Abstract
Although Pichia pastoris is a popular protein expression system, it exhibits limitations in its ability to secrete heterologous proteins. Therefore, a REMI (restriction enzyme mediated insertion) strategy was utilized to select mutant beta-galactosidase supersecretion (bgs) strains that secreted increased levels of a β-galactosidase reporter.Many of the twelve BGS genes may have functions in intracellular signaling or vesicle transport. Several of these strains also appeared to contain a more permeable cell wall. Preliminary characterization of four bgs mutants showed that they differed in the ability to enhance the export of other reporter proteins. bgs13, which has a disruption in a gene homologous to Saccharomyces cerevisiae protein kinase C(PKC1), gave enhanced secretion ofmost recombinant proteins that were tested, raising the possibility that it has the universal super-secreter phenotype needed in an industrial production strain of P. pastoris.
Keywords: Enhanced secretion, Mutagenesis, Pichia pastoris, Recombinant proteins, Secretion
Introduction
Over 1,000 recombinant proteins have been produced using the methlotrophic yeast Pichia pastoris. Heterologous expression is usually driven by the alcohol oxidase I (AOX1) promoter, allowing for 1,000-fold transcriptional induction when cells are shifted to a medium containing methanol. Additionally, proteins can be produced intracellularly or secreted into the growth medium. Secreted recombinant proteins are expressed using their native signal or the Saccharomyces cerevisiae alpha mating factor (MATα) secretion leader. Because P. pastoris secretes few of its own proteins, foreign proteins can represent up to 80 % of total extracellular protein, acting as a valuable purification step (Cereghino and Cregg 2000).
Despite these attributes, P. pastoris has limitations, with inefficient secretion being the most serious obstacle (Idiris et al. 2010). Expression levels of different secreted proteins vary from 1 mg/l to 10 g/l even though identical strains, promoters, and secretion leaders are used. Instead of being exported, recombinant proteins may: (1) accumulate in ER, Golgi or endosomal compartments as either folded or misfolded; (2) undergo ER-associated protein degradation (ERAD), where proteins are translocated from the ER to the cytosol and degraded in proteosomes; or, (3) be degraded in the vacuole (Idiris et al. 2010; Hou et al. 2012).
Saccharomyces cerevisiae also has problems with the secretion of recombinant peptides (Idiris et al. 2010; Hou et al. 2012). One approach used in baker’s yeast to solve secretion issues has been to isolate ‘supersecreter strains’ (Moir 1989), which feature the overexpression, modification, or decreased function of a secretory trans-acting factor. For example, elevated levels of the ER chaperone, protein disulfide isomerase (PDI), increases the export of some peptides (Idiris et al. 2010) while decreasing the activity of other genes, such as PMR1 (Plasma Membrane Related ATPase) and certain VPS (Vacuolar Protein Sorting) genes, can enhance secretion as well (Hou et al. 2012; Rudolph et al. 1989). However, due to the complex relationship between factors, modification of one step can lead to rate limitation of following steps, causing a bottleneck in the secretory system. The effects appear protein specific: the strategy improves secretion of only one or a few proteins (Idiris et al. 2010). Thus, a strain of baker’s yeast engineered for the oversecretion of one peptide cannot be used as a general cell factory platform for many different proteins (Hou et al. 2012).
Although S. cerevisiae is a model to understand P. pastoris, significant functional differences exist between the secretory systems of the two yeasts (Delic et al. 2013), leaving much to be understood about the secretory pathway of P. pastoris. Though the complete genome was released in 2009 (Mattanovich et al. 2009; De Schutter et al. 2009) only a few P. pastoris gene products affecting secretion have been identified. These include Pmr1p, Pdip, the transcription factor Hac1p, and glycophospholipid-anchored surface protein (Gas1p) (Hou et al. 2012; Idiris et al. 2010). Manipulation of these gene products has increased the secretion of heterologous proteins, but the effects are protein specific (Idiris et al. 2010).
This work describes a novel selection strategy to identify gene products involved in the secretion process of P. pastoris. The disruption of 12 genes increased the secretion efficiency of a β-galactosidase reporter. Preliminary characterization of three genes revealed the combination of trans-acting factors used in secretion varies, depending on the structural features of the specific protein traveling through secretory pathway. However, one mutant, bgs13 affects the secretion of a wide range of recombinant proteins, suggesting that it may play a more general role in protein export.
Materials and methods
Strains, plasmids and reagents
yJC100 (wild type) and yGS115 (his4) have been described previously (Lin-Cereghino et al. 2006). yDT39 is a his4 met2 derivative of yGS115, and culturing methods for bacteria and yeasts have been described (Thor et al. 2005), The pAM1 (Li et al. 2009), pCH3K, and pKanB (Lin-Cereghino et al. 2008) plasmids were previously generated. pCH1, encoding horseradish peroxidase, and pPpT4_CalB, containing Candida antarctica lipase B gene were supplied by Anton Glieder, Technical University of Graz, Austria (Morawski et al. 2000; Krainer et al. 2012). pREMI-Z was provided by Benjamin Glick (University of Chicago, Chicago, IL). James Cregg (Keck Graduate Institute, Claremont, CA) supplied pHSA413 (Prevatt and Sreekrishna 1994).
