Pc and P·S (Scheme 4). The binding behavior of the polymers to LUVs depends on both the hydrophobic content of the polymers and the chemical structures of lipids. Polymer D0 displayed higher Kd″ values for LUVs containing an anionic POPG lipid than those for their neutral controls (Table 3). This result indicates that the affinity of the polymer to neutral lipid bilayers is higher than that to negatively charged bilayers. On the other hand, the LUVs containing a POPE lipid showed relatively larger Kd″ values than those for LUVs of POPC and POPC/POPG. Interestingly, the difference between neutral lipids (POPC and POPE), which are the major components of LUVs, seems to be a primary factor in the binding of cationic polymers rather than the population of negative charges in the membrane surfaces, even though the effect of charge coupling between cationic side chains of polymer and anionic lipids is expected to positively contribute to the formation of polymer–lipid complex. These data suggest that the properties of lipids play an important role in the affinity of polymers as well as their specificity.
Figure 6.
Binding isotherms of dansyl-labeled polymers A) D0, B) D27, and C) D49.
Table 3.
Dissociation constants of the membrane binding of polymers
| Polymer | POPC | POPC/POPG | Kd’’ [μm] POPE/POPC |
POPE/POPG |
|---|---|---|---|---|
| D0 | 2.27 | 3.30 | 5.00 | 5.59 |
| D27 | 0.83 | 3.27 | 15.85[a] | 1.76 |
| D49 | 1.08 | 0.87 | n.d.[a],[b] | n.d.[b] |
The isotherms displayed at least two different modes of binding at lower (0–20 μm) and higher lipid concentrations.
Not determined due to low quality of fitting.
In general, the Kd″ values for all lipid types except POPC/ POPG decrease as the mole fraction of butyl groups of polymers increases, indicating that hydrophobicity of polymers enhances the binding of polymers to lipid bilayers. The isotherms of D27 and D49 for POPE/ POPC displayed at least two different modes of binding in the low (0–20 µm) and high (>20 µm) lipid concentrations that yield a relatively large Kd″ value of 15.85 for D27, which deviates from that of other polymers. We were unable to obtain good fitting to the data for D49 due to the multiple modes of binding. The polymers displayed lower Kd″ values for POPC than that for POPE/POPG, indicating that these polymers bind RBC-type membranes in preference to bacterial cell membranes. In addition, the differences in Kd″ diminishes as hydrophobicity of the polymers increases, implying that their binding to lipids is no longer dependent on the lipid contents when the hydrophobic polymer–lipid interaction is dominant in the complex formation. Polymers D27 and D49 displayed similar Kd″ values for their binding to POPC, suggesting a further increase in hydrophobic content does not affect the affinity of polymers to lipid bilayers. This is likely due to a hydrophobic collapse of the polymer chain in aqueous media or irreversible aggregation, preventing interactions with lipid layers. This binding behavior of polymers is consistent with the trends in the antimicrobial and hemolytic activities and suggests that the membrane-disruption mechanism correlates to the hydrophobic binding of polymers to lipid bilayers. It is interesting that the Kd″ displayed a maximum increase of only four-fold as the fraction of hydrophobic (butyl) groups of polymers increased to from 0 to 0.49, whereas the HC50 varies by up to 100-fold. This indicates that the activity of polymers should be interpreted as the result of a combination of both the binding of polymers and pore formation, a mechanism that also likely reflects the amount of polymers bound to the membranes as well as the hydrophobic nature of polymers.
We also examined the effect of the formation of the polymer– lipid complex on fluorescence intensity by comparing emission-intensity values in the absence of lipid and once the polymer was fully bound to LUVs. The level of enhancement in fluorescence intensity shows a dependence on lipid composition (Figure 6), although the fluorescence spectra of polymers in high lipid concentrations are similar (Figure 5). In all cases the polymers bound to POPE/POPG vesicles exhibited the greatest enhancement, and those bound to POPC LUVs showed the least enhancement. These results indicate that the microenvironments around the dansyl groups in the polymer–lipid complex seem to reflect the membrane-bound conformation of polymers, that is, how the polymers physically interact with the membrane surface (P·Sbound) and insert into the bilayer (P·Sinserted) (Scheme 3). The presence or absence of charged groups on both the membrane surface and the polymer chain can affect the exact location at which the polymer and, therefore, the dansyl group is oriented in the membrane. Small changes in fluorophore depth have been shown to affect fluorescence properties; for example, a dansyl residing in the headgroup region could show slightly different emission properties to a dansyl group either oriented at the interfacial region or that becomes buried in the nonpolar core of the bilayer upon binding the vesicle. Additionally, as the polymers become hydrophobic, the value of Finf/F0 decreases (see Table S11 in the Supporting Information). It could be possible that for more hydrophobic polymers F0 is larger than Finf, resulting in a decrease in Finf/F0.
