Abstract
In many plants, major developmental transitions such as the initiation of flowering are synchronized to the changing seasons. Day length provides one of the environmental cues used to achieve this. We describe the molecular mechanisms that measure day length and control flowering in Arabidopsis. Also, we compare these mechanisms with those that control flowering time in rice. This comparison suggests that components of the Arabidopsis regulatory network are conserved in other species, but that their regulation can be altered to generate different phenotypic responses.
Keywords: Arabidopsis, circadian clock, flowering, photoperiodism, vernalization
Introduction
The life cycle of many plants is synchronized to the changing seasons. This pattern of behaviour ensures that developmental transitions, such as the onset of flowering, occur under the most appropriate environmental conditions and in many locations is an essential aspect of the sessile growth habit of plants. Fluctuations in day length (or photoperiod) and temperature provide the information used to synchronize these developmental decisions to the seasons. The mechanism underlying photoperiodic responses has been of interest since they were first described in detail in the 1920s (Garner and Allard, 1920). A conceptual breakthrough was the realization that a circadian clock, an endogenous timing mechanism with a cycle time or period length of approximately 24 h, is the time-keeping mechanism required to measure day length (Bünning, 1936; Thomas and Vince-Prue, 1997). This was later refined as a coincidence model in which exposure of a plant to light at a particular phase of a circadian rhythm would trigger or repress a developmental transition (Pittendrigh and Minis, 1964). Such a system would consist of two parts: a circadian rhythm in a component that regulates the developmental response and whose activity is controlled by exposure of the plant to light, and a light signalling pathway that activates or represses the activity of this component. Genetic analysis of the control of flowering has identified genes that confer a photoperiodic response on Arabidopsis and suggested a molecular basis for the coincidence between circadian rhythms and light (Hayama and Coupland, 2003; Yanovsky and Kay, 2003). In this review, we describe the mechanisms that underlie the response to day length in Arabidopsis, and how these are modified in other plant species.
A regulatory pathway that induces flowering of Arabidopsis in response to photoperiod
Arabidopsis shows a strong photoperiod response in the onset of flowering, and most strains (or accessions) flower in spring or early summer as the days become longer. In laboratory conditions, flowering occurs much earlier under long days of 16 h light than under short days of 10 h light. Mutations that disrupt these responses were isolated by identifying mutants with a reduced response to day length (Redei, 1962; Koornneef et al, 1991). These mutants fell into two classes, those that flower later than wild-type plants under long days but are unaffected under short days or, alternatively, early-flowering mutants under short days.
Some of the mutations that cause early flowering under short days also cause a general disruption of circadian rhythms. In Arabidopsis, behaviours such as leaf movements or the elongation of cells in the hypocotyl (Dowson-Day and Millar, 1999) as well as the expression of around 6% of genes (Harmer et al, 2000; Schaffer et al, 2001) are under circadian clock control. The mutations that reduce day-length responses by causing early flowering under short days also cause a general disruption of these circadian rhythms. These include mutations in the EARLY FLOWERING3 (ELF3), TIMING OF CHLOROPHYLL A/B BINDING PROTEIN1 (TOC1), LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED1 (CCA1) genes (). The expression of these genes is also regulated by the circadian clock, so that their mRNAs only accumulate in the morning (LHY and CCA1) or the evening (TOC1 and ELF3).
LHY, CCA1 and TOC1 may be part of the central mechanism that generates circadian rhythms in plants. CCA1 and LHY are similar in sequence and expression pattern (Schaffer et al, 1998; Wang and Tobin, 1998), and are genetically partially redundant (Alabadi et al, 2002; Mizoguchi et al, 2002). In the lhy cca1 double mutant or the toc1 single mutant, circadian rhythms cycle faster and the plants flower earlier under short days than wild-type plants (Somers et al, 1998; Mizoguchi et al, 2002). LHY and CCA1 were proposed to act along with TOC1 in a transcriptional feedback loop in which TOC1, which is expressed only in the evening, promotes the expression of LHY/CCA1 at dawn, and in turn LHY/CCA1 repress the expression of TOC1 (Alabadi et al, 2001). In contrast, ELF3 does not appear to encode a central component of the circadian clock but modulates light signalling to the oscillator, so that exposure of elf3 mutants to continuous light or long photoperiods stops circadian rhythms (McWatters et al, 2000; Hicks et al, 2001; Liu et al, 2001b).
