Abstract
Manipulation of gene expression is one of the most informative ways to study gene function. Genetic screens have been an informative method to identify genes involved in developmental processes. In the zebrafish, loss-of-function screens have been the primary approach for these studies. We sought to complement loss-of-function screens using an unbiased approach to overexpress genes with a Gal4-UAS based system, similar to the gain-of-function screens in Drosophila. Using MMLV as a mutagenic vector, a cassette containing a UAS promoter was readily inserted in the genome, often at the 5′ end of genes, allowing Gal4-dependent overexpression. We confirmed that genes downstream of the viral insertions were overexpressed in a Gal4-VP16 dependent manner. We further demonstrate that misexpression of one such downstream gene gucy2F, a membrane-bound guanylate cyclase, throughout the nervous system results in multiple defects including a loss of forebrain neurons. This suggests proper control of cGMP production is important in neuronal survival. From this study we propose that this gain-of-function approach can be applied to large-scale genetic screens in a vertebrate model organism and may reveal previously unknown gene function.
Keywords: insertional mutagenesis, forward genetic screen, Gal4-VP16, guanylate cyclase
INTRODUCTION
One of the most informative ways to study gene function has been to manipulate its expression in a model organism either through loss-of-function or overexpression. While loss-of-function studies can be performed in a non-biased manner, genes with redundant functions may be overlooked. Overexpression studies in vertebrates commonly examine the effect of a predetermined set of genes when expressed in a tissue of interest. We sought to develop, in a vertebrate model organism, an unbiased approach to overexpress genes for genetic analysis, similar to the gain-of-function screens that have been done in Drosophila.
Zebrafish is a well established model system to study vertebrate development and other processes and is amenable to genetic manipulation. One of the key advantages of zebrafish is the ability to perform large-scale forward genetic screens to identify genes involved in developmental processes. This has been achieved primarily using ENU to mutate genes (Driever et al. 1996; Haffter and Nusslein-Volhard 1996; Muto et al. 2005), although insertional mutagenesis mediated by retrovirus (Amsterdam et al. 1999; Golling et al. 2002; Wang et al. 2007) or Tol2 based transposons (Kawakami et al. 2004; Kotani et al. 2006; Asakawa et al. 2008) is gaining wider use. While highly informative, forward genetic screens based on recessive phenotype cannot identify mutations in genes with redundant function. In addition, these mutations impact the entire organism, making determination of tissue-specific function of the affected gene very challenging. A complementary approach to the loss-of-function screen is to overexpress genes in a specific tissue or cell type and determine phenotypic consequences. In Drosophila, large-scale gain-of-function screens through overexpression of random genes have been used to identify genes involved in specific developmental processes (Hay et al. 1997; Rorth et al. 1998; Staudt et al. 2005; Molnar et al. 2006). Because these mutations are “dominant” and can be controlled spatially and temporally, the gain-of-function screens have identified genes that had been unnoticed in loss of function screens.
While used extensively in Drosophila, the Gal4-UAS system has not been widely applied in zebrafish. More recently, Gal4-UAS is being incorporated as a component of gene-trap and enhancer trap screens (Davison et al. 2007; Scott et al. 2007; Asakawa et al. 2008). However, these approaches rely primarily on a UAS driven fluorescent protein as an indicator of the expression pattern of the gene tagged by a Gal4 cassette and often does not significantly impact the expression of the trapped gene itself. In order to examine the function of a gene, its expression would need to be altered. In the Drosophila gain-of-function screens, a Gal4 responsive promoter (UAS) is randomly inserted in the genome by EP elements and genes downstream of these insertions are expressed when the Gal4 activator is present (Rorth 1996; Hay et al. 1997). For zebrafish, retrovirus is an appealing vector to deliver the UAS element into the zebrafish genome. The mouse Moloney leukemia virus (MMLV) has been used by several groups as an insertional mutagen in zebrafish (Amsterdam et al. 1999; Golling et al. 2002; Ellingsen et al. 2005; Wang et al. 2007), facilitating loss-of-function screens and providing a tool to quickly identify essential genes in development. MMLV is also uniquely poised to facilitate gain-of-function screens since the insertions occurring in genes have been shown to be primarily located at the 5′ end (Wu et al. 2003; Wang et al. 2007). The gain-of-function approach increases the efficiency of phenotype-based screens since the analysis can be performed in the F1 generation, in contrast to the F3 generation used for loss-of-function screens. Furthermore the effect of all germline insertions in a mosaic founder can be examined, instead of propagating a fraction in selected F1 progeny for the loss-of-function screens.
We show here the application of a MMLV mediated gain-of-function strategy with overexpression of genes that lie downstream of the viral insertions. Overexpression of some of these genes in specific tissues can lead to a detectable phenotype, showing this approach is valid for a forward genetic screen in a vertebrate system. Additionally, many of the insertions are also expected to inactivate the host gene in the absence of Gal4-based transcription factors, allowing loss-of-function effects to also be determined.
MATERIALS AND METHODS
Fish husbandry
Zebrafish were raised in Aquatic Habitats systems (Apopka, FL) on a 14 to 10 light dark cycle as described previously (Chen and Casey Corliss 2004). Tubingen fish were acquired from the Zebrafish International Resource Center (ZIRC). Tg(ath5:Gal4VP16) fish were genotyped for the transgene and maintained as homozygous adults. Embryos were reared in 0.3x Danieau’s solution at 28°C.
Generation of gene-activating virus
The pClnZ plasmid (Chen et al. 2002) was modified to contain the gene-activation cassette in the reverse orientation of LacZ. The gene activating cassette was generated using 14xUAS obtained from Dr. Scott Fraser (California Institute of Technology), the synthetic element subcloned from the Gene-Switch plasmid (Invitrogen) and the BGH pA subcloned from pCDNA3.1Myc-His (Invitrogen). The viral packaging cell line 293 gp/bsr (Amsterdam et al. 1999) was grown in Dulbecco’s modified Eagle medium with 10% fetal calf serum, penicillin and streptomycin. Transfection of 293 gp/bsr cells, transduction and selection of individual cells by FACS was performed as previously described (Chen et al. 2002). When individual clones became confluent, the viral titer from these clones was assessed by QPCR (Chen et al. 2002). In this assay, viral titer is determined by examining Ct values for viral DNA compared to the Ct values for the endogenous Rag1 gene (ΔCt = Ct(virus) − Ct(Rag1)). In this comparison a more negative number indicates more viral copies are present. To compare virus producing clones a ΔΔCt value is determined by comparing the ΔCt of a selected clone to the control GT186 (ΔΔCt = ΔCt(clone)−ΔCt(control)). Once again, a negative number indicates more viral copies are present in cells infected with the test clones than with the control. Clones producing the highest titer in the initial assay were scaled up to 10 cm dishes. For large scale virus preparation, viral producing cells were plated on poly-L-lysine coated 15 cm dishes for 90% confluency. Cells were transfected overnight with CMVG and Lipofectamine 2000 using conditions yielding the optimal viral titer. For clone 10D11, optimal transfection condition was 5μg CMVG and 10:1 Lipofectamine 2000; for clone 9D4, 10μg CMVG with 10:1 Lipofectamine 2000. Following 16 hours of transfection, the medium was changed to normal growth medium. At day 2 post transfection, medium was changed to 12.5 ml of growth medium supplemented with 10mM HEPES (collection medium) and conditioned medium collected 24 hours later at 3 days post transfection. To maximize the amount of virus obtained from a single transfection, when day 3 conditioned medium was removed it was replaced with an additional 12.5 ml of collection medium and collected at day 4. To concentrate the virus, medium was filtered through 0.2 μM filters and concentrated by centrifugation at 58,000 RCF for 45 min in an SW28 rotor at 4°C. Viral pellets were resuspended in 30μl of calcium and magnesium free PBS supplemented with 10 mM HEPES. Zebrafish embryos were injected with concentrated virus from 9D4, 10D11 or GT186 clones as previously described (Lin et al. 1994) and the number of proviral inserts determined using QPCR as described (Amsterdam et al. 1999). To determine the number of proviral inserts, the ΔCt values are calculated as outlined above and compared to the ΔCt values of a standard curve generated using genomic DNA containing a known number of proviral copies.