In summary, the hydrophobicity of polymers enhances binding to lipid bilayers and induces a collapse of polymer chains in solution, causing a decrease in the apparent affinity of polymers to the LUVs. In addition, the binding of polymers to lipid bilayers depends on the overall hydrophobicity of polymers as well as the chemical properties of the lipid headgroups. The activity of polymers is due to both membrane binding of polymers and pore formation induced by polymers.
Membrane binding and antimicrobial activity of polymers
According to the results of membrane binding of polymers, the hydrophobic interaction between polymers and lipid bilayers drives the binding of polymers to both bacteria and erythrocytes. However, the affinity of polymers to POPC LUVs is higher than that to POPE/POPG LUVs (Table 3), indicating bacteria would be more resistant than erythrocytes to the binding of polymers. Because the antimicrobial activity of polymers levels off for highly hydrophobic polymers, displaying a MIC of 5–10 µm (Figure 2), the antimicrobial action also likely depends on the balance between the membrane binding and collapsing of polymers driven by the polymer hydrophobicity as shown in the hemolysis. Therefore, the bacterial resistance to the binding of polymers could cause a turnover in MIC at lower hydrophobicity of polymers, which is seen in the HC50 for more hydrophobic polymers (Figure 4, panels C and D). This earlier turnover in the MIC of polymers increases the minimum MIC values (decreases the maximum efficacy) and reduces the selectivity of activity towards bacteria over erythrocytes. Considering these results, the rational design of polymer structures and a fine tuning of their properties are necessary for the further development of non-toxic synthetic polymers.
MIC and HC50 of polymers
Although the hemolytic activity is driven by the hydrophobic nature of polymers, the amphiphilic balance seems to be important in their antimicrobial activity (Figure 2) and selectivity towards bacteria over RBCs.[40] To examine a relationship between these activities of the polymers, the MIC and HC50 values were plotted in Figure 7. The MIC values are mostly within the range of 5–20 µm for all polymers, whereas the HC50 values range from about 0.03 to 100 µm. This is reflected in the different dependencies of MIC and HC50 on the fHB values of the polymers: the HC50 values for most of the polymer series decrease as fHB increases, whereas the MICs of all polymers reached approximately the same value at high fHB (Figures 2 and 3). Thus, polymers with methyl side chains show selectivity towards E. coli over RBCs with a maximum selectivity index (HC50/MIC) of 10 and a high antimicrobial activity (MIC=5–10 µm).
Figure 7.
Relationship between HC50 and MIC of copolymers. Selectivity index (SI) is defined as HC50/MIC. Polymers with MIC<10 µm and SI<1 are suitable for preparation of biocides due to their high activity against bacteria and human cells. Those with MIC<10 µm and SI>1 display efficacy selective towards bacterial cells and are useful as non-toxic antimicrobials.
Important properties of synthetic antimicrobials are not only high activity against bacteria but also the lack of toxicity to human cells. To this end, the activity profile in Figure 7 provides general information on the optimal composition of polymers for a given application, which is useful for the future design and manufacturing of polymers. In the case of preparation of polymeric biocides for cleaning or sanitizing surfaces, polymers are required to have high antimicrobial activity (lowest MIC). Because their toxicity to human cells is not crucial, the polymers at the bottom left region (MIC < 10 µm and selectivity <1), which contain mainly C2- and C4- polymer series, will be useful for this purpose. On the other hand, for polymers used as antibacterial agents in cosmetics, food, and house paints, their toxicity to human cells should be avoided. To this end, polymers displaying possibly lowest MIC and high HC50 or high selectivity (>1) will be available in the polymer series with methyl side chains (top left region).
