A systematic screen of the entire human Rab GTPase family for interactions with myosin Va identified 10 novel Rab partners for myosin Va, all of which belong to the endocytic recycling and post-Golgi secretory membrane network. However, Rab10 and Rab11 appear to be the major determinants of its recruitment to intracellular membranes.
Abstract
Myosin Va is a widely expressed actin-based motor protein that binds members of the Rab GTPase family (3A, 8A, 10, 11A, 27A) and is implicated in many intracellular trafficking processes. To our knowledge, myosin Va has not been tested in a systematic screen for interactions with the entire Rab GTPase family. To that end, we report a yeast two-hybrid screen of all human Rabs for myosin Va-binding ability and reveal 10 novel interactions (3B, 3C, 3D, 6A, 6A′, 6B, 11B, 14, 25, 39B), which include interactions with three new Rab subfamilies (Rab6, Rab14, Rab39B). Of interest, myosin Va interacts with only a subset of the Rabs associated with the endocytic recycling and post-Golgi secretory systems. We demonstrate that myosin Va has three distinct Rab-binding domains on disparate regions of the motor (central stalk, an alternatively spliced exon, and the globular tail). Although the total pool of myosin Va is shared by several Rabs, Rab10 and Rab11 appear to be the major determinants of its recruitment to intracellular membranes. We also present evidence that myosin Va is necessary for maintaining a peripheral distribution of Rab11- and Rab14-positive endosomes.
INTRODUCTION
Class V myosins are versatile actin-based motor proteins that have been implicated in organelle transport and dynamic organelle tethering (Woolner and Bement, 2009; Hammer and Sellers, 2011). They are large homodimers consisting of an amino-terminal actin-binding motor domain, a lever arm, and a carboxy-terminal globular-tail domain (GTD) that mediates binding to cargo. Mammals have three myosin V isoforms: myosin Va, which is widely expressed but is enriched in brain, testes, and skin (Mercer et al., 1991); myosin Vb, which is ubiquitously expressed; and myosin Vc, which is epithelial specific (Vale, 2003).
An increase in calcium concentration triggers a structural rearrangement in myosin Va by which it switches from an inactive, “closed” conformation in which the GTD folds back and binds to the motor domain near its ATP-binding site to an unfolded and thus extended active conformation (Li et al., 2006; Liu et al., 2006; Thirumurugan et al., 2006; Trybus et al., 2007). Another layer of myosin Va regulation is maintained by members of the Rab family of GTPases. Rabs make up the largest family of small GTPases, with >60 members in humans (Kelly et al., 2012), which can be subdivided into ∼39 subfamilies based on sequence and functional similarity (Pereira-Leal and Seabra, 2001). They reversibly associate with membranes via hydrophobic geranylgeranyl groups attached to their carboxy termini and undergo a guanine nucleotide–dependent molecular switch (Kelly et al., 2012). When GTP bound, Rabs recruit a diverse range of downstream “effectors,” which include coat proteins, motor proteins, tethering factors, and soluble N-ethylmaleimide–sensitive factor attachment protein receptors to specific intracellular compartments (Kelly et al., 2012). In doing so, Rabs serve to organize the four main steps in membrane transport: vesicle budding, delivery, tethering, and fusion with target compartments (Hutagalung and Novick, 2011; Kelly et al., 2012).
Myosin Va has been well characterized in melanocytes and neuronal cells, where it plays a key role in melanosome and secretory vesicle transport, respectively (Hammer and Sellers, 2011). However, although myosin Va is known to directly bind Rab3A (Wollert et al., 2011), Rab8A/Rab10, and Rab11A (Roland et al., 2009) and indirectly bind Rab27A (Fukuda et al., 2002; Hume et al., 2002; Strom et al., 2002; Wu et al., 2002a, b; Desnos et al., 2003), relatively little is known about its function or mechanism of regulation in other cell types. Furthermore, no systematic evaluation of the ability of all human Rabs to bind myosin Va has been reported. We describe here such a study and reveal several new Rab partners for myosin Va. In addition, we define three distinct Rab-binding domains on myosin Va and examine the functional significance of novel Rab/myosin Va interactions.
RESULTS
Myosin Va interacts with multiple Rab GTPases of the endosomal and secretory pathways
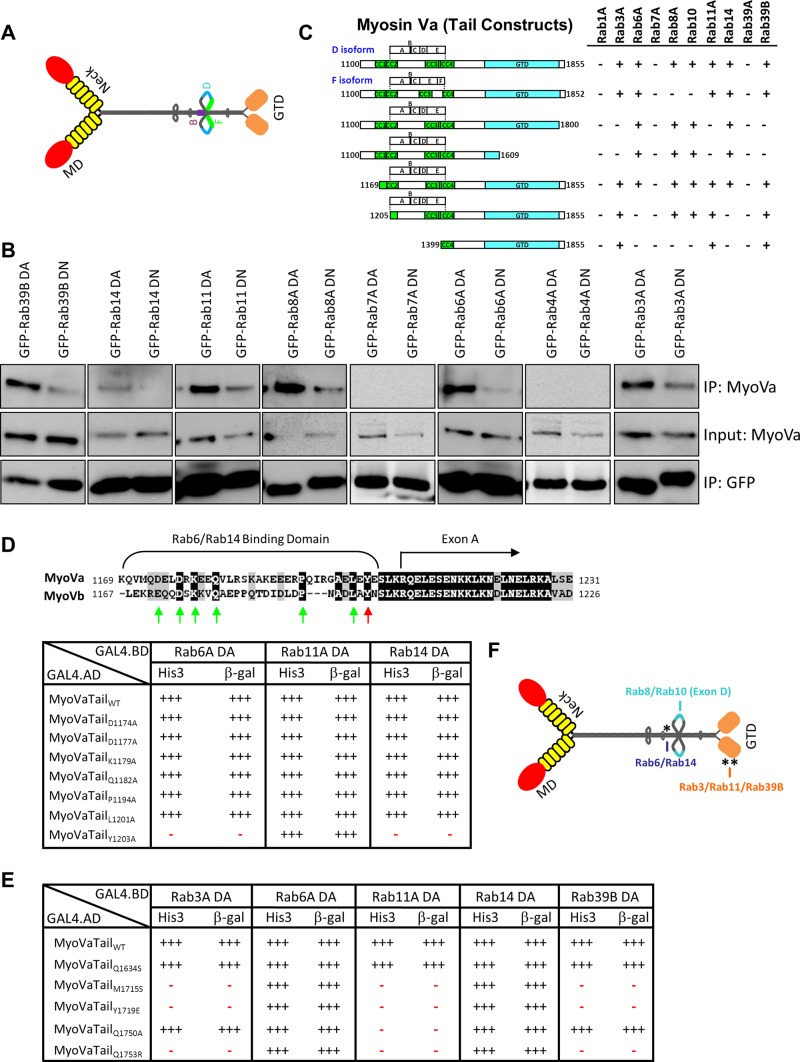
In a systematic approach to investigate the ability of the entire complement of human Rab GTPases to interact with myosin Va, we performed a yeast two-hybrid “living chip” assay (see Materials and Methods) in which a library of the Y187 yeast strain transformed with the wild-type (wt), constitutively active (DA; GTP bound), and constitutively inactive (DN; GDP bound) forms of each human Rab in the pLexA bait vector was mated with the L40 yeast strain transformed with a prey vector expressing the tail region of the F isoform of myosin Va (see later discussion). Colonies containing interacting bait and prey constructs were selected for by growth on synthetic medium lacking histidine (Supplemental Figure S1) and further assayed for β-galactosidase activity (Supplemental Table S1). We observed the previously reported interactions with active Rab3A and Rab11A (Roland et al., 2009; Wollert et al., 2011), which validated our screen (Supplemental Table S1). In addition, we also found interactions between myosin Va and the activated forms of Rabs 3B, 3C, 3D, 6A, 6A′, 