PCR analysis of viral insertions
For inverse PCR, genomic DNA was digested with Taq1α (NEB) and diluted twofold for ligation. PCR was performed with primers either for the left side LTR or right side LTR of the provirus using Phusion polymerase (Finnzymes) to ensure amplification of large products. For linker-mediated PCR, genomic DNA was digested with Tsp509I followed by incubation with 10 μg/ml Proteinase K to digest the enzyme. Linkers were subsequently ligated onto the fragments and nested PCR was performed using primers for the left side or the right side of the viral insertion. Products were separated on a 2% agarose gel, purified from the gel, and directly sequenced. To determine location of the viral insertion, BLAST searches were performed for the resulting sequences to the zebrafish assembly version 7 and only unambiguous alignments are reported. To genotype for a specific insertion, a primer for the genomic location was used in conjunction with a primer for the LTR of the virus.
Expression analysis
For RT-PCR, pools of embryos were collected from appropriate matings. RNA was isolated using Trizol (Invitrogen) and reverse transcription performed using Transcriptor Reverse Transcriptase (Roche Applied Science) with oligo(dT) containing a T7 adaptor. Two rounds of PCR using Phusion polymerase (Finnzymes) were performed with primers for the viral tag and T7 based step-out primers for the 3′ end. Products were separated on 1.2% agarose, purified and directly sequenced. The sequences were compared using BLAST against the zebrafish assembly database (Zv7).
For In Situ Hybridization, when possible products from the RT-PCR analysis were used as templates for anti-sense probes as they contain a T7 promoter. For Gal4-VP16, cDNA was cloned into pBluescript II (Stratagene). Antisense probes were generated using the appropriate RNA polymerase (T7 or T3, Promega). Hybridization was performed as previously described (Chen et al. 2001).
Guanylate cyclase construct expression in zebrafish embryos and cGMP assay
For gucy2F, the full-length cDNA was amplified by PCR using Phusion polymerase to reduce the possibility of errors being introduced. For gucy1a3, the full-length cDNA was subcloned from an available IMAGE clone. The cDNAs were placed behind 14x UAS and a synthetic element containing an intron. These expression cassettes were then inserted into the pBS-I-Sce meganuclease vector (Grabher et al. 2004). For injection into one-cell stage embryos, plasmid DNA was prepared at a concentration of 30 ng/μl with I-Sce I meganuclease and one nanoliter injected into embryos.
To assess cGMP levels, appropriate samples were homogenized in 0.1M HCl. All samples were acetylated and cGMP levels assessed using a direct cGMP kit (Assay Designs/Stressgen) according to manufacturer’s instructions.
RESULTS
Generation of the gene-activating virus
To facilitate a gain-of-function screen in zebrafish, we constructed a MMLV based virus that contained a UAS responsive promoter. As shown in Figure 1, the parent GT vector (Chen et al. 2002; Golling et al. 2002) was modified to include a polyadenylation signal (pA), a tandem UAS-based promoter, and a downstream synthetic element that includes a splice donor. Also included in the construct are the viral LTRs, viral packaging signals and a LacZ gene to allow isolation of virus producing cells. When the provirus is inserted in the correct orientation the pA, in conjunction with a preceding cryptic splice acceptor in the viral vector, is expected to terminate transcription that may be initiated upstream of the insertion. This provides an additional mechanism leading to a loss-of-function insertion and allows these viral insertions to serve a dual purpose. Where the Gal4-VP16 fusion protein is present, transcription should be initiated from the UAS promoter within the provirus and the splice donor of the synthetic element should be joined to the nearest exon, leaving a portion of the viral construct on the transcript which acts as a tag for identification. No initiator ATG is present within the viral tag, therefore, if the insertion occurs upstream of the first coding exon, a full-length protein should be produced. If the viral insertion occurs within an intron, the preceding exons will not be contained in the resulting RNA and may result in a gene product that is constitutively active or dominant negative.
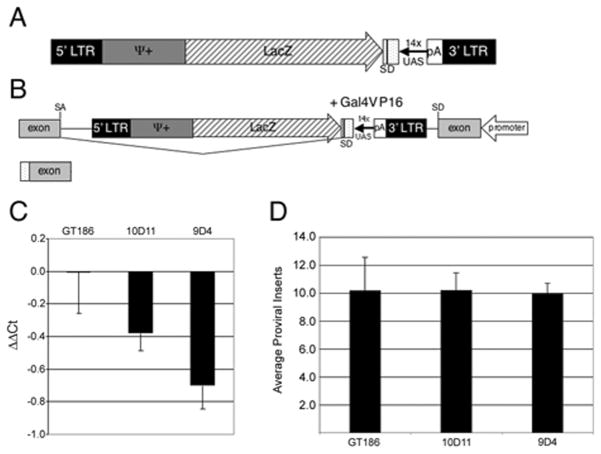
Figure 1. Generation of the gene-activation virus.
A – Schematic of the elements in the gene activation virus. The gene-activation cassette includes a polyadenylation signal from bovine growth hormone (pA), Gal4 responsive promoter (UAS) and a synthetic element with a splice donor (SD). Additional elements are the viral long terminal repeats (LTR), Ψ+ viral packaging signals and LacZ. B – integration of the virus in the intron of a gene. In the presence of Gal4-VP16 transcription is initiated from the UAS and splicing should occur from the splice donor to the splice acceptor (SA) of the following exon. The resulting RNA will have a portion of the gene-activation cassette on the 5′ end. C – Infection of Pac2 zebrafish cells with virus from two clones, 10D11 and 9D4 determined using QPCR. Negative ΔΔCt values indicate more proviral inserts per cell compared to the GT186 virus. D - Infection of zebrafish embryos with virus from two clones, 10D11 and 9D4. Number of proviral inserts determined using QPCR.
