Abstract
The cauliflower mosaic virus reinitiation factor TAV interacts with host translation initiation factor 3 (eIF3) and the 60S ribosomal subunit to accomplish translation of polycistronic mRNAs. Interaction between TAV and eIF3g is critical for the reinitiation process. Here, we show that eIF4B can preclude formation of the TAV/eIF3 complex via competition with TAV for eIF3g binding; indeed, the eIF4B- and TAV-binding sites on eIF3g overlap. Our data indicate that eIF4B interferes with TAV/eIF3/40S ribosome complex formation during the first initiation event. Consequently, overexpression of TAV in plant protoplasts affects only second initiation events. Transient overexpression of eIF4B in plant protoplasts specifically inhibits TAV-mediated reinitiation of a second ORF. These data suggest that TAV enters the host translation machinery at the eIF4B removal step to stabilize eIF3 on the translating ribosome, thereby allowing translation of polycistronic viral RNA.
Keywords: caulimovirus, initiation, ribosome, translation reinitiation, translational transactivator (TAV)
Introduction
Cap- and scanning-dependent translation initiation—the main initiation pathway of protein synthesis in eucaryotes—involves numerous factors and the interplay of a succession of protein–protein and protein–RNA complexes (Hershey and Merrick, 2000). mRNA is recruited to ribosomes through recognition of the capped 5′-end by eIF4F (central adaptor eIF4G, cap-binding protein eIF4E and helicase eIF4A). eIF4A, supported by eIF4B (Jaramillo et al, 1990) and ATP, is thought to melt RNA secondary structure, facilitating binding of a 43S preinitiation complex consisting of the 40S ribosomal subunit carrying eIF3, eIF1, eIF1A and the ternary complex (eIF2, GTP and Met-tRNAi). The resulting 48S preinitiation complex is strengthened by multiple interactions between RNA and the 40S ribosomal subunit mediated by eIF4F, eIF4B and eIF3. The multisubunit factor eIF3 thus plays a major role in mediating linkage between the cap-binding complex and the complex centered around the 40S ribosomal subunit (Hershey and Merrick, 2000). eIF3 interacts with the central domain of mammalian eIF4G (Lamphear et al, 1995) and with eIF4B in yeast (via eIF3g; Vornlocher et al, 1999) and mammals (via eIF3a; Méthot et al, 1996a). Once assembled, the 48S preinitiation complex scans the 5′-untranslated region (5′-UTR) until a suitable initiation codon is encountered (Pestova et al, 1998), whereupon the ORF is translated. Other than reinitiation after short open reading frames (sORFs; Fütterer and Hohn, 1992; Hinnebusch, 2000; Morris and Geballe, 2000), reinitiation of translation is suppressed in eucaryotes. To date, the only known exception is reinitiation mediated by the cauliflower mosaic virus (CaMV) reinitiation factor TAV (transactivator/viroplasmin; reviewed in Hohn et al, 2002).
CaMV is a plant pararetrovirus that produces a terminally redundant 35S RNA that acts both as replicative intermediate and as polycistronic mRNA. Translation initiation of the 35S polycistronic RNA is strongly cap dependent, and expression of all major ORFs absolutely requires TAV. The transactivation activity of TAV does not require cis-specific RNA sequences and is not much affected by the distance between two ORFs (Fütterer and Hohn, 1991). However, the presence of an sORF upstream of the long ORFs strongly enhances TAV-mediated reinitiation. In addition to mediating polycistronic translation from the 35S RNA, TAV is implicated in the regulation of post-translational events such as stabilization of viral proteins (Kobayashi et al, 1998), and virion assembly (Himmelbach et al, 1996).
Recently, we have shown that TAV interacts with the host translational machinery via eIF3 and the 60S ribosomal subunit, influencing its behavior in favor of reinitiation (Park et al, 2001). Complexes between TAV and eIF3 subunit g (Park et al, 2001), as well as between TAV and 60S ribosomal proteins L24 (Park et al, 2001) and L18 (Leh et al, 2000), have been characterized. The TAV–eIF3g interaction is critical for TAV-mediated reinitiation of translation both in vivo and in vitro. The significant accumulation of eIF3 in polysomal fractions isolated from CaMV-infected cells suggests that TAV might prevent removal of eIF3 from the host translation machinery during translation of viral polycistronic mRNA, thus sequestering it for subsequent reinitiation events (Park et al, 2001).
Here, we identify a new player in the TAV-mediated reinitiation process, initiation factor 4B, and demonstrate that the complex formed between the canonical initiation factors eIF4B and eIF3 on the 40S ribosome can preclude the binding of TAV to 40S-bound eIF3 prior to the 60S subunit-joining step. Thus TAV can interact with ribosome-bound eIF3 only after disruption of the 48S initiation complex, explaining why it affects exclusively the second (and following) round of initiation on polycistronic mRNA.
Results
The eIF4B- and TAV-binding sites on eIF3g overlap
We used the yeast two-hybrid assay to investigate whether plant eIF3 can interact with eIF4B via subunit g of eIF3 as proposed in yeast (Vornlocher et al, 1999). Our analysis also included eIF3i, which is linked to eIF3g in yeast (Verlhac et al, 1997). Complete or truncated fragments encoding these three proteins from Arabidopsis thaliana were fused to the DNA binding (BD) and activation (AD) domains of the transcription activator Gal4, and the resulting constructs were tested in all combinations for two-hybrid interactions in yeast. We detected interactions between AD-eIF3g and either BD-eIF4B or BD-eIF3i, but not with the reciprocal combinations (i.e. BD-eIF3g versus AD-eIF4B or AD-eIF3i) (data not shown). Observing false-negative interactions between full-length proteins in two-hybrid assays is not uncommon and can be caused by steric hindrance or other factors. To confirm that the full-length proteins do in fact interact, GST pull-down experiments were performed. GST-eIF3g and GST were expressed in bacteria and purified by immobilization on glutathione–sepharose beads. 35S-labeled eIF4B, eIF3i and luciferase (as a control) were produced by in vitro translation. The labeled proteins were incubated with GST, or GST-eIF3g, bound to glutathione beads. After removal of the supernatant containing the unbound proteins and extensive washing, proteins bound to glutathione beads were eluted, separated by SDS–PAGE and visualized by autoradiography. After incubation with GST-eIF3g, 3i and eIF4B were present in the bound fraction (Figure 1, lanes 5 and 7), while they remained in the unbound fraction after incubation with GST (lanes 10 and 12). The 35S-labeled luciferase control always remained in the unbound fraction (lanes 8 and 14). These results demonstrate specific binding of full-length eIF3g to eIF3i and eIF4B in vitro.
Figure 1.
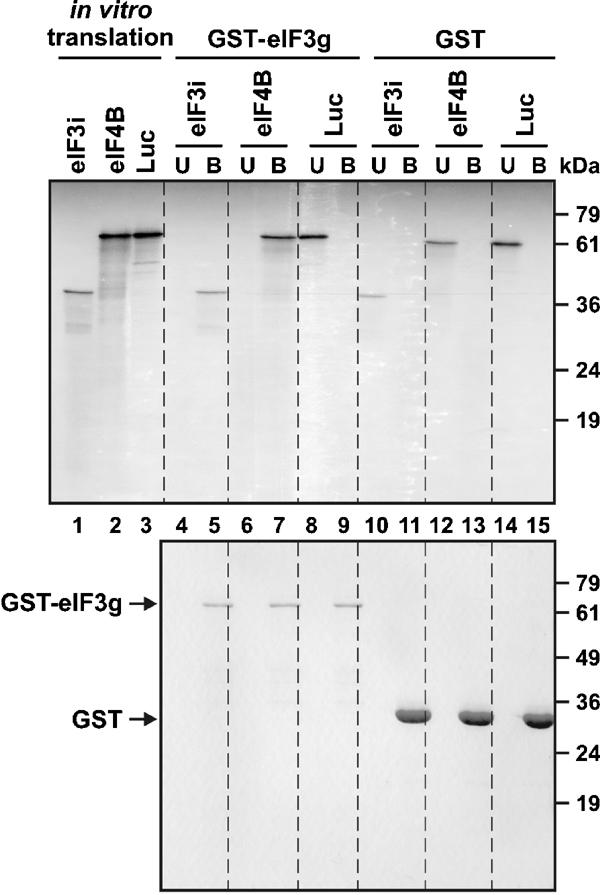
eIF3g interacts with eIF4B and eIF3i in vitro. Upper panel: Autoradiograph of SDS–PAGE gel of 35S-labeled proteins produced by in vitro translation of eIF3i, eIF4B or luciferase mRNAs (lanes 1, 2 and 3, respectively). Lanes 4–15: interaction of 14.5 pmol GST-eIF3g or 48 pmol GST bound to glutathione–sepharose 4B beads with approximately 3–4 pmol 35S-labeled eIF3i, eIF4B or luciferase. U, unbound; B, bound fractions. Lower panel: Same gel stained with Coomassie blue. The positions of size markers are indicated on the right.