Plasmid constructions
The primers 5′GAPNheI: GGCGCTAGCTTTTTGTAGAAATGTC and LeuBgalNotI: CAAGCGGCCGCTTTTTGACACCAGACCAA were used to amplify the GAP1-MATα-lacZ expression cassette from pJGBG (Lin-Cereghino et al. 2006). This fragment was digested with NheI and NotI, and inserted into the same sites of pBLMETIX (Thor et al. 2005) to create pBLMET-βgal. AOX1 promoter-lipase was amplified from pPpT4_CalB using primers AOX1promlipaseB-Sac1rhobeforw: CGGAGCTCGCTCATTCCAATTCCTTC and lipaseBSac2stoprhoberev: CGCCGCGG TTATGGGGTCACGATACCG, digestedwith SacI and SacII and inserted into the same sites in pKanB to create pKanB-lipase. All plasmid constructions were confirmed by restriction digestions and sequencing. Restriction enzymes were purchased from MBI Fermentas (Hanover, MD). The QiaQuick PCR Cleanup Kit (Qiagen) was used to purify all PCR reactions, probes and digestions. Plasmid purification from E. coli was performed with the QIAprep Spin Miniprep Kit (Qiagen). Restriction enzyme-mediated integration (REMI) and selection
pREMI-Z (Schroder et al. 2007) was linearized with BamH1. 50 µl of electrocompetent yDT39: pBLMET-pGAP-βgal (containing ~1.25 × 108 cells) were mixed with 1 µg purified pREMI-Z and BamH1 (1U) and electroporated using standards conditions (1.5 kV, 50 µF, 200 Ω). After adding 1 ml of cold 1 M sorbitol, the cells were plated in 100 µl aliquots onYNLplates supplemented with histidine (50 µg/ml). Transformants were single streaked twice on YNL+HIS plates to confirm positive growth phenotype and onto YPD-zeocin plates to confirm that they contained the pREMI-Z vector. Finally, the colonies were grown on YNM+HIS plates supplemented with Xgal (80 µg/ml) to confirm β-galactosidase secretion, as evidenced by the formation of a blue halo around the colony (Staley et al. 2012).
Preparation of pREMI-Z probe and Southern analysis of genomic DNA
Using primers REMIprobeForw:5′TCATAGCTGTTTCCGATCCC3′ and REMIprobeRev: 5′CCAGCTTGCAAATTAAAGCC3′, a 1.2 kb biotinylated probe of the pREMI-Z plasmid was prepared using the Bioprime DNA Labeling System (Life Technologies, Carlsbad, CA). Southern analysis of EcoRI-digested genomic DNA isolated using the Yeast Geno-DNA-Template DNA Extraction Kit (G-Biosciences, St. Louis, MO) was performed using the North2 South Chemiluminescent Hybridization and Detection Kit (Pierce, Rockford, IL). Images were captured using the ChemiImager 5500 (Alpha Innotech, San Leandro, CA.)
Plasmid rescue from bgs strains
Genomic DNA (3 µg) was digested with either EcoR1 or HindIII. 1 µg of the purified, digested DNA was incubated with T4 DNA ligase (Fermentas, Hanover, MD) overnight at room temperature. One Shot MAX Efficiency DH10B competent E. coli cells (Life Technologies, Carlsbad, CA) were transformed with ligation mixes and plated on LB-zeocin. Flanking genomic DNA in the rescued plasmids, from resulting transformants was sequenced with M13 forward and reverse primers by Geneway Research, LLC (Hayward, CA).
Bioinformatic analysis
The National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) and the Bioinformatics Online Genome Annotation System (BOGAS, http://bioinformaticspsb.ugent.be/webtools/bogas) were used to determine the gene disrupted in the mutant strains by the pREMI-Z sequence. Amino acid sequence homology to other cloned genes was analyzed with the NCBI BLAST program.
Transmission electron microscopy
Transmission electron microscopy was performed as described previously (Yount et al. 2006).
Small-scale P. pastoris methanol induction and cell-free extract preparation
The amount of reporter protein (β-galactosidase, horseradish peroxidase, lipase, secretory leukocyte protease inhibitor, and human serum albumin) secreted was determined by performing small-scale expression assays and generating cell-free extracts as described previously (Staley et al. 2012).
Enzyme assays
Functional activity levels of secreted or intracellular β-galactosidase were measured using protocols described in (Lin-Cereghino et al. 2006). Horseradish peroxidase and lipase activities were determined using previously described methods (Lin-Cereghino et al. 2013).
Alkaline phosphatase (ALP) plate assay
BCIP [5-bromo-4-chloro-3-indolyl phosphate p-toluidine salt, (Sigma)] at 50 mg/ml in N,N-dimethylformamide was added to YPD agar plates to give 60 µg/ml (Molina et al. 1998). P. pastoris wild type and bgs strains, as well as S. cerevisiae wild-type yDL100 and temperature sensitive pkc1 mutant yDL523 (Denis and Cyert 2005) were spotted onto plates and incubated at 30 °C for 48 h. Plates were then either kept at 30 °C or shifted to 37 °C for an additional 48 h.