6B, 9B, 11B, 14, 29, 30, and 39B, as well as with wild-type Rab25, which naturally contains a leucine residue at position 71 (Supplemental Table S1). The Rab29 and Rab30 interactions were dismissed as false positives because they scored positive with the majority of preys tested (Supplemental Table S1 and unpublished data). The remaining interactions were retested individually in the yeast two-hybrid system (Figure 1C), and only Rab9B was eliminated in this more stringent round of assays (Supplemental Figure S3). Cells expressing green fluorescent protein (GFP)–fused DA or DN mutants of representative members from each of its partner Rab subfamilies were lysed and the fusion proteins immunoprecipitated with an anti-GFP antibody. Coimmunoprecipitating myosin Va proteins were then revealed with an anti-myosin Va antibody. The data revealed that endogenous myosin Va forms a complex with each of the DA mutants of Rabs 39B, 14, 11A, 8A, 6A, and 3A, whereas only weak binding was observed with their DN mutants (Figure 1B). Of importance, no binding was observed with the DA or DN mutants of Rab7A or Rab4A, which were used as negative controls (Figure 1B). These results are consistent with our yeast two-hybrid studies, which demonstrated that myosin Va preferentially associates with a subset of GTP-bound Rabs.
FIGURE 1:
Myosin Va interacts with multiple Rab GTPases. (A) Schematic diagram depicting the extended “active” structure of myosin Va. The location of the alternatively spliced exons B, D, and F are indicated. GTD, globular-tail domain; MD, motor domain. (B) HeLa cells expressing dominant-active (DA) or dominant-negative (DN) mutants of the indicated Rab GTPases fused to GFP were lysed, and the fusion proteins were immunoprecipitated with an anti-GFP antibody. The ability of myosin Va to form a complex with these Rabs was tested by Western blot analysis using anti-myosin Va antibody. Input represents 10% of the starting material used in all conditions. (C) Regions of the myosin Va tail tested using the yeast two-hybrid technique. The constitutively active mutant of each Rab GTPase was used. CC, coiled coil; GTD, globular tail domain. (D) Identification of a critical amino acid for binding to Rab6 and Rab14. ClustalW alignment of the Rab6/Rab14-binding regions of human myosin Va and myosin Vb. Arrows indicate the conserved amino acids that were mutated to alanine, and the red arrow indicates tyrosine 1203, which, when mutated, abolishes the Rab6 and Rab14 interaction. The table indicates the strength of interaction of each mutant with constitutively active Rab6A, Rab11A, and Rab14 using the yeast two-hybrid HIS3 and LacZ reporter assays. (E) Identification of critical amino acids that mediate binding to Rab3A, Rab11A, and Rab39B. The amino acids in myosin Va that correspond to those identified by Lipatova et al. (2008) that mediate the binding of Myo2 to Ypt31/32 were mutated and the mutants tested for interaction with the constitutively active mutants of the indicated Rab GTPases by yeast two-hybrid assay. (F) Schematic diagram of myosin Va indicating the identified Rab-binding domains. Asterisk indicates the location of the Y1203A mutation; double asterisks indicate the location of the Q1753R mutation.
Myosin Va has several splice variants, which are expressed in a tissue-specific manner (Seperack et al., 1995; Huang et al., 1998; Lambert et al., 1998). The alternative splicing occurs in a coiled-coil region of the central stalk domain (Figure 1A). This region contains six exons (A–F), with exons B, D, and F subject to alternative splicing (Seperack et al., 1995). Exon D mediates the interaction with Rab8A and Rab10 (Roland et al., 2009), and exon F mediates an indirect interaction with Rab27A (Fukuda et al., 2002; Hume et al., 2002; Strom et al., 2002; Wu et al., 2002a, b). Throughout this study, we examined in parallel two myosin Va splice isoforms (D and F), both of which are expressed in the HeLa cells used in this work (Supplemental Figure S2A). The D isoform has the exon structure ABCDE, and the F isoform has the structure ABCEF (Figure 1C). Of note is the presence of the three-residue exon B in both splice isoforms. Exon B was reported to be a brain-specific exon (Seperack et al., 1995), although it has since been found to be present in myosin Va splice isoforms expressed in melanocytes and leukocytes (Lambert et al., 1998).
Myosin Va has three distinct Rab-binding domains
To identify the Rab-binding domain(s) on myosin Va, we tested truncation mutants of the tail regions of the D isoform in the yeast two-hybrid system. Deleting the GTD (MyoVa1100-1800 and MyoVa1100-1609) abolished the Rab11A interaction, but it also abolished the interactions with Rab3A and Rab39B (Figure 1C). A series of deletions of the D isoform tail (MyoVa1169-1855, MyoVa1205-1855, and MyoVa1399-1855) identified a short, 35–amino acid region (residues 1169–1204) that is necessary for the myosin Va interaction with Rab6A and Rab14 (Figure 1C).
Myosin Vb also interacts with Rab6 (unpublished data), and alignment of the Rab6-binding regions of myosin Va and myosin Vb revealed several conserved amino acids (Figure 1D). Seven of these residues were individually mutated to alanine, and the mutants were tested for their ability to interact with Rab3A, Rab6A, Rab11A, Rab14, and Rab39B. Mutation of the tyrosine at position 1203 in myosin Va to alanine (Y1203A) abolished the interactions with Rab6A and Rab14 (Figure 1D) but had no effect on the interactions with Rab11A (Figure 1D), Rab3A, or Rab39B (unpublished data).