We next identified the domains in eIF4B, eIF3g and eIF3i that mediate their mutual interactions in vivo. The central part of eIF3g—F2, a fragment spanning amino-acid residues 66–173 and including a zinc-finger motif—interacted strongly with the eIF4B central domain including the G-rich domain (residues 121–275 (4B-2); Figure 2A and B). eIF3g-F2 is also involved in interaction with CaMV TAV (Park et al, 2001). eIF3g-F1 (N-terminal residues 1–65) interacted strongly with the N-terminal 162 amino acids of 3i (3i-1; Figure 2B and C). In addition, weak interactions were observed between eIF3g-F2 and 3i-1, and between eIF3g-F1 and eIF4B-2 (Figure 2B), showing that the two interaction domains of eIF3g might overlap. The C-terminal part of eIF3g contributes to neither eIF4B nor eIF3i binding (Figure 2B). Likewise, the N- and C-terminal domains (1–120 and 276–545) of eIF4B, and the C-terminal domain (163–328) of eIF3i seem to be not involved in binding to eIF3g (Figure 2A and C).
Figure 2.
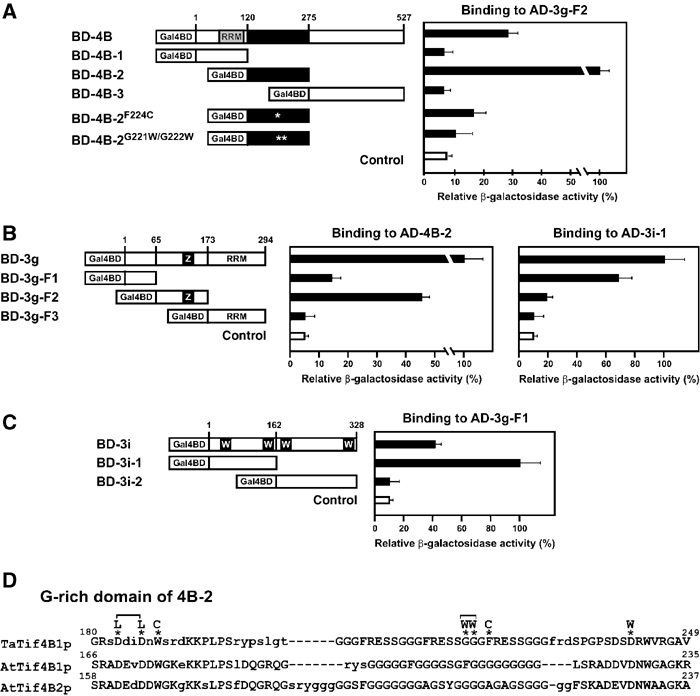
Mapping interacting regions of eIF4B, eIF3g and eIF3i in the yeast two-hybrid system. Interactions were scored by measuring β-galactosidase activity in liquid assays. In each set of experiments, the highest value of β-galactosidase activity in the diploid yeast transformed with the corresponding constructs was set to 100% (8 Miller units for BD-4B-2 interaction with AD-eIF3g-F2, 5 and 4 Miller units for BD-3g interaction with AD-4B-2 and AD-eIF3i-1, respectively, and 7 Miller units for BD-3i-1 interaction with AD-eIF3g-F1). Results in (A–C) represent the mean values from triplicate experiments±standard deviation. (A) eIF4B constructs fused to the Gal4 binding domain (BD), and their binding activities to eIF3g fused to Gal4AD (AD-3g-F2). RRM, RNA recognition motif; asterisk, amino-acid substitutions. (B) eIF3g constructs fused to Gal4BD and their binding activities to the central domain of eIF4B fused to the Gal4 activation domain (AD) (AD-4B-2) and to the N-terminal part of eIF3i fused to Gal4AD (AD-3i-1). Z, zinc-finger domain. (C) eIF3i constructs fused to Gal4BD and their binding activities to AD-3g-F1. W, WD40-repetitive motifs. (D) Alignment of the central region of domain 4B-2 with that of wheat eIF4B (TaTif4B1p) and Arabidopsis eIF4B1 and 2 (AtTif4B1p and AtTif4B2p) generated using MACAW v. 2.0.5. Blocks of similarity between the deduced amino-acid sequences are indicated by capital letters in agreement with Blossom 62 and Jonson amino-acid substitution matrices. Dashes, gaps in the sequence introduced to maximize alignment; asterisks, mutations.
Mammalian eIF4B exists as a homodimer in vitro, the self-dimerization motif mapping to the centrally situated DRYG-rich domain (Méthot et al, 1996a). This motif is also present in Saccharomyces cerevisiae eIF4B. The comparable region in plant eIF4B is rich only in glycine (G) and aspartic acid (D). A. thaliana eIF4B is a monomer with an extended shape (L Mayberry and KS Browning, unpublished). The central domain of wheat eIF4B (TaTif4B1p) has four repeats of a GGGFRESS motif, and the equivalent Arabidopsis sequence has four repeats of 4–9 glycine residues (see Figure 2D). Analysis of the TaTif4B1p GGGFRESS motif revealed its significance for the eIF4B–eIF3g interaction. eIF4B derivatives F224C or G221W/G222W failed to interact with eIF3g (Figure 2A), while other mutations (D183L/D186L, W188C and D242W; Figure 2D) did not affect the interaction.
eIF4B outcompetes TAV for binding to intact eIF3
As residues 66–173 of eIF3g interact with both eIF4B and CaMV TAV (Park et al, 2001), we tested whether eIF3g can interact with eIF4B and TAV simultaneously. GST-TAV, purified recombinant Arabidopsis eIF4B and intact purified wheat germ eIF3 were used in GST pull-down assays. GST-TAV pulled down eIF3, confirming earlier results, but not eIF4B (Figure 3A). No binding was observed when GST was used as a control (Figure 3A). When a mixture of eIF3 and eIF4B (1:1 molar ratio) was incubated with GST-TAV, almost no eIF3 was bound to GST-TAV (Figure 3A). This was the first indication that eIF4B outcompetes TAV for binding to eIF3. To exclude that the bulky GST tag might be causing specific binding, the experiment was also performed using the small His tag, with similar results (Figure 3B). In this case, two different ratios of eIF4B to eIF3 were used. At a ratio of 0.5:1, some eIF3 was still found in the bound fraction, while at a ratio of 1:1 it was seen only in the unbound fraction (Figure 3B).
Figure 3.