The viral construct was transfected into 293gp cells, and individual cells selected based on the level of β-galactosidase production using FACS. The viral titer produced from each clone was determined by infecting PAC2 zebrafish fibroblast cells and determining the proviral inserts by QPCR as described (Chen et al. 2002). As shown in Figure 1B and C, although the modifications increased the overall size of the viral vector, there was no negative impact on viral titer. When compared to a previously used viral clone, GT186 (Chen et al. 2002), two clones, 9D4 and 10D11, consistently produced more viral insertions in the zebrafish cell line shown as a negative ΔΔCt value due to the normalization to the endogenous rag1 gene(Fig 1B). To analyze the ability of these virus producing clones to mutagenize the zebrafish genome, purified, concentrated retrovirus was injected into zebrafish embryos. The 9D4 and 10D11 clones produced similar numbers of proviral insertions in zebrafish embryos as the GT186 clone (Fig 1C), with an average of 10 proviral insertions per cell. While both clones produced virus of similar titer, the 9D4 clone was used for subsequent experiments since 70% of 9D4 injected embryos, compared to 30% of 10D11 injected embryos, survived after 16 hours post injection. Large scale preparations of the 9D4 virus were generated and injected into Tubingen embryos at the 1000-cell stage and surviving embryos were raised to maturity resulting in approximately 6000 adult virus injected F0 fish.
Germline integration of the gene-activating virus in the zebrafish genome
To determine whether the UAS-based promoter in the provirus functions as predicted, we characterized germline insertions in a subset of founders. Twenty one founders were outcrossed and the insertions in the F1 fish identified using inverse PCR and linker-mediated PCR. Inverse PCR on randomly selected F1 embryos (Figure 2A) indicated the number of unique viral insertions carried varied from a single insertion (Family 5 embryo 1) to multiple insertions (Family 1). These results are consistent with the data obtained by QPCR and Southern blot analysis (data not shown). From these 21 founders, a subset of F1 fish were raised to maturity and the insertions carried by these F1 adults were analyzed using linker mediated PCR (Figure 2B). A total of 70 insertions were characterized but represent only a fraction of the total insertions carried by the 21 founder fish. Table 1 summarizes the insertions where a known gene or EST may be a target for Gal4-VP16 mediated overexpression. Several of the viral insertions are likely too distant from the nearest gene to result in Gal4-VP16-dependent overexpression. In agreement with data previously observed for the MMLV retrovirus, when predicted genes are included for the collection, the majority of the insertions occur at the 5′ end of genes: 25 (36%) of the insertions occur in the 1st intron of a gene, 15 (21%) occur in the proximal region within 1 kb of the 1st intron, 13 (19%) insertions occurred in introns other than the first. The remaining 16 insertions (25%) occur at least 1 kb away from the 1st exon as identified in the ENSEMBL database. However, many of the initial exons annotated in the zebrafish database are the first coding exon and non-coding exons may exist near the viral insertion. This is the case for family pa008 (Portland, activating) where an insertion on chromosome 3 occurs 12.3 kb upstream of LOC557147, a gene related to Pax6. However, transcript analysis indicated previously unidentified 5′ untranslated exons with the virus insertion occurs 1 kb upstream from an untranslated exon and this gene can be overexpressed in the presence of Gal4-VP16.
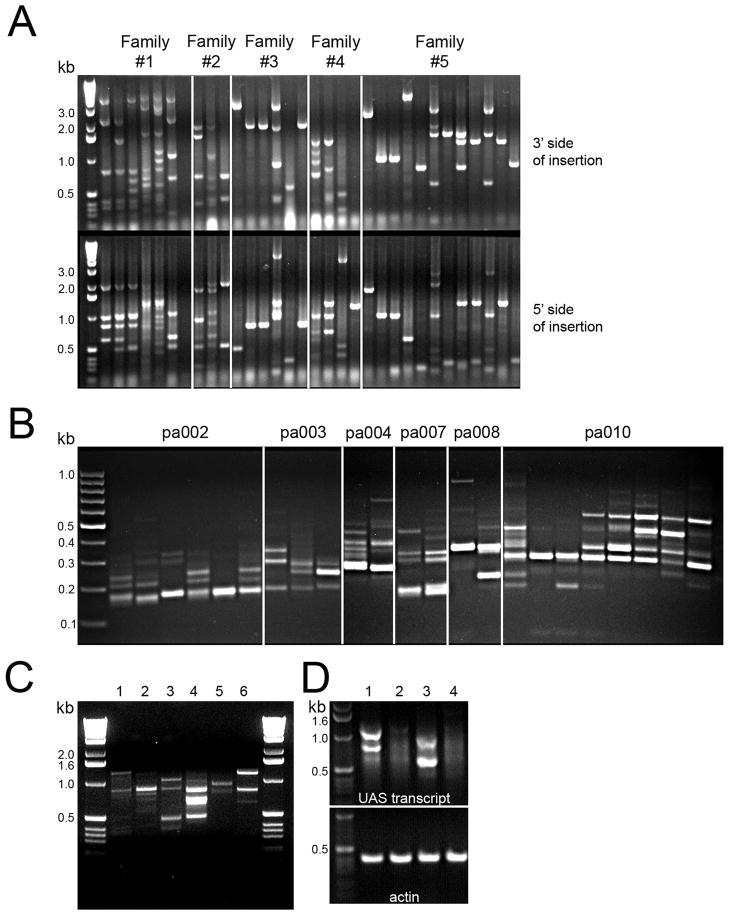
Figure 2. The gene-activation virus integrates throughout the zebrafish genome and downstream genes are targeted for overexpression.
A – inverse PCR for F1 embryos. Each lane is DNA for an individual embryo with 5 different families represented. Each band is a unique viral insertion with both flanking genomic regions analyzed. B – linker-mediated PCR using genomic DNA from F1 fish previously genotyped for a selected insertion. Each lane is a separate fish representing 6 different families. The number of stable insertion in these fish varies from 1 to 7 insertions. C – overexpression of genes downstream of viral insertions. Each lane represents an individual family with embryos obtained by crossing to Tg(ath5:Gal4VP16) and pooled at 48 hours post fertilization. Each band is a unique gene induced by Gal4-VP16. The number of overexpressed genes can vary from one (lane 5) to multiple genes (lane 4). D- Two families of F1 embryos were analyzed after crossing to wild-type (lanes 2 and 4) or Tg(ath5:Gal4VP16) lanes 1 and 3) using RT-PCR. Gal4 induced transcripts are detected only in the presence of Gal4-VP16.
TABLE 1.
Genomic location of viral insertions in F1 families maintained with a selected insertion. The genomic location of the viral insertion in these fish, strand impacted by the insertion and putative gene targeted for overexpression in the presence of Gal4-VP16 are indicated.