Conclusion
We have investigated the optimal balance of hydrophobic and cationic side chains for the antimicrobial and hemolytic activities of amphiphilic polymethacrylate copolymers. The polymers vary systematically in the nature of their hydrophobic groups, composition, and length. Analysis of membrane binding and efficacy indicates that the hydrophobic nature of polymers is a key factor in controlling the antimicrobial and hemolytic activities. This reflects the balance between the polymers bound to lipid bilayers, which are membrane- active and cause lysis, and hydrophobically collapsed polymers, which are not interacting with membranes. In addition, the membrane binding of polymers strongly depends on the lipid properties, and bacteria could be more resistant to the binding of polymers than erythrocytes, resulting in a lack of selective activity towards bacteria over erythrocytes. This may be an inherent property of these synthetic polymers and reflects the need for careful engineering in the design of non-toxic antimicrobial polymers.
Experimental Section
Materials
The bee-venom toxin melittin (85%) was purchased from Sigma–Aldrich. All other chemicals and solvents were obtained from Sigma–Aldrich and Fisher Scientific and used without further purification.
Polymer synthesis
The polymers, except those having methyl (C1-3.3, 2.0, and 1.6) and ethyl groups (C2-2.9(2), and 1.8) were prepared according to procedures reported previously,[40] in which the Boc-protected precursor polymers were isolated by precipitation in diethyl ether, followed by Boc-deprotection by TFA, yielding cationic polymers. For the polymers with methyl (C1-3.3, 2.0, and 1.6) and ethyl groups (C2-2.9(2) and 1.8), the crude polymers were deprotected by treatment with TFA without isolation by precipitation in diethyl ether of the Boc-protected precursor polymers. The experimental procedure is described briefly as follows: N-(tert-butoxycarbonyl)aminoethyl methacrylate and alkyl or benzyl methacrylate (0.435 mmol total) were polymerized in acetonitrile (0.5 mL) containing 2,2′-azobisisobutyronitrile (AIBN) (0.716 mg, 4.35µmol) and methyl 3-mercaptopropionate (MMP) as a chain-transfer agent at 60–70 °C. The solvents were removed under reduced pressure, and the obtained crude polymer was dissolved in TFA and stirred for 30 min. After removing TFA by N2 gas perfusion, the oily product was dissolved in a small amount of methanol and added into diethyl ether. The resultant precipitate was collected by centrifugation and lyophilized to give cationic copolymers as a white powder. Note that polymers having high MWs and a high percentage of cationic groups selectively precipitate in diethyl ether. As a result, the MWs of the collected polymers are higher than those of the Boc-protected precursors, and the hydrophobic content is less than 60–70% in general, whereas the hydrophobic content of the Boc-protected precursors is greater. 1H NMR analysis provided determination of the degree of polymerization (DP) and the mole percentage of alkyl methacrylate, fHB, of the polymers (see Supporting Information for more details).
Synthesis of a chain-transfer agent with a dansyl group
Dansyl chloride (500 mg, 1.85 mmol) was added to cystamine dihydrochloride (209 mg, 0.97 mmol) in DMF (15 mL) and triethylamine (0.78 mL, 5.6 mmol). The reaction mixture was stirred at RT overnight. After the solvent was evaporated under reduced pressure, the residue was taken up by ethylacetate and washed in water. The aqueous layer was back-washed in ethylacetate. The combined organic layer was washed in 1n NaOH, 10% citric acid, sat. NaHCO3, and brine. The organic layer was dried over Na2SO4, and the solvent was evaporated under reduced pressure. The product was purified by silica-gel column chromatography (EtOAc/hexane 1:1) to give product 1 (450 mg, 75%); 1H NMR (500 MHz, CDCl3): δ=8.56 (d, 1H), 8.25 (m, 2H), 7.53 (m, 2H), 7.19 (d, 1H), 5.10 (brm, 1 H), 3.11 (m, 2H), 2.89 (s, 6 H), 2.50 ppm (t, 2H).
Tris-(2-carboxyethyl)phosphine (TCEP) (38 mg, 0.13 mmol) was added to 1 (60 mg, 0.097 mmol) in methanol (1 mL). After addition of 3 drops of water with a glass pipette, the reaction mixture was stirred at RT overnight. The solvent was evaporated under reduced pressure, and the residue was dissolved in CH2Cl2 and washed by water and brine. The organic layer was dried over Na2SO4, and the solvent was evaporated under reduced pressure. The crude product was purified by silica-gel column chromatography (EtOAc/hexane 1:1) to give product 2. 1H NMR (500 MHz, CDCl3): δ=8.57 (d, 1H), 8.28 (m, 2 H), 7.59 (t, 1H), 7.53 (t, 1H), 7.21 (d, 1 H), 5.13 (brm, 1H), 3.10 (m, 2 H), 2.90 (s, 6H), 2.53 ppm (m, 2H); HRMS (ESI): calcd for C14H18N2O4S2 : 310.0810 [M+]; found: 310.0805.