It was reported that the GTD of myosin Va interacts with Rab11A (Roland et al., 2009), and in yeast, the class V myosin, Myo2, interacts with the Rab11 orthologues Ypt31/32 (Lipatova et al., 2008). A Ypt/Rab-binding pocket in the Myo2 GTD, as well as individual amino acids that mediate the Ypt31/32 interaction within this pocket, have been identified (Lipatova et al., 2008). Each of the corresponding residues in myosin Va were individually mutated, and we found that four of these mutants (M1715S, Y1719E, Q1750A, Q1753R) abolished binding to Rab11A, whereas the Q1634S mutation did not (Figure 1E). In addition, three of these myosin Va mutants (M1715S, Y1719E and Q1753R) failed to interact with Rab3A and Rab39B (Figure 1E). Of interest, although the Q1750A mutant abolished the Rab11A interaction, it did not affect the Rab3A or Rab39B binding (Figure 1E). None of these myosin Va GTD mutations affected the Rab6A or Rab14 binding (Figure 1E). These data suggest that Rab3A, Rab11A, and Rab39B bind to overlapping residues in a pocket in the GTD.
Taken together, these results demonstrate that myosin Va has three distinct Rab-binding domains: a Rab6/Rab14-binding domain in the central stalk just upstream of the alternatively spliced region, Rab8- and Rab10-binding sites on exon D, and Rab3-, Rab11-, and Rab39B-binding sites in a small pocket in the GTD (Figure 1F).
Myosin Va displays varying degrees of colocalization with its Rab partners
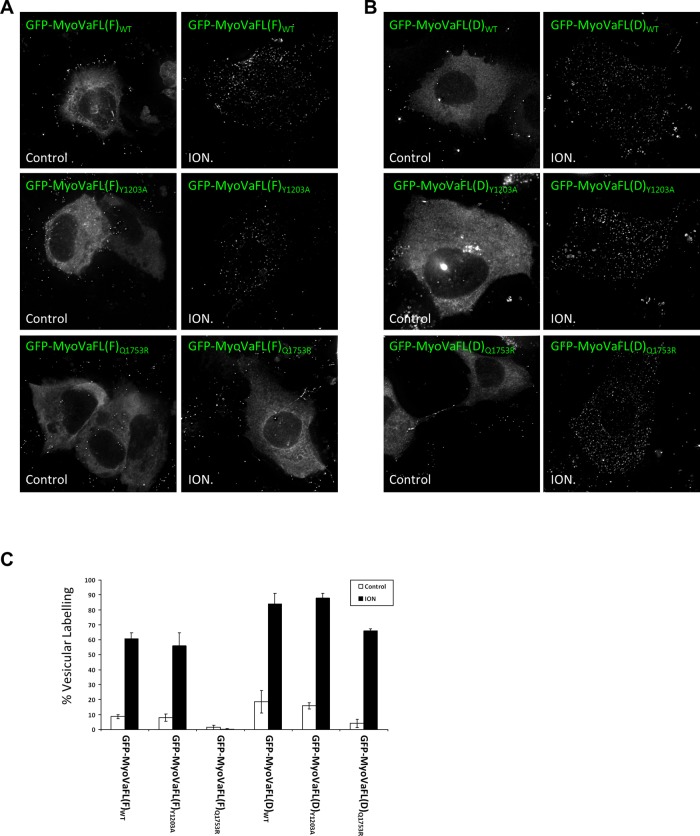
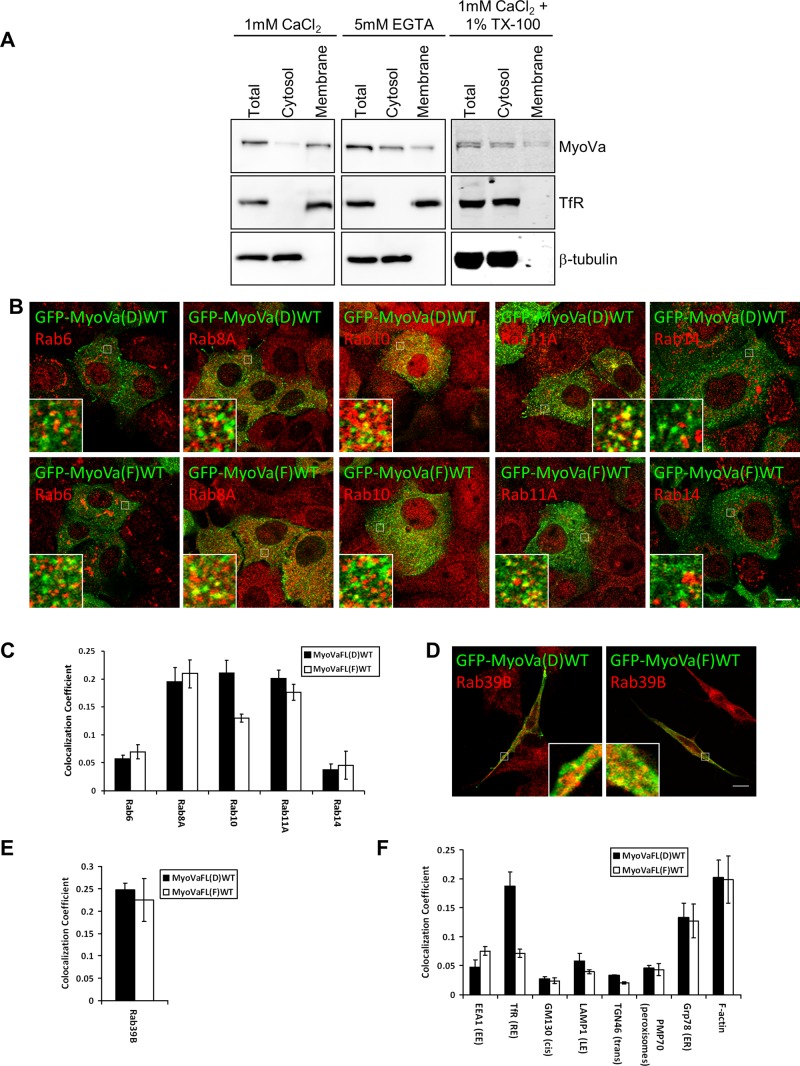
Many of the identified Rab partners of myosin Va have been the focus of considerable research efforts and reported to function on overlapping transport pathways, largely in the endocytic recycling and post-Golgi secretory systems (Chen et al., 1998; Darchen and Goud, 2000; Junutula et al., 2004; Larance et al., 2005; Miserey-Lenkei et al., 2007; Stenmark, 2009; Grigoriev et al., 2011; Hutagalung and Novick, 2011). Rab39B, which is less well characterized, is neuronal specific and has been reported to localize to the Golgi complex (Giannandrea et al., 2010). To evaluate the degree of overlap between myosin Va and its Rab partners, we established stable cell lines expressing the GFP-fused D and F isoforms of full-length myosin Va in A431 cells. Under basal conditions, both isoforms display a predominantly cytosolic distribution, likely due to the fusion protein adopting the closed, “inactive” conformation (see later discussion of Figure 5, A and B). Micromolar levels of calcium and/or cargo binding induce myosin Va to undergo a conformational switch to its open, active conformation (Liu et al., 2006; Thirumurugan et al., 2006). Indeed, subcellular fractionation experiments demonstrated that the majority of endogenous myosin Va partitioned in the membrane fraction when A431 cell lysates were prepared with CaCl2, whereas when calcium was chelated by ethylene glycol tetraacetic acid (EGTA), the major pool of myosin Va was found in the cytosolic fraction (Figure 2A). To rule out the possibility that calcium results in myosin Va becoming insoluble, thus accounting for its recovery in the high-speed pellet, we pretreated A431 lysates with 1% TX-100 to solubilize the membranes before centrifugation. Under these conditions the majority of myosin Va was found in the high-speed supernatant (Figure 2A). This indicates that calcium allows myosin Va to adopt a conformation that is capable of binding to membranes.