| Family | Insertion(Chr;bp) | Strand | Target gene | Distance | Accession # |
|---|---|---|---|---|---|
| pa001 | 16; 25040525 | minus | ENSDARESTG00000016651 | 1st exon | BC134041 |
| pa002 | 13; 547721 | plus | ENSDART00000029051 (AS) | downstream | NM_001109731 |
| pa003 | 8; 3258272 | plus | cxxc1l (zgc:63512) | 765 bp upstream | NM_200599.1 |
| 4; 4157290 | plus | si:dkey14d8.5 | 1st intron | NM_001044841 | |
| 25; 24786581 | plus | zgc:162316 | 627 bp upstream | NM_001089524 | |
| 9; 35404108 | plus | LOC564612 | 12kb upstream | XM_687945 | |
| 21; 20464166 | plus | zgc:63532 | 13 kb upstream | NM_200451 | |
| 1; 14743223 | minus | ENSDARESTG00000017576 | 15 kb upstream | BC163858 | |
| 19; 37538331 | plus | gngt1 | 10kb upstream | NM_199967 | |
| pa004 | 23; 5325388 | plus | gata5 | 34 kb upstream | NM_131235 |
| 9; 6372551 | plus | Q0V969_DANRE | 1st intron | XM_686856 | |
| 10; 1548701 | plus | ENSDARESTG00000013253 | 1st intron | XR_045177 | |
| pa005 | 17; 8060679 | plus | sec23a | 68 bp upstream | NM_213465 |
| pa007 | 21; 330010041 | plus | zgc:92775 | 100 bp upstream | NM_001003424 |
| 19; 28445980 | plus | zgc:153389 | 100 bp upstream | NM_001045380 | |
| 25; 24786583 | minus | Serdin1 | 4.5 kb upstream | NM_001009559 | |
| pa008 | 3; 30287910 | plus | LOC557147 | 12.3 kb upstream | XM_680152 |
| 22; 1567618 | plus | ENSDARG00000071738 | 180bp upstream | XM_001922442 | |
| pa010 | 12; 17866096 | minus | zgc:64155 | 5 kb upstream | NM_200049 |
| 19; 12118629 | minus | no target within 100 kb | |||
| pa011 | 23; 19856166 | plus | ENSDART00000104210 | 2nd intron | BC075740 |
| 22; 3247549 | plus | LOC568321 | 4 kb upstream | XM_691644 | |
| pa013 | 20; 48008187 | minus | zgc:56518 | 600 bp upstream | NM_213427 |
| 17; 18866024 | plus | ENSDARESTG00000001086 | 3rd intron | XM_001341307 | |
| 21; 4743363 | minus | ENSDARESTG00000008827 | 2nd intron | BC134922 | |
| pa014 | 24; 12685262 | plus | no target within 100kb | ||
| 14; 4889047 | minus | ENSDARESTG00000016928 | 1st intron | XM_001922888 | |
| 15; 26281200 | plus | foxn1 | 16 kb upstream | NM_212573 | |
| 6; 21336874 | plus | ENSDARESTG00000003321 | 1st intron | NM_199526 | |
| 8; 54135254 | plus | FEM1 | 28 kb upstream | AY249190 | |
| pa015 | 5; 13747065 | minus | sox19a | 1.6 kb upstream | NM_130908 |
| pa016 | 5; 59290951 | plus | ENSDARESTG00000019052 | 29 kb upstream | XM_684978 |
| 23; 20977122 | plus | ENSDARESTG00000023027 | 1st intron | CR388207 | |
| pa019 | 9; 33247346 | minus | zgc:77231 | 50 bp upstream | NM_200000 |
| pa020 | 12; 23590340 | minus | ENSDARESTG00000009439 | 276 bp upstream | XM_683728 |
| pa025 | Zv7_scaffold2487 | plus | ENSDARESTG00000024884 | 7kb upstream | BX927103 |
| 6; 9608673 | minus | dnah9l | 4th intron | XM_001340261 | |
| 8; 19746052 | minus | ENSDARESTG00000017700 | 4.2 kb upstream | BC075974 | |
| pa026 | 15; 3362892 | minus | smc4 | 42kb upstream | NM_173253 |
| pa027 | 13; 34015197 | minus | txndc1 | 1st intron | XM_001336514 |
| pa028 | 22; 14723856 | minus | nfia | 66 kb upstream | NM_001079962 |
| 24; 22419219 | plus | pol1a | 1kb upstream | XP_001342845 | |
| 15; 30463249 | minus | gucy2f | 17 kb upstream | XM_684538 | |
| 17; 1838398 | plus | zgc:56248 | 2nd intron | NM_201215 | |
| pa030 | 19; 20906119 | minus | ENSDARESTG00000003278 | 6 kb upstream | BC152302 |
| pa031 | Zv7_NA2685 | minus | ENSDARESTG00000018886 | 1st intron | NM_131081 |
Gal4-VP16 induces expression of genes downstream of the viral insertion
There is an orientation dependence to the gene-activating virus; therefore, while the insertions occur within or just upstream of genes, not all of these will be expressed in the presence of Gal4-VP16. When the genomic location of the viral insertions is examined, the nearest gene that may be regulated by Gal4-VP16 can be predicted. However, in some cases, such as with pa010 or pa014, the closest gene on the same strand is more than 20kb away and may not be overexpressed in the presence of Gal4-VP16. To examine expression of Gal4 induced transcripts, we used a procedure similar to 3′ RACE that uses the viral tag on the 5′ end of the transcript and an adaptor for the 3′ tail to specifically amplify the transcripts initiated from the viral insertion. No transcript is identified in a pool of embryos obtained by outcrossing founders to wild-type fish (Figure 2D, lanes 2 and 4). In contrast, multiple transcripts are identified in a pool of embryos from founder fish crossed to Tg(ath5:Gal4VP16) (Figure 2C), supporting that Gal4-VP16 efficiently induces transcription of genes downstream from the proviral insertion. Genes induced by Gal4-VP16 were examined for 15 of the 21 families maintained as F1 adults (Table 2). The majority (18/21 (86%)) of the identified transcripts lie downstream of the viral insertions identified in the F1 fish, although 3 (14%) transcripts were from insertions not identified by LM-PCR or inverse PCR. As expected, where the viral insertion occurs in the first intron, the corresponding transcript begins at the second exon. In many cases, the first exon of the mutated gene is noncoding, and a full length protein is expected from these viral-initiated transcripts. Interestingly, some of the identified transcripts contain noncoding exons that have not been previously annotated. Two (9.5%) transcripts identified contain spliced exonic sequences that do not correspond to an annotated gene in the database; the sequence of these transcripts have been deposited into the EMBL/GenBank data library. These may be novel genes that have not been previously identified or may be a result of splicing of cryptic exonic sequences and are artificial transcripts initiated from the insertions. Two additional transcripts were identified that lie downstream of a viral insertion and contains a single exon but without an ORF and but do not align with an annotated gene;. These are most likely artificial transcripts and are not included in Table 2. In one case, pa002, such an artificial transcript aligns with a portion of a known gene on the antisense strand and may act as antisense RNAs to decrease the expression of the known gene.
TABLE 2.