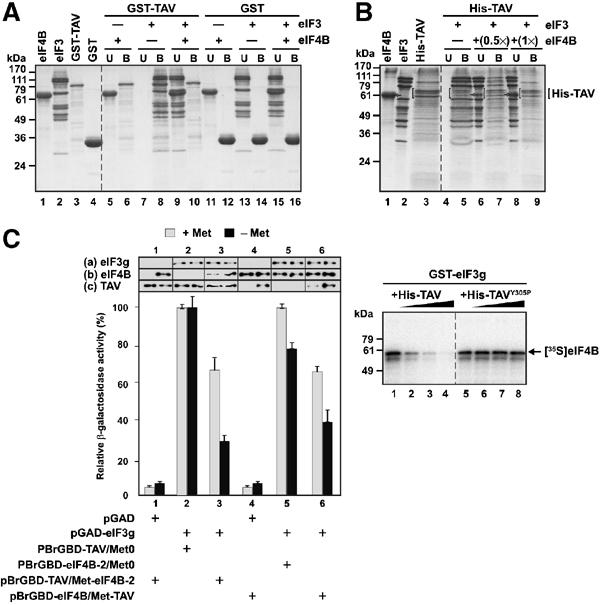
eIF4B outcompetes TAV for binding to eIF3. (A) GST-TAV and GST were overexpressed in E. coli and purified by affinity chromatography; eIF3 and eIF4B were purified from WGE. Lanes 1–4: Purified components. For pull-down experiments, GST-TAV or GST bound to glutathione–sepharose beads were incubated with the components to be tested. After washing, unbound (U) and bound (B) fractions were separated by SDS–PAGE and stained with Coomassie blue (note that TAV is less well stained by Coomassie blue than eIF4B and eIF3 in equimolar amounts). In all, 20 pmol GST-TAV or GST was incubated with 20 pmol eIF4B (lanes 5, 6 and 11, 12), 15 pmol eIF3 (lanes 7, 8 and 13, 14) or 15 pmol eIF3 followed by incubation with 31 pmol eIF4B (lanes 9, 10 and 15, 16). The positions of size markers are indicated on the left. (B) His-TAV was overexpressed in E. coli and purified by affinity chromatography. Lanes 1–3: Purified components. Bracketed triple bands, His-TAV (confirmed by Western blotting with anti-TAV antibodies); arrow, eIF4B. For pull-down experiments, His-TAV bound to Ni-NTA agarose beads was incubated with the components to be tested. After washing the beads, unbound (U) and bound (B) fractions were analyzed by SDS–PAGE. A measure of 20 pmol His-TAV was incubated with 15 pmol eIF3 (lanes 4 and 5), or 15 pmol eIF3 followed by incubation with 8 pmol (lanes 6 and 7) or 15 pmol eIF4B (lanes 8 and 9). The positions of size markers are indicated on the left. (C) Quantification of β-galactosidase activity in yeast cells cotransformed with the plasmid pairs indicated. The highest values of β-galactosidase activity transformed with pGAD-eIF3g and either pBrGBD-TAV/Met0 or pBrGBD-eIF4B-2/Met0±inhibitor (Met) are set to 100%. Expression levels of eIF3g (a), eIF4B (b) and TAV (c) in yeast cells±Met were analyzed by immunoblotting using polyclonal antibodies against eIF3g, eIF4B and TAV. (D) GST-eIF3g, His-TAV and His-TAVY305P were overexpressed in E. coli and purified by affinity chromatography. 35S-labeled eIF4B was produced by in vitro translation. The effect of increasing concentrations of His-TAV (lanes 2–4) or His-TAVY305P (lanes 6–8) on the binding affinity of [35S]eIF4B (2 pmol) to GST-eIF3g (3.5 pmol) was tested by adding 7, 28 and 56 pmol TAV (or mutant TAV) to the binding reaction.
To investigate competition between TAV and eIF4B for eIF3 binding in vivo, we used a three-hybrid system (Tirode et al, 1997) in which, besides the pair of two-hybrid fusion proteins, a third protein is conditionally expressed under the control of the Met25 promoter, which is inhibited by methionine (Met). We analyzed β-galactosidase expression in yeast cells transformed with pGAD-eIF3g expressing eIF3g and plasmid pBrGBD-TAV/Met-eIF4B-2 expressing TAV and, in the absence of Met, the central peptide of eIF4B. Overexpression of eIF4B decreased reconstitution of the Gal4 transcriptional activator complex three-fold, indicating that the level of interaction between TAV and eIF3g was significantly decreased in vivo (Figure 3C, cf. columns 2 and 3, −Met). An ∼45% reduction in the strength of interaction between eIF4B and eIF3g was observed when TAV was overexpressed from plasmid pBrGBD-eIF4B-2/Met-TAV (Figure 3C, cf. columns 5 and 6, −Met). The presence of the inhibitor (Met) in the medium still allowed a low level of expression of the third partner (eIF4B, see Figure 3C, cf. lanes 2 and 3, +Met; and TAV, cf. lanes 5 and 6, +Met), resulting in an ∼30% reduction in the interaction strength (Figure 3C, cf. lanes 2 and 3, +Met and 5 and 6, +Met). In control experiments, cells cotransformed with pGAD-eIF3g and either pBrGBD-TAV/Met0 or pBrGBD-eIF4B-2/Met0 showed a LacZ+ phenotype both in the presence and absence of Met (Figure 3C, lanes 2 and 5). These results show that TAV and eIF4B compete for eIF3g binding in vivo, with the eIF3g–eIF4B interaction again being somewhat stronger than that between eIF3g and TAV.
To further analyze the ability of TAV to compete with eIF4B bound to eIF3, polyhistidine-tagged wild-type TAV (His-TAV) and a TAV mutant devoid of eIF3g affinity (His-TAVY305P; Park et al, 2001) were tested for their ability to compete with 35S-eIF4B for binding to GST-eIF3g. Addition of a two-fold molar excess of wild-type His-TAV to GST-eIF3g significantly reduced the amount of 35S-eIF4B bound to GST-eIF3g, and an eight-fold molar excess abolished the interaction (Figure 3D, lanes 2–4). His-TAVY305P had no effect (Figure 3D, lanes 6–8).
Taken together, these results suggest that eIF3g cannot simultaneously interact with molecules of eIF4B and TAV, that the same domain in eIF3g is responsible for both interactions, and that eIF4B can outcompete TAV for eIF3g binding.
Both eIF4B and TAV can form stable complexes with eIF3 and 40S ribosomal subunits
As eIF4B interacts with the TAV-binding site of eIF3 and, in mammalian systems, also with the 18S RNA component of the 40S ribosomal subunit (Méthot et al, 1996b), we investigated whether TAV interferes with the eIF4B interaction network.
First, we analyzed the physical association of GST-eIF4B with purified wheat germ 40S subunits and eIF3 in vitro. GST-eIF4B was incubated with 40S subunits alone, or with a preformed complex of 40S subunit and eIF3. This indeed led to the formation of a GST-eIF4B/40S complex (Figure 4A, lane 6), GST-eIF4B/eIF3 complex (Figure 4B, lane 6) and a higher-order complex GST-eIF4B/eIF3/40S (Figure 4B, lane 8). Addition of His-TAV in molar ratio did not change either of these complexes, with His-TAV remaining in the unbound fraction (Figure 4A, lane 8 and Figure 4B, lane 10). Furthermore, the GST-eIF4B/eIF3 ternary complex with the 40S ribosomal subunit was not disrupted even by a five-fold molar excess of His-TAV (Figure 4B, lanes 11 and 12). In a control experiment, no binding of eIF3, 40S and His-TAV was detected (data not shown; see also Figure 3A, lanes 14 and 16 and Figure 4A, lane 12). The reciprocal experiment, in which a GST-TAV/eIF3/40S complex was challenged with purified eIF4B, gave similar results, with the challenger remaining in the unbound fraction (Figure 4C, lanes 9–12), showing that the preformed GST-TAV/eIF3 complex bound to the 40S ribosome (Figure 4C, lane 8) strongly resists eIF4B binding. Interestingly, eIF3 complexed with TAV, or TAV alone, interferes with direct binding of eIF4B to the 40S ribosomal subunit (Figure 4C, lanes 9 and 11).
Figure 4.
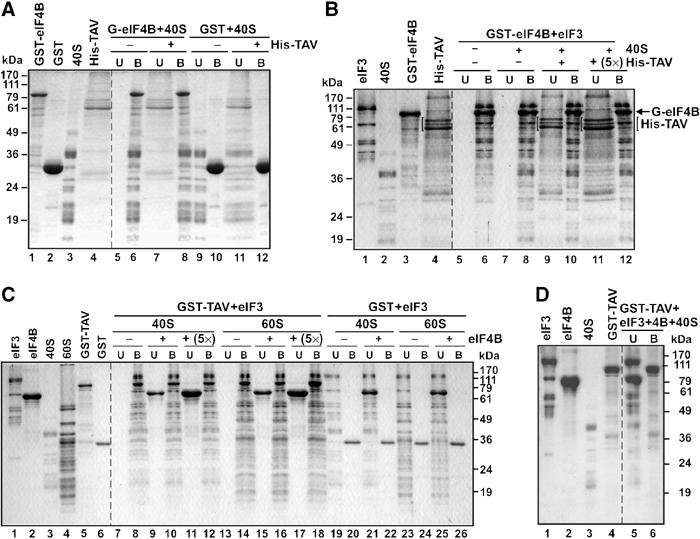
Interaction network between eIF4B, TAV, eIF3 and ribosomal subunits. GST, GST-eIF4B, GST-TAV and His-TAV were overexpressed in E. coli and purified by affinity chromatography. 60S, 40S, eIF3 and eIF4B were purified from WGE. The left panel of each gel shows the purified components. For pull-down experiments, GST or GST fusion proteins were bound to glutathione–sepharose beads and incubated with the components to be tested. After washing, unbound (U) and bound (B) fractions were analyzed by SDS–PAGE. The positions of size markers are indicated. (A) Binding of GST-eIF4B to 40S. A measure of 20 pmol GST-eIF4B, or GST alone, was incubated with 40S ribosomal subunits (22 pmol; lanes 5–8 and 9–12, respectively). A subset was further incubated with 21.5 pmol His-TAV (lanes 7, 8 and 11, 12). (B) GST-eIF4B interactions with eIF3 and 40S subunits. GST-eIF4B (20 pmol) was incubated with 20 pmol eIF3 (lanes 5–12). A subset of GST-bound fractions was then further incubated with 22 pmol 40S subunits without (lanes 7–8) or with increasing concentrations of His-TAV—14 pmol (lanes 9 and 10) and 70 pmol (lanes 11 and 12). (C) eIF4B does not displace eIF3 from eIF3/GST-TAV/60S or GST-TAV/eIF3/40S complexes. In all, 20 pmol GST-TAV or GST was incubated with 20 pmol eIF3 and 22 pmol 40S (lanes 7–12 and 19–22, respectively) or 20 pmol eIF3 and 24 pmol 60S (lanes 13–18 and 23–26, respectively). A subset of GST-bound fractions was further incubated with 21 pmol (lanes 9–10, 15–16, 21–22 and 25–26) or 100 pmol (lanes 11–12 and 17–18) eIF4B. (D) eIF4B interferes with the formation of the GST-TAV/eIF3/40S complex. GST-TAV (20 pmol) was incubated with 20 pmol eIF3, 22 pmol 40S and 100 pmol eIF4B (lanes 5 and 6) simultaneously.
