Abstract
The extracellular portions of cell surface receptor proteins are often comprised of independently folding protein domains. As they are translated into the endoplasmic reticulum (ER), some of these domains require protein chaperones to assist in their folding. Members of the low-density lipoprotein receptor (LDLR) family require the chaperone called Boca in Drosophila or its ortholog, Mesoderm development, in the mouse. All LDLRs have at least one six-bladed β-propeller domain, which is immediately followed by an epidermal growth factor (EGF) repeat. We show here that Boca is specifically required for the maturation of these β-propeller/EGF modules through the secretory pathway, but is not required for other LDLR domains. Protein interaction data suggest that as LDLRs are translated into the ER, Boca binds to the β-propeller. Subsequently, once the EGF repeat is translated, the β-propeller/EGF module achieves a more mature state that has lower affinity for Boca. We also show that Boca-dependent β-propeller/EGF modules are found not only throughout the LDLR family but also in the precursor to the mammalian EGF ligand.
Keywords: β-propeller, chaperone, EGF, LDLR, LRP
Introduction
The low-density lipoprotein receptor (LDLR) family includes a diverse group of cell surface proteins that carry out a broad range of cellular functions. Most family members are endocytic cargo receptors. For example the founding member of the family, the human LDL receptor (hLDLR), mediates the endocytosis of cholesterol-containing lipoprotein particles. Other members of the family, such as LDLR-related protein 1 (LRP1) and Megalin, have multiple ligand binding domains and accordingly endocytose a wide variety of ligands including proteases and protease inhibitor complexes, vitamin carrier proteins, and lipoproteins (see for reviews Hussain et al, 1999; Strickland et al, 2002). In addition, some members of this receptor family have signaling capacity. For example, in Wnt/Wingless (Wg) signaling, the mammalian LDLR-related proteins 5 and 6 (LRP5/LRP6) and their Drosophila homolog Arrow (Arr) are essential co-receptors. Other recently characterized examples are very low-density lipoprotein receptor (VLDLR) and apolipoprotein E receptor 2 (ApoER2), which serve as redundant receptors for the secreted protein Reelin and are required for neuronal migration during mammalian brain development (Willnow et al, 1999; Nimpf and Schneider, 2000; Herz, 2001; Howell and Herz, 2001).
As with many cell surface and secreted proteins, LDLRs have a modular structure. The extracellular domains of all LDLRs are composed of three types of repeated sequences: the complement-type repeats, also called type A repeats, epidermal growth factor (EGF) repeats, and YWTD repeats (Hussain et al, 1999) (Figure 1). Each type A repeat and EGF repeat fold largely autonomously as shown by the structures of receptor fragments containing only type A or EGF domains (Herz, 2001). In contrast, six contiguous YWTD repeats fold together into a six-bladed β-propeller conformation (Springer, 1998; Jeon et al, 2001; Rudenko et al, 2002). Each β-propeller is also thought to pack against an adjacent, C-terminal EGF repeat. Although these domains can be found individually in other proteins, only in the LDLR family do they coexist in the same proteins (Bork et al, 1996).
Figure 1.
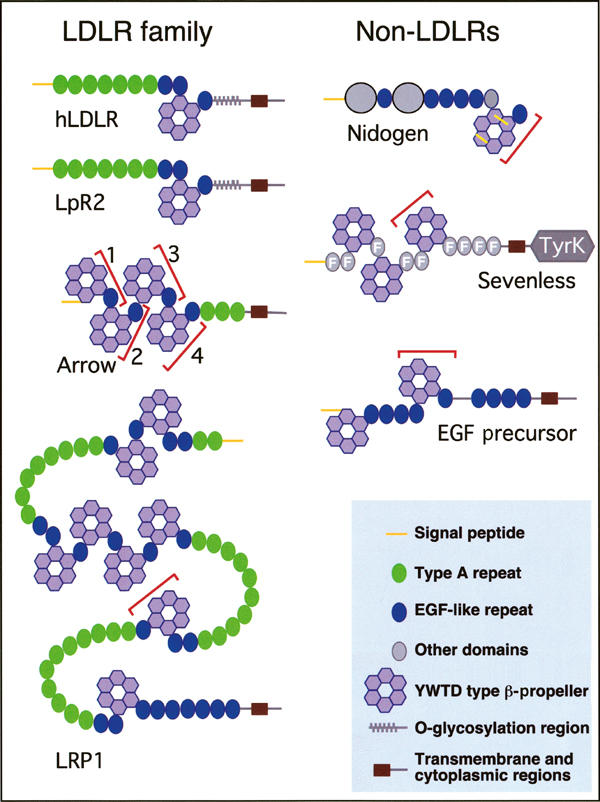
Schematic representation of YWTD β-propeller-containing proteins. Most proteins with this type of β-propeller belong to the LDLR family, which also have type A and EGF repeats in their extracellular domains. Shown here are Drosophila LpR2 and Arrow and human LDLR and LRP1. The non-LDLR proteins with YWTD β-propellers used in this work are Drosophila Nidogen, an extracellular matrix protein, the tyrosine kinase receptor Sevenless, and mouse EGFP. The yellow bars in the Nidogen β-propeller represent two disulfide bonds. Sevenless has a cytoplasmic tyrosine kinase domain (TyrK) and fibronectin type III modules (F) in the extracellular region. The red brackets indicate the domains analyzed in this work. Arrow β-propeller/EGF pairs are numbered from N to C. The domain structure for Drosophila Nidogen was tentatively derived from a predicted open reading frame.
Like other cell surface and secreted proteins, LDLR family members enter the secretory pathway as they are translated by endoplasmic reticulum (ER)-associated ribosomes. In the ER, proteins often fold and mature with the help of molecular chaperones (Stevens and Argon, 1999; Parodi, 2000; Frydman, 2001). Some of these chaperones have a broad specificity and are required for the trafficking of many different proteins. On the other hand, a few other molecular chaperones have been identified that are specific for a single protein or for a restricted subset of related proteins (Ellgaard et al, 1999; Ellgaard and Helenius, 2003; Sitia and Braakman, 2003). We recently identified a novel ER chaperone in Drosophila, named Boca, that is specifically required for the correct folding and trafficking of several members of the LDLR family, including the Wg co-receptor Arrow (Culi and Mann, 2003). In cells that do not express Boca, LDLR proteins cannot traffic to the cell membrane because they fail to exit the ER. Significantly, the murine homolog of Boca, Mesoderm development (Mesd), was shown to play a similar role during the synthesis of LRP5/6 (Hsieh et al, 2003). Consequently, flies or mice with a nonfunctional boca or mesd gene die during embryogenesis and display phenotypes that are consistent with an inability to transduce a Wg/Wnt signal. Additionally, during Drosophila oogenesis, boca mutant eggs accumulate abnormally low levels of yolk proteins because the endocytic receptor for yolk proteins (Yolkless, a member of the LDLR family) fails to exit the ER (Culi and Mann, 2003). The early embryonic lethality of mesd mutant mice makes it difficult to determine if this gene is required for other LDLR-dependent functions in addition to Wnt signaling (Hsieh et al, 2003).