Transcripts overexpressed in the presence of Gal4-VP16. F2 embryos were analyzed using 3′ RACE and all transcripts sequenced. For each transcript the genomic location is identified with start as the first nucleotide of the alignment. The direction of the transcript is identified as well as relevant structural information and accession numbers.
| Family | Chromosome, 1st bp | Strand | Gene | Transcript features | Accession # |
|---|---|---|---|---|---|
| pa001 | 16, 25039037 | minus | ENSDARESTG00000016651 | missing 1st exon | BC134041 |
| pa002 | 13, 551588 | plus | AS for ENSDARG00000039833 | BX323800 | |
| pa003 | 4, 4158304 | minus | si:dkey-14d8.5 | missing 1st exon | NM_001044841 |
| 8, 3260547 | plus | cxxc1l | missing 1st exon | NM_200599.1 | |
| pa004 | 23, 5329961 | plus | novel-unidentified transcript | 3 exons, short ORF | FJ151013 |
| pa005 | 17, 8064176 | plus | sec23a | missing 1st exon | NM_213465.1 |
| pa007 | 17, 39393740 | minus | zgc:77004 | extra 5′UTR exons | NM_213316.1 |
| 21, 33011784 | plus | zgc:92775 | missing 1st exon | NM_001003424 | |
| pa008 | 3, 30289479 | plus | LOC557147 | extra 5′UTR exons | XM_680152.2 |
| 22, 1569032 | plus | ENSDARG00000071738 | full length | BC155813 | |
| pa010 | 7, 14879344 | plus | nedd8 | missing 1st exon | NM_201321.1 |
| 12, 14879344 | minus | zgc:64155 | full-length | NM_200049.1 | |
| pa013 | 19, 4725956 | minus | tpsn | missing 1st exon | NM_130974.1 |
| pa014 | 14, 4884769 | minus | ENSDARESTG00000016928 | missing 1st exon | BC122283 |
| 24, 12695317 | plus | novel transcript | 3 exons, short ORF | FJ151014 | |
| pa020 | 12, 23589829 | minus | ENSDARESTG00000009439 | missing 1st exon | XM_683728.2 |
| pa025 | 6, 9709034 | minus | dnah9l | missing exons 1–4 | XM_001340261 |
| pa028 | 17, 1840419 | plus | zgc:56248 | missing exons 1–2 | NM_201215.1 |
| 24, 22421077 | plus | pola1 | full-length | XM_682004.2 | |
| 15, 30462034 | minus | gucy2f | full-length | XM_684538.1 | |
| pa031 | 2, 39795141 | plus | CDH2 | extra 5′UTR | XM_693056.2 |
We examined the expression pattern of several of these genes in both a wild-type and Tg(ath5:Gal4VP16) background. The ath5 promoter has been demonstrated to direct expression to the retinal ganglion cells, as well as ectopic expression in the olfactory bulb (Kay et al. 2005) In these transgenic fish, onset of Gal4-VP16 expression is at 24hpf (Figure 3A) with expression in a few cells in the ventral retina as well as slight expression in the olfactory bulb. At 50 hpf, there is a marked increase in expression which encompasses the entire retina (Figure 3B), but the expression in the olfactory bulb is much weaker. Genes downstream of viral insertions are expressed in a similar pattern. For example, at 48hpf nedd8, a gene downstream of a viral insertion identified in pa010, is not normally expressed in the retina in embryos that carry the viral insertion but not ath5:Gav4VP16 (Figure 3C). In embryos that carry both ath5:Gal4VP16 and the viral insertion, the expression of nedd8 is prominent in the retina (Figure 3D). At 48hpf, the expression pattern of the nedd8 is primarily in the ventral retina. This slight restriction in expression domains suggests that the Gal4-VP16 fusion protein may need to accumulate to a sufficient level to induce expression from the viral insertion, which in this Gal4-VP16 transgenic line appears to occur predominantly in the ventral retina at this stage. We also examined expression of another candidate gene in embryos at a later stage. This target gene, a Pax6-like gene identified in family pa008, was found to be expressed in neurogenic regions at early stages of development in wild type embryos (data not shown). At 54 hpf expression is barely detectable (Figure 3E). However in embryos that carry the upstream viral insertion as well as ath5:Gal4VP16, expression in the retina continues to be detected at 54 hpf and is restricted to the retinal ganglion cell layer (Figure 3F). Using a separate Gal4-VP16 diver line that is more fully discussed in later sections, we also observe upregulation of an additional target gene in cells where Gal4-VP16 is expressed (Figure 4B and C). These data support the Gal4-VP16 dependent activation of genes downstream of viral insertions.
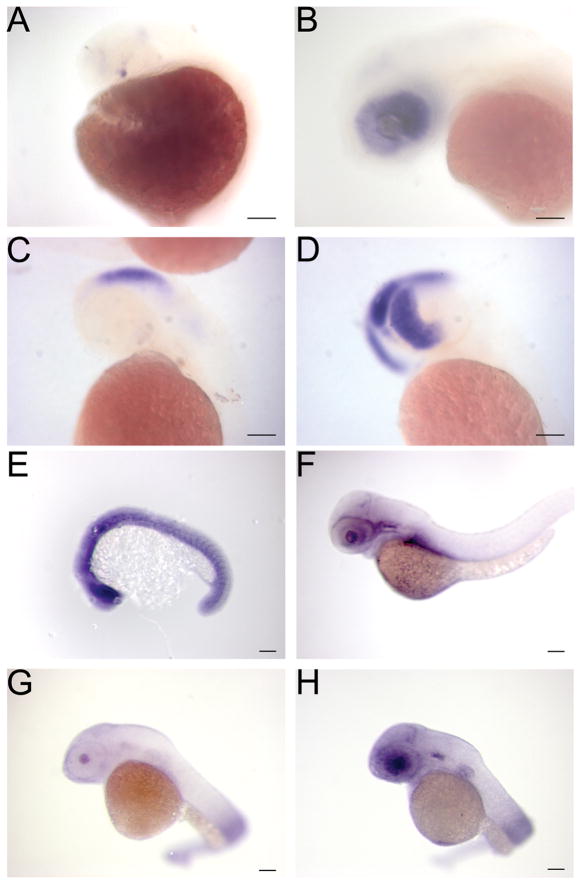
Figure 3. Genes downstream of viral insertions are overexpressed in the presence of Gal4-VP16.
A, B - Expression of Gal4-VP16 in Tg(ath5:Gal4VP16) transgenic embryos. At 24 hpf a few positive cells in the retina are detected (arrow in A). This expands to encompass the entire retina by 50 hpf (B). C, D - Expression of a gene, nedd8, downstream of a viral insertion at 48 hpf. Embryos without the viral insertion have normal expression in the brain (C) and embryos carrying both the viral insertion and Tg(ath5:Gal4VP16) show overexpression in the retina (D). E, F Overexpression of a Pax6-like gene. At 54 hpf there is little expression (E) but embryos that carry the upstream viral insertion and Tg(ath5:Gal4VP16) continue to express the Pax6-like gene primarily in the retinal ganglion cell layer (F). Scale Bars are 50 μM
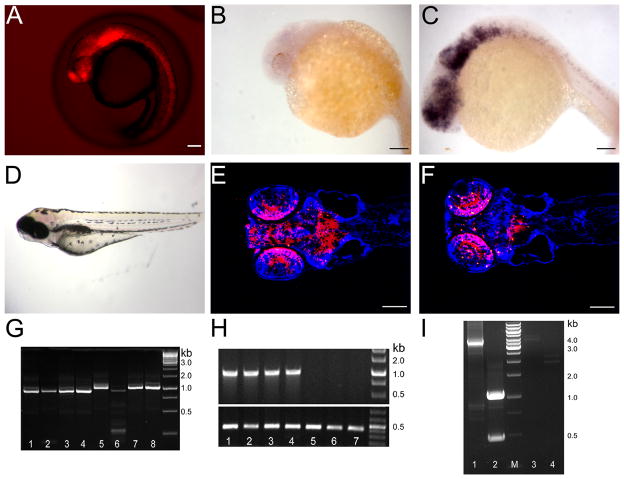
Figure 4. Overexpression of a guanylate cyclase downstream of a viral insertion results in morphological defects.