Creation of reporter strains
pAM1 (AOX1:SLPI), pCH3K(AOX1:HRP), pKanB-lipase (AOX1:lipase) and pKanB (control) were linearized with MssI or Mph1103I and transformed into electrocompetent mutant (bgs2, bgs3, bgs13) or wild type cells (yDT39:pBLMET-βgal) and selected on medium containing 500 µg G418/ml (Lin-Cereghino et al. 2013). pHSA413 was linearized with SalI and transformants were selected on YND. Colony PCR (Thor et al. 2005) was performed to confirm that transformants harbored the reporter genes.
Spot western analysis
Spot westerns were used to analyze supernatants by loading samples standardized to equivalent cell numbers. Westerns were performed with the SNAP i.d. Protein Detection System (Millipore, Billerica, MA) as previously described (Lin-Cereghino et al. 2013) and using the following amounts of antibodies: 40 µl rabbit anti-SLPI (Santa Cruz Biotechnology, Santa Cruz, CA) and 2 µl secondary goat anti-rabbit IgG (Applied Biosystems, Foster City, CA) or 20 µl mouse anti-HSA antibodies (Santa Cruz Biotechnology) and 2 µl secondary goat anti-mouse IgG (Applied Biosystems, Foster City, CA).
Results and discussion
Rationale for the selection strategy
Wild type P. pastoris cannot grow on medium containing lactose as a sole carbon source because it lacks β-galactosidase, that converts lactose to glucose and galactose. P. pastoris strains transformed with E. coli β-galactosidase (lacZ) fused to the MATα secretion leader under the control of the strong, constitutive GAP promoter also cannot grow on medium containing lactose as they do not secrete the enzyme. We reasoned that we could use this inability of wild type P. pastoris to secrete β-galactosidase as a basis to select mutants with defects in the secretion process that enabled them to export enzyme and thus grow on lactose medium. We generated the mutant strains by utilizing the restriction enzyme-mediated integration (REMI) method that randomly inserts a zeocin-resistant vector (pREMI-Z) into genomic DNA (Schroder et al. 2007) to disrupt gene expression. Mutants that secrete significant levels of β-galactosidase should grow on lactose medium.
Selection to isolate supersecreter mutants
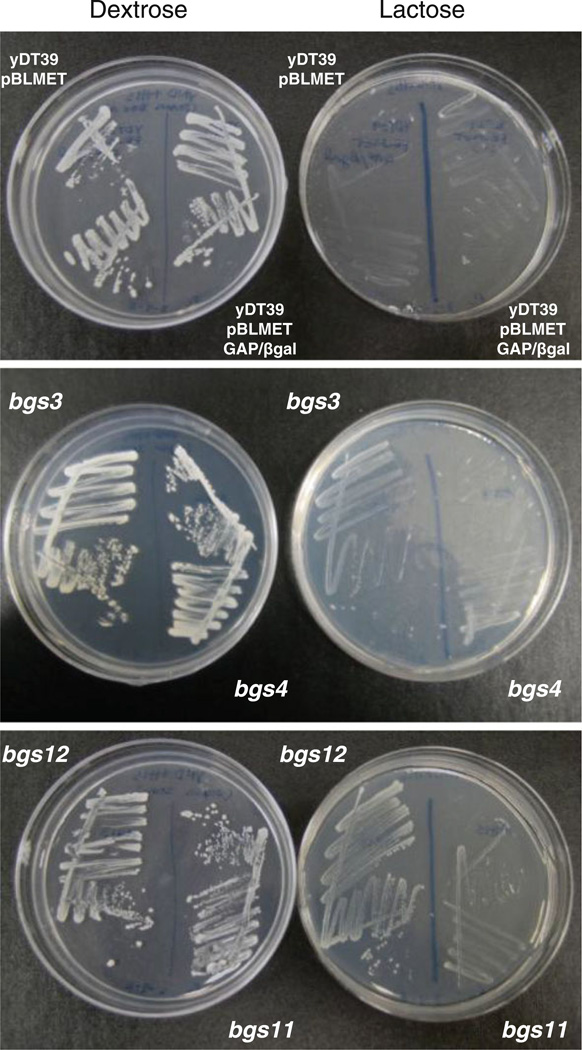
The parent strain used for mutagenesis was yDT39 (met2 his4) transformed with the vector, pBLMET-βgal, encoding a MATα-β-galactosidase fusion under the control of the P. pastoris GAP promoter. As expected, this strain was unable to grow on lactose medium (Fig. 1). Electroporation of yDT39: pBLMET-pGAP-βgal with pREMI-Z resulted in insertions of pREMI-Z at random locations in the yeast genome. Selection on lactose medium identified mutants that secreted significant levels of β-galactosidase. Furthermore, the colonies were grown on YNM plates containing X-gal to confirm that the colonies were secreting β-galactosidase, indicated by the formation of a blue halo around the colony. Of the original 2 × 109 cells that were electroporated with pREMI-Z, which corresponds to ~20–25,000 zeocin-resistant transformants, 15 supersecreter mutants were ultimately isolated from this selection and screening. These mutants were designated bgs (beta-galactosidase supersecretion) and are shown in Table 1. These zeocin resistant bgs strains expressed β-galactosidase from the GAP promoter, which is a strong, constitutive promoter on glucose and methanol (Waterham et al. 1997). These strains demonstrated different growth rates on lactose plates (Fig. 1) and secreted different levels of β-galactosidase (Table 1).