FIGURE 5:
Calcium is required for membrane binding of myosin Va. GFP-MyoVaFL(F)WT (A) or GFP-MyoVaFL(D)WT (B) and their Y1203A or Q1753R mutants were treated with 5 μM ionomycin, or solvent alone, for 4 min at 37°C. The cells were then fixed immediately and processed for fluorescence microscopy. (C) Percentage of cells displaying vesicular labeling of the GFP–myosin Va fusion proteins for each condition (mean ± SEM; n = 150 cells from three independent experiments).
FIGURE 2:
Characterization of the intracellular localization of myosin Va. (A) A431 (epidermoid carcinoma) cells were separated into cytosol and membrane fractions in the presence of CaCl2, EGTA, or CaCl2 and 1% TX-100. The presence of myosin Va in these fractions was determined by Western blot analyses using anti–myosin Va antibody. The distribution of TfR and β-tubulin was used to determine the accuracy of the fractionation procedure. (B) A431 cells stably expressing GFP-MyoVaFL(D)WT or GFP-MyoVaFL(F)(WT) were treated with 5 μM ionomycin for 4 min before fixation, permeabilization, and labeling with antibodies to the indicated endogenous Rab GTPases. Insets contain zoomed images of the boxed regions. (C, E, F) Quantitative analysis of the colocalization coefficients of each GFP–myosin Va splice variant with the indicated Rab GTPase or organelle marker (mean ± SEM; n = 15–20 cells). (D) SH-SY5Y cells expressing the indicated GFP–myosin Va splice variant treated with 5 μM ionomycin for 5 min before fixation and labeling with an antibody that detects endogenous Rab39B. Each immunofluorescence image is a single confocal section. Bar, 10 μm.
To assess the distribution of myosin Va with respect to that of membrane-associated endogenous Rab GTPases, we first treated cells with ionomycin for 4 min at 37°C, to increase the intracellular calcium concentration and allow GFP-myosin Va to associate with membranes, before processing for immunofluorescence microscopy. In cells stably expressing the GFP–myosin Va isoforms and treated with ionomycin, both isoforms were found to be present on membranous structures that were distributed throughout the cell (Figure 2B). This localization pattern resembles the distribution of endogenous myosin Va in both HeLa (Supplemental Figure S2C) and A431 cells (unpublished data). Both myosin Va isoforms displayed their highest degree of overlap with Rab8A and Rab11A, and there was little difference in colocalization coefficients between either isoform and these Rabs (Figure 2, B and C). This was somewhat surprising, given that Rab8A does not interact with the F isoform. Rab10, which, like Rab8A, does not bind the F isoform, also colocalized with both isoforms, but, as would be expected, it displayed considerably greater overlap with the D isoform (Figure 2, B and C). The degree of colocalization observed between Rab6 or Rab14 and either myosin Va isoform was much lower than that observed for the other Rabs tested (Figure 2, B and C). In the case of Rab6, colocalization was evident on Rab6-positive post-Golgi carriers rather than on the Rab6-positive membranes of the Golgi complex (Figure 2B). Similarly, the myosin Va D and F isoforms colocalized with Rab14-positive punctate structures (Figure 2B).
Because Rab39B is predominantly expressed in neuronal tissue, the SH-SY5Y human neuroblastoma cell line was used to examine myosin Va distribution with respect to that of Rab39B. Thus SH-SY5Y cells transiently expressing the GFP-fused D or F isoforms of myosin Va were treated with ionomycin for 5 min and then processed and immunolabeled for endogenous Rab39B. In these cells, Rab39B localized to the Golgi and to vesicles distributed throughout the cell body, as well as to the neurites that extended from the cell body (Figure 2D). Both myosin Va isoforms displayed considerable overlap with Rab39B-positive vesicles in the neurites (Figure 2, D and E). We also investigated the distribution of endogenous Rab3A with respect to that of endogenous myosin Va in SH-SY5Y cells and found that they also displayed considerable overlap (colocalization coefficient of 0.42 ± 0.031; Supplemental Figure S2, D and E). Endogenous Rab3A also displayed substantial colocalization with Rab39B in SH-SY5Y cells, indicating that these two Rabs may function along the same transport pathway in neurons (Supplemental Figure S2, D and E).
A pool of myosin Va localizes to the endocytic recycling system
The observation that the greatest overlap is with Rab8A, Rab10, and Rab11A suggests that a significant pool of myosin Va localizes to recycling endosomes. This was confirmed by measuring the colocalization coefficient between both myosin Va isoforms and markers of various intracellular compartments. We found that the D isoform displayed substantial overlap with the transferrin receptor (TfR; 0.19 ± 0.025), a marker of the endosomal recycling pathway, whereas the overlap observed between the F isoform and TfR was much lower (0.07 ± 0.007; Figure 2F and Supplemental Figure S4, A and B). In contrast, the F isoform displayed more, albeit modest, overlap with the early endosomal marker EEA1 than the D isoform (Figure 2F and Supplemental Figure S4, A and B). Limited overlap was observed with markers of late endosomes/lysosomes (LAMP1) and peroxisomes (PMP70; Figure 2F and Supplemental Figure S4, A and B). Neither was there strong overlap with markers of the cis-Golgi (GM130) and trans-Golgi (TGN46; Figure 2F and Supplemental Figure S4, A and B), but both myosin Va isoforms displayed considerable colocalization with the endoplasmic reticulum (ER) marker protein Grp78 (Figure 2F and Supplemental Figure S4, A and B). This is not entirely surprising, as the yeast class V myosin Myo4 is required for ER inheritance in budding yeast (Estrada et al., 2003), and myosin Va is required for transport of the ER into the spines of Purkinje neurons in mammals (Wagner et al., 2011). Of interest, Rab10 was recently reported to localize to the ER (English and Voeltz, 2013), which raises the possibility that it could link myosin Va to this organelle. In addition, both myosin Va isoforms were found to overlap considerably with phalloidin-labeled actin filaments (see later discussion; Figure 2F and Supplemental Figure S4, A and B).