A- Gal4-VP16 expression in a gene-trap line where mCherry is under the control of UAS and serves as a marker for expression. Expression is present in all areas of neurogenesis including the retina, brain and spinal cord. B–C Expression of gucy2F at 24 hpf in normal (B) and embryos with the viral insertion upstream of gucy2F and Gal4-VP16 (C). D – Phenotype of mutant larvae at 5 dpf presenting with a shortened trunk, hemorrhaging in the brain and abnormal head morphology particularly in the area of the forebrain. E–F- Coronal sections of a phenotypically normal (E) and abnormal (F) 5 dpf larvae. Many neurons are absent in the forebrain of the abnormal larvae shown as a decrease in mCherry+ neurons. Sections were counterstained in Hoechst 33342 to show nuclei in blue G - Linker-mediated PCR for viral insertion. Lanes 1–4 are abnormal larvae, lanes 5–8 are normal siblings showing one viral insertion appears only in the abnormal larvae. The smaller band in lane 6 is due to incomplete enzymatic digestion and shares no sequence similarity to those bands in lanes 1–4. H - Genomic PCR for the gucy2F viral insertion. Lanes 1–4 are abnormal larvae, lanes 5–7 are normal siblings. The viral insertion upstream of gucy2F is present only in the abnormal larvae. Lower panel is β-actin showing DNA is present in all samples. I – analysis of the overexpressed gucy2F transcript at 24hpf. RNA from mCherry + embryos (lanes 1&2) and mCherry − embryos (lane 3&4) was analyzed. Analysis of the 3′ end reveals a 3 kb band is present only mCherry + embryos. Analysis of the 5′ end indicates two splice variants in mCherry+ embryos that are not present in the mCherry − embryos. Scale Bars are 50 μM.
Misexpression of a Gal4-VP16 induced gene results in morphological defects
To determine whether virus-mediated overexpression can lead to a phenotype, we performed two pilot screens using Gal4-VP16 expression lines. The first screen used Tg(ath5:Gal4VP16) as a driver to overexpress tagged genes in progenitors of retinal ganglion cells and individuals were screened at 7-day postfertilization for impaired vision using the vision mediated background adaptation (VBA) response (Goldsmith and Harris 2003). This assay screens for a change in pigmentation when larvae are exposed to a bright light, a known vision mediated process. Because the germline of founders is mosaic, an individual insertion occurs in between 1 to 40% of a clutch with an average of 15% (unpublished observation). To take into account this mosaicism, 100 larvae from each clutch were screened and only clutches with >5% but <40% phenotypic embryos were regarded as carrying a putative phenotype-inducing mutation. A total of 1500 founder fish were screened in this manner. However, linkage analysis of the 21 families with a putative phenotype using both Southern blot and linker-mediated PCR did not identify any insertion with linkage to the phenotype. We concluded that the screen using Tg(ath5:Gal4VP16) as a driver line was not productive because of several factors. In our hands the VBA assay was not sufficient to detect what may be a subtle phenotype, since this assay identifies fish with gross defects in retinal function. We also determined that the Gal4-VP16 expression in the Tg(ath5:Gal4VP16) is too limited and occurs too late to impact retinal development as a whole. Assays to determine more subtle changes in the retina were beyond the scope of this study.
As an alternative, a second screen used a Gal4-VP16 gene/enhancer trap line. This gene/enhancer line was generated using Tol2 transgenesis to deliver a cassette consisting of a UAS-driven mCherry and the hsp70 basal promoter to drive Gal4-VP16 in conjunction with transcriptional elements near the insertion. This gene/enhancer trap line carries an insertion in the 14-3-3 epsilon gene and displays strong mCherry throughout the CNS (Figure 4A) beginning at 12 somites. We crossed these Gal4-VP16 expressing fish to the F1s carrying viral insertions from the 21 families that were maintained from the pilot retinal screen. After screening 10 of the 21 F1 families, one interesting phenotype was identified. In one family, when compared to a normal sibling (Figure 4E), the F2 larvae at 5 dpf displayed a shortened body, hemorrhage in the brain (Figure 4D and Figure 5A, B, D) and a loss of neurons in the forebrain (Figure 4F). This family, pa0028, carries multiple insertions, however only one insertion showed linkage with the phenotype (Figure 4G). This insertion is upstream of gucy2F, a Type 1 membrane-bound guanylate cyclase normally expressed exclusively in the retina (Yang et al. 1995; Yang and Garbers 1997; Baehr et al. 2007). Additional analysis of mutant embryos from multiple crosses supports linkage of this insertion and the phenotype (Figure 4H).
Figure 5. Morphological defects in larvae with overexpression of guanylate cyclases.

A–C- F2 progeny from a cross of pa0028 virus insertion fish to the 14-3-3epsilon Gal4-VP16 gene/enhancer trap line. The upper larva has Gal4-VP16 expression but does not carry the gucy2F viral insertion and is phenotypically normal. The lower larva has both Gal4-VP16 and the gucy2F virus insertion. Dorsal (A) view highlights the forebrain defect (arrowhead) and the shortened trunk, lateral view (B) highlights the hemorrhaging in the brain (arrow). Higher magnification of normal larvae (C) and mutant larvae with hemorrhaging in the brain (D, arrow). E, F-transient expression of gucy2F. Normal, Gal4-VP16 negative (top) and abnormal, Gal4-VP16 positive (bottom) larvae showing shortened trunk and hemorrhaging in the brain.
We examined the transcript generated from this virus insertion. We used a primer specific to the viral tag and a primer specific to gucy2F to ensure expression was a result of Gal4 induction. We also examined the 3′ end of the transcript, which gives an indication of gucy2F expression but is not exclusive to a Gal4 induced transcript. In 24hpf embryos that contain both the viral insertion and Gal4-VP16, we readily detected the UAS directed transcript of gucy2F (Figure 4I, lane1 and 2). No transcript was identified using RNA from 24hpf embryos that carried the viral insertion but no Gal4-VP16 (Figure 4I, lane 3,4) suggesting low levels of expression at this stage. This analysis also indicated two alternative splice sites within the first exon and both products should be able to produce a protein although the shorter transcript will lack 200 amino acids at the N-terminus of the protein.
We examined the pattern of expression for gucy2F in normal embryos and in those with expression from the upstream viral insertion. At 24 hour post fertilization, there is no expression of gucy2F in embryos without Gal4-VP16 (Figure 4B) consistent with the data obtained from the RT-PCR analysis. This is not surprising since gucy2F has been described as restricted to photoreceptors (Yang et al. 1995) which have yet to develop at this stage. In contrast, in embryos that carry the viral insertion and Gal4-VP16 expression from the gene-trap cassette, strong expression of gucy2F is detected in the same pattern as mCherry (Figure 4C). Since these embryos go on to develop abnormally, this provides a good correlation between overexpression of a gene downstream of a viral insertion and a phenotype and supports the gain-of-function approach in zebrafish.
To further tie overexpression of gucy2F and the development of this phenotype, embryos carrying the gene-trap cassette resulting in Gal4-VP16 expression in neurons were injected with a UAS driven gucy2F cDNA. At four days post-fertilization, we observed a phenotype similar to that of gucy2F expression from the viral insertion. Specifically, injected larvae with Gal4-VP16 expression have a shortened body and visible hemorrhaging in the brain (Figure 5E, F – lower larvae). These phenotypes were not observed in larvae that were injected with UAS driven gucy2F cDNA but lack Gal4-VP16 expression (Figure 5E, F – upper panels). This supports that the phenotype observed in the family of pa0028a F2 fish is a result of gucy2F overexpression and not a gene downstream from another viral insertion.