Together, these experiments indicate that high activation energy separates the two types of ternary complexes, giving either one a quasistability in the presence of the competitive challenger. However, incubating a mixture of eIF4B, eIF3, 40S and GST-TAV in a ratio of 5:1:1:1 did not result in the formation of a complex between GST-TAV, eIF3 and 40S (Figure 4D, lane 6); eIF4B, eIF3 and 40S were found in the unbound fraction (Figure 4D, lane 5).
eIF3 associates with 60S subunits only in the presence of TAV, and in CaMV-infected plants both TAV and eIF3 cosediment with plant polysomes (Park et al, 2001). We asked whether eIF4B could displace eIF3 from the eIF3/TAV/60S complex. Addition of a molar or ∼5-fold molar excess of eIF4B to the preformed eIF3/GST-TAV/60S complex did not lead to disruption of the complex; the added eIF4B was found mainly in the unbound fraction (Figure 4C, lanes 15 and 17).
Thus, in vitro, eIF3/TAV or eIF3/eIF4B are able to form stable supercomplexes with the 40S ribosome, and eIF3/TAV with the 60S ribosome. Any exchange between TAV and eIF4B within these already preformed complexes is restricted under our conditions (i.e. with the limited set of initiation factors used).
60S ribosomal subunit joining disrupts the eIF4B/eIF3/40S complex
We investigated if 60S subunit joining triggers removal of eIF4B from the eIF4B/eIF3/40S complex, and of TAV/eIF3 from the TAV/eIF3/40S complex, by challenging with addition of purified 60S ribosomal subunits in vitro. Both complexes were preformed and incubated with 60S ribosomal subunits at a ratio of 1:1. Addition of 60S to the GST-eIF4B/eIF3/40S complex led to destabilization of the complex, and 60S, 40S and eIF3 were found in the unbound fraction (Figure 5A, lane 6).
Figure 5.
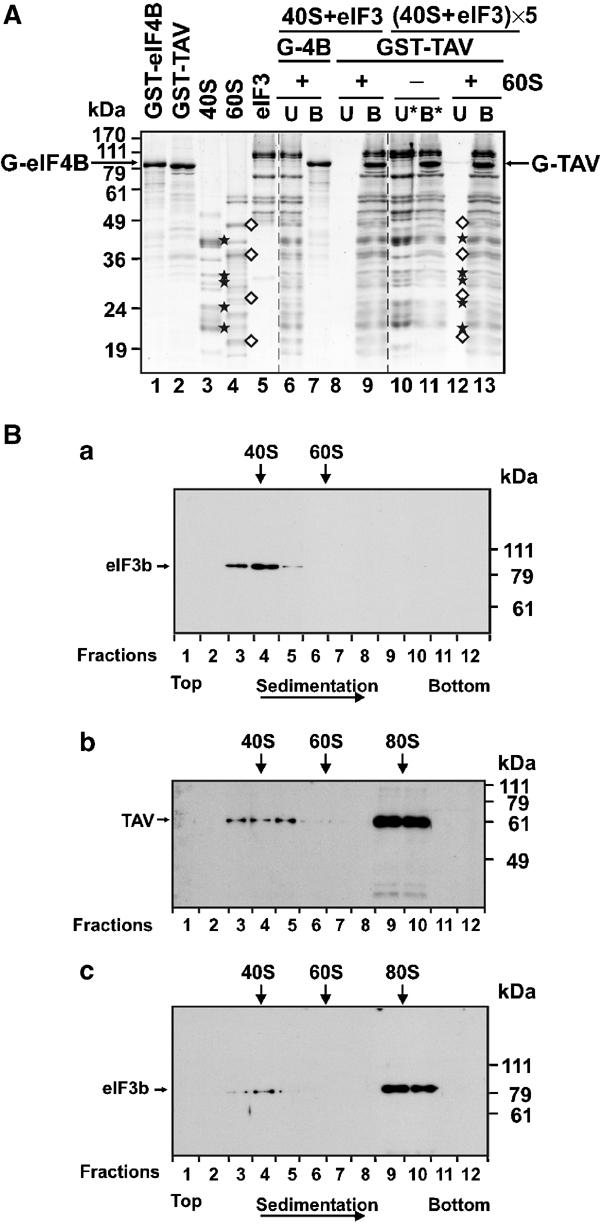
TAV and eIF3 can form a complex with 80S ribosomes. (A) GST, GST-eIF4B and GST-TAV were overexpressed in E. coli and purified by affinity chromatography. 60S, 40S and eIF3 were purified from WGE. The left panel shows the purified components (lanes 1–5). For pull-down experiments, 20 pmol GST-eIF4B and GST-TAV was bound to glutathione–sepharose beads and incubated with a 1:1:1 molar ratio of eIF3:40S:60S (lanes 6, 7 and 8, 9, respectively). To saturate TAV with the eIF3/40S complex, GST-TAV bound to glutathione beads was incubated with a five-fold molar excess of eIF3:40S (lanes 10 and 11). After removal of the unbound fraction (U*, lane 10) the GST-TAV-bound fraction (B*, lane 11) was further incubated with 60S ribosomal subunits at a 1:1 molar ratio (GST-TAV:60S). The beads were then washed and the unbound (U) and bound (B) fractions were analyzed by SDS–PAGE (lanes 12 and 13, respectively). Asterisks, characteristic 40S proteins; diamonds, characteristic 60S proteins specifically co-precipitated with GST-TAV. (B) GST-TAV was overexpressed in E. coli, purified by affinity chromatography, and TAV released from GST-TAV by protease cleavage. Ribosomal subunits and eIF3 were purified from WGE. 40S ribosomal subunits were incubated with eIF3 alone (panel a) and with TAV (panels b and c), followed by incubation with 60S ribosomal subunits and then subjected to velocity sedimentation through sucrose density gradients. Gradients were fractionated with optical scanning at 254 nm. The sedimentation positions of 40S, 60S and 80S ribosomes are shown. Aliquots (150 μl) of each fraction were precipitated with 20% TCA and analyzed by SDS–PAGE and immunoblotting using polyclonal antibodies against TAV and eIF3b.
In contrast, upon addition of 60S to the GST-TAV/eIF3/40S complex, all three components (60S, 40S and eIF3) were found in the GST-TAV-bound fraction (Figure 5A, lane 9). As GST-TAV can interact individually with both eIF3 and the 60S ribosome, in a separate experiment GST-TAV was presaturated with a five-fold molar excess of the eIF3/40S complex (Figure 5A, lane 11). After removal of unbound eIF3 and 40S ribosomal subunits (U*, Figure 5A, lane 10), the glutathione-bound fraction with the immobilized GST-TAV/eIF3/40S complex (B*, Figure 5A, lane 11) was further incubated with 60S ribosomal subunits at a 1:1 molar ratio (60S:GST-TAV). Again, 60S, 40S and eIF3 were all found in the GST-TAV-bound fraction (Figure 5, lane 13), suggesting that a TAV/eIF3/40S/60S complex indeed forms (note that eIF3, 40S and 60S were not found in the unbound fraction). This supports the hypothesis that the TAV/eIF3/40S complex is not disrupted at the 60S-joining step in the absence of Met-tRNAi and other initiation factors.