Mutations that reduce day-length responses by delaying flowering under long days define a set of circadian-clock-regulated genes. These include the CONSTANS (CO), GIGANTEA (GI) and FLOWERING LOCUS T (FT) genes, which were initially placed in the same genetic pathway based on their mutant phenotypes and the genetic interactions between the mutations (Koornneef et al, 1991). All of these genes have now been cloned (Table I), and are circadian clock regulated. Transgenic overexpression of each of the genes in this group causes early flowering (Kardailsky et al, 1999; Kobayashi et al, 1999; Borner et al, 2000; Lee et al, 2000; Onouchi et al, 2000; Samach et al, 2000).
Table 1.
Proteins involved in the response to day length in Arabidopsis
| Proteins | Putative biochemical function | References |
|---|---|---|
| EARLY FLOWERING3 (ELF3) | Nuclear protein proposed to act as a transcriptional activator | McWatters et al (2000), Hicks et al (2001) and Liu et al (2001b) |
| TIMING OF CHLOROPHYLL A/B BINDING PROTEIN1 (TOC1) | N-terminus is similar to the receiver domain of bacterial response regulators; C-terminus is the plant-specific CCT domain | Strayer et al (2000) |
| LATE ELONGATED HYPOCOTYL (LHY) | Myb domain DNA binding | Schaffer et al (1998) and Wang and Tobin (1998) |
| CIRCADIAN CLOCK ASSOCIATED1 (CCA1) | Myb domain DNA binding | Schaffer et al (1998) and Wang and Tobin (1998) |
| CONSTANS (CO) | Nuclear protein containing two B-box zinc fingers; C-terminus CCT domain | Putterill et al (1995) and Robson et al (2001) |
| GIGANTEA (GI) | Nuclear protein of unknown function | Fowler et al (1999) and Park et al (1999) |
| FLOWERING LOCUS T (FT) | Homology to RAF kinase inhibitor | Kardailsky et al (1999) and Kobayashi et al (1999) |
| SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1) | MADS box transcription factor | Borner et al (2000), Lee et al (2000) and Samach et al (2000) |
| CRYPTOCHROME 2 (CRY2) | Blue light photoreceptor involved in the post-transcriptional regulation of CO | El-Assal et al (2001), Yanovsky and Kay (2003) and Valverde et al (2004) |
| PHYTOCHROME A (PHYA) | Red/far-red light photoreceptor involved in the post-transcriptional regulation of CO | Yanovsky and Kay (2002), Johnson et al (1994) and Valverde et al (2004) |
| PHYTOCHROME B (PHYB) | Red light photoreceptor regulating the degradation of CO protein at dawn | Guo et al (1998), Yanovsky and Kay (2002), Cerdan and Chory (2003) and Valverde et al (2004) |
| FLAVIN-BINDING, KELCH REPEAT, F-BOX (FKF1) | Photoreceptor required to increase CO transcription at dusk | Imaizumi et al (2003) |
Analysis of the effects of mutant alleles or transgenes on the expression of genes within the pathway allowed their order of action to be determined (Figure 1). Mutations in LHY/CCA1 and TOC1 affect the temporal pattern of expression of later-acting genes such as GI, CO and FT (Suarez-Lopez et al, 2001; Blazquez et al, 2002; Mizoguchi et al, 2002; Yanovsky and Kay, 2002). The expression of GI is regulated by LHY/CCA1, so that in the lhy cca1 double mutant the timing of expression of GI occurs 4 h earlier under long-day conditions (Mizoguchi et al, 2002). The major effect of gi mutations on flowering appears to be through the regulation of CO mRNA levels, because in gi mutants these are reduced and the overexpression of CO in a gi mutant overcomes the late-flowering phenotype (Suarez-Lopez et al, 2001). However, gi mutations cause additional defects inducing circadian