Here we determine which domains within LDLRs require Boca for their trafficking through the secretory pathway. We identify the β-propeller and its associated C-terminal EGF repeat as a Boca-dependent module shared by all LDLRs. Interestingly, this module is also present in the precursor to the mammalian epidermal growth factor (EGFP). Consistent with Boca's proposed specificity, we find that EGFP's β-propeller/EGF module also requires Boca for its trafficking through the secretory pathway. In contrast, other YWTD β-propellers, such as those in the basement membrane protein Nidogen and the tyrosine kinase receptor Sevenless, do not require Boca. The distinction between those β-propellers that require Boca and those that do not is that the trafficking of Boca-dependent β-propellers also requires a C-terminal EGF domain.
Results and discussion
The β-propeller and its associated C-terminal EGF repeat constitute the minimal unit in LpR2 that requires Boca
The modular organization of LDLR proteins suggests the possibility that Boca may be required for the maturation and/or trafficking of only one of the three different repeats present in these proteins (A repeats, EGF repeats, and YWTD type β-propellers). To address this question, we undertook a deletion analysis of the Drosophila lipophorin receptor 2 (LpR2) protein. We selected LpR2 for this study because it requires Boca during its maturation, has a relatively small size (861 amino acids), and its domain organization is analogous to the human LDL receptor (Figure 1), whose trafficking in Drosophila S2 cells is also Boca dependent (Culi and Mann, 2003).
We used two different assays to measure the trafficking of LpR2 and its derivatives (Figure 2). In the first assay, the subcellular localization of epitope-tagged versions of these proteins was directly examined by immunofluorescence and confocal microscopy following their expression in wild-type and boca− Drosophila S2 cells. boca− S2 cells were generated by RNA interference (RNAi) as described previously (Culi and Mann, 2003). For full-length LpR2, the protein was detected at the cell membrane in wild-type S2 cells, but not in boca− S2 cells (Figure 2). In the second assay, epitope-tagged LpR2 derivatives in which the TM and cytoplasmic domains had been deleted were expressed in wild-type and boca− S2 cells (Figure 2). In the case of an otherwise intact LpR2 (LpR2ΔTM), expression in wild-type S2 cells resulted in its secretion into the media, and could be readily detected by immunoblotting (Figure 2). In contrast, in boca− S2 cells, secretion of LpR2ΔTM was nearly eliminated. In both wild-type and boca− cells, LpR2ΔTM could be detected at comparable levels in total cell lysates (Figure 2). Thus, both assays agree that Boca is required for the trafficking of LpR2 through the secretory pathway, but is not required for its synthesis or stability.
Figure 2.
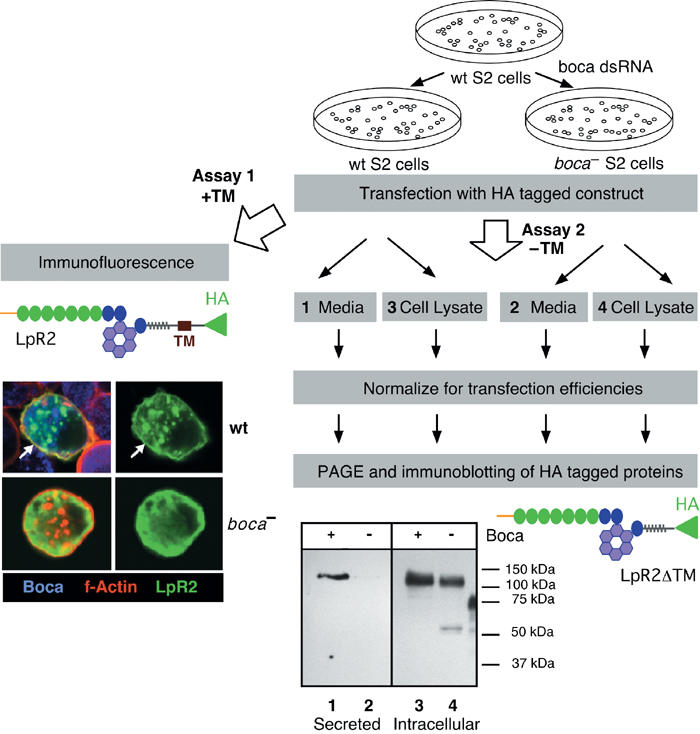
Flow diagram of the two assays used to analyze the trafficking of LpR2 derivatives. Both assays begin with expression of HA-tagged LpR2 derivatives into wild-type (wt) and boca− S2 cells. In assay 1, the cells were transfected with LpR2 derivatives that contain a TM and examined by immunofluorescence and confocal microscopy. The cells were stained to detect Boca (blue), which is a marker for the ER (Culi and Mann, 2003), the LpR2 derivative (green, using the HA epitope), and F-actin (red), which serves as a marker for the cell membrane. For clarity, the green channel is shown to the right of the merged image. In wt S2 cells, LpR2 is found in intracellular vesicles and at the cell membrane, which appears yellow due to colocalization with F-actin (arrows). In boca− S2 cells, LpR2 is found diffusely throughout the cytoplasm and not at the cell membrane, which appears red due to lack of colocalization with F-actin. This staining is consistent with LpR2 accumulating in the ER, which is diffuse in S2 cells as shown by the distribution of the ER marker Boca in wild-type cells (blue) (see also Culi and Mann, 2003). In assay 2, the cells were transfected with LpR2 derivatives that do not have a TM. Transfection efficiencies were normalized by cotransfecting with a lacZ expression plasmid and comparing β-galactosidase activities in the cell extracts. The amount of the HA-tagged protein in the media and in total cell lysates was examined by immunoblotting with an anti-HA antibody. Although LpR2 was detected intracellularly in both wild-type and boca− S2 cells, it was only secreted from wild-type cells.
We next measured the Boca dependency of a series of LpR2 derivatives to determine the minimal fragment that requires Boca to traffic through the secretory pathway. In all cases in which both the immunofluorescence (Figure 3A) and secretion (Figure 3B) assays were used, they were in agreement with each other. LpR2 contains a signal peptide at its N-terminus followed by a string of seven complement type A repeats (A), two EGF repeats (E1 and E2), a single β-propeller (β), a third EGF repeat (E3), a putative O-glycosylation sequence, a transmembrane (TM) domain, and a short cytoplasmic domain (Figure 3A). Deletion of the type A repeats, all three EGF repeats, and the β-propeller resulted in a small protein (LpR2_Gly) with an extracellular domain containing only the O-glycosylation segment. This protein was able to traffic through the secretory pathway in a Boca-independent manner, indicating that Boca is not required for the maturation of the O-glycosylation or cytoplasmic domains of LpR2 (Figure 3A). This result is consistent with the fact that LDLRs are class I TM proteins that during synthesis have their extracellular domains in the ER lumen where Boca resides. Unexpectedly, additional deletion of the O-glycosylation segment resulted in a protein (LpR2_∅) that was retained in the ER both in wild-type and boca− cells (Figure 3A). Thus, in this assay, the O-glycosylation segment is essential for LpR2 to mature properly even in wild-type cells. For this reason we included this region of the protein, as well as the LpR2 signal sequence, in all of the remaining constructs.