Rab10 and Rab11 recruit myosin Va to membranes
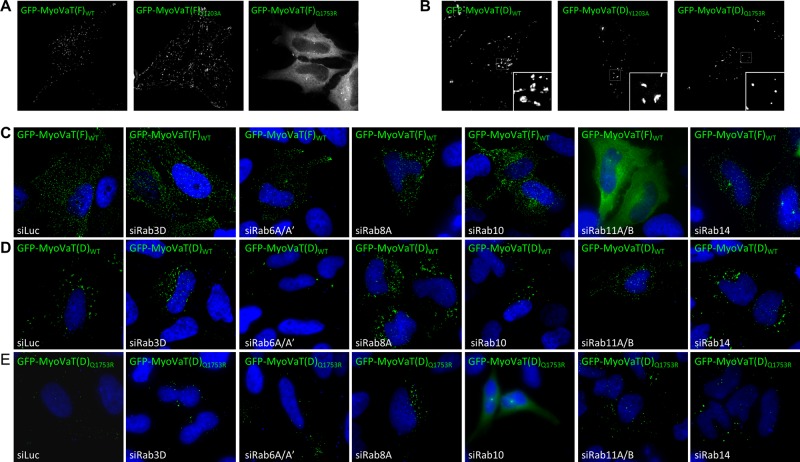
Although it is believed that Rab GTPases recruit class V myosins to membranes, to our knowledge this has not yet been proven in mammalian cells. To address this, we expressed the tail regions of the D isoform (residues 1100–1855; henceforth referred to as the D isoform tail or MyoVaT(D)) and the F isoform (residues 1100–1852; henceforth referred to as the F isoform tail or MyoVaT(F)) of myosin Va bearing the Y1203A (abolishes Rab6 and Rab14 binding) or Q1753R (abolishes Rab3, Rab11, and Rab39B binding) mutations as GFP fusions in HeLa cells. At low levels of expression the F isoform displayed a punctate pattern reminiscent of the endogenous protein (Figure 3A). The Y1203A mutation does not affect this localization pattern, but introduction of the Q1753R mutation renders the F isoform mostly cytosolic (Figure 3A). This suggests that one or all of the Rabs that bind at this site are involved in the recruitment of the F isoform to membranes. To confirm this and determine which Rabs are important for myosin Va recruitment, we expressed MyoVaT(F)WT in HeLa cells in which many of its interacting Rabs had been individually depleted by RNA interference (RNAi). MyoVaT(F)WT was found to be cytosolic in cells in which Rab11A and Rab11B had been depleted (Figure 3C), whereas depletion of the other Rabs, including Rab3D, which is expressed in our HeLa cells (Supplemental Figure S2F), had no such effect (Figure 3C). To rule out the possibility that the level of overexpression of GFP-MyoVaT(F)WT was so high as to obscure any punctate staining, we recorded confocal images of siRab11A/B–transfected cells expressing low, medium, or high levels of the fusion protein, and in each case the myosin Va fusion was found to be predominantly cytosolic (Supplemental Figure S5A). A second small interfering RNA (siRNA) targeting each Rab was used in an independent series of experiments, and the same results were observed (Supplemental Figure S5, B, C, and E). To further verify these results, we cotransfected DN mutants of each GFP-Rab with mCherry-MyoVaT(F)WT (Supplemental Figure S6A). In agreement with the RNAi data, only Rab11A DN resulted in the redistribution of myosin Va into the cytosol (Supplemental Figure S6A).
FIGURE 3:
Myosin Va is recruited to membranes by Rab10 and Rab11. HeLa cells expressing GFP-MyoVaTail(F)WT (A) or GFP-MyoVaTail(D)WT (B) or their mutants were fixed and imaged by fluorescence deconvolution microscopy. HeLa cells were transfected for 72 h with control siRNA or siRNA targeting the indicated Rab GTPase, and GFP-MyoVaTail(F)WT (C), GFP-MyoVaTail(D)WT (D), or GFP-MyoVaTail(D)Q1753R (E) was transfected into these cells for the final 18 h. The cells were fixed and the nuclei labeled with DAPI and imaged by fluorescence deconvolution microscopy. (See Figure S5D for Western blots analyzing the knockdown efficiency of the siRNA duplexes.)
The D isoform tail was found to localize to several large cytoplasmic aggregates (Figure 3B), and electron microscopy revealed that these aggregates are composed of multiple small, membrane-bound, TfR-positive vesicles (Supplemental Figure S4C). It remains membrane bound when the Q1753R mutation is introduced, although the mutant localizes to smaller and more uniformly round structures than the wild type (Figures 3B and Supplemental Figure S4C). This raises the possibility that the Q1753R mutation may have disengaged Rab11-positive recycling endosomes from aggregation and suggests that Rab8 and/or Rab10, in addition to Rab11, can facilitate the binding of the D isoform to membranes. To investigate whether this is the case, HeLa cells expressing MyoVaT(D)WT and MyoVaT(D)Q1753R were depleted of each of the interacting Rabs (Figure 3, D and E). MyoVaT(D)WT displayed membrane localization under all conditions, even after depletion of Rab11A and Rab11B or Rab8A (Figure 3D); however, MyoVaT(D)Q1753R was mostly cytosolic in cells in which Rab10 had been depleted (Figure 3E). This suggests that a combination of Rab10 and Rab11, but not Rab8, is required to recruit myosin Va to membranes and thus to its cargo.