Synthesis of copolymers labeled with a dansyl group
Cationic homo-/copolymers labeled with a dansyl group at the polymer end were prepared by the procedure used for the synthesis of copolymers previously described. The monomers (0, 30, 50 mol% of butyl methacrylate relative to total amount of monomers, 0.161 mmol) were polymerized by using AIBN in the presence of a chain-transfer agent 2 with a dansy group (5 mg, 0.016 mmol). The resultant polymers were treated by TFA to remove Boc groups. After removing TFA by N2 gas perfusion, the oily residue was dissolved in a small amount of methanol and added into diethyl ether. The resultant precipitate was collected by centrifugation and lyophilized to give cationic copolymers as a white powder. 1H NMR analysis provided determination of the degree of polymerization (DP) and the mole fraction of alkyl methacrylate, fHB, of the polymers. 1H NMR (500 MHz, CD3OD): for the representative polymer (fHB=49 and DP= 11.8): δ=8.62 (brs, 1H), 8.35 (brs, 1H), 8.24 (brs, 1H), 7.63 (brs, 2H), 7.32 (brs, 1H), 3.67 (brs, 12.07 H), 4.02 (brs, 11.53 H), 3.01 (brs, 1.46 H), 2.92 (s, 6H), 2.3–1.8 (brm, 20.32 H), 1.8–0.8 ppm (brm, 85.73 H).
Antimicrobial testing
A protocol for an antimicrobial test was reported previously.[40] A bacterial strain, Escherichia coli D31 (ampicillin- and streptomycin-resistant) was grown in Mueller–Hinton broth (MH broth) at 37 °C overnight. Turbidity of the E. coli culture was measured as optical density at λ=600 nm (OD600) in a 1-mL plastic disposable cuvette (1-cm path length) using an Eppendorf BioPhotometer. This cell culture was diluted with MH broth to give a cell suspension (20 mL) with OD600=0.1, which was incubated at 37 °C for 1.5 h. Healthy cell growth was confirmed by measuring OD600 (valued between 0.5 and 0.6). This cell suspension was diluted to give a bacterial stock solution with OD600=0.001, and this E. coli stock (90 µL) was added to each well in a 96-well, sterile assay palate (Costar Clear Polystyrene 3370, Corning). The assay plate was incubated at 37°C for 18 h without shaking. Bacterial growth was detected at OD595 using ThermoLabsystems Multiskan Spectrum and was compared to that of MH broth without copolymers and E. coli. MIC was defined as the lowest polymer concentration to completely inhibit bacterial growth in at least two samples of triplicate measurements.
Hemolysis assay
A hemolysis-assay protocol was reported previously.[40] Human red blood cells (RBCs) were obtained by centrifuging a whole-blood sample and removing plasma and white blood cells. The RBCs (1 mL) were diluted with TBS buffer (9 mL, 10 mm Tris buffer, pH 7.0, 150 mm NaCl) and this suspension (0.75 mL) was further diluted by TBS buffer (33 mL) to give a RBC stock suspension. This RBC stock (90 µL), and the polymer stock solutions (10 µL, [polymer]=10–0.0003 mgmL−1) were added to a 96-well, round-bottomed microplate and incubated at 37°C for 1 h with shaking. The range of final polymer concentrations in the microplate was 1 mgmL−1–0.03 µgmL−1. The microplate was centrifuged at 4000 ppm for 5 min. Supernatant (30 µL) was diluted with TBS buffer (100 µL), and OD414 of the solution was measured as hemoglobin concentration. Melittin was used as a positive control, and the most concentrated sample (100 µgmL−1) was used as a reference for 100% hemolysis. Percentage of hemolysis (P) was calculated from Equation (11):
| (11) |
HC50 was obtained as the polymer concentration at 50% hemolysis, which was estimated by curve fitting using Equation (12):
| (12) |
in which P(Cp) and K form a hemolysis curve for a given polymer concentration (Cp), and HC50, respectively. K and n are variable parameters in the curve fitting. The polymers with a low percentage of hydrophobic groups displayed relatively low hemolytic activity, and provided HC50 values around or larger than the highest polymer concentration used here (1 mgmL−1), which is not presented in Figure 3 because the limited data points did not present reliable intra- or extrapolate estimation. Under these assay conditions, the HC50 of melittin was 1.25±0.64 µgmL−1 (n=10). The HC50 reported is the average value of at least two independent experiments, except the data for C2-2.9(2), for which only one experiment was performed.