rhythms to cycle faster under constant conditions, and impairing red-light signalling from phytochrome B (Park et al, 1999; Huq et al, 2000), and the relationship of these effects to the flowering phenotype is unclear. CO activates the expression of the downstream genes FT and SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO 1) (Kardailsky et al, 1999; Kobayashi et al, 1999; Borner et al, 2000; Lee et al, 2000; Onouchi et al, 2000; Samach et al, 2000). The hierarchy of gene action in the pathway suggested that the early-flowering phenotypes caused by loss-of-function mutations of elf3, lhy, cca1 and toc1 may be largely due to alterations in the timing of expression of these circadian-clock-regulated genes that control flowering time, particularly CO (discussed later) (Strayer et al, 2000; Suarez-Lopez et al, 2001; Yanovsky and Kay, 2002).
Figure 1.
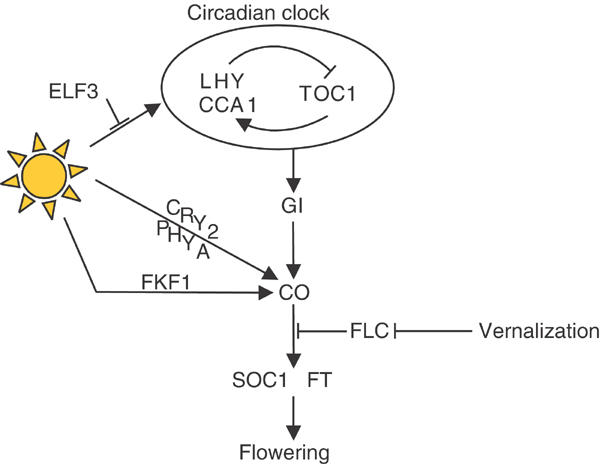
Molecular hierarchy that controls flowering of Arabidopsis in response to photoperiod. Arrows between genes represent promotive effects, whereas perpendicular lines represent repressive effects.
FT and SOC1 are among the most potent activators of flowering, so that they cause extreme early flowering when overexpressed (Kardailsky et al, 1999; Kobayashi et al, 1999; Borner et al, 2000; Lee et al, 2000; Samach et al, 2000). Furthermore, in addition to the response to photoperiod, these genes are also regulated by other environmental conditions that influence flowering time, such as exposure of plants to low temperatures for extended durations that mimic winter conditions (vernalization). Therefore, FT and SOC1 are at the point of convergence of several flowering-time pathways and are often therefore described as floral integrators (Mouradov et al, 2002; Simpson and Dean, 2002).
A coincidence model for CONSTANS activation by photoreceptors
CO plays a central role in the photoperiod response pathway by mediating between the circadian clock and the floral regulators FT and SOC1 (Suarez-Lopez et al, 2001). Furthermore, CO mRNA shows a striking temporal pattern of expression that was proposed to provide a basis for the regulation of the pathway by day length. Under long days the mRNA peaks in the evening and stays high until the following dawn, whereas under short days the mRNA peaks during the night (Figure 2) (Suarez-Lopez et al, 2001). This suggested that post-transcriptional regulation of CO by light specifically under long days might be responsible for the activation of CO, and thereby the response to long days. That the exact timing of CO expression is important in distinguishing between long and short days was also suggested by experiments in which the temporal pattern of CO expression was altered using mutants or by altering the length of the daily cycle from 24 h (Roden et al, 2002; Yanovsky and Kay, 2002). The toc1-1 mutant causes circadian rhythms to cycle faster under constant light, and under short days causes CO mRNA abundance to peak earlier. This earlier peak in CO mRNA under short days occurs during the photoperiod rather than during the night. Surprisingly, this effect of toc1-1 appears to be specific to CO expression, and does not affect expression of the upstream genes GI and LHY (Yanovsky and Kay, 2002). Nevertheless, expression of CO mRNA during the photoperiod correlates with increased FT expression and early flowering under short days, and these effects require CO function since they are largely abolished in a co mutant.