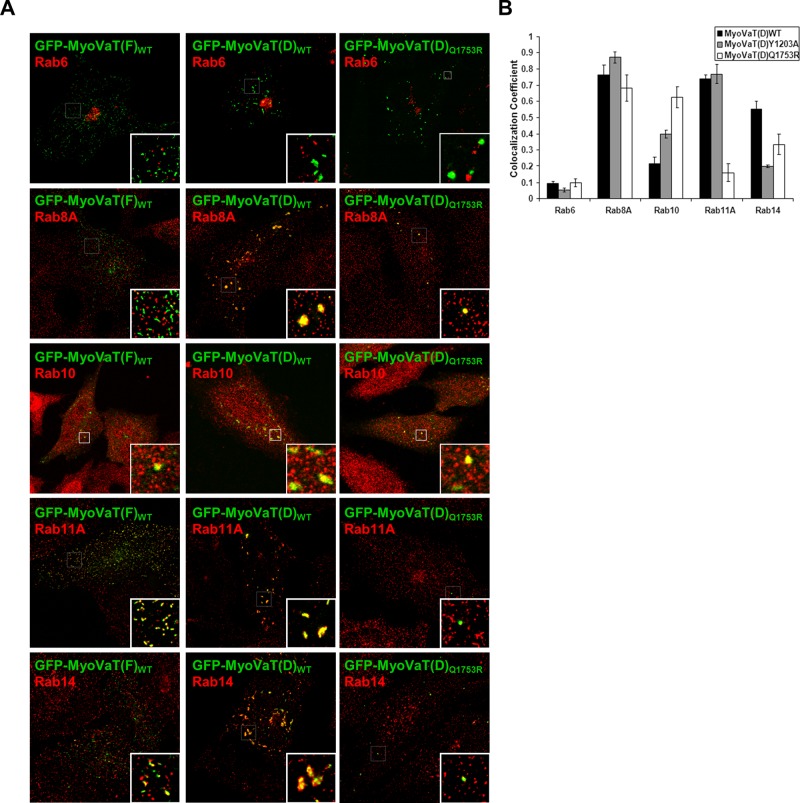
Immunofluorescence microscopy revealed that the large D isoform tail aggregates are positive for Rab8A, Rab10, Rab11A, and Rab14, indicating that they are accumulations of Rab-positive endosomal membranes (Figure 4A). The smaller MyoVaT(D)Q1753R-induced structures also contain Rab8A, Rab10, and Rab14 but lack Rab11A (Figure 4, A and B), confirming that Rab11-positive vesicles disengaged due to their inability to associate with the mutant protein.
FIGURE 4:
Expression of the myosin Va tail induces the aggregation of Rab-positive vesicles. (A) HeLa cells expressing the indicated myosin Va tail fused to GFP were fixed and labeled with antibodies that detect endogenous Rab GTPases. Insets show zoomed images of the boxed areas. (B) Quantitative analysis of the colocalization coefficients of each of the GFP–myosin Va tail constructs in HeLa cells with the indicated Rab GTPase (mean ± SEM; n = 20–40 cells).
To further investigate the hypothesis that Rab10 and Rab11 can recruit myosin Va to membranes, we performed experiments involving the full-length motor protein. Exogenous full-length myosin Va is predominantly cytosolic, with a fraction localizing to the tips of filopodia (Figure 5, A and B). Treatment of A431 cells with ionomycin results in the rapid redistribution of GFP-MyoVaFL from the cytosol to vesicles (Figure 5). The Y1203A mutation did not affect the vesicular association of either isoform (Figure 5). However, abolishing the Rab11 interaction by introducing the Q1753R mutation into the F isoform completely inhibited its membrane association upon ionomycin treatment (Figure 5, A and C), whereas this mutation had only a slight effect on the vesicular localization of the D isoform (Figure 5, B and C). These results indicate that Rab11 is crucial for the recruitment of the F isoform of myosin Va to membranes, whereas both Rab10 and Rab11 can fulfill this role for the D isoform.
Myosin Va plays a role in maintaining organelle distribution
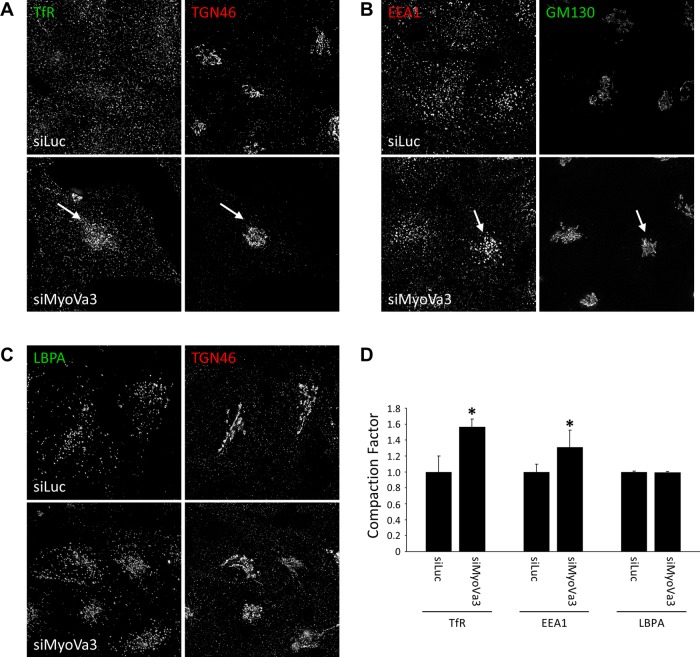
We next investigated the effects of myosin Va knockdown on organelle distribution. RNAi-mediated knockdown of myosin Va resulted in the accumulation of TfR-positive membranes in the perinuclear region of the cell (Figure 6A). Quantification revealed an almost 60% increase in TfR fluorescence near the Golgi in comparison with the control cells (Figure 6D). Myosin Va depletion also caused a similar accumulation of EEA1-labeled early endosomes in the perinuclear region (Figure 6, B and D), whereas the distribution of late endosomes/lysosomes was unaffected (Figure 6, C and D). This juxtanuclear vesicle accumulation is reminiscent of the post-Golgi vesicle clustering observed in yeast cells when Myo2 binding to Sro7 is disrupted (Rossi and Brennwald, 2011).
FIGURE 6:
Myosin Va depletion induces perinuclear clustering of endocytic recycling pathway membrane compartments. HeLa cells transfected with control siRNA or myosin Va siRNA for 72 h were fixed, permeabilized, and colabeled with antibodies to the TfR and TGN46 (A), EEA1 and GM130 (B), or LBPA and TGN46 (C). (D) Quantification of the fluorescence intensity of TfR, EEA1, or LBPA in the pericentrosomal region relative to the total cellular fluorescence intensity and expressed as a “compaction factor.” Data were normalized to the control (mean ± SEM; n = 30–50 cells). *p <10−4, Student's t test.