One potential mechanism leading to this abnormal phenotype is an overall increase in the levels of cGMP. To investigate if cGMP regulation may be altered in these larvae, we assayed cGMP levels in the mutant larvae arising from gene-activation from the upstream viral insertion as well as examining cGMP levels in the larvae injected with the expression constructs. At 5 dpf the pa0028 mutant larvae had a 9-fold increase levels in cGMP when compared to normal siblings (Figure 6). Similarly, the larvae injected with the gucy2F construct showed a 10-fold increase in cGMP at 24hpf, a time where there should be expression of gucy2F from the injected plasmid. The increase in cGMP levels suggests that aberrant production of cGMP due to gucy2F expression plays a central role in the development of the mutant phenotype.
Figure 6. Increased cGMP in embryos with gucy2F overexpression.
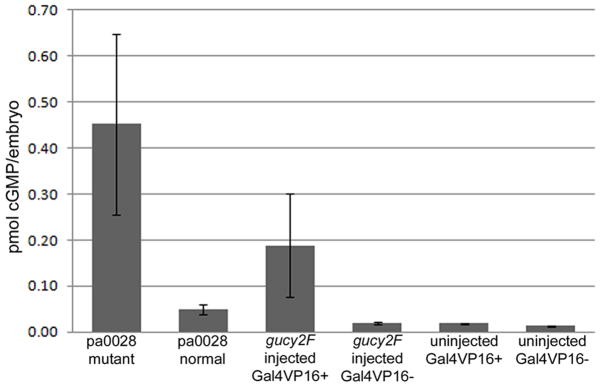
cGMP levels assayed at 5 dpf for pa0028 mutant larvae and normal siblings and 24 hpf embryos injected with the gucy2F expression construct. Increased cGMP levels are detected in those embryos that have overexpression of gucy2F. No difference in cGMP levels due to the presence of Gal4-VP16.
These studies indicate that this gain-of-function approach is a viable method in a vertebrate model in order to identify new genes involved in developmental processes. Using MMLV as a tool to deliver a Gal4 responsive cassette into the genome, thousands of genes can easily be targeted. In conjunction with a Gal4-VP16 expression in tissues of interest, genes can be overexpressed and the resultant phenotypes observed. This approach may uncover genes that have been previously overlooked in loss-of-function screens.
DISCUSSION
In Drosophila, gain-of-function screens using gene-activation have been useful to identify genes involved in a number of developmental processes (Hay et al. 1997; Staudt et al. 2005; Molnar et al. 2006). Here we sought to use an analogous approach in a vertebrate system to demonstrate the utility of gain-of-function in zebrafish and the MMLV retrovirus as an agent for cassette delivery into the genome. As has been highlighted by others (Wu et al. 2003; Wang et al. 2007) and further supported by this work, the MMLV mediated viral integrations that occur within genes are primarily at the 5′ end of the gene. For the UAS containing virus, this allows the simultaneous production of a loss-of-function insertion and the ability to overexpress the downstream gene when Gal4-VP16 is present. In a screen of F1 fish, the loss-of-function insertion should have minimal impact since only one allele is altered. However, in subsequent generations, the loss-of-function phenotype can be examined for genes of interest. When insertions occur in 5′ non-coding regions, the protein resulting from Gal4-VP16 activation should function normally. If the viral insertion occurs within an intron in the coding regions, this could yield a truncated protein with dominant negative or aberrant function. During the analysis of transcripts induced by Gal4-VP16, we discovered there may be cryptic splice acceptor sites throughout the zebrafish genome. Many of these artificial transcripts likely have no function although the possibility exists that some of them represent previously unidentified genes. It is also important to consider that due to the expression of Gal4-VP16, the gene downstream of the viral insertion may now be present in cells or tissues where it would not normally be expressed. This is the case for gucy2F expression in the CNS. Although it is simple to discount this as merely an artifact, a phenotype induced by misexpression may reveal important information regarding the function of a related gene or biological process.
In this study only a small number of insertions were examined in detail compared to the number generated. However, this study provides important information about the gain-of-function approach. The virus readily integrates into the zebrafish genome and at least one insertion is transmitted to each progeny. This suggests that a large number of insertions can be analyzed. On average, each founder carries insertions that can produce at least two unique Gal4-inducible transcripts (Fig 2C). Therefore the 6,000 founders we generated may contain at least 12,000 activating mutations, likely covering a significant portion of the zebrafish genome.
During the course of this study we discovered in order to impact a developmental process, it is necessary to have a robust assay and to have Gal4-VP16 induced expression at the appropriate time, place and at an adequate level. This is highlighted by our initial phenotype based screen using Tg(ath5:Gal4VP16) fish as the driver line. This screen was designed to identify gross changes in retinal development using the VBA assay and was unable to identify more subtle phenotypic changes. The Gal4-VP16 expression in the transgenic line begins just as cells in the retina begin to differentiate and is likely unable to impact the overall function of the retina. However, using the 14-3-3 epsilon gene/enhancer trap line revealed that phenotype based screens using this gain-of-function approach is a viable approach. Although we used a small sample size, we found a phenotype in one of ten founders screened, which are estimated to carry about 20 activating mutations. This phenotype is easily identifiable and linked with a specific virus insertion. While the range of gain-of-function screens in Drosophila (Staudt et al. 2005; Molnar et al. 2006), the true frequency would need to be determined with a larger collection of founders carrying viral insertions.
Overexpression of gucy2F in the CNS clearly leads to an abnormal phenotype. The significance of this observation can be debated since gucy2F is normally restricted to the retina, however it does yield useful information about cGMP in neurons. The increase in cGMP levels likely plays a central role in this phenotype, which suggests that precise control of cGMP in neurons is necessary. This process would be controlled by a guanylate cyclase other than GUCY2F. The transient expression of gucy1a3, a guanylate cyclase unrelated to gucy2F, only slightly increased cGMP levels and did not fully recapitulate the phenotype from gucy2F overexpression was (data not shown). The unremarkable increase of cGMP by gucy1a3 overexpression may be related to differences in cyclase activation since GUCY1A3 is dependent on nitric oxide for stabilization and activation (Zabel et al. 1998). Interestingly, at early stages embryos with gucy2F overexpression appear morphologically normal and proliferation and apoptosis appear to be unchanged (data not shown) suggesting that patterning and early development is not impacted by gucy2F overexpression. The abnormal phenotype begins at 4 dpf and severe defects evident by 7 dpf. This suggests that the phenotype may be due to neuronal degeneration rather than a developmental defect.
The observed brain hemorrhaging may be related to neurodegeneration phenotype. It may be a consequence of high concentration of extracellular cGMP released from dying neurons. High levels of cGMP can induce apoptosis of endothelial cells (Suenobu et al, 1999). However, it is not known how much extracellular cGMP in the brain can enter endothelial cells. Alternatively, the hemorrhaging may induce neurodegeneration, The hemorrhaging suggests defects in the blood-brain barrier, which has been suggested to be an element in neuronal degeneration with such diseases as Parkinson’s disease (Desai et al. 2007; Monahan et al. 2008). Nevertheless, the two phenotypes may be independent.