Figure 2.
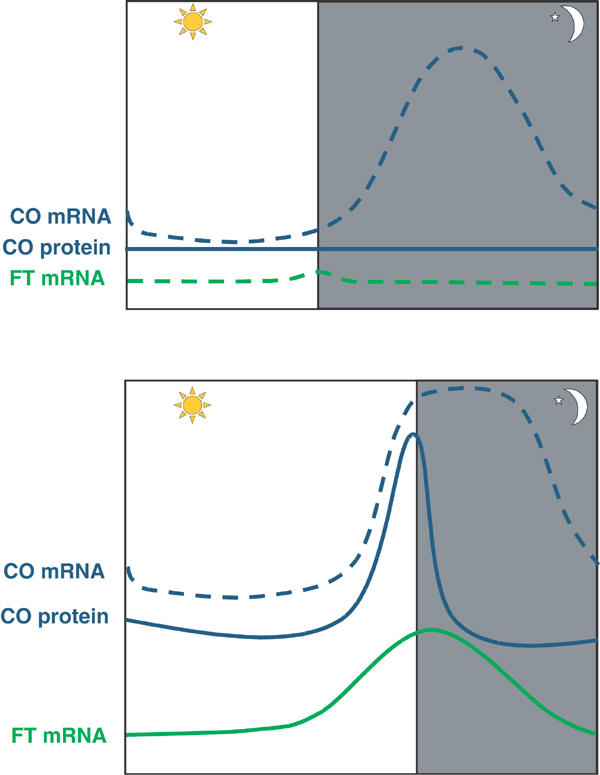
Expression patterns of the mRNAs of circadian-clock-controlled genes CO and FT under long and short days. Under short days (8 h light:16 h dark), CO mRNA expression peaks during the night (upper panel), CO protein does not accumulate and the downstream gene FT is not expressed. Under long-day conditions (16 h light:8 h dark), the peak of CO mRNA expression partly coincides with light (lower panel), the protein accumulates in the nucleus and the expression of FT mRNA is activated. FT promotes early flowering.
A second approach to altering the phase of CO expression involved changing the duration of the 24 h daily cycle. The timing of expression of CO relative to the light–dark transitions could be altered by maintaining the ratio of light to dark within the daily cycle, but extending or shortening the cycle from 24 h to 21 or 30 h (Roden et al, 2002; Yanovsky and Kay, 2002). This demonstrated a strong correlation between the expression of CO in the light, increased expression of the downstream gene FT and early flowering. Although the toc1-1 mutation and the alteration in cycle duration are likely to affect the timing of expression of many clock-controlled genes, the striking correlation between CO expression during the photoperiod, upregulation of FT and early flowering strongly suggested that post-transcriptional regulation of CO by exposure to light is at least one mechanism by which flowering of Arabidopsis is activated in response to long days.
Two molecular mechanisms underlying this activation of CO by light were recently described. The stability of the CO protein was shown to be regulated by light, so that in plants exposed to blue or far-red light the protein accumulates in the nucleus, but in darkness or red light the protein is absent (Valverde et al, 2004). This correlates with blue and far-red light being the most effective in promoting flowering. Also genetic experiments demonstrate that the blue light photoreceptors cryptochrome 1 and cryptochrome 2 as well as the far-red photoreceptor phytochrome A both promote flowering and stabilize the CO protein, whereas phytochrome B, which is activated by red light, delays flowering and promotes the degradation of CO protein (Johnson et al, 1994; Guo et al, 1998; El-Assal et al, 2001; Yanovsky and Kay, 2002; Cerdan and Chory, 2003; Valverde et al, 2004). This post-transcriptional regulation of CO stability by light provides a basis for the original proposal that the coincidence between CO mRNA and exposure to light is required to promote flowering.