These results suggest that the 40S/eIF3/TAV complex is stable in the presence of 60S ribosomal subunits and can form an 80S/eIF3/TAV complex. Formation of such a complex was tested directly by preincubating isolated 40S subunits with eIF3 or eIF3+TAV followed by addition of 60S ribosomal subunits at 3 mM Mg(OAc)2; products were analyzed by sucrose gradient centrifugation and the positions of ribosomes and subunits were determined by UV scanning (Figure 5B). The amount of TAV and eIF3 in gradient fractions was analyzed by Western blot with polyclonal purified anti-TAV (Nakayashiki et al, 1993) or anti-human eIF3b (PRT1) (Lin et al, 2001) antisera (Figure 5B). The addition of TAV leads to the formation of an 80S complex. The 80S complex formed under these conditions (i.e. a two-fold molar excess of isolated ribosomal subunits over eIF3 and TAV) cosediments with about 70% of the TAV and eIF3 detected in the gradient (Figure 5B, panels b and c, fractions 9 and 10), the remaining ∼30% eIF3 and TAV being found in the 40S ribosomal subunit fraction, with not more than 5% TAV in the 60S ribosomal fraction.
In contrast, if 40S ribosomes were preincubated only with eIF3 and 60S, most eIF3b was found in the 40S ribosomal fraction (Figure 5B, panel a, fractions 3–5). In the absence of ribosomal subunits, a mixture of eIF3 and recombinant TAV was found at the top of the gradient (data not shown).
Overexpression of TAV and eIF4B in plant protoplasts exclusively affects reinitiation
The effect of TAV, eIF3g and eIF4B on expression of first and second ORFs in mono- and bicistronic constructs was tested in Nicotiana plumbaginifolia protoplasts. Expression from monoGUS, where β-glucuronidase (GUS) is the first and only ORF, was unaffected by the presence of TAV, even in large excess (Figure 6A, cf. lanes 1 and 3–6). In contrast, expression of GUS from the dicistronic construct biGUS (with CaMV ORF VII as a first ORF and GUS as a second ORF) was strongly enhanced if a TAV-expressing plasmid was provided (Figure 6B, cf. lanes 1 and 3), confirming earlier results (Bonneville et al, 1989). Thus, TAV is inert for first ORF initiation but efficiently supports reinitiation at a second ORF.
Figure 6.
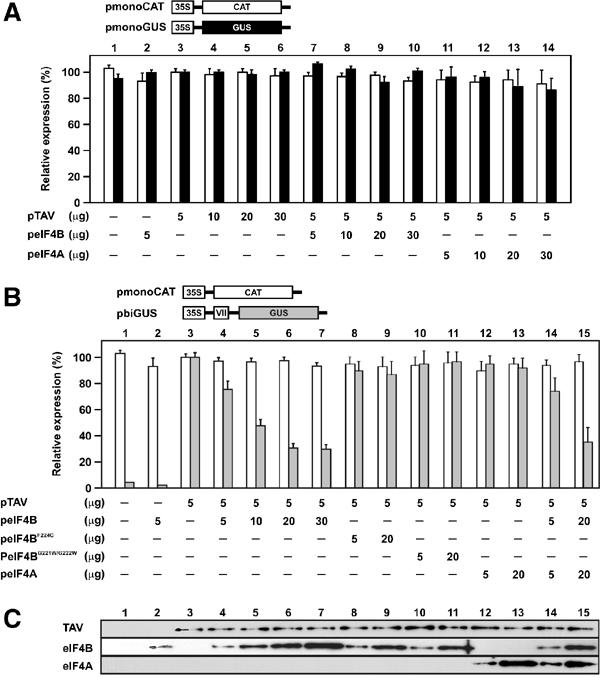
eIF4B suppresses TAV-mediated reinitiation in plant protoplasts. All transfection experiments in N. plumbaginifolia protoplasts included the two reporter plasmids shown, as well as effector plasmids in the amounts indicated below the charts. The results represent the means obtained in three independent experiments. In lane 3, GUS (20 000 units for monoGUS and 8500 units for biGUS measured under comparable conditions) and CAT levels were defined as 100%. (A) Effect of eIF4B and eIF4A overexpression on the first initiation event. CAT (white bars) and GUS (black bars) expression levels measured in protoplast extracts are indicated. (B) Effect of eIF4B, eIF4B mutants and eIF4A on TAV activity. CAT (white bars) and GUS (gray bars) expression levels measured in protoplast extracts are indicated. (C) Expression levels of TAV, eIF4B, their mutants and eIF4A in plant protoplasts were analyzed by immunoblotting using polyclonal antibodies against TAV, eIF4B and eIF4A.
Overexpression of eIF3g in this system diminishes TAV-dependent translation transactivation of the second ORF (Park et al, 2001). As eIF4B competes with TAV for eIF3 binding, we predicted it to be an inhibitor of TAV-mediated second-ORF translation, while not affecting translation of the first ORF. Overexpression of eIF4B did not affect GUS expression from monoGUS (Figure 6A, lanes 7–10), but led to significant inhibition of TAV-mediated GUS expression from pbiGUS (Figure 6B, lanes 4–7). This negative effect of eIF4B on TAV-mediated reinitiation efficiency is most likely due to competition with TAV for eIF3 binding, leading to increased loss of eIF3 from the translation machinery. Indeed, eIF4B mutants unable to interact with eIF3 (F224C and G221W/G222W) do not affect TAV-mediated transactivation (Figure 6B, lanes 8–11). (Expression levels as determined by Western blotting with antibodies against TAV, eIF4B and 4A are shown in Figure 6C.) Increased eIF4B concentrations did not stimulate the first initiation event, which is apparently well enough supported by endogenous eIF4B.
Unlike in mammals, plant eIF4B stimulates, but is not essential for, helicase activity (Altmann et al, 1995 and references therein). Nevertheless, it remains possible that an RNA melting activity of eIF4B plays a role in TAV-mediated reinitiation in plant protoplasts. If so, it might be expected that this function could be replaced by the helicase eIF4A. We tested the effect of eIF4A alone and in combination with eIF4B on TAV-mediated transactivation in vivo. Overexpression of eIF4A had no effect (Figure 6A, lanes 11–14 and Figure 6B, lanes 12 and 13), and coexpression of eIF4A with eIF4B had no additional effect over eIF4B alone, on TAV-mediated reinitiation (Figure 6B, lanes 14 and 15). Thus, the eIF4B RNA unwinding function is not involved in TAV-mediated reinitiation, and eIF4B has functions other than just being a helicase coactivator.
Discussion
In principle, polycistronic translation is possible in eucaryotes if the first ORF is short. However, reinitiation efficiency decreases rapidly with the increasing length of the sORF (Kozak, 1987; Fütterer and Hohn, 1992). It is assumed that certain initiation factors remain bound to the translation machinery during translation of an sORF, while they are successively lost during translation of a longer one. The CaMV TAV protein modifies the translation machinery in such a way that reinitiation occurs independently of the length of the first-translated ORF and the intercistronic distance between two ORFs. Accumulation of eIF3 in the polysomal fraction isolated from CaMV-infected cells (Park et al, 2001) suggested that TAV might prevent loss of eIF3 from the translating ribosome after the first initiation event, thereby allowing it to be used for subsequent reinitiation events.
This study reveals participation of another initiation factor, eIF4B, in TAV-mediated reinitiation. Our data indicate that eIF4B precludes binding of TAV to eIF3 by occupying the same eIF3g-based binding site. We demonstrate, both in vitro and in vivo, that interactions between plant eIF4B and intact eIF3 and/or the 40S ribosome exist, and are not disrupted by TAV (up to at least a five-fold molar excess). Under the conditions used, 60S subunit joining might trigger removal of eIF4B from the eIF4B/eIF3/40S complex. Following eIF4B removal, TAV can participate in higher-order complex formation consisting of (at least) eIF3 and 80S ribosomes.