Figure 3.
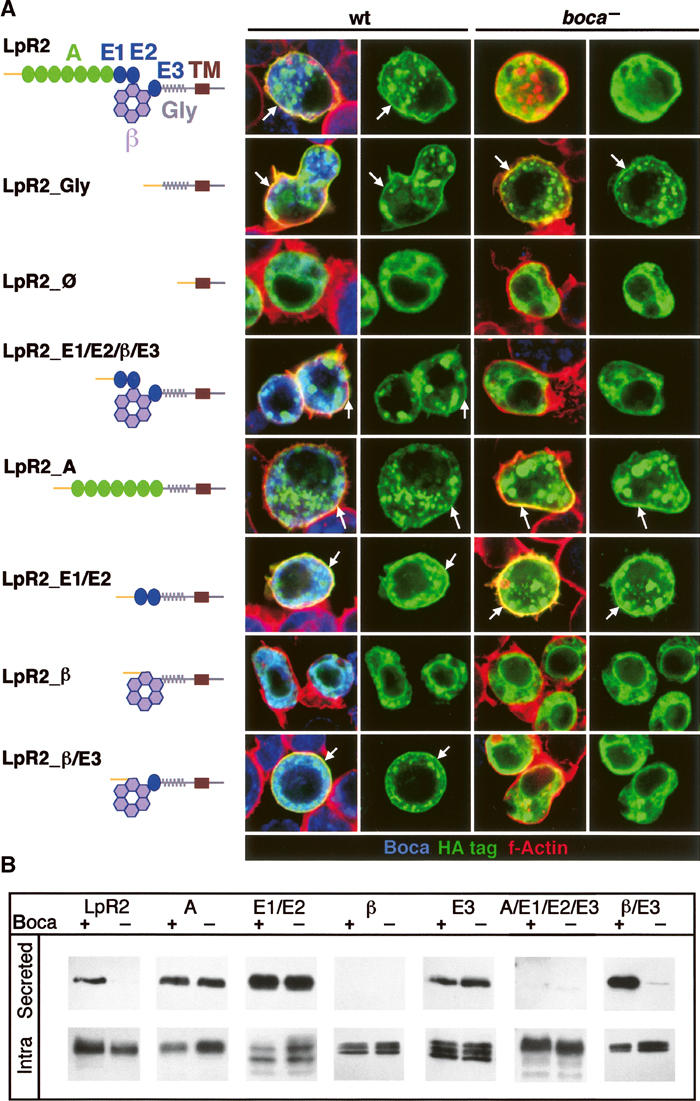
The β-propeller/E3 module constitutes the minimal Boca-dependent fragment of LpR2. (A) Assay 1: Full-length LpR2 and several deletion derivatives were transfected into wild-type (wt) and boca− S2 cells. A schematic representation of the various LpR2 derivatives is shown on the left. The seven A type repeats (A), three EGF repeats (E1, E2, and E3), β-propeller (β), O-glycosylation sequence (Gly), and TM are indicated. The domains listed refer to those present in the protein being analyzed. Cells were stained as described in Figure 2. In these images, the white arrows point to membrane localization of the HA-tagged protein. Of these proteins, only LpR2_Gly, LpR2_A, and LpR2_E1/E2 show membrane localization in the absence of Boca. (B) Assay 2: Proteins without a TM domain were expressed in wild-type (+) and boca− (−) cells. Secreted and intracellular fractions were analyzed by immunoblotting with anti-HA antibody. The domains listed refer to those present in the protein being analyzed. Of these proteins, only LpR2 and LpR2_β/E3 require Boca for secretion into the media.
Deletion of all seven type A repeats from LpR2 resulted in a protein (LpR2_E1/E2/β/E3) that still required Boca for its maturation (Figure 3A). Conversely, a protein comprised only of type A repeats (LpR2_A) trafficked through the secretory pathway in a Boca-independent manner (Figure 3A and B). Thus, type A repeats do not require Boca for their maturation. Similarly, a derivative containing only the two contiguous N-terminal EGF repeats (LpR2_E1/E2) trafficked through the secretory pathway in both wild-type and boca− cells (Figure 3A and B). Thus, EGF repeats E1 and E2 do not require Boca. However, an LpR2 derivative that is comprised only of the β-propeller (LpR2_β) was retained in the ER both in wild-type and boca− cells (Figure 3A and B). This suggests that even in wild-type cells, LpR2_β cannot fold properly and is therefore retained in the ER. Interestingly, crystallographic studies of the hLDLR revealed that the third EGF repeat (E3) packs against the β-propeller such that part of the surfaces of the β-propeller and this EGF repeat become buried and solvent inaccessible (Jeon et al, 2001). In LpR2_β, the β-propeller surface that is normally masked by E3 is probably exposed to the solvent. This surface includes several hydrophobic residues that may result in misfolding and failure to exit the ER due to the activity of quality control systems that block the trafficking of misfolded proteins (Ellgaard et al, 1999). Consistent with the structural studies by Jeon et al (2001), an LpR2 derivative that contains both the β-propeller and the E3 repeat, but is missing the other repeats (LpR2_β/E3), trafficked through the secretory pathway in wild-type S2 cells. However, this protein failed to exit the ER in boca− cells (Figure 3A and B). These results suggest that the module comprised of the β-propeller and E3 is the minimal and only region of LpR2 that requires Boca for its trafficking through the secretory pathway.
The importance of the E3 repeat prompted us to characterize two additional proteins. LpR2_E3, which contains only the E3 repeat, was secreted into the media both in wild-type and boca− cells (Figure 3B). Thus, as with E1 and E2, E3 does not require Boca function. Surprisingly, however, deletion of the β-propeller from an otherwise intact LpR2 resulted in a protein (LpR2_A/E1/E2/E3) that was secreted poorly both in wild-type and boca− cells (Figure 3B). The juxtaposition of E2 next to E3 apparently resulted in a protein that was unable to fold properly even in wild-type cells.
These results indicate that the β-propeller requires an adjacent E3 domain to mature through the secretory pathway. To further explore the requirement for an E3 domain, we tested if the β-propeller and E3 must be present in the same polypeptide or, alternatively, if E3 can be provided in trans. We expressed in S2 cells two hemagglutinin (HA)-tagged proteins, one containing a signal sequence plus the β-propeller and the second containing a signal sequence plus the E3 domain. To avoid interference from the HA tags, we tagged the β-propeller at the N-terminus and E3 at the C-terminus (see Materials and methods). Consistent with our previous results, the β-propeller failed to be secreted when expressed in the absence of E3, but E3 was secreted when expressed in the absence of the β-propeller (Figure 4A). However, when the two proteins were coexpressed, secretion of E3 was strongly reduced (Figure 4A). This observation suggests that these two proteins can associate with each other in the ER, but that they do so in a manner that is inconsistent with trafficking. Identical results were obtained in boca− S2 cells, suggesting that Boca is not required for the β-propeller and E3 domains to interact with each other as measured by this assay (Figure 4A).