Steady-state fluorescence spectroscopy
Fluorescence spectra and intensity measurements were recorded by using an Aviv Automated Titrating Differential/Ratio Spectrofluorometer (Aviv Biomedical, Lakewood, NJ). For liposome-binding experiments, the excitation and emission bandpass were both set to 2 nm, whereas for emission-spectrum measurements, the excitation bandpass was set to 3 nm and the emission bandpass was set to 1.5 nm. For liposome-binding experiments the excitation and emission wavelengths were 353 and 500 nm, respectively, for dansyl-labeled polymers. Dansyl-labeled-polymer fluorescence-emission spectra were recorded by using an excitation wavelength of 353 nm and monitoring the emission spectra from 450–600 nm. All experiments were performed in 1×1- cm quartz semi-microcuvettes with constant stirring by a magnetic stir-flea. Background intensities or spectra were subtracted from all samples.
Liposome preparation
Lipids were purchased from Avanti Polar Lipids, Inc. (Alabaster, Al) and used without further purification. Liposomes were composed of either 100% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC); POPC and 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho- rac-(1-glycerol)] (POPG) (8:2 mol/mol); 1-palmitoyl-2-oleoyl-sn-glycero- 3-phosphoethanolamine (POPE) and POPC (8:2 mol/mol); or POPE and POPG (8:2 mol/mol). Lipids dissolved in chloroform were mixed in the appropriate ratios and first dried under a stream of N2, then further dessicated under vacuum for at least 3 h to remove any remaining chloroform. The dried lipid film was then stored under N2 at −20°C until used. To form liposomes, the lipid film was rehydrated in HBS buffer (10 mm HEPES, 100 mm NaCl, pH 7.1) to obtain a final lipid concentration of 10 mm (a final volume of 0.5 mL was typically used). The sample was vortexed for at least 3 min to ensure complete resuspension of the lipid film. The sample was then subjected to five rounds of freezing in a dry-ice/acetone bath and thawing at 37 °C in a heated water bath. The sample was then passed 21 times through a Liposofast extruder (Avestin, Inc., Ottowa, Canada) with two stacked polycarbonate membranes, each with a pore size of 400 nm. POPC and POPC/POPG liposomes were stored at 4 °C and used for up to 1 week after preparation, whereas POPE/POPG and POPE/POPC liposomes were used immediately after extrusion.
Polymer-binding assay
Polymer binding to liposomes was assayed by monitoring changes in the emission spectrum of dansyl. An aliquot of polymer from a concentrated stock in water was added to HBS buffer to a final concentration of 2 µm polymer in 800 L, and was allowed to equilibrate for 5 min at which point the dansyl fluorescence-emission intensity was recorded. The sample was then titrated with aliquots of liposomes (typically 1–5 µL), allowed to equilibrate for 5 min with constant mixing by means of a microstir bar, and fluorescence emission was recorded again. Intensities were corrected for dilution and for the inner filter effect at the highest lipid concentrations. For binding-constant analysis, fluorescence-emission data were fit to a single site-binding isotherm [Eq. (13)] by using the KaleidaGraph software package:
| (13) |
in which [P]T and [L]T are total polymer and lipid concentrations, respectively, F0 is fluorescence intensity at [L]=0, and n is the number of lipids per polymer-binding site, which was held fixed at n=6. This was chosen to allow good data fitting for all tested polymers, but similarly good fits were obtained using n=4–9. ΔF is defined as ΔF=Finf–F0, in which Finf is the fluorescence intensity at [L]=infinite, which corresponds to all polymers being completely bound to LUVs.