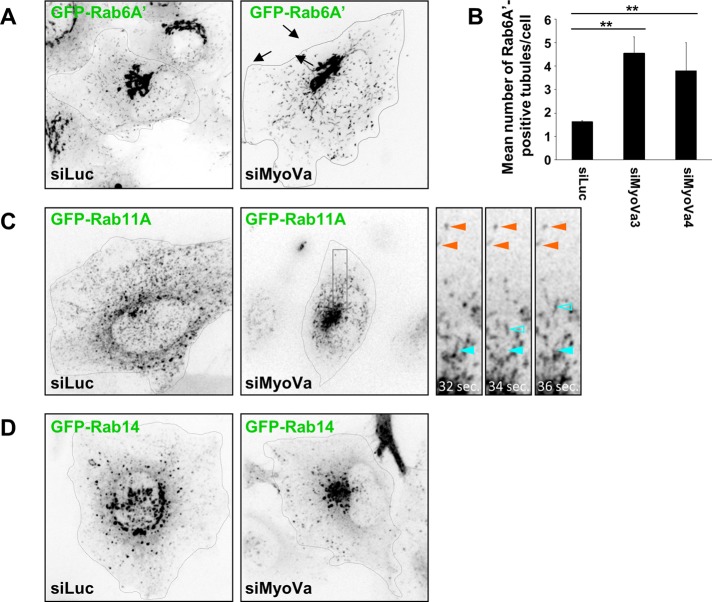
To investigate the functional relationship(s) between myosin Va and its partner Rabs, we performed live-cell spinning-disk confocal microscopy in control or myosin Va-depleted HeLa cells expressing GFP fusions of each Rab. Myosin Va knockdown had no observable effect on the distribution/motility of GFP-Rab3D, GFP-Rab8A, or GFP-Rab10 (unpublished data); however, effects were observed in the distribution of Rab6-, Rab11A-, and Rab14-positive structures (Figure 7 and Supplemental Movies S1–S3).
FIGURE 7:
Myosin Va depletion affects Rab motility and organelle distribution. (A) A HeLa cell line stably expressing GFP-Rab6A′ was transfected with control or myosin Va siRNA for 72 h. Two-minute time-lapse movies were recorded on a spinning disk confocal microscope. Representative frames from Supplemental Movie S1. Black arrows indicate tubules emerging from the Golgi complex. (B) Quantification of the number of Rab6A′-positive tubules connected to the Golgi for each condition (mean ± SEM; n = 11–15 cells). Two independent siRNAs targeting myosin Va were used. **p < 10−3. (C) HeLa cells were transfected with control or myosin Va siRNA for 72 h and plasmid DNA encoding GFP-Rab11A for the final 18 h. Two-minute time-lapse movies were recorded on a spinning disk confocal microscope. Representative frames from Supplemental Movie S2. Insets depict zoomed images of the boxed region. Indicated are examples of immobile vesicles (orange arrowheads) and rapidly moving vesicles (blue arrowheads). Closed arrowheads indicate the position of the vesicle in the first frame (recorded at 32 s), and the open arrowheads indicate the position of the same vesicle in the given frame. (D) HeLa cells transfected with control or myosin Va siRNA for 72 h and plasmid DNA encoding GFP-Rab14 for the final 18 h. Two-minute time-lapse movies were recorded on a spinning disk confocal microscope. Representative frames from Supplemental Movie S3.
Myosin Va knockdown had no obvious effect on Golgi distribution or morphology, but in myosin Va–depleted cells, short, Rab6A′-positive tubules emanating from the Golgi complex could be readily observed (Figure 7A and Supplemental Movie S1). Quantification revealed a greater than twofold increase in the number of tubules extending from the Golgi in comparison to control siRNA–transfected cells (Figure 7B). In addition, the Rab6-positive transport carriers that had detached from the Golgi were slightly longer and traveled in a more chaotic and less-directed manner. These tubules were different from the Rab6-positive tubules induced upon the inhibition of myosin II function (Miserey-Lenkei et al., 2010), in that they tended to be shorter and could occasionally be seen to undergo fission from the donor membrane (Supplemental Figure S6B).
In control cells GFP-Rab11A displayed a broadly dispersed distribution with some concentration in the perinuclear area, whereas when myosin Va was knocked down Rab11A-positive vesicles were heavily clustered in the perinuclear region of the cell (Figure 7C and Supplemental Movie S2). Analysis of the time-lapse movies revealed that in both control and myosin Va–knockdown cells GFP-Rab11A localizes to two types of transport carriers: those that are continuously moving, and those that remain static for short periods of time before bursts of rapid movement (Figure 7C, inset, and Supplemental Movie S2). This effect on Rab11 localization is reminiscent of the phenotype observed in melanocytes derived from dilute (myosin Va-null) mice (Wu et al., 1998). In those cells, the melanosomes fail to be captured at the actin-rich periphery, leading to their accumulation in the perinuclear region of the cell (Wu et al., 1998).
Knockdown of myosin Va was also found to result in the clustering of GFP-Rab14–labeled endosomes in the perinuclear region of live cells (Figure 7D and Supplemental Movie S3). In control cells, Rab14-positive endosomes underwent frequent short-range movements, whereas in the myosin Va–depleted cells these endosomes were clustered in the perinuclear region and their motility was reduced. However, the smaller, Rab14-positive structures that undergo rapid and long-range movement appear to be unaffected by myosin Va knockdown (Figure 7D and Supplemental Movie S3).
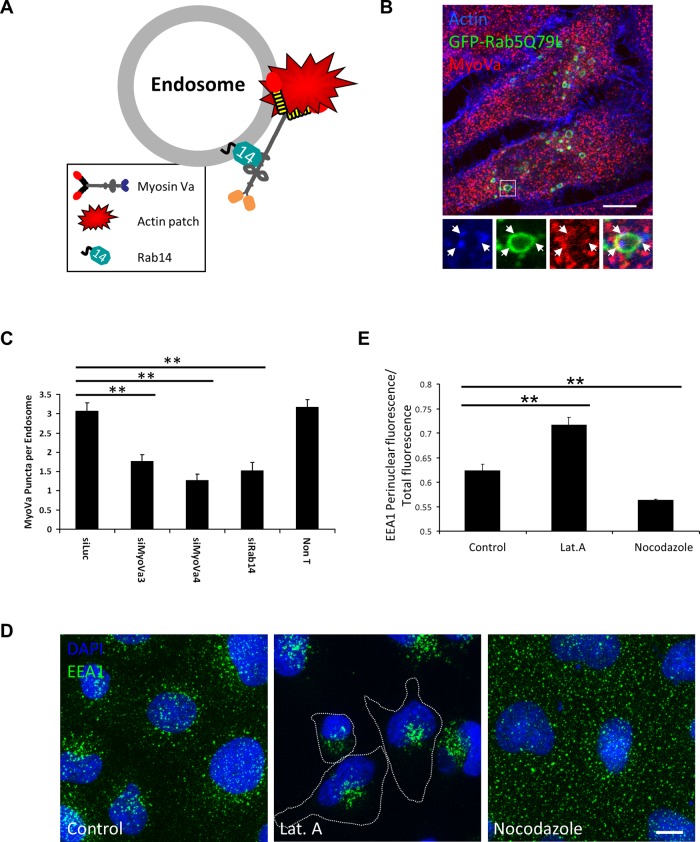
The foregoing findings led us to speculate that myosin Va may tether Rab14-positive endosomes to peripheral actin patches in order to maintain their correct intracellular distribution. To investigate this hypothesis further, we examined the localization of endogenous myosin Va to endosomal structures in cells in which Rab14 had been depleted (Figure 8, B and C). Because the size of these endosomal organelles is at the resolution limit of optical microscopy, we induced their enlargement by expressing a DA mutant of Rab5 (GFP-Rab5AQ79L), which promotes endosomal fusion (Stenmark et al., 1994). Cells expressing GFP-Rab5AQ79L were labeled with an antibody that specifically recognizes endogenous myosin Va (Supplemental Figure S2, B and C) and with coumarin-labeled phalloidin. These enlarged endosomal structures possessed an average of three myosin Va puncta, which were present on the endosomal membrane, where they frequently overlapped with actin patches (Figure 8, B and C). This is reminiscent of the findings of Morel et al. (2009), which demonstrate the presence of actin patches on endosomal membranes that are important for endosome biogenesis and remodeling. These puncta are indeed myosin Va rather than nonspecific antibody labeling, as depletion of myosin Va with two independent siRNAs resulted in ∼50% reduction in the number of puncta per endosome (Figure 8C). In addition, the puncta remaining after siRNA treatment had much lower fluorescence intensity. Depletion of Rab14 also significantly reduced the average number of myosin Va puncta per endosome (Figure 8C), raising the possibility that Rab14 is involved in the targeting of myosin Va to these enlarged endosomes.