In this study, we demonstrate that the gene-activation approach can be applied in a forward genetic screen in zebrafish by using MMLV as a method to deliver a UAS driven cassette into the genome. This is a versatile approach since once fish with viral insertions are generated, they can be used in a variety of screens assaying effects in different tissues or cell types. However, for this to be a robust approach it requires appropriate Gal4-VP16 expression in tissues or cell types of interest, and appropriate assays to identify mutant phenotypes. By applying this gain-of-function approach, we identified that appropriate control of cGMP production is needed for neuronal survival. The availability of this gain-of-function approach in a vertebrate system should facilitate the identification of new genes or previously unknown roles of known genes in development and diseases.
Acknowledgments
We would like to thank Emily Janega for assistance with PCR and In situ hybridization. This work is supported by NIH EY016092 to (WC), NIH Training Grant 1T32HD049309-01A1 to Jan Christian (LAM) and P40 RR012546 (ZIRC).
Footnotes
LITERATURE CITED
- Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A. 2008;105:1255–1260. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehr W, Karan S, Maeda T, Luo DG, Li S, Bronson JD, Watt CB, Yau KW, Frederick JM, Palczewski K. The function of guanylate cyclase 1 and guanylate cyclase 2 in rod and cone photoreceptors. J Biol Chem. 2007;282:8837–8847. doi: 10.1074/jbc.M610369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Burgess S, Golling G, Amsterdam A, Hopkins N. High-throughput selection of retrovirus producer cell lines leads to markedly improved efficiency of germ line-transmissible insertions in zebra fish. J Virol. 2002;76:2192–2198. doi: 10.1128/jvi.76.5.2192-2198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Burgess S, Hopkins N. Analysis of the zebrafish smoothened mutant reveals conserved and divergent functions of hedgehog activity. Development. 2001;128:2385–2396. doi: 10.1242/dev.128.12.2385. [DOI] [PubMed] [Google Scholar]
- Chen W, Casey Corliss D. Three modules of zebrafish Mind bomb work cooperatively to promote Delta ubiquitination and endocytosis. Dev Biol. 2004;267:361–373. doi: 10.1016/j.ydbio.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai BS, Monahan AJ, Carvey PM, Hendey B. Blood-brain barrier pathology in Alzheimer’s and Parkinson’s disease: implications for drug therapy. Cell Transplant. 2007;16:285–299. doi: 10.3727/000000007783464731. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Ellingsen S, Laplante MA, Konig M, Kikuta H, Furmanek T, Hoivik EA, Becker TS. Large-scale enhancer detection in the zebrafish genome. Development. 2005;132:3799–3811. doi: 10.1242/dev.01951. [DOI] [PubMed] [Google Scholar]
- Goldsmith P, Harris WA. The zebrafish as a tool for understanding the biology of visual disorders. Semin Cell Dev Biol. 2003;14:11–18. doi: 10.1016/s1084-9521(02)00167-2. [DOI] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin SY, Nissen RM, Hopkins N. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- Grabher C, Joly JS, Wittbrodt J. Highly efficient zebrafish transgenesis mediated by the meganuclease I-SceI. Methods Cell Biol. 2004;77:381–401. doi: 10.1016/s0091-679x(04)77021-1. [DOI] [PubMed] [Google Scholar]
- Haffter P, Nusslein-Volhard C. Large scale genetics in a small vertebrate, the zebrafish. Int J Dev Biol. 1996;40:221–227. [PubMed] [Google Scholar]
- Hay BA, Maile R, Rubin GM. P element insertion-dependent gene activation in the Drosophila eye. Proc Natl Acad Sci U S A. 1997;94:5195–5200. doi: 10.1073/pnas.94.10.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kay JN, Link BA, Baier H. Staggered cell-intrinsic timing of ath5 expression underlies the wave of ganglion cell neurogenesis in the zebrafish retina. Development. 2005;132:2573–2585. doi: 10.1242/dev.01831. [DOI] [PubMed] [Google Scholar]
- Kotani T, Nagayoshi S, Urasaki A, Kawakami K. Transposon-mediated gene trapping in zebrafish. Methods. 2006;39:199–206. doi: 10.1016/j.ymeth.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Lin S, Gaiano N, Culp P, Burns JC, Friedmann T, Yee JK, Hopkins N. Integration and germ-line transmission of a pseudotyped retroviral vector in zebrafish. Science. 1994;265:666–669. doi: 10.1126/science.8036514. [DOI] [PubMed] [Google Scholar]
- Molnar C, Lopez-Varea A, Hernandez R, de Celis JF. A gain-of-function screen identifying genes required for vein formation in the Drosophila melanogaster wing. Genetics. 2006;174:1635–1659. doi: 10.1534/genetics.106.061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan AJ, Warren M, Carvey PM. Neuroinflammation and peripheral immune infiltration in Parkinson’s disease: an autoimmune hypothesis. Cell Transplant. 2008;17:363–372. [PubMed] [Google Scholar]
- Muto A, Orger MB, Wehman AM, Smear MC, Kay JN, Page-McCaw PS, Gahtan E, Xiao T, Nevin LM, Gosse NJ, Staub W, Finger-Baier K, Baier H. Forward genetic analysis of visual behavior in zebrafish. PLoS Genet. 2005;1:e66. doi: 10.1371/journal.pgen.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci U S A. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milan M, Benes V, Ansorge W, Cohen SM. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Staudt N, Molitor A, Somogyi K, Mata J, Curado S, Eulenberg K, Meise M, Siegmund T, Hader T, Hilfiker A, Bronner G, Ephrussi A, Rorth P, Cohen SM, Fellert S, Chung HR, Piepenburg O, Schafer U, Jackle H, Vorbruggen G. Gain-of-Function Screen for Genes That Affect Drosophila Muscle Pattern Formation. PLoS Genet. 2005;1:e55. doi: 10.1371/journal.pgen.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Jao LE, Zheng N, Dolan K, Ivey J, Zonies S, Wu X, Wu K, Yang H, Meng Q, Zhu Z, Zhang B, Lin S, Burgess SM. Efficient genome-wide mutagenesis of zebrafish genes by retroviral insertions. Proc Natl Acad Sci U S A. 2007;104:12428–12433. doi: 10.1073/pnas.0705502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Yang RB, Foster DC, Garbers DL, Fulle HJ. Two membrane forms of guanylyl cyclase found in the eye. Proc Natl Acad Sci U S A. 1995;92:602–606. doi: 10.1073/pnas.92.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RB, Garbers DL. Two eye guanylyl cyclases are expressed in the same photoreceptor cells and form homomers in preference to heteromers. J Biol Chem. 1997;272:13738–13742. doi: 10.1074/jbc.272.21.13738. [DOI] [PubMed] [Google Scholar]
- Zabel U, Weeger M, La M, Schmidt HH. Human soluble guanylate cyclase: functional expression and revised isoenzyme family. Biochem J. 1998;335 (Pt 1):51–57. doi: 10.1042/bj3350051. [DOI] [PMC free article] [PubMed] [Google Scholar]