Supplementary Material
Acknowledgements
This work was funded primarily by AI74866 and partially by GM54616. We thank NIH and PolyMedix, Inc for financial support. We also acknowledge use of a microplate reader at Dr. Dewey McCafferty’s laboratory at the Department of Biochemistry and Biophysics at the University of Pennsylvania.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/chem.200800xxx.
References
- 1.Levy SB, Marshall B. Nat. Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 2.Nathan C. Nature. 2004;431:899–902. doi: 10.1038/431899a. [DOI] [PubMed] [Google Scholar]
- 3.Lewis K. Nat. Rev. Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 4.Wright GD. Nat. Rev. Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 5.Zasloff M. Proc. Natl. Acad. Sci. USA. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 7.Brogden KA. Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 8.Hancock REW, Sahl HG. Nat. Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 9.Hancock REW, Rozek A. FEMS Microbiol. Lett. 2002;206:143–149. doi: 10.1111/j.1574-6968.2002.tb11000.x. [DOI] [PubMed] [Google Scholar]
- 10.Powers JPS, Hancock REW. Peptides. 2003;24:1681–1691. doi: 10.1016/j.peptides.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 11.Hilpert K, Volkmer-Engert R, Walter T, Hancock REW. Nat. Biotechnol. 2005;23:1008–1012. doi: 10.1038/nbt1113. [DOI] [PubMed] [Google Scholar]
- 12.Shai Y. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 13.Dathe M, Wieprecht T. Biochim. Biophys. Acta Biomembr. 1999;1462:71–87. doi: 10.1016/s0005-2736(99)00201-1. [DOI] [PubMed] [Google Scholar]
- 14.Mecke A, Lee DK, Ramamoorthy A, Orr BG, Holl MMB. Biophys. J. 2005;89:4043–4050. doi: 10.1529/biophysj.105.062596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porcelli F, Buck-Koehntop BA, Thennarasu S, Ramamoorthy A, Veglia G. Biochemistry. 2006;45:5793–5799. doi: 10.1021/bi0601813. [DOI] [PubMed] [Google Scholar]
- 16.Goodman CM, Choi S, Shandler S, DeGrado WF. Nat. Chem. Biol. 2007;3:252–262. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 18.Porter EA, Wang X, Lee HS, Weisblum B, Gellman SH. Nature. 2000;405:298–298. doi: 10.1038/35007145. [DOI] [PubMed] [Google Scholar]
- 19.Hamuro Y, Schneider JP, DeGrado WF. J. Am. Chem. Soc. 1999;121:12200–12201. [Google Scholar]
- 20.Liu DH, DeGrado WF. J. Am. Chem. Soc. 2001;123:7553–7559. doi: 10.1021/ja0107475. [DOI] [PubMed] [Google Scholar]
- 21.Patch JA, Barron AE. J. Am. Chem. Soc. 2003;125:12092–12093. doi: 10.1021/ja037320d. [DOI] [PubMed] [Google Scholar]
- 22.Tew GN, Liu DH, Chen B, Doerksen RJ, Kaplan J, Carroll PJ, Klein ML, DeGrado WF. Proc. Natl. Acad. Sci. USA. 2002;99:5110–5114. doi: 10.1073/pnas.082046199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu DH, Choi S, Chen B, Doerksen RJ, Clements DJ, Winkler JD, Klein ML, DeGrado WF. Angew. Chem. 2004;116:1178–1182. doi: 10.1002/anie.200352791. Angew. Chem. Int. Ed.2004, 43, 1158–1162. [DOI] [PubMed] [Google Scholar]
- 24.Arnt L, Nusslein K, Tew GN. J. Polym. Sci. Polym. Chem. Ed. 2004;42:3860–3864. [Google Scholar]
- 25.Nusslein K, Arnt L, Rennie J, Owens C, Tew GN. Microbiology-Sgm. 2006;152:1913–1918. doi: 10.1099/mic.0.28812-0. [DOI] [PubMed] [Google Scholar]
- 26.Tashiro T. Macromol. Mater. Eng. 2001;286:63–87. [Google Scholar]