The role of eIF4B in the translation initiation process is not yet well defined. It has been shown that eIF4B can catalyze the hybridization of two complementary single-stranded RNAs via an RNA annealing activity (Altmann et al, 1995), and stimulate the RNA helicase and ATPase (Bi et al, 2000) activities of eIF4A, although no physical association of eIF4A and eIF4B has been observed. eIF4B might also play a role in internal initiation because it interacts with poliovirus IRES in an energy-dependent manner (Ochs et al, 2002). What role is played by eIF4B in the TAV-mediated reinitiaton process? According to our in vitro data, eIF4B is occluded in the binary complex with the 40S ribosome and in the ternary complex with eIF3 and the 40S ribosome, and cannot be replaced by TAV (Figure 4B and C), thereby ensuring that TAV has no effect on primary translation initiation events. Furthermore, when simultaneously supplied with eIF4B, TAV could not form a complex with eIF3 and 40S (Figure 4D). Conversely, once TAV had formed a complex with 40S ribosome-bound eIF3, not even a five-fold excess of eIF4B was able to release it. This supports our hypothesis that within the 48S preinitiation complex, 40S-bound eIF3 forms a stable complex with eIF4B, while after release of eIF4B during the translation elongation process, TAV takes over the eIF4B-binding site of the ribosome-bound eIF3. Reconstruction of the 48S preinitiation complex with ternary complex will be required to test this hypothesis.
Release of factors from the 48S initiation complex
The release of cap-binding initiation factors from the 48S initiation complex is not well understood. eIF5 stimulates GTP hydrolysis, promoting the removal of GDP/eIF2 from the 48S preinitiation complex, but other participants, for example, eIF3, eIF1 and eIF1A, remain associated with 40S ribosomes (Pestova et al, 2000). eIF5B promotes the 60S subunit-joining step but has no apparent role in displacing eIFs 3, 1 and 1A from the 48S complex; rather, displacement of these factors may involve the 60S ribosome. eIFs 1, 2 and 3 were detected in 48S but not in 80S complexes with a complete set of initiation factors and GTP (Pestova et al, 2000). It is not known whether eIF4F, eIF4A and eIF4B are removed before the 60S subunit-joining step.
In addition to addressing specifically the interplay of TAV and eIF4B in the reinitiation process, our experiments also reveal an important role for eIF4B in the factor release cascade. Addition of the 60S subunit to the eIF4B/eIF3/40S complex caused a dramatic release of eIF4B (Figure 5A, lane 6). In contrast, TAV is not released upon addition of the 60S subunit to the TAV/eIF3/40S complex. Also, in the presence of TAV, eIF3 is not necessarily removed from the 40S subunit during the 60S subunit-joining step, and can remain as part of the 80S ribosome complex (Figure 5A). That TAV stabilizes binding of eIF3 to the 80S ribosome is also seen in vivo; eIF3 is found in the polysomal fraction isolated from CaMV-infected, but not healthy, plants.
Results from overexpression studies in vivo also support our in vitro data: TAV does not affect the efficiency of first initiation events even at elevated concentrations (Figure 6A), thus confirming that TAV does not affect the eIF3 and eIF4B interaction network at this stage. TAV affects exclusively reinitiation events on dicistronic mRNA (Figure 6B). eIF4B overexpression did not affect the efficiency of the first initiation event, but diminished TAV-dependent reinitiation to a level of 15–20%, most likely by outcompeting TAV for eIF3 binding.
Role of TAV in the viral context
In CaMV-infected cells, eIF3 remains associated with polysomes due to its interaction with TAV. A likely scenario (Figure 7) is that following, or concomitant with, disruption of the eIF3–eIF4B interaction, TAV is sequestered by eIF3, thus strengthening the otherwise unstable complex, which remains attached to the translation machinery. This would allow second and subsequent eIF3-dependent reinitiation events at downstream viral ORFs. The affinity of TAV for the 60S ribosomal subunit might further enhance TAV entry and might lead to deposition of eIF3 on the solvent surface of 60S via L18, where it would not interfere with the elongation process.
Figure 7.
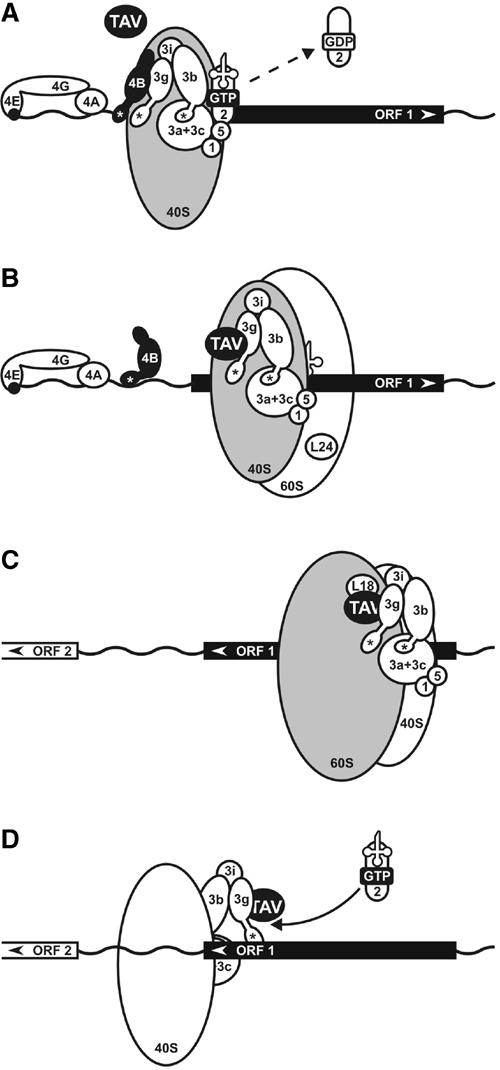
Model of TAV entry into the host translational machinery. Note that in (A) and (B), the model is viewed from the ‘front', with movement of the initiation complex from left to right, and in (C) and (D) from the ‘back' (movement from right to left). The solvent side of 40S and 60S ribosomal subunits is depicted in gray, with the internal interface side in white. Asterisks, RNA recognition domains (RRM) within eIF4B and eIF3g. (A) eIF4B, in concert with eIF4F (4E/4G/4A) or eIFiso4F, interacts with eIF3 bound to the 48S preinitiation complex. The complex scans until it encounters the first suitable start codon. During the codon–anticodon recognition step, eIF5-dependent GTP hydrolysis occurs and eIF2 is removed from 40S. eIF3 and its subunits g (3g), i (3i), b (3b), a and c (3a+3c); eIF1 (1), eIF2 (2), eIF5 (5), tRNA and TAV are depicted. (B) During the 60S subunit-joining step eIF4B is displaced from the ribosome, while eIF3 is still associated with the solvent side of the 40S subunit. TAV binds to the eIF3/40S complex via eIF3 subunit g. ORF1 elongation begins. L24 is positioned on the interface side of the 60S subunit. (C) During the elongation process, the TAV/eIF3/80S complex is stabilized, possibly by transfer of TAV/eIF3 to the 60S subunit through TAV interaction with L18. (D) The TAV/eIF3 complex is relocated back to the 40S subunit during ORF 1 termination. The TAV/eIF3/40S complex re-acquires the ternary complex and scans for ORF 2.
At a later stage of infection, when TAV accumulates to higher concentrations and becomes a main component of viral inclusion bodies, it would inhibit translation by binding to L24 (Park et al, 2001), which is located on the ribosome interface surface of the 60S ribosomal subunit, or interfere with eIF3 and eIF4B at the initiation phase of viral RNA translation, thereby liberating the CaMV RNA for packaging and reverse transcription.
Experimental procedures
Protein purification
eIF4B was purified from wheat germ extract (WGE) as described in Metz et al (1999). Wheat germ eIF3, 40S and 60S ribosomes were isolated from WGE according to Lax et al (1986) and Spremulli et al (1977).
pGST-TAV, pHis-TAV and the mutant pHis-TAV(Y305P) (Park et al, 2001) were transformed into Escherichia coli BL21 (DE3). pGST6P-TAV was constructed by cloning a TAV fragment obtained by PCR from pHelp7 (Bonneville et al, 1989) into the Xma–XhoI sites of pGEX6P-1 (Amersham Pharmacia). Bacterial growth, lysis, protein Ni-NTA affinity column and glutathione–sepharose 4B bead purifications were performed as described in the manufacturer's protocols (Qiagen, Amersham Pharmacia). Purified proteins were quantified by Bradford assay.