Fig. 1.
Growth characteristics of selected pREMI-induced mutants. Colonies were streaked onto YND +histidine (dextrose) and YNL +histidine (lactose)
Table 1. Cloned bgs mutants.
Bioinformatic analysis suggested that the predicted proteins have similarity to characterized proteins with the indicated structural or functional properties. β;galactosidase activity was measured in the extracellular fractions of cells induced on methanol growth medium. Residual β-galactosidase activity of the wild type parent strain containing the lacZ expression plasmid was subtracted, and the adjusted values are reported as units/ L OD600. Doubling times were calculated on YPD medium. The values are averages of three trials
| Mutant | Gene locus disrupted |
Comments/putative function | β-gal secretion |
Doubling time (min) |
|---|---|---|---|---|
| bgs1 | PAS_chr1-1_0268 | Subunit of dynactin | 8.7 ± 0.4 | 130 |
| bgs2 | PAS_chr3_1240 | SKIP_SNW family | 0.7 ± 0.2 | 240 |
| bgs3 | PAS_chr2-1_0743 | Unknown function | 2.9 ± 0.1 | 110 |
| bgs4 | PAS_chr3_1240 | Same as bgs2 | – | – |
| bgs5 | PAS_chr2-1_0744 | Heavy chain dynein | 13 ± 0.7 | 120 |
| bgs6 | PAS_chr2-2_0224 | TRAPP (transport protein particle) | 4.2 ± 0.1 | 140 |
| bgs7 | PAS_chr1-4_0031 | Pleckstrin-like, nuclear transport | 39 ± 5.6 | 110 |
| bgs8 | PAS_chr4_0054 | Nuclear transport protein | 1.5 ± 0.2 | 140 |
| bgs9 | PAS_chr1-4_0456 | Polyphosphatidy-linositol phosphatase | 1.0 ± 0.1 | 120 |
| bgs10 | PAS_chr3_0754 | Aminoacyl tRNA synthetase | 0.6 ± 0.1 | 140 |
| bgs11 | PAS_chr1-1_0068 | Staphylococcal nuclease-like (SNase-like) | 4.6 ± 0.2 | 110 |
| bgs12 | PAS_chr1-4_0443 | Cytoplasmic dynein, intermediate chain | 8.6 ± 0.6 | 110 |
| bgs13 | PAS_chr2-1_0124 | Protein kinase C | 13 ± 0.5 | 160 |
| bgs14 | PAS_chr2-1_0124 | Same as bgs13 | – | – |
| bgs15 | PAS_chr2-1_0124 | Same as bgs13 | – | – |
Cloning of supersecreter genes by plasmid rescue
Next, Southern blot analysis using a probe generated from pREMI-Z was performed to confirm that the pREMI-Z insert was integrated into the genome of each bgs strain at only a single site. The Southern result suggests that the genomic DNA of each mutant had only one copy of the pREMI-Z inserted into its genome, since each lane contained only one band (data not shown). A variety of genes were disrupted because most bands appeared to be of different sizes.
To identify the disrupted gene in each strain, the genomic DNA was digested, allowed to self-ligate, and then transformed into competent E. coli cells. The rescued plasmids, which contained the incorporated pREMI-Z insertion and flanking genomic DNA, were isolated, characterized by extensive restriction digestion, and sequenced. Sequences were aligned with P. pastoris genome to identify the disrupted gene using BLAST and BOGAS. Twelve unique genes in total were identified (Table 1).
Bioinformatics were used to determine the putative sequence motifs and function of these P. pastoris genes. While some BGS genes appear to be novel genes with unknown function, most have domains similar to proteins with known functions in other eukaryotes (Table 1). Many of the genes (BGS1, 5, 6, 7, 8, 12) encode proteins that are associated with vesicular movement and expected to be identified in a search for genes involved in secretion. For example, dynactin (BGS1) has been shown to bind dynein (BGS5, BGS12) to intracellular organelles to promote movement in endosome trafficking (Flores-Rodriguez et al. 2011). The predicted proteins of other genes, such as BGS9 (polyphosphatidylinositol phosphatase) and BGS13 (protein kinase C), are similar to proteins known to function in intracellular signaling. In addition, some of the identified gene products can potentially bridge both processes. For instance, S. cerevisiae polyphosphatidylinositol phosphatase/INP53 (BGS9) plays a role in the trans-Golgi network to early endosome pathway (Ha et al. 2003; Yu et al. 2004).