Figure 4.
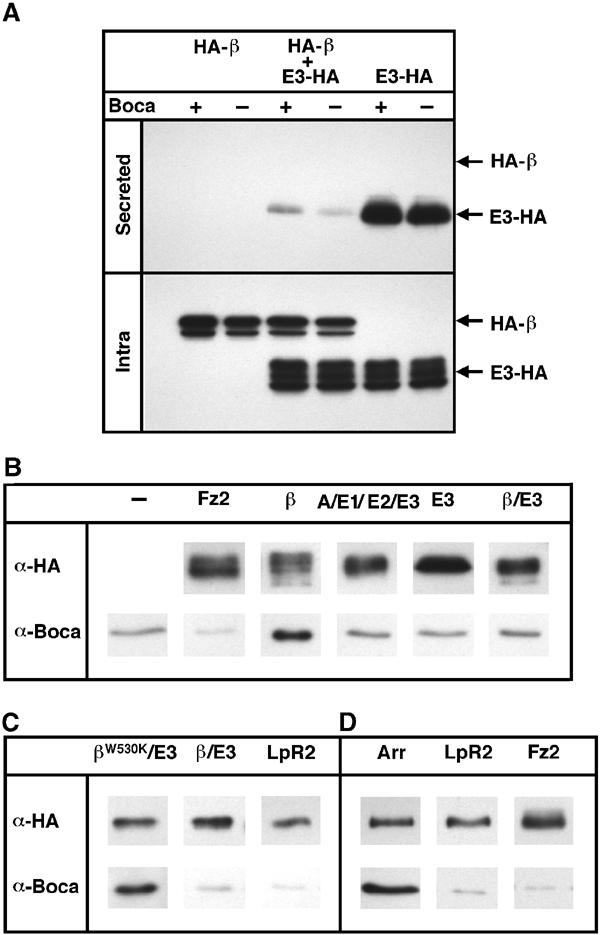
Boca interacts with the β-propeller. (A) The LpR2ΔTM derivatives HA-β, E3-HA, and mock DNA were cotransfected into wild-type (+) and boca− (–) S2 cells as indicated. Twice the amount of E3-HA DNA was used compared to HA-β. Secreted and intracellular fractions were analyzed by immunoblotting with anti-HA antibody. The HA-β and E3-HA bands are indicated at the right of the figure. Note that for the intracellular fraction, multiple glycosylation bands can be detected for each protein. Secretion of E3-HA is strongly impaired by the coexpression of HA-β, both in wild-type and boca− cells. (B) The indicated LpR2ΔTM derivatives were expressed in wild-type S2 cells, the cells treated with a crosslinking reagent, and cell extracts immunoprecipitated with anti-HA antibody. An experiment carried out with untransfected cells (–) represents the level of background observed in this experiment. The TM protein Frizzled2 (Fz2), whose trafficking is Boca independent (Culi and Mann, 2003), served as another negative control. The immunoprecipitates were immunoblotted with anti-HA and anti-Boca in parallel as indicated. Of these proteins, only LpR2_β co-immunoprecipitated Boca above background. (C) LpR2_βW530K/E3, which has a mutation in Trp530 of LpR2, co-immunoprecipitated Boca. In contrast, wild-type LpR2_β/E3 and LpR2 failed to co-immunoprecipitate Boca. All proteins used in this experiment are ΔTM derivatives. (D) Arrow (Arr), but not LpR2 or Fz2, co-immunoprecipitated Boca. All proteins used in this experiment contain a TM.
Boca interacts with the β-propeller
The experiments described above suggest that Boca is required for the trafficking of the β-propeller/E3 module through the secretory pathway. Boca could carry out this function by interacting with the β-propeller, E3, or both. To address this question, we carried out co-immunoprecipitation (co-IP) experiments. These experiments suggest that Boca's primary site of interaction is the β-propeller. Most significantly, Boca was co-immunoprecipitated with LpR2_β, which has a β-propeller but no E3 domain. In contrast, Boca did not interact above background with LpR2_A/E1/E2/E3, a protein that does not have a β-propeller (Figure 4B). In multiple experiments, the amount of Boca that was co-immunoprecipitated with LpR2_β was consistently above that observed in two different negative controls (untransfected cells and cells coexpressing the Boca-independent receptor Fz2). Furthermore, both LpR2_β and LpR2_A/E1/E2/E3 were present at similar levels when expressed in S2 cells and both failed to exit the ER (Figure 3 and data not shown). Thus, the difference in Boca's ability to interact with them is unlikely to be due to differences in their levels within the ER. Moreover, a protein containing only the E3 repeat (LpR2_E3) also failed to interact with Boca (Figure 4B). The interaction with LpR2_β was only observed in the presence of a crosslinking reagent, suggesting, as is often the case for chaperones, that it may be weak and/or transient (see Materials and methods) (Kuznetsov et al, 1994; Ellgaard et al, 1999).
Surprisingly, we were unable to detect an interaction between Boca and full-length LpR2 or with LpR2_β/E3, even though both proteins have a β-propeller and require Boca for their trafficking (Figure 4B–D). The observation that LpR2_β, but not LpR2_β/E3, could be co-immunoprecipitated with Boca suggests that in the absence of E3, LpR2_β exists in an immature state that binds to Boca, but when E3 is present, as in LpR2_β/E3, the β-propeller reaches a more mature state that has a lower affinity for Boca. Based on this idea, we reasoned that if we disrupt the E3–β-propeller interaction, we might stabilize the immature state and the interaction between Boca and the β-propeller. To this end, we mutated the hydrophobic β-propeller residue Trp530 to a Lys, a residue with a high propensity for surface exposure. In the hLDLR, the equivalent residue (Phe509) is also aromatic and is buried by and makes van der Waals contacts with the EGF domain (Jeon et al, 2001) (see Materials and methods). Consistent with the idea that Boca binds to an immature form of the β-propeller, Boca was co-immunoprecipitated with the mutant protein (LpR2_βW530K/E3) but not with wild-type LpR2_β/E3 (Figure 4C). We also found that Boca was co-immunoprecipitated when the β-propeller and E3 domains were expressed as separate proteins, suggesting that E3 and Boca do not compete for an interaction with the β-propeller (data not shown). LpR2_βW530K/E3, like LpR2_β, also failed to exit the ER when expressed in wild-type S2 cells (data not shown). The interaction between these proteins and Boca may contribute to their ER retention, as has been observed for more general chaperone–substrate interactions (Ellgaard et al, 1999; Jorgensen et al, 2000).