- 27.Gabriel GJ, Som A, Madkour AE, Eren T, Tew GN. Mater. Sci. Eng. R. 2007;57:28–64. doi: 10.1016/j.mser.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenawy ER, Worley SD, Broughton R. Biomacromolecules. 2007;8:1359–1384. doi: 10.1021/bm061150q. [DOI] [PubMed] [Google Scholar]
- 29.Gelman MA, Weisblum B, Lynn DM, Gellman SH. Org. Lett. 2004;6:557–560. doi: 10.1021/ol036341+. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda T, Tazuke S. Makromol. Chem.-Rapid. Commun. 1983;4:459–461. [Google Scholar]
- 31.Chen Y, Worley SD, Huang TS, Weese J, Kim J, Wei CI, Williams JF. J. Appl. Polym. Sci. 2004;92:363–367. [Google Scholar]
- 32.Tiller JC, Liao CJ, Lewis K, Klibanov AM. Proc. Natl. Acad. Sci. USA. 2001;98:5981–5985. doi: 10.1073/pnas.111143098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sellenet PH, Allison B, Applegate BM, Youngblood JP. Biomacromolecules. 2007;8:19–23. doi: 10.1021/bm0605513. [DOI] [PubMed] [Google Scholar]
- 34.Varun Sambhy BRPAS. Angew. Chem. 2008;120:1270–1274. Angew. Chem. Int. Ed.2008, 47, 1250–1254. [Google Scholar]
- 35.Kenawy ER, Abdel-Hay FI, El-Raheem A, El-Shanshoury R, El-Newehy MH. J. Controlled Release. 1998;50:145–152. doi: 10.1016/s0168-3659(97)00126-0. [DOI] [PubMed] [Google Scholar]
- 36.Lenoir S, Pagnoulle C, Detrembleur C, Galleni M, Jerome R. J. Polym. Sci. Polym. Chem. 2006;44:1214–1224. [Google Scholar]
- 37.Lu GQ, Wu DC, Fu RW. React. Funct. Polym. 2007;67:355–366. [Google Scholar]
- 38.Ilker MF, Nusslein K, Tew GN, Coughlin EB. J. Am. Chem. Soc. 2004;126:15870–15875. doi: 10.1021/ja045664d. [DOI] [PubMed] [Google Scholar]
- 39.Mowery BP, Lee SE, Kissounko DA, Epand RF, Epand RM, Weisblum B, Stahl SS, Gellman SH. J. Am. Chem. Soc. 2007;129:15474–15476. doi: 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- 40.Kuroda K, DeGrado WF. J. Am. Chem. Soc. 2005;127:4128–4129. doi: 10.1021/ja044205+. [DOI] [PubMed] [Google Scholar]
- 41.Matsuzaki K, Nakamura A, Murase O, Sugishita K, Fujii N, Miyajima K. Biochemistry. 1997;36:2104–2111. doi: 10.1021/bi961870p. [DOI] [PubMed] [Google Scholar]
- 42.Hansch CL, Leo A. Exploring QSAR. Washington DC: American Chemical Scoiety; 1995. [Google Scholar]
- 43.Sangster J. J. Phys. Chem. Ref. Data. 1989;18:1111–1229. [Google Scholar]
- 44.Verkleij AJ, Zwaal RFA, Roelofse B, Comfuriu P, Kastelij D, Van Deene L. Biochim. Biophys. Acta. 1973;323:178–193. doi: 10.1016/0005-2736(73)90143-0. [DOI] [PubMed] [Google Scholar]
- 45.Zwaal RFA, Comfurius P, Vandeenen LLM. Nature. 1977;268:358–360. doi: 10.1038/268358a0. [DOI] [PubMed] [Google Scholar]
- 46.Cronan JE. Annu. Rev. Microbiol. 2003;57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- 47.Wilkinson SG. In: Microbiral Lipids. Ratledge C, Wilkinson SG, editors. Vol. 1. London: Acadmic Press; 1988. p. 299. [Google Scholar]
- 48.Li YH, Chan LM, Tyer L, Moody RT, Himel CM, Hercules DM. J. Am. Chem. Soc. 1975;97:3118–3126. [Google Scholar]
- 49.Caputo GA, London E. Biochemistry. 2003;42:3265–3274. doi: 10.1021/bi026696l. [DOI] [PubMed] [Google Scholar]
- 50.Kachel K, Asuncion-Punzalan E, London E. Biochim. Biophys. Acta Biomembr. 1998;1374:63–76. doi: 10.1016/s0005-2736(98)00126-6. [DOI] [PubMed] [Google Scholar]
- 51.Kaiser RD, London E. Biochim. Biophys. Acta Biomembr. 1998;1375:13–22. doi: 10.1016/s0005-2736(98)00127-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.