Cell wall integrity analysis of the bgs strains
Because of its apparent homology with S. cerevisiae protein kinase C (PKC1), which is involved in regulating the cell wall integrity pathway (Denis and Cyert 2005), we hypothesized that a mutation in BGS13 would generate a yeast strain with defects associated with the cell wall. Because changes in cell wall structure have been associated with enhanced secretion (Marx et al. 2006), we decided to examine the permeability of the cell wall in bgs13 along with all the other bgs strains, using an alkaline phosphatase (ALP) colorimetric assay. ALP is a vacuolar protein which is a commonly used marker for qualitative analysis of cell wall integrity (Molina et al. 1998). Cells with abnormally permeable cell walls release ALP, permitting the hydrolysis of the chromogenic substrate BCIP (5-Bromo-4-chloro-3-indolyl phosphate p-toluidine salt), turning the colonies blue.
As expected, the S. cerevisiae temperature sensitive (ts) pkc1 yDL523 strain appeared white at the permissive temperature but turned blue after a shift to 37 °C (Fig. 2a), indicating loss of Pkc1p function and release of ALP while the wild type strain remained white at both temperatures, indicating an intact cell wall. As hypothesized, at 30 °C, the P. pastoris bgs13 colony turned blue (Fig. 2b), suggesting a leaky cell wall that released ALP. Interestingly, the same phenotype was exhibited by bgs2 and to a lesser degree by bgs6 and bgs9 (Fig. 2b).
Fig. 2.
Alkaline phosphatase (ALP) plate assays. Both S. cerevisiae (a) and P. pastoris (b) strains were spotted onto YPD medium containing BCIP. The plates were either maintained at 30 °C or shifted to the restrictive temperature (37 °C) either in the absence or presence of 1 M sorbitol
Because Pkc1p plays a role in many cellular responses in baker’s yeast (Krause et al. 2008; Lin et al. 2001), it is possible that ALP release may not have resulted necessarily from a more permeable cell wall. To examine this possibility, the same colonies were spotted onto YPD-BCIP medium containing the osmotic stabilizer sorbitol. S. cerevisiae cells with deletions of PKC1 are lethal under standard growth conditions but are viable and do not release ALP in the presence of sorbitol (Denis and Cyert 2005). As shown in Fig. 2, the ts pkc1 strain remained white after being shifted to the restrictive temperature in the presence of sorbitol, demonstrating that the sorbitol restored cell wall function. In the same way, the presence of osmotic stabilizer prevented the release of ALP from bgs13, bgs2 and the other bgs mutants, which remained white (Fig. 2b). These results suggest that the ALP release found in some bgs strains was the result of a change in cell wall permeability.
Effects of pREMI-Z mutations on the secretion of different reporters
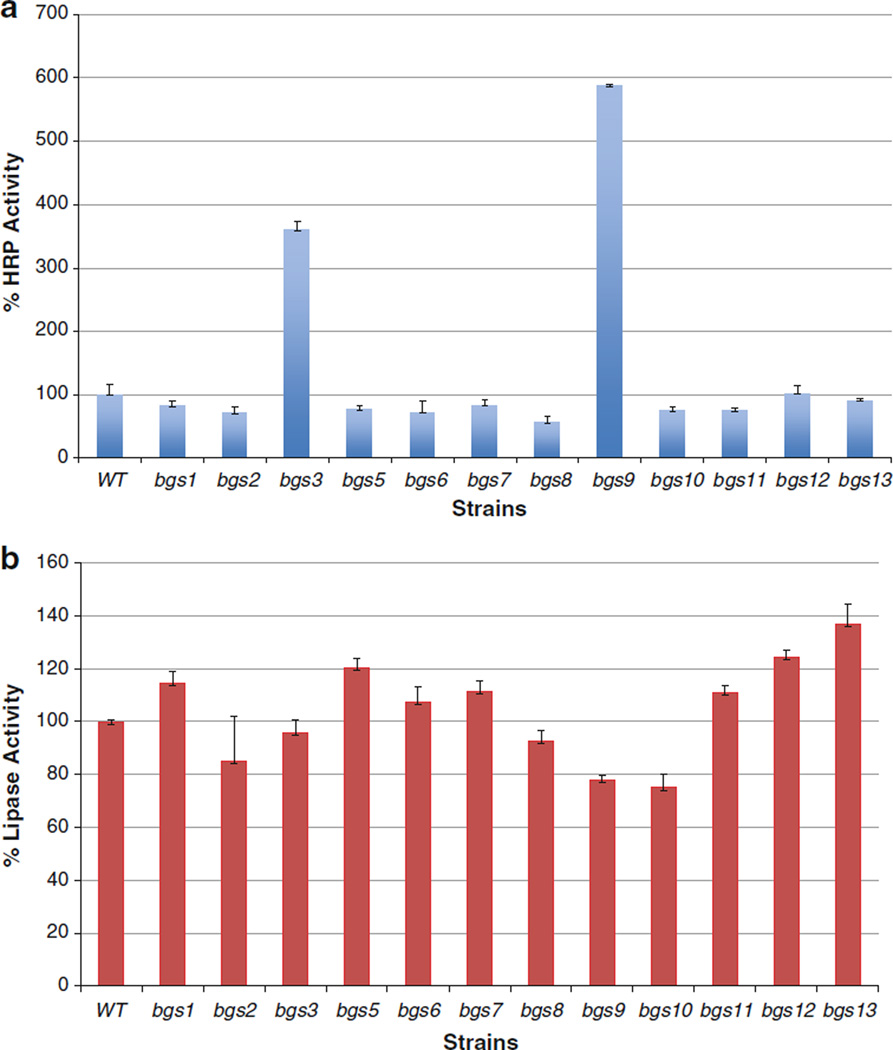
To comprehend the extent of the supersecretion phenotype, we investigated the effects of all the pREMI-Z disruptions on the secretion of two reporter genes. Horseradish peroxidase (HRP) and Candida antarctica lipase B are enzymes of ~44 and 34 kDa, respectively, that are secreted with moderate efficiency (10–20 mg/l) (Morawski et al. 2000; Rotticci-Mulder et al. 2001). HRP has a structure rich in α-helices containing a ferric heme, two calcium ions, and four disulfide bridges while lipase is a globular enzyme with an α/β hydrolase fold structure containing three disulfide bridges. These reporters present different challenges to the secretory system of a eukaryotic cell because of their diverse physical structures. pCH3K (HRP) and pKanB-lipase were transformed into the mutant as well as wild type strains, their expression was induced in methanol medium, and the reporter levels were measured in the extracellular medium. The bgs mutants differ in the efficiency by which they secrete HRP and lipase (Fig. 3a, b). Some strains, such as bgs3, showed enhanced secretion of one reporter but not the other while some gene disruptions, such as bgs8, significantly reduced secretion in comparison to wild type.