Although we were unable to detect an interaction between Boca and full-length LpR2, the murine Boca ortholog Mesd has been reported to interact with LRP5 and LRP6 (Hsieh et al, 2003). In contrast to LpR2, which has only a single β-propeller, LRP5/6 and their fly ortholog Arrow have four β-propellers (Figure 1). We considered the possibility that if the β-propeller was the primary site of Boca interaction, we might be able to detect an interaction between Boca and LDLRs, such as Arrow, which have multiple β-propellers. In agreement with this idea, we detected an interaction between Boca and full-length Arrow (Figure 4D). Thus, the interaction between Boca and LDLRs may be increased by the presence of multiple β-propellers. In addition to having more sites of Boca interaction, it is also possible that the presence of multiple β-propellers in Arrow complicates the folding process and thereby increases the amount of time Boca interacts with this LDLR, which would increase our ability to detect an interaction.
Taken together, these results suggest that Boca is necessary for the folding and assembly of the β-propeller/E3 module present in LpR2. In order to carry out its function, the co-IP experiments suggest that Boca interacts, either directly or indirectly, with the β-propeller. Further, our results suggest that the β-propeller–Boca interaction only occurs when the β-propeller is in an immature state. In proteins containing a native β-propeller/E3 module, the presence of E3 allows the module to achieve a mature state that has a low affinity for Boca.
β-Propeller/EGF modules from other LDLRs also require Boca
The results described above show that the β-propeller/E3 module from LpR2 is the minimal fragment of this receptor that requires Boca for its trafficking through the secretory pathway. In addition to LpR2, Boca is required for the maturation of other LDLR family members (Culi and Mann, 2003). Thus, we wanted to determine if the conclusions from the analysis of LpR2 could be extrapolated to other LDLR proteins. There are several observations that support this idea. First, in all LDLR family members, both in mammals and Drosophila, each β-propeller is immediately followed by an adjacent C-terminal EGF repeat (Springer, 1998; Hussain et al, 1999) (Figure 1 and data not shown). Second, sequence comparison of the EGF repeats present in the Drosophila LDLRs shows that EGF repeats C-terminal to the β-propellers are more similar to each other than they are to those EGF repeats occurring at other positions (data not shown). Third, a crystal structure of the β-propeller/E3 module from the human LDL receptor shows that this EGF repeat packs against the β-propeller (Jeon et al, 2001), suggesting that the mature structures of these two domains depend on each other.
Based on these considerations, we hypothesized that the maturation of β-propeller/EGF modules from other LDLRs would also be Boca dependent. As a test of this hypothesis, we used the secretion assay to analyze five different β-propeller/EGF modules. Four of these were derived from Drosophila Arrow and one from human LRP1 (Figure 1). In all cases in which the β-propeller/EGF module was secreted in wild-type cells, trafficking was Boca dependent (Figure 5A). The second β-propeller/EGF module from Arrow failed to be secreted in wild-type cells, preventing our ability to assess Boca dependency. Furthermore, an isolated β-propeller from LRP1 failed to be secreted from wild-type or boca− cells (Figure 5B), emphasizing the integral role played by the EGF repeats during the assembly of β-propeller/EGF modules.
Figure 5.
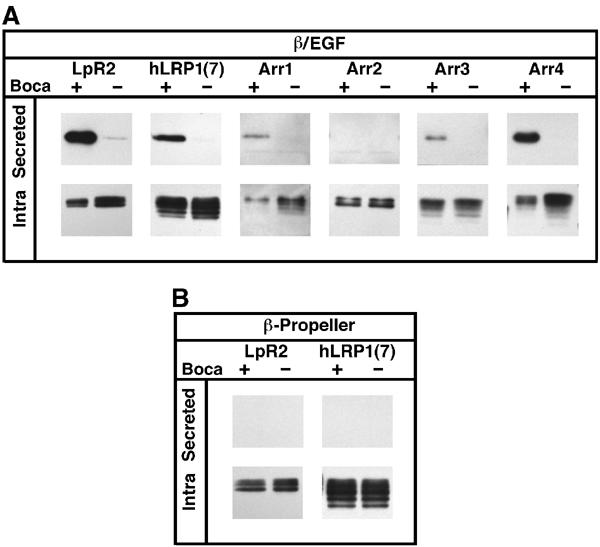
β-Propeller/EGF modules from other LDLRs also require Boca. (A) All four β-propeller/EGF pairs from Arrow and the seventh β-propeller/EGF pair from human LRP1 (hLRP1(7)) were tested for Boca dependency using the secretion assay. LpR2_β/E3 was included as a positive control. All proteins required Boca except for Arr2, for which Boca dependency could not be assessed because it failed to be secreted even in wild-type (+) cells. (B) Without their C-terminal EGF repeats, the β-propellers from LpR2 and hLRP1(7) failed to be secreted in wild-type and boca− cells.
A β-propeller/EGF module from a non-LDLR family member also requires Boca
Outside the LDLR family, only a few proteins contain β-propellers of the YWTD type. These are the EGFP, the basement membrane proteins Nidogen and Osteonidogen, and the tyrosine kinase receptor c-ros and its Drosophila homolog Sevenless (Springer, 1998). Of these, Nidogen and EGFP have a β-propeller/EGF module, whereas Osteonidogen and c-ros/Sevenless have YWTD type β-propellers but lack a C-terminal EGF repeat (Figure 1). To test if Boca is required for the maturation of these non-LDLR proteins, we first analyzed two proteins containing β-propeller/EGF modules from murine EGFP and Drosophila Nidogen. (Note that there are no orthologs of EGFP or Osteonidogen in Drosophila.) All of these domains were tested in the secretion assay, after inserting them into the LpR2_Gly vector (Figure 3A). Strikingly, the β-propeller/EGF module from EGFP required Boca for its proper maturation and secretion into the media (Figure 6A). Thus, EGFP conforms to the rule that β-propeller/EGF modules require Boca.
Figure 6.
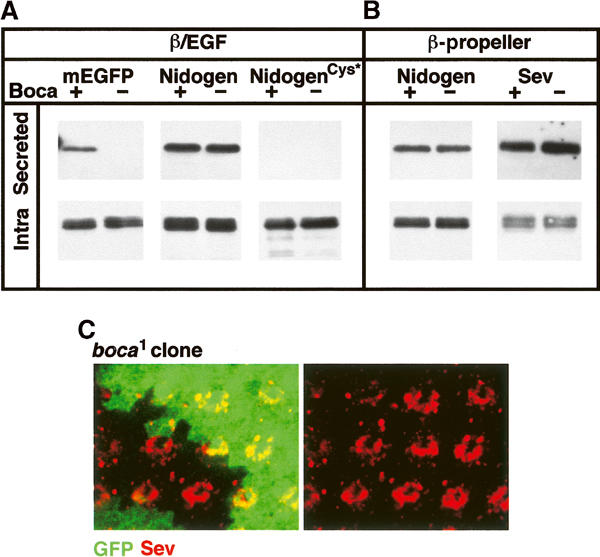
A β-propeller/EGF pair from EGFP requires Boca. (A, B) β-Propellers/EGF pairs (β/EGF) (A) and β-propellers from Sevenless (Sev), murine EGF precursor (mEGFP), and Nidogen were tested in S2 cells for Boca-dependent maturation using the secretion assay. NidogenCys* denotes a β/EGF protein that is unable to form its two intrapropeller disulfide bridges. Of these proteins, only the β-propeller/EGF pair from EGFP required Boca for secretion. (B) Clones of homozygous boca1 cells were induced in the eye imaginal disc and stained for Sevenless (red) and GFP (green). The absence of GFP marks the boca1 mutant cells. The focal plane was limited to the apical surface of these cells. Sevenless was localized at the apical cell membrane in both boca+ and boca1 cells.