Ribosome complex formation and density centrifugation
GST6P-TAV was overexpressed in E. coli, and purified by affinity chromatography followed by TAV release from the GST6P-TAV fusion by cleavage with Prescission protease (Amersham). 80S ribosome complexes were formed as follows: reaction mixture a (100 μl) in buffer A (20 mM Tris–HCl (pH 7.5), 100 mM KCl, 3 mM MgAc2, 1 mM DTT) contained 0.6 A260 units 40S ribosomal subunits and 7 μg eIF3; reaction mixture b contained in addition 1 μg purified TAV. After incubation for 20 min at 30°C, 1.2 A260 units 60S ribosomal subunits were added to a and b. After incubating for an additional 10 min at 30°C, reactions a and b were chilled, then layered onto 10–30% sucrose gradients in reaction buffer A and centrifuged at 45 000 rpm for 2.5 h in an SW 50.1 rotor. Gradients were fractionated using an ISCO gradient fractionator, and the absorbance profile at 254 nm was monitored.
Western blots
Proteins were separated on 12.5% polyacrylamide gels and transferred to nitrocellulose membranes by electroblotting (1 h, 1 mA/cm2 gel). TAV and eIF3b in sucrose density gradient fractions were detected using a purified polyclonal rabbit anti-TAV antiserum (Nakayashiki et al 1993) or an anti-human eIF3b (PRT1) antiserum (Lin et al, 2001). In plant protoplast extracts, TAV and its mutants were detected using rabbit anti-TAV antiserum (De Tapia et al, 1993). eIF4B and its mutants and eIF4A were detected using rabbit anti-eIF4B and -eIF4A polyclonal antisera.
Yeast two-hybrid analysis
Two-hybrid analysis of interacting proteins was performed using a cotransformation procedure. Diploid strain GC1945xY187 was cotransformed with plasmids expressing BD fused with full-length or truncated TAV sequences and AD fused to full-sized or truncated wheat eIF4B (TaTif4B1; Metz et al, 1999) or Arabidopsis eIF3i sequences. Plasmid BD-eIF3g was produced by PCR from AD-eIF3g (Park et al, 2001) and cloned between the SmaI and PstI sites of pAS2ΔΔ. AD-eIF4B and AD-eIF3i were produced by ligating EcoRI–BamHI fragments prepared by PCR from peIF4B/pET3d (Metz et al, 1999) and from the expressed sequence tag (EST) from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University (Columbus, OH), into EcoRI–BamHI-digested pGAD424 (Clontech). eIF4B and eIF3i deletion mutants fused to AD were produced by PCR and inserted between the EcoRI and BamHI sites of pGAD424. Protein expression levels were controlled by Western blotting; no significant variations in the amount of protein expressed were observed between wild-type and truncated fusion proteins (data not shown).
In vitro transcription and translation
pT7eIF4B and pT7eIF3i were produced by ligating BamHI–PstI fragments prepared by PCR from pAD-eIF4B and pAD-eIF3i, respectively, into BamHI–PstI-digested pBluescript II (Stratagene). pT7 Luciferase was supplied by Promega. T7-directed transcripts were transcribed in the presence of the cap analog 7mGpppG and translated in WGE as described by Ryabova and Hohn (2000).
In vitro GST pull-down assays
Constructs pGST-TAV and pGST-eIF3g were described previously (Park et al, 2001). pGST-eIF4B was made by cloning EcoRI–SalI fragments from pLex4B—constructed by cloning EcoRI–BamHI fragments from peIF4B/pET3d (Arabidopsis eIF4B, AtTif4B2p; Metz et al, 1999)—into pGEX6P-1 (Pharmacia) as in-frame fusions with the GST domain. Expression of GST fusion proteins was induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. In vitro GST pull-down assays were performed essentially as described by Park et al (2001), as detailed below. In all cases, bound and unbound fractions were separated by 12.5% SDS–PAGE and [35S]-labeled proteins were visualized by autoradiography.
GST-eIF3g interaction with [35S]eIF4B or [35S]eIF3i. [35S]eIF4B or [35S]eIF3i were produced by in vitro translation using [35S]Met. Aliquots (300 μl) of GST or GST-eIF3g bound to beads were incubated with 10 μl WGE translation mixture containing [35S]-labeled proteins for 1 h at 4°C with gentle shaking prior to separation of bound (60 μl) and unbound fractions by centrifugation. Aliquots of 5 μl (bound fraction) or 25 μl (unbound fraction) were analyzed.
Competition between [35S]eIF4B and TAV for eIF3 binding. Fractions of GST-eIF3g (3.5 pmol) bound to beads and 10 μl of in vitro translation mixtures containing [35S]-labeled eIF4B (about 1–2 pmol) were incubated with increasing concentrations of purified His-TAV or His-TAVY305P (0.5, 10 and 20 μg) for 1 h at 4°C with gentle shaking. After extensive washing, proteins bound to beads were eluted and analyzed by SDS–PAGE.
In the following sections, unless stated otherwise, binding reactions were carried out by incubating the components indicated in a final volume of 300 μl buffer R (10 mM Tris–HCl, pH 7.5, 50 mM NaCl, 3 mM MgCl2 and 2 mM β-mercaptoethanol) for 30 min at 30°C with gentle shaking, followed by centrifugation to separate unbound and bound (60 μl) fractions. Aliquots of 10 μl (bound fractions) or 25 μl (unbound fractions) were separated by 12.5% SDS–PAGE.
Competition between eIF4B and TAV for eIF3 binding. Aliquots of 20 pmol GST TAV (or GST alone) bound to beads were incubated with 1.2 μg eIF4B or 10.2 μg eIF3. Bound fractions of GST-TAV (or GST) and eIF3 were further incubated with 1.8 μg purified eIF4B. His-TAV (20 pmol) bound to Ni-resin was incubated with 10 μg eIF3. Bound fractions of His-TAV and eIF3 were further incubated with 0.6 or 1.2 μg purified eIF4B.
Interactions between GST-eIF4B, 40S and His-TAV. GST-eIF4B (20 pmol) (or GST) bound to beads was incubated with 0.5 A260 units 40S. After washing, the GST-bound fraction was further incubated with 1.5 μg purified His-TAV.
Interactions between GST-TAV, 40S, eIF3 and eIF4B. GST-TAV (20 pmol) bound to beads was incubated with 20 pmol eIF3, 22 pmol 40S and 100 pmol eIF4B.
Interactions between GST-eIF4B, 40S, eIF3 and His-TAV. GST-eIF4B or GST (20 pmol) bound to beads was incubated with 20 pmol eIF3. After washing, GST-bound fractions were further incubated with 0.5 A260 units 40S or with a mixture of 0.5 A260 units 40S and 1 or 5 μg purified His-TAV.
Interactions between GST-TAV, eIF3, eIF4B and ribosomal subunits. GST-TAV or GST (20 pmol) bound to beads was incubated with 13.3 μg eIF3 and 0.5 A260 units 40S subunits or 0.9 A260 60S subunits. GST-bound fractions containing eIF3, 40S or 60S ribosomes were further incubated with 1.2 or 5.8 μg eIF4B.
Complexes between GST-TAV or GST-eIF4B with eIF3 and both ribosomal subunits. About 20 pmol each of GST-TAV, GST-eIF4B or GST alone was incubated with a mixture of 13.3 μg eIF3, 0.5 A260 units 40S subunits (22 pmol) and 20 pmol tRNA in 300 μl buffer R for 5 min at 30°C with gentle shaking. After addition of 0.9 A260 units 60S subunits (24 pmol), the mixture was further incubated for 30 min at 30°C with gentle shaking.
To saturate TAV with the eIF3/40S complex, in a parallel experiment GST-TAV was incubated with a five-fold molar excess of the eIF3/40S subunit complex. After washing to remove unbound eIF3 and 40S ribosomal subunits, the GST-TAV/eIF3/40S complex was incubated with 60S ribosomal subunits at a 1:1 molar ratio.
Yeast three-hybrid system
The three-hybrid system was performed according to standard Clontech protocols using the yeast strain HF7c. The eIF3g ORF was fused to AD behind the ADH promoter of vector pGAD424 carrying the LEU 2 selection gene (pGAD-eIF3g). BglII–NotI eIF4B-2 and TAV fragments obtained by PCR from pBD-eIF4B and pBD-TAV (Park et al, 2001) were introduced into the BglII–NotI site of the Bridge vector (pBr, Clontech). In addition, EcoRI–BamHI fragments from pGAD-TAV and pGAD-eIF4B-2 were fused to BD in the Bridge vector with or without eIF4B-2 and TAV to produce pBrBD-TAV/Met-eIF3g and pBrBD-TAV/Met-0 and pBrBD-eIF4B-2/Met-TAV and pBrBD-eIF4B-2/Met-0, respectively.