An independent mechanism based on transcriptional regulation was also recently shown to regulate CO in response to light (Imaizumi et al, 2003). Under long days, the broad peak in CO mRNA is biphasic with one peak occurring in the light prior to dusk and a second during the night. This first peak in mRNA abundance, which facilitates the coincidence between CO expression and light, requires exposure to light. The photoreceptor FKF1 (Table I) is required for the expression of this peak, and mutations in FKF1 both delay flowering and reduce CO expression at dusk (Imaizumi et al, 2003).
The responsiveness of CO activity to day length therefore depends on regulation at several levels. Circadian clock control of CO transcription underlies the system and restricts CO expression to the later part of the day/night cycle. The presence of light during the evening both enhances CO transcription and stabilizes the protein in the nucleus ensuring activation of the floral regulator FT. This requirement for light ensures that CO activation and flowering only occur under long days.
Interactions between the vernalization and photoperiod responses
Natural accessions of Arabidopsis differ in their responses to seasonal cues of day length and temperature. Summer annual accessions germinate in spring or early summer and rapidly flower in response to the long-day photoperiod. In contrast, winter annuals typically germinate in summer, grow vegetatively through the winter until the following spring and then flower in response to exposure to long photoperiods the following summer. Thus winter annuals do not respond to inductive photoperiods in the first summer, but require exposure to cold winter temperatures before they can respond to long days the following summer. Winter and summer annuals typically differ at one of two loci, FLOWERING LOCUS C (FLC) and FRIGIDA (FRI), and dominant alleles at these loci in the winter annual are required to confer a vernalization requirement (Simpson and Dean, 2002).
FLC encodes a MADS box transcription factor that is expressed at high levels in winter annuals before vernalization, and at lower levels when plants are exposed to cold temperatures for several weeks (Michaels and Amasino, 1999; Sheldon et al, 1999). In addition, overexpression of FLC in summer annual varieties causes a dramatic late-flowering phenotype. Therefore, FLC encodes a repressor of flowering, and high FLC levels correlate with the vernalization requirement of winter annual varieties (Michaels and Amasino, 1999; Sheldon et al, 1999). FRI encodes a protein with unknown biochemical function (Johanson et al, 2000) and is required to increase FLC mRNA abundance (Michaels and Amasino, 1999; Sheldon et al, 1999). This effect is dependent on functional FLC alleles as loss-of-function flc mutations suppress the effect of FRI on flowering time.
The photoperiod and vernalization pathways respond to different environmental signals, but these pathways converge to regulate the expression of the same downstream genes, FT and SOC1 (Borner et al, 2000; Lee et al, 2000; Samach et al, 2000; Michaels and Amasino, 2001). Transcription of FT and SOC1 is activated by CO and repressed by FLC, which represses SOC1 transcription by directly binding to its promoter (Hepworth et al, 2002). Therefore, in winter annual accessions, response to long days is prevented during the first summer, at least in part because high FLC levels block the capacity of CO to activate downstream genes.