In contrast to EGFP, the β-propeller/EGF module from Drosophila Nidogen was secreted into the media in both wild-type and boca− cells (Figure 6A), indicating that Nidogen does not require Boca for its maturation. Nidogen is the only exception we have found in which a β-propeller/EGF pair does not require Boca for its trafficking. However, the Nidogen β-propeller is unusual because it contains two disulfide bonds that may help to stabilize its structure. Such disulfide bonds are not present in the β-propellers of LDLRs or EGFP. Also, several of the hydrophobic residues that make up the β-propeller–EGF interface in the LDLR β-propeller (including Trp530) are hydrophilic in Nidogen's β-propeller. To test the significance of these differences, we determined if the Nidogen β-propeller could traffic through the secretory pathway in the absence of the EGF repeat. Unlike LpR2's β-propeller, the isolated β-propeller from Nidogen was secreted from both wild-type and boca− S2 cells (Figure 6B) (see also Takagi et al, 2003). We also mutated the four cysteines that form the two disulfide bridges in the Nidogen β-propeller to the equivalent amino acids present in the LpR2 β-propeller. This mutated β-propeller/EGF module was not secreted from wild-type or boca− cells. Although it is possible that these substitutions disrupt the structure of Nidogen's β-propeller, this result is consistent with the idea that these disulfide bonds are required for this β-propeller to fold properly. In further agreement with these results, Osteonidogen has a β-propeller very similar to Nidogen's (including the cysteines that form the disulfide bonds), but does not have a C-terminal EGF repeat (Kimura et al, 1998). Thus, Osteonidogen's β-propeller is also able to fold without an EGF repeat. In sum, these results reveal a correlation in which Boca-dependent β-propellers, such as those in LDLRs, also require an adjacent EGF repeat to mature properly, whereas Boca-independent β-propellers, such as Nidogen's, do not require an adjacent EGF repeat to traffic through the secretory pathway.
To provide additional support for this correlation, we examined a YWTD type β-propeller from Sevenless, which is not associated with an EGF repeat (Figure 1). From our previous results, we would predict that the folding and trafficking of this β-propeller would also be Boca independent. Consistent with this prediction, the Sevenless β-propeller did not require Boca to be secreted after its expression in S2 cells (Figure 6B). We also directly visualized the subcellular localization of full-length Sevenless protein in Drosophila tissues. Sevenless is normally expressed in a subset of photoreceptors in the eye imaginal disc where it accumulates at their apical cell membranes (Banerjee et al, 1987; Tomlinson et al, 1987). We induced clones of cells homozygous for the allele boca1, which codes for a nonfunctional Boca protein (Culi and Mann, 2003). In boca1 clones, the apical localization of Sevenless was still observed (Figure 6C), indicating that full-length Sevenless also does not require Boca to mature through the secretory pathway to reach the cell membrane.
Mesd can substitute for Boca in S2 cells
To begin to extend our results to the mammalian proteins, we analyzed if mouse Mesd can rescue the absence of Boca in S2 cells. Boca and its mammalian homolog Mesd have a high degree of sequence conservation, except that Mesd has 34 amino acids close to its C-terminus that are not present in Boca (Culi and Mann, 2003; Hsieh et al, 2003). To address the significance of this insertion, we tested if full-length Mesd could substitute for Boca in the secretion assay. As described above, LpR2ΔTM or its β-propeller/E3 module is not secreted in boca− S2 cells. However, secretion of these proteins was rescued by coexpressing Mesd (Figure 7). Thus, mouse Mesd can substitute for Drosophila Boca to allow the maturation of LpR2 in S2 cells.
Figure 7.
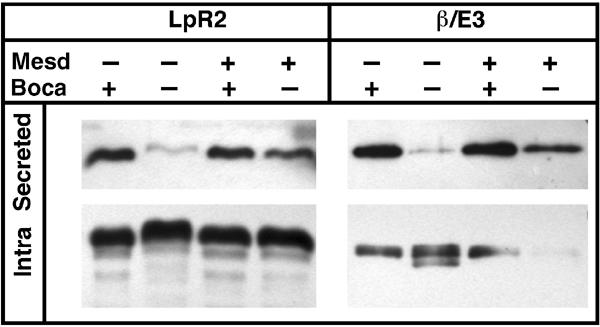
Boca and Mesd are functionally equivalent in S2 cells. Constructs encoding a secreted version of LpR2 or the β/E3 module from LpR2 were transfected into wild-type (+) and boca− S2 cells with a Mesd expression construct or mock DNA as indicated. Secreted and intracellular levels of the HA-tagged LpR2 proteins were measured using assay 2. For both proteins, secretion was rescued in boca− cells by expressing Mesd.
Conclusions
Taken together, our results demonstrate that Boca is an unusually specialized chaperone that is dedicated to the assembly of β-propeller/EGF modules. Boca-dependent β-propeller/EGF modules are present in all LDLR family members, consistent with Boca's requirement for the maturation of these receptors through the secretory pathway (Culi and Mann, 2003). Outside the LDLR family, Boca-dependent β-propeller/EGF modules are only present in EGFP. Thus, Boca is required for the trafficking of a highly select subset of eucaryotic proteins. Other YWTD type β-propellers, such as those from Nidogen and Sevenless, do not require Boca. These results can be generalized as follows: if the β-propeller, such as the one from LpR2, requires an EGF repeat to traffic through the secretory pathway, then it also requires Boca. Conversely, if the β-propeller can fold independently of any other domains, such as Nidogen's and Sevenless' β-propellers, then it does not require Boca. This correlation suggests that, for a subset of YWTD type propellers, Boca may be required to promote the correct association between the β-propeller and its C-terminal EGF repeat.
For those β-propeller/EGF modules that require Boca, our data further suggest that Boca transiently associates with the β-propeller as it is being translated in the ER. By binding to the β-propeller, Boca might maintain it in an interaction-competent state. This interaction must be transient because we only detect Boca binding to β-propeller/EGF modules when this module is prevented from reaching its mature state by either removing the E3 domain, as in LpR2_β, or by interfering with the association between the β-propeller and E3, as in the LpR2_βW530K/E3 mutant.