Transient expression
The eIF4B coding sequence from pAD-eIF4B and wheat eIF4A from pETeIF4A (Metz and Browning, 1993) was subcloned under the control of the CaMV 35S promoter in pTAV (p35S-P6; Kobayashi et al 1998). pmonoCAT, pmonoGUS and pbiGUS have been described previously (Bonneville et al, 1989).
Leaf protoplasts derived from Nicotiana plumbaginifolia were prepared and samples of 6 × 105 protoplasts were used for polyethylene glycol-mediated transfection as described previously (Fütterer and Hohn, 1992). A monocistronic (pmonoGUS; 5 μg) or dicistronic (pbiGUS; 5 μg) GUS-expressing reporter plasmid was cotransfected with a CAT-expressing plasmid (pmonoCAT; 2 μg) as an internal standard of transfection efficiency and protoplast quality. In addition, 5 μg pTAV with and without increasing concentrations of peIF4B and/or peIF4A (as indicated) was cotransformed. CAT and GUS activities were determined in protein extracts prepared after overnight incubation as described previously (Pooggin et al, 2000). Values given are the means from more than three independent experiments.
Acknowledgments
We are grateful to S Fumagalli, T Pestova and V Boyko for their expertise. We thank B Hohn, K Kobayashi, M Pooggin and H Rothnie for helpful suggestions and critical reading of the manuscript, and S Oakeley and M Rothnie for help in preparation of the figures. We thank the Arabidopsis Biological Resource Center (ABRC) at Ohio State University for providing an EST clone. This work was supported by the Novartis Research Foundation and INTAS to H-SP, TH and LR, the Roche Research Foundation to H-SP, and by DOE (DE-FG03-97ER20283), NSF (MCB-0214996) and the Welch Foundation (F1339) to KSB.
References
- Altmann M, Wittmer B, Méthot N, Sonenberg N, Trachsel H (1995) The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF-4B, have RNA annealing activity. EMBO J 14: 3820–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Ren J, Goss DJ (2000) Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry 39: 5758–5765 [DOI] [PubMed] [Google Scholar]
- Bonneville J-M, Sanfaçon H, Fütterer J, Hohn T (1989) Posttranscriptional transactivation in cauliflower mosaic virus. Cell 59: 1135–1143 [DOI] [PubMed] [Google Scholar]
- De Tapia M, Himmelbach A, Hohn T (1993) Molecular dissection of the cauliflower mosaic virus translational transactivator. EMBO J 12: 3305–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J, Hohn T (1991) Translation of a polycistronic mRNA in the presence of the cauliflower mosaic virus transactivator protein. EMBO J 10: 3887–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J, Hohn T (1992) Role of an upstream open reading frame in the translation of polycistronic mRNAs in plant cells. Nucleic Acids Res 20: 3851–3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC (2000) The pathway and mechanism of initiation of protein synthesis. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB, Mathews MB (eds) pp 33–88. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Himmelbach A, Chapdelaine Y, Hohn T (1996) Interaction between cauliflower mosaic virus inclusion body protein and capsid protein: implications for virus assembly. Virology 217: 147–157 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG (2000) Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB, Mathews MB (eds) pp 185–243. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hohn T, Park H-S, Guerra-Peraza O, Stavolone L, Pooggin MM, Kobayashi K, Ryabova LA (2002) Shunting and controlled reinitiation. The encounter of cauliflower mosaic virus with the translational machinery. Cold Spring Harb Symp Quant Biol 66: 269–276 [DOI] [PubMed] [Google Scholar]
- Jaramillo M, Browning K, Dever TE, Blum S, Trachsel H, Merrick WC, Ravel JM, Sonenberg N (1990) Translation initiation factors that function as RNA helicase from mammals, plants and yeast. Biochim Biophys Acta 1050: 134–139 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Tsuge S, Nakayashiki H, Mise K, Furusawa I (1998) Requirement of cauliflower mosaic virus open reading frame VI product for viral gene expression and multiplication in turnip protoplasts. Microbiol Immunol 42: 377–386 [DOI] [PubMed] [Google Scholar]
- Kozak M (1987) Effects of intercistronic length on the efficiency of reinitiation by eukaryotic ribosomes. Mol Cell Biol 7: 3438–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE (1995) Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral protease. Implications for cap-dependent and cap-independent translation initiation. J Biol Chem 270: 21975–21983 [DOI] [PubMed] [Google Scholar]
- Lax SR, Lauer SJ, Browning KS, Ravel JM (1986) Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol 118: 109–128 [DOI] [PubMed] [Google Scholar]
- Leh V, Yot P, Keller M (2000) The cauliflower mosaic virus translational transactivator interacts with the 60S ribosomal subunit protein L18 of Arabidopsis thaliana. Virology 266: 1–7 [DOI] [PubMed] [Google Scholar]
- Lin L, Holbro T, Alonso G, Gerosa D, Burger MM (2001) Molecular interaction between human tumor marker protein p150, the largest subunit of eIF3, and intermediate filament protein K7. J Cell Biochem 80: 483–490 [DOI] [PubMed] [Google Scholar]
- Méthot N, Pickette G, Keene JD, Sonenberg N (1996b) In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA recognition motif. RNA 2: 38–50 [PMC free article] [PubMed] [Google Scholar]
- Méthot N, Song MS, Sonenberg N (1996a) A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol 16: 5328–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz AM, Browning KS (1993) Sequence of a cDNA encoding wheat eukaryotic protein synthesis initiation factor 4A. Gene 131: 299–300 [DOI] [PubMed] [Google Scholar]
- Metz AM, Wong KCH, Malmström SA, Browning KS (1999) Eukaryotic initiation factor 4B from wheat and Arabidopsis thaliana is a member of a multigene family. Biochem Biophys Res Commun 266: 314–321 [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 23: 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki H, Tsuge S, Kobayashi K, Okuno T, Furusawa I (1993) Reasons for the low accumulation level of aphid transmission factor protein in infected leaves with an aphid-non-transmissible cauliflower mosaic virus isolate, CM 1841. J Gen Virol 74: 2469–2472 [DOI] [PubMed] [Google Scholar]
- Ochs K, Saleh L, Bassili G, Sonntag VH, Zeller A, Niepmann M (2002) Interaction of translation initiation factor eIF4B with the poliovirus internal ribosome entry site. J Virol 76: 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-S, Himmelbach A, Browning KS, Hohn T, Ryabova LA (2001) A plant viral ‘reinitiation' factor interacts with the host translational machinery. Cell 106: 723–733 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CUT (1998) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Pooggin MM, Hohn T, Fütterer J (2000) Role of a short open reading frame in ribosome shunt on the cauliflower mosaic virus RNA leader. J Biol Chem 275: 17288–17296 [DOI] [PubMed] [Google Scholar]
- Ryabova LA, Hohn T (2000) Ribosome shunting in the cauliflower mosaic virus 35S RNA leader is a special case of reinitiation of translation functioning in plant and animal systems. Genes Dev 14: 817–829 [PMC free article] [PubMed] [Google Scholar]
- Spremulli LL, Walthall BJ, Lax SR, Ravel JM (1977) Purification and properties of a Met-tRNA binding factors from wheat germ. Arch Biochem Biophys 178: 565–575 [DOI] [PubMed] [Google Scholar]
- Tirode F, Malaguti C, Romero F, Attar R, Camonis J, Egly JM (1997) A conditionally expressed third partner stabilizes or prevents the formation of a transcriptional activator in a three-hybrid system. J Biol Chem 272: 22995–22999 [DOI] [PubMed] [Google Scholar]
- Verlhac M-H, Chen R-H, Hanachi P, Hershey JWB, Derynck R (1997) Identification of partners of TIF34, a component of the yeast eIF3 complex, required for cell proliferation and translation initiation. EMBO J 16: 6812–6822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vornlocher HP, Hanachi P, Ribeiro S, Hershey JWB (1999) A 110-kilodalton subunit of translation initiation factor eIF3 and an associated 135-kilodalton protein are encoded by the Saccharomyces cerevisiae TIF32 and TIF31 genes. J Biol Chem 274: 16802–16812 [DOI] [PubMed] [Google Scholar]