Many other plant species show similar genetic variation between summer and winter annual forms (Laurie, 1997), and must also block the day-length response until they have been exposed to winter conditions. CO function is conserved (Yano et al, 2000; Griffiths et al, 2003) in distantly related species, raising the possibility that antagonism between CO and FLC orthologues may be the general basis of the winter annual form. No FLC orthologues, however, have been identified outside the Cruciferae, suggesting that the role of FLC in the vernalization response may not be widely conserved. VRN1 is required in winter wheat varieties to confer a vernalization response. A candidate for the VRN1 gene was recently cloned and encodes a MADS box protein most similar to APETALA1 (AP1) from Arabidopsis (Yan et al, 2003). vrn1 mutants contain a deletion in the promoter region, suggesting that a negative regulator can no longer repress VRN1 expression prior to vernalization and this lack of repression promotes flowering. This is consistent with the observation of low VRN1 mRNA levels before vernalization in winter wheat varieties and increases in its mRNA levels after vernalization (Yan et al, 2003). Therefore, no repressor analogous to FLC has been described in monocotyledonous plants, and how the photoperiod response is prevented prior to vernalization remains unclear.
Diversity in photoperiodic responses
Control of flowering by photoperiod is widespread in the plant kingdom, but the type of response can vary widely between species (Thomas and Vince-Prue, 1997). For example, short-day plants flower early under short days and late under long days, and therefore show the reverse response to Arabidopsis. The distinction between long- and short-day response types has evolved independently in different families of flowering plants. The grasses include the long-day response plants wheat and barley as well as the short-day response plants maize and rice, while in Nicotiana, a single genus of dicotyledonous plants, long- and short-day response types occur. Therefore, whether the molecular pathway described in Arabidopsis is conserved in species showing responses to short days is of importance, since this would enable analysis of how the pathway is modified to generate a short-day response and whether these modifications are the same in different branches of the Angiosperm phylogeny.
Genetic analysis of photoperiod response in rice has provided evidence that even in short-day plants distantly related to Arabidopsis, the same components regulate photoperiod response. By identifying natural allelic variation affecting photoperiodic control of flowering, the rice orthologues of CO (Hd1 in rice) and FT (Hd3a in rice) were shown to be required for flowering in response to short days (Yano et al, 2000; Kojima et al, 2002). Hd3a is expressed at higher levels under short days, which induce flowering, and therefore is similar to the transcriptional upregulation of FT detected under long days in Arabidopsis. Thus, transcriptional upregulation of FT/Hd3a specifically under day lengths that induce flowering is conserved in both species (Izawa et al, 2002; Hayama et al, 2003). Furthermore, Hd1 and the rice orthologue of GI (OsGI) regulate the expression of Hd3a mRNA in rice. However, the relationship between Hd1 and Hd3a appears to be reversed, with elevated Hd1 causing a reduction in Hd3a expression. This relationship between Hd1 and Hd3a suggests that the role of Hd1 is to repress Hd3a under long days and that this repression is relieved in short days leading to an upregulation of Hd3a and early flowering (Hayama et al, 2003). Therefore, although both Arabidopsis and rice CO/Hd1 regulate FT/Hd3a expression, their relationships are reversed with CO activating FT whereas Hd1 represses Hd3a.
Whether a similar mechanism operates in other short-day plants remains to be tested, but at least the components have been shown to occur in other species. CO-like genes have been cloned from the short-day plant Pharbitis nil (Liu et al, 2001a; Kim et al, 2003), which is closely related to the Nicotiana species, and show a similar pattern of diurnal regulation to CO from Arabidopsis. Also, Maryland Mammoth tobacco, which shows an absolute requirement for exposure to short days to flower, will flower under long days if it carries a transgene driving constitutive expression of the Sinapis alba orthologue of SOC1. This suggests that related MADS box proteins act downstream of the photoperiod response in Arabidopsis and short-day tobacco varieties (Borner et al, 2000).
In addition to flowering, other developmental transitions including tuberization in potato and the onset of dormancy in the buds of perennial plants such as deciduous trees are controlled by day length (Thomas and Vince-Prue, 1997). Some of the proteins identified as regulating photoperiodic control of flowering in Arabidopsis may also regulate these other responses. Tuberization of potato is induced by short days, and transgenic plants overexpressing the Arabidopsis CO gene showed delayed tuberization, suggesting that potato orthologues of CO may negatively regulate tuberization (Martinez-Garcia et al, 2002).