Recent results suggest that a different family of β-propellers, the WD40 type, also requires chaperones to fold properly. In this case, it is thought that the chaperonin TRiC/CCT, which also folds actin and tubulin, collaborates with upstream chaperones to fold a subset of WD40-containing proteins (Ho et al, 2002; Camasses et al, 2003; Craig, 2003; Siegers et al, 2003). Thus, β-propellers may be inherently difficult domains to fold, and specialized chaperones such as Boca have apparently evolved to assist in the folding process. Chaperones may be especially important when the folding of β-propellers also requires additional domains, as is the case in LDLRs and EGFP.
Materials and methods
Expression constructs
Constructs to express in Drosophila S2 cells, HA-tagged versions of Arr, Fz2, and full-length LpR2 have been described previously (Culi and Mann, 2003). Two sets of LpR2 deletion constructs were used in this work. In the first set, all proteins contained a TM domain, whereas in the second set the TM domains were deleted. In both cases, the C-terminal ends were fused to three copies of the HA tag followed by a stop codon. The different deletion constructs were cloned into pAc5.1 (Invitrogen) for expression in Drosophila S2 cells. All constructs were generated by standard cloning, PCR amplification, and inverted PCR mutagenesis techniques. Fragments generated by PCR were confirmed by sequencing. LpR2 residues were numbered as before (Culi and Mann, 2003).
LpR2 constructs with a TM domain are as follows: LpR2 (Culi and Mann, 2003); LpR2_∅: amino acids (aa) Cys34 to Tyr780 were substituted by a NotI site (GCG GCC GCN, coding for three Ala); LpR2_Gly: Cys34 to Glu717 were substituted by a NotI site; LpR2_E1/E2/β/E3: Cys34 to Cys319 were substituted by a NotI site; LpR2_A: Ile321 to Glu717 were substituted by a NotI site; LpR2_E1/E2: Gly320 to His406 were flanked by NotI sites and cloned into the NotI site of LpR2_Gly; LpR2_β: Glu404 to Gly670 were flanked by NotI sites and cloned into the NotI site of LpR2_Gly; LpR2_β/E3: Cys34 to Ser403 were substituted by a NotI site. LpR2 constructs without the TM domain were identical to those described above, except that they were truncated at Tyr780. Additional constructs without a TM domain are as follows: LpR2_E3: Cys34 to Val671 were substituted by a NotI site; LpR2_A/E1/E2/E3: Glu404 to Pro664 were substituted by a NotI site. Constructs shown in Figure 4A are as follows: HA-β: a signal peptide and three copies of the HA tag were placed N-terminal to the β-propeller (Gly405 to Gly670), followed by a stop codon; E3-HA: Ala24 to Gly670 were substituted by a NotI site. Cleavage of the signal peptide is predicted just before Val671. The following constructs contain sequences other than LpR2. The indicated aa were flanked by NotI sites and cloned into LpR2_Gly. hLRP1_β/EGF(7): Asp3026 to Ser3332 of human LRP1 (Q07954; GI:1708865); hLRP1_β(7): Asp3026 to Asp3288; Arr_β/EGF(1): Asn84 to Asn399 of Drosophila Arrow (NP_524737.1; GI:17864330); Arr_β/EGF(2): Gly400 to Val705; Arr_β/EGF(3): from Val704 to Pro1012; Arr_β/EGF(4): Pro1012 to Ala1315; Sev_β(3): Glu1397 to Pro1675 of Sevenless (P13368; GI:14424434); Nidogen_β/EGF: His1041 to the end of Drosophila predicted protein NP_610575.1, GI:24652408; Nidogen_β: His1041 to Gln1304; Nidogen_βCys*/EGF: Cys1081, 1220, 1231, and 1302 in Nidogen_β/EGF were mutated to the structurally equivalent positions occurring in LpR2 β-propeller (Val, Tyr, Ser, and Thr, respectively); mEGFP_β/EGF: Ser485 to Asp790 of murine EGFP (P01132; GI:119227).
LpR2_βW530K/E3 was identical to LpR2_β/E3 except that Trp530 in the β-propeller was changed to Lys. Trp530 of LpR2 was targeted for mutagenesis based on the following considerations. Quantitation of buried, solvent-accessible surface area in the β-propeller–EGF domain interface of the LDLR (Jeon et al, 2001) using the areaimol program of the CCP4 suite (Collaborative Computational Project, 1994) identified Phe509 as the most buried hydrophobic residue that was conserved in LDLR proteins but not in Nidogen. Phe509 makes van der Waals contacts with EGF domain residues Pro664, Gln665, and Ile666 but does not appear to be required for the structural integrity of the β-propeller. Therefore, Trp530 of LpR2, which is equivalent to Phe509 in hLDLR, was mutated to Lys, a residue with a high propensity for surface exposure, to disrupt the β-propeller–E3 interface.
Trafficking assays and Mesd rescue
Drosophila S2 cells were cultured at room temperature in serum-free media Sf-900 II SFM (Gibco) containing penicillin (50 IU/ml) and streptomycin (50 μg/ml). boca− cells were generated by incubation with dsRNA as described in Culi and Mann (2003). For immunostaining, wild-type and boca− S2 cells were seeded into eight-well slides (Falcon) at 300 000 cells/well and then transfected with 0.5 μg of the expression construct per well using cellfectin (Invitrogen). After 24 h, cells were fixed with 4% paraformaldehyde in PBS for 20 min, washed with PBS–0.3% triton and incubated with the primary antibodies rat anti-HA (Roche) and anti-Boca (Culi and Mann, 2003), followed by incubation with the corresponding secondary antibodies and Texas-red-conjugated phalloidin. Finally, cells were analyzed by confocal microscopy. For the secretion assay, wild-type and boca− cells were seeded in 48-well plates at 300 000 cells/well and 300 μl media/well. They were transfected with a mixture of the expression construct and pAc5.1-lacZ at a ratio of 3:1. The total amount of transfected DNA was 0.5 μg/well. After 24 h, the medium was collected and the cells were lysed in 200 μl of lysis buffer (reporter lysis buffer, Promega). β-Galactosidase activity of the lysate was analyzed using ONPG as substrate. After normalizing for transfection efficiencies, media and cell lysate fractions were analyzed by PAGE and immunoblotting with anti-HA antibody.
For the Mesd rescue experiment, the mesd coding sequence (Hsieh et al, 2003) was cloned into pAc5.1 for its expression in S2 cells. Wild-type and boca− S2 cells were transfected as described above with a mixture of DNAs as shown in Figure 7 and also containing pAc5.1-lacZ. Nonspecific DNA was added when necessary. After normalization for transfection efficiencies, media and cell lysate fractions were analyzed as described above.