Fig. 3.
Secretion of HRP and lipase by bgs and wild type strains. Strains carrying expression plasmids for HRP (a) and lipase (b) were induced on methanol, and the activity of each reporter, standardized to equal cell numbers, was measured in the extracellular fractions. Enzyme activity of wild type was defined as 100 % for HRP (58 U/l OD600) and lipase (3.5 U/l OD600). The values represent averages at least three trials containing each reporter plasmid
Effects of bgs2, bgs3, bgs7 and bgs13 on cell growth, morphology and secretion
The different supersecreting strains were divided into three groups based on their relative level of β-galactosidase secretion (Table 1). We decided to perform a preliminary analysis on representatives of each group: low (bgs2), medium (bgs3) and high (bgs7 and bgs13) to determine if there were major differences in the behavior of the strains.
The doubling time of the wild type strain was ~110 min. As seen in Table 1, bgs2 and bgs13 strains displayed slower growth rates, suggesting that the REMI mutations affected cell viability to varying degrees. bgs3 and bgs7, however, grew at wild type rates. Second, transmission electron microscopy indicated that the mutants looked nearly identical to the wild type strain in terms of cell size, appearance of organelles and overall architecture (data not shown).
Furthermore, the secretion of a protein that posed a challenge to the secretory system was evaluated. This reporter, human SLPI (secretory leukocyte protease inhibitor), is a 12 kDa non-glycosylated, cysteine-rich enzyme inhibitor peptide that contains eight disulfide bonds and is secreted at low efficiency (~2 mg/l), as measured by spot westerns (Li et al. 2009). The SLPI expression of a strain harboring pAM1 was induced in methanol medium and measured in the extracellular medium by spot westerns. The secretion levels of SLPI along with HRP and lipase were compared for the selected strains in Table 2.
Table 2.
Expression of different reporter proteins secreted by bgs and wild type strains
| Reporter protein | Wild type | bgs2 | bgs3 | bgs7 | bgs13 |
|---|---|---|---|---|---|
| HRP | 100 | 71 | 360 | 84 | 99 |
| Lipase | 100 | 85 | 96 | 112 | 137 |
| SLPI | 100 | 93 | 103 | 218 | 214 |
The values are from enzyme activity and spot western data and represent averages of at least three trials of the strains containing each reporter plasmid. Wild type activity was defined as 100 % for HRP (59 U/L OD600) and lipase (3.5 U/L OD600). For SLPI expression, supernatant volumes corresponding to equivalent numbers of cells were spotted onto nitrocellulose and probed with anti-myc antibodies. Spot western images captured with a ChemiImager 5500 were subjected to Integrated Densitometry Values (IDV) analysis. 100 % wild type SLPI secreted corresponds to ~2 mg/l
bgs2, which is mutated in a gene that contains a domain with significant similarity to the SNW family of transcriptional activators, secretes the three reporters at levels lower than or equal to wild type. These data suggest that its enhanced secretion phenotype is fairly specific to the β-galactosidase protein. On the other hand, bgs3, whose mutant gene shows no significant homology to other genes in the searched databases, secretes higher levels of HRP than the wild type but this supersecretion phenotype does not extend to SLPI or lipase. Opposite of bgs3, bgs13 exports greater amounts of SLPI and lipase than the wild type and secretes normal levels of HRP. bgs7 secretes normal levels of HRP and lipase but has approximately twice as much SLPI in its extracellular region.