At present our understanding of these processes is largely at the genetic level, there is a need to extend this to include the biochemical basis of the response in Arabidopsis, and how the biochemical function of the constituent proteins is altered to generate other photoperiodic response types. This will provide an understanding of how the activity of this pathway has been modified during evolution and provide insights into how the flowering time of crop or horticultural plants could be modified to generate novel flowering phenotypes.
Acknowledgments
Work in the laboratory of GC is supported by a core grant from the Max Planck Society. IS is supported by EC grant (QLK5-CT-2001-01412) within FP5.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Alabadi D, Yanovsky MJ, Mas P, Harmer SL, Kay SA (2002) Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr Biol 12: 757–761 [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Trenor M, Weigel D (2002) Independent control of gibberellin biosynthesis and flowering time by the circadian clock in Arabidopsis. Plant Physiol 130: 1770–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24: 591–599 [DOI] [PubMed] [Google Scholar]
- Bünning E (1936) Die endogene Tagesrhythmik als Grundlage der photoperiodischen Reaktion. Ber Dtsch Bot Ges 54: 590–607 [Google Scholar]
- Cerdan PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ (1999) Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 17: 63–71 [DOI] [PubMed] [Google Scholar]
- El-Assal SED, Alonso-Blanco C, Peeters AJM, Raz V, Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet 29: 435–440 [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner WW, Allard HA (1920) Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J Agric Res 18: 553–606 [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131: 1855–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HW, Yang WY, Mockler TC, Lin CT (1998) Regulations of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch LB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hayama R, Coupland G (2003) Shedding light on the circadian clock and the photoperiodic control of flowering. Curr Opin Plant Biol 6: 13–19 [DOI] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Albertson TM, Wagner DR (2001) EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Tepperman JM, Quail PH (2000) GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci USA 97: 9789–9794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16: 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC (1994) Photoresponses of light-grown phyA mutants of Arabidopsis: phytochrome A is required for the perception of daylength extensions. Plant Physiol 105: 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Moon J, Lee I, Maeng J, Kim SR (2003) Molecular cloning and expression analysis of a CONSTANS homologue, PnCOL1, from Pharbitis nil. J Exp Bot 54: 1879–1887 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Van Der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Laurie DA (1997) Comparative genetics of flowering time. Plant Mol Biol 35: 167–177 [PubMed] [Google Scholar]
- Lee H, Suh S-S, Park E, Cho E, Ahn JH, Kim S-G, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JY, Yu JP, McIntosh L, Kende H, Zeevaart JAD (2001a) Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol 125: 1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wanger DR (2001b) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Virgos-Soler A, Prat S (2002) Control of photoperiod-regulated tuberization in potato by the Arabidopsis flowering-time gene CONSTANS. Proc Natl Acad Sci USA 99: 15211–15216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar AJ (2000) The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14 (Suppl): S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Minis DH (1964) The entrainment of circadian oscillations by light and their role as photoperiodic clocks. Am Nat 98: 261–322 [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Redei GP (1962) Supervital mutants of Arabidopsis. Genetics 47: 443–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F, Costa MMR, Hepworth S, Vizir I, Pineiro M, Reeves PH, Putterill J, Coupland G (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J 28: 619–631 [DOI] [PubMed] [Google Scholar]
- Roden LC, Song HR, Jackson S, Morris K, Carre IA (2002) Floral responses to photoperiod are correlated with the timing of rhythmic expression relative to dawn and dusk in Arabidopsis. Proc Natl Acad Sci USA 99: 13313–13318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AAR, Pearson M, Kay SA (1998) The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125: 485–494 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue B (1997) Photoperiodism in Plants, 2nd edn. San Diego, CA: Academic Press [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein and the mechanism of photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2003) Living by the calendar: how plants know when to flower. Nat Rev Mol Cell Biol 4: 265–275 [DOI] [PubMed] [Google Scholar]