Co-immunoprecipitation
S2 cells were grown in 60 mm tissue culture dishes at 450 000 cells/dish and transfected with 5 μg of the expression construct. After 24 h, cells were collected and washed twice in 1 ml of cold D-PBS. The membrane-permeable crosslinking reagent DSP (Pierce) was added to the last wash at 100 μg/ml, in a 1 ml volume (∼0.5 × 106 cells) and incubated with rotation at room temperature for 30 min. The nonreacted crosslinker reagent was inactivated by the addition of Tris buffer (pH 7.4) at a final concentration of 100 mM and allowed to react for 5 min at room temperature with rotation. Cells were then pelleted and lysed with 0.5 ml of lysis buffer (1% triton, 10 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, 1% BSA, protease inhibitor cocktail (Roche), PMSF 100 μg/ml) at 4°C for 15 min. After eliminating the debris by centrifugation, the lysate was incubated with 20 μl of anti-HA affinity matrix (Roche) for 2 h at 4°C with rotation. The beads were extensively washed with buffer (0.1% triton, 50 mM Tris (pH 7.4), 300 mM NaCl, 5 mM EDTA) five times at room temperature for a total time of 45 min. The HA-tagged proteins bound to the beads were eluted with 20 μl of 2 × PAGE loading buffer (200 mM DTT, 4% SDS, 100 mM Tris (pH 6.8), 20% glycerol, 0.1% bromophenol blue) at 100°C for 4 min. This treatment also cleaves DSP and reverses the crosslink. The eluted fractions were analyzed by PAGE and immunoblotting with the anti-HA and anti-Boca antibodies. In the gels shown in Figure 4, 1% of the sample was loaded for the anti-HA blots and the remainder was loaded for the anti-Boca blots. Note that there is a low level of nonspecific binding of Boca to the anti-HA matrix seen in cells that do not express any HA-tagged proteins (−).
Drosophila stocks and clonal analysis
Clones of cells homozygous for the boca1 allele were generated as described (Culi and Mann, 2003). Eye imaginal discs were stained with anti-Sevenless antibody (Tomlinson et al, 1987) following standard techniques and analyzed by confocal microscopy.
Acknowledgments
We thank J Herz, B Holdener, G Struhl, the Berkeley Drosophila Genome Project, and the IMAGE consortium for materials, E Gouaux, B Konforti, MJ García-García, and B Gebelein for comments on the manuscript, and members of the lab for discussions. This work was supported by grants from the NIH and The American Heart Association to RSM.
References
- Banerjee U, Renfranz PJ, Hinton DR, Rabin BA, Benzer S (1987) The sevenless+ protein is expressed apically in cell membranes of developing Drosophila retina; it is not restricted to cell R7. Cell 51: 151–158 [DOI] [PubMed] [Google Scholar]
- Bork P, Downing AK, Kieffer B, Campbell ID (1996) Structure and distribution of modules in extracellular proteins. Q Rev Biophys 29: 119–167 [DOI] [PubMed] [Google Scholar]
- Camasses A, Bogdanova A, Shevchenko A, Zachariae W (2003) The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol Cell 12: 87–100 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Craig EA (2003) Eukaryotic chaperonins: lubricating the folding of WD-repeat proteins. Curr Biol 13: R904–R905 [DOI] [PubMed] [Google Scholar]
- Culi J, Mann RS (2003) Boca, an endoplasmic reticulum protein required for wingless signaling and trafficking of LDL receptor family members in Drosophila. Cell 112: 343–354 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A (2003) Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4: 181–191 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A (1999) Setting the standards: quality control in the secretory pathway. Science 286: 1882–1888 [DOI] [PubMed] [Google Scholar]
- Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Herz J (2001) The LDL receptor gene family: (un)expected signal transducers in the brain. Neuron 29: 571–581 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Howell BW, Herz J (2001) The LDL receptor gene family: signaling functions during development. Curr Opin Neurobiol 11: 74–81 [DOI] [PubMed] [Google Scholar]
- Hsieh JC, Lee L, Zhang L, Wefer S, Brown K, DeRossi C, Wines ME, Rosenquist T, Holdener BC (2003) Mesd encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell 112: 355–367 [DOI] [PubMed] [Google Scholar]
- Hussain MM, Strickland DK, Bakillah A (1999) The mammalian low-density lipoprotein receptor family. Annu Rev Nutr 19: 141–172 [DOI] [PubMed] [Google Scholar]
- Jeon H, Meng W, Takagi J, Eck MJ, Springer TA, Blacklow SC (2001) Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD–EGF domain pair. Nat Struct Biol 8: 499–504 [DOI] [PubMed] [Google Scholar]
- Jorgensen MM, Jensen ON, Holst HU, Hansen JJ, Corydon TJ, Bross P, Bolund L, Gregersen N (2000) Grp78 is involved in retention of mutant low density lipoprotein receptor protein in the endoplasmic reticulum. J Biol Chem 275: 33861–33868 [DOI] [PubMed] [Google Scholar]
- Kimura N, Toyoshima T, Kojima T, Shimane M (1998) Entactin-2: a new member of basement membrane protein with high homology to entactin/nidogen. Exp Cell Res 241: 36–45 [DOI] [PubMed] [Google Scholar]
- Kuznetsov G, Chen LB, Nigam SK (1994) Several endoplasmic reticulum stress proteins, including ERp72, interact with thyroglobulin during its maturation. J Biol Chem 269: 22990–22995 [PubMed] [Google Scholar]
- Nimpf J, Schneider WJ (2000) From cholesterol transport to signal transduction: low density lipoprotein receptor, very low density lipoprotein receptor, and apolipoprotein E receptor-2. Biochim Biophys Acta 1529: 287–298 [DOI] [PubMed] [Google Scholar]
- Parodi AJ (2000) Protein glucosylation and its role in protein folding. Annu Rev Biochem 69: 69–93 [DOI] [PubMed] [Google Scholar]
- Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J (2002) Structure of the LDL receptor extracellular domain at endosomal pH. Science 298: 2353–2358 [DOI] [PubMed] [Google Scholar]
- Siegers K, Bolter B, Schwarz JP, Bottcher UM, Guha S, Hartl FU (2003) TRiC/CCT cooperates with different upstream chaperones in the folding of distinct protein classes. EMBO J 22: 5230–5240 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sitia R, Braakman I (2003) Quality control in the endoplasmic reticulum protein factory. Nature 426: 891–894 [DOI] [PubMed] [Google Scholar]
- Springer TA (1998) An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J Mol Biol 283: 837–862 [DOI] [PubMed] [Google Scholar]
- Stevens FJ, Argon Y (1999) Protein folding in the ER. Semin Cell Dev Biol 10: 443–454 [DOI] [PubMed] [Google Scholar]
- Strickland DK, Gonias SL, Argraves WS (2002) Diverse roles for the LDL receptor family. Trends Endocrinol Metab 13: 66–74 [DOI] [PubMed] [Google Scholar]
- Takagi J, Yang Y, Liu JH, Wang JH, Springer TA (2003) Complex between nidogen and laminin fragments reveals a paradigmatic beta-propeller interface. Nature 424: 969–974 [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Bowtell DD, Hafen E, Rubin GM (1987) Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell 51: 143–150 [DOI] [PubMed] [Google Scholar]
- Willnow TE, Nykjaer A, Herz J (1999) Lipoprotein receptors: new roles for ancient proteins. Nat Cell Biol 1: E157–E162 [DOI] [PubMed] [Google Scholar]