Because bgs7 and bgs13 exported the highest amounts of β-galactosidase and showed the broadest phenotype in terms of the number of reporters supersecreted, human serum albumin (HSA) was also tested in these strains. HSA is one of the most efficiently secreted, recombinant proteins by P. pastoris, reaching the g/l range using its native secretion signal (Cereghino and Cregg 2000). Structurally, HSA is comprised of three domains but lacks glycosylation and requires no post-translational modifications. Our western data revealed that bgs7 and bgs13 secrete about three times the level of HSA protein compared to the yDT39 parent, suggesting that the effect is not limited to proteins with the MATα leader (Fig. 4).
Fig. 4.
Secretion of HSA by bgs and wild type strains. Strains, either untransformed or carrying expression plasmids for HSA, were induced on methanol, and the volumes of extracellular medium, standardized to equal cell numbers, were probed with anti-HSA antibody. Spot western images captured with a ChemiImager 5500 were quantified with integrated densitometry values (IDV) analysis. Negative refers to the untransformed parent strain. WT refers to the parental wild type strain containing the HSA expression plasmid
Therefore, besides releasing higher levels of endogenous alkaline phosphatase, bgs13 shows increased secretion of four of the five heterologous proteins tested. The fact that a mutation of BGS13 can improve the secretion of so many diverse peptides raises the possibility that modification of its activity could be a step toward developing an industrial strain for recombinant expression. Although modification of Bgs13p results in a defect in cell wall integrity just as with S. cerevisiae protein kinase C (Denis and Cyert 2005), it remains unclear, however, whether (a) a leaky cell wall alone is sufficient to generate the bgs13 supersecretion phenotype since bgs2 also had a permeable cell wall, or (b) whether a mutation in BGS13 affects a downstream pathway that indirectly impacts secretion. For instance, in S. cerevisiae, Pkc1p plays a role in regulating ER membrane fusion (Lin et al. 2001). Interestingly, some proteins with a pleckstrin homology domain, which is found in Bgs7p, interact with intracellular signaling factors, such as protein kinase C in other systems (Ha et al. 2003; Yu et al. 2004). To our knowledge, our study is the first report of a potential Pkc1p ortholog being involved in regulating secretion efficiency. Furthermore, the identification of this and the other BGS genes, none of which had been associated with enhanced secretion in P. pastoris prior to this study, presents opportunities for improving this heterologous expression system and providing insights into eukaryotic secretion.
Acknowledgments
This study was supported by National Institutes of Health Academic Research Enhancement Award GM65882-03 to Joan Lin-Cereghino and Geoff P. Lin-Cereghino and a University of the Pacific Summer Undergraduate Research Fellowship to Amy Huang.
References
- Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, Rouze P, Van de Peer Y, Callewaert N. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;27:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- Delic M, Valli M, Graf AB, Pfeffer M, Mattanovich D, Gasser B. The secretory pathway: exploring yeast diversity. FEMS Microbiol Rev. 2013 doi: 10.1111/1574-6976.12020. [DOI] [PubMed] [Google Scholar]
- Denis V, Cyert MS. Molecular analysis reveals localization of Saccharomyces cerevisiae protein kinase C to sites of polarized growth and Pkc1p targeting to the nucleus and mitotic spindle. Eukaryot Cell. 2005;4:36–45. doi: 10.1128/EC.4.1.36-45.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rodriguez N, Rogers SS, Kenwright DA, Waigh TA, Woodman PG, Allan VJ. Roles of dynein and dynactin in early endosome dynamics revealed using automated tracking and global analysis. PLoS One. 2011;6(9):e24479. doi: 10.1371/journal.pone.0024479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SA, Torabinejad J, DeWald DB, Wenk MR, Lucast L, De Camilli P, Newitt RA, Aebersold R, Nothwehr SF. The synaptojanin-like protein Inp53/Sjl3 functions with clathrin in a yeast TGN-to-endosome pathway distinct from the GGA protein-dependent pathway. Mol Biol Cell. 2003;14:1319–1333. doi: 10.1091/mbc.E02-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Tyo K, Liu Z, Petranovic D, Nielsen J. Engineering of vesicle trafficking improves heterologous protein secretion in Saccharomyces cerevisiae. Metabol Eng. 2012;14:120–127. doi: 10.1016/j.ymben.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Idiris A, Tohda H, Kumagai H, Takegawa K. Engineering of protein secretion in yeast: strategies and impact on protein production. Appl Microbiol Biotechnol. 2010;86:403–417. doi: 10.1007/s00253-010-2447-0. [DOI] [PubMed] [Google Scholar]
- Krainer FW, Dietzsch C, Hajek T, Herwig C, Spadiut O, Glieder A. Recombinant protein expression in Pichia pastoris strains with an engineered methanol utilization pathway. Microb Cell Fact. 2012;11:22. doi: 10.1186/1475-2859-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause SA, Xu H, Gray JV. The synthetic genetic network around PKC1 identifies novel modulators and components of protein kinase C signaling in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:1880–1887. doi: 10.1128/EC.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Moy A, Sohal K, Dam C, Kuo P, Whittaker J, Whittaker M, Duzgunes N, Konopka K, Franz AH, Lin-Cereghino J, Lin-Cereghino GP. Expression and characterization of recombinant human secretory leukocyte protease inhibitor (SLPI) protein from Pichia pastoris. Protein Expr Purif. 2009;67:175–181. doi: 10.1016/j.pep.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Patel S, Latterich M. Regulation of organelle membrane fusion by Pkc1p. Traffic. 2001;2:698–704. doi: 10.1034/j.1600-0854.2001.21004.x. [DOI] [PubMed] [Google Scholar]
- Lin-Cereghino GP, Godfrey L, de la Cruz BJ, Johnson S, Khuongsathiene S, Tolstorukov I, Yan M, Lin-Cereghino J, Veenhuis M, Subramani S, Cregg JM. Mxr1p, a key regulator of the methanol utilization pathway and peroxisomal genes in Pichia pastoris. Mol Cell Biol. 2006;26:883–897. doi: 10.1128/MCB.26.3.883-897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Cereghino J, Hashimoto MD, Moy A, Castelo J, Orazem CC, Kuo P, Xiong S, Gandhi V, Hatae CT, Chan A, Lin-Cereghino GP. Direct selection of Pichia pastoris expression strains using new G418 resistance vectors. Yeast. 2008;25:293–299. doi: 10.1002/yea.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Cereghino GP, Stark CM, Kim D, Chang J, Shaheen N, Poerwanto H, Agari K, Moua P, Low LK, Tran N, Huang AD, Nattestad M, Oshiro KT, Chang JW, Chavan A, Tsai JW, Lin-Cereghino J. The effect of alpha-mating factor secretion signal mutations on recombinant protein expression in Pichia pastoris. Gene. 2013;519:311–317. doi: 10.1016/j.gene.2013.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx H, Sauer M, Resina D, Vai M, Porro D, Valero F, Ferrer P, Mattanovich D. Cloning, disruption and protein secretory phenotype of the GAS1 homologue of Pichia pastoris. FEMS Microbiol Lett. 2006;264:40–47. doi: 10.1111/j.1574-6968.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- Mattanovich D, Callewaert N, Rouze P, Lin YC, Graf A, Redl A, Tiels P, Gasser B, De Schutter K. Open access to sequence: browsing the Pichia pastoris genome. Microb Cell Fact. 2009;8:53. doi: 10.1186/1475-2859-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir DT. Barr PJ, Brake AJ, Valenzuela P, editors. Yeast mutants with increased secretion efficiency. Yeast genetic engineering. Butterworths, Boston. 1989:215–231. [PubMed] [Google Scholar]
- Molina M, Martín H, Sánchez M, Nombela C. 20 MAP kinase-mediated signal transduction pathways. Methods Microbiol. 1998;26:375–393. [Google Scholar]
- Morawski B, Lin Z, Cirino P, Joo H, Bandara G, Arnold FH. Functional expression of horseradish peroxidase in Saccharomyces cerevisiae and Pichia pastoris. Prot Eng. 2000;13(5):377–384. doi: 10.1093/protein/13.5.377. [DOI] [PubMed] [Google Scholar]
- Prevatt W, Sreekrishna K. Expression of human serum albumin in Pichia pastoris. 5330901. US Patent. 1994
- Rotticci-Mulder JC, Gustavsson M, Holmquist M, Hult K, Martinelle M. Expression in Pichia pastoris of Candida antarctica lipase B and lipase B fused to a cellulose-binding domain. Protein Exp Purif. 2001;21:386–392. doi: 10.1006/prep.2000.1387. [DOI] [PubMed] [Google Scholar]
- Rudolph HK, Antebi A, Fink GR, Buckley CM, Dorman TE, LeVitre J, Davidow LS, Mao J, Moir DT. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell. 1989;58:133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- Schroder LA, Glick BS, Dunn WA. Identification of pexophagy genes by restriction enzyme-mediated integration. Methods Mol Biol. 2007;389:203–218. doi: 10.1007/978-1-59745-456-8_15. [DOI] [PubMed] [Google Scholar]
- Staley CA, Huang A, Nattestad M, Oshiro KT, Ray LE, Mulye T, Li ZH, Le T, Stephens JJ, Gomez SR, Moy AD, Nguyen JC, Franz AH, Lin-Cereghino J, Lin-Cereghino GP. Analysis of the 5′; untranslated region (5′;UTR) of the alcohol oxidase 1 (AOX1) gene in recombinant protein expression in Pichia pastoris. Gene. 2012;496:118–127. doi: 10.1016/j.gene.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor D, Xiong S, Orazem CC, Kwan AC, Cregg JM, Lin-Cereghino J, Lin-Cereghino GP. Cloning and characterization of the Pichia pastoris MET2 gene as a selectable marker. FEMS Yeast Res. 2005;5:935–942. doi: 10.1016/j.femsyr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Waterham HR, Digan ME, Koutz PJ, Lair SV, Cregg JM. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene. 1997;186:37–44. doi: 10.1016/s0378-1119(96)00675-0. [DOI] [PubMed] [Google Scholar]
- Yount BA, Lin-Cereghino J, Lin-Cereghino GP, Fox MM. Preparation of the yeast Pichia pastoris for transmission electron microscopy. Microsc Today. 2006;14:36–37. [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]