FIGURE 8:
Endosomes are tethered to the actin cytoskeleton by Rab14 and myosin Va. (A) Model depicting the tethering of endosomes to the actin cytoskeleton by myosin Va, which is associated with the endosomal membrane via a direct interaction with Rab14. (B) HeLa cells expressing GFP-Rab5Q79L for ∼24 h were fixed and labeled with coumarin–phalloidin and an antibody to myosin Va. Inset contains a zoomed image of the indicated enlarged endosome. (C) Quantification of the number of endogenous myosin Va puncta on GFP-Rab5AQ79L enlarged endosomes in cells transfected with the indicated siRNA for 72 h (mean ± SEM; n ≥ 60 endosomes per condition). **p <10−3. (D) A431 cells treated with solvent alone, 0.5 μM latrunculin A, or 20 μM nocodazole for 1 h at 37°C before fixation and labeling with anti-EEA1 (green) and DAPI (blue). (E) The fluorescence intensity in the perinuclear region of the cell was expressed as a ratio of the total cellular fluorescence using the Radial Profile plug-in in ImageJ (mean ± SEM; n ≥ 100 cells per condition). **p <10−3.
These data are consistent with a model in which Rab14 and myosin Va cooperate to tether endosomes to peripheral actin patches (Figure 8A). If this is the case, then disruption of the actin cytoskeleton should also result in the perinuclear clustering of these endosomal compartments. Indeed, treatment with the actin polymerization inhibitor latrunculin A results in perinuclear clustering of EEA1-positive endosomes, whereas disruption of microtubules caused a small but significant dispersion of EEA1-positive endosomes toward the cell periphery (Figure 8, D and E).
The hypothesis that myosin Va may function as a dynamic tether to anchor membranes onto the actin cytoskeleton is further supported by the observation that full-length myosin Va is relatively immobile once it is recruited to membranes. Time-lapse images of GFP-MyoVaFL(D)WT–expressing cells revealed that the fusion protein was rapidly recruited to membrane structures after addition of 5 μM ionomycin to the culture medium (recruitment could be observed as soon as 40 s post-addition; Supplemental Movie S4). However, these structures were relatively static (GFP-MyoVaFL(D)WT-positive membranes had an average velocity of 0.036 ± 0.013 μm/s; GFP-MyoVaFL(F)WT-positive membranes had an average velocity of 0.030 ± 0.011 μm/s) and did not undergo the directional movements characteristic of a point-to-point transporter. To rule out the possibility that the ionomycin has an inhibitory effect on motility, we investigated the distribution and movement of a myosin Va mutant (D136A) that is constitutively active owing to its inability to adopt the folded inactive conformation (Li et al., 2008). We found that GFP-MyoVaFL(D)D136A (Supplemental Movie S5) and GFP-MyoVaFL(F)D136A (unpublished data) localized to vesicles in the absence of ionomycin. Time-lapse imaging revealed that both of these mutant proteins displayed movement patterns akin to those of the corresponding wild-type isoforms with mean velocities of 0.038 ± 0.016 and 0.037 ± 0.019 μm/s, respectively.
Taken together, our data suggest that myosin Va functions to tether distinct membrane organelles to the actin cytoskeleton via interactions with its partner Rab GTPases.
DISCUSSION
Here we report the first systematic screen of the human Rab GTPase family for interactions with myosin Va. In addition to the previously identified direct interactions (Rab3A, see Wollert et al., 2011; Rab8A, Rab10, and Rab11A, see Roland et al., 2009) and the indirect interaction with Rab27A (Hume et al., 2002; Nagashima et al., 2002; Strom et al., 2002; Wu et al., 2002b), we found that myosin Va also directly interacts with Rabs 3B, 3C, 3D, 6A, 6A′, 6B, 11B, 14, 25, and 39B. These data identified three additional Rab subfamilies (Rab6, Rab14, Rab39B) as myosin Va–binding proteins and demonstrate that myosin Va interacts with all members of each of these subfamilies, with the exception of the Rab39 subfamily. Remarkably, myosin Va interacts only with a subset of the Rabs associated with the endocytic recycling and post-Golgi secretory systems. For example, myosin Va interacts with Rab14 but not with other endosomal Rabs such as Rab4, Rab5, or Rab22A or with Rab22B/Rab31, which, like Rab14, is present on both the TGN and endosomes (Ng et al., 2007; Kelly et al., 2009). Neither does myosin Va interact with Rab35, which operates along the same transport pathway as Rab8 (Rahajeng et al., 2012).
Despite the fact that the total pool of myosin Va is shared by many Rabs and different membrane compartments, our data indicate that it is Rab11 (and Rab10 for the D isoform) that is chiefly responsible for the recruitment of myosin Va to membranes. Without ruling out the possibility that other proteins and/or lipids play a role in facilitating the membrane targeting of myosin Va or stabilizing its membrane association, this suggests that membrane-bound myosin Va could “exchange/convert” Rab11 (or Rab10) for another Rab, similar to the Rab conversion events that take place in the transition between early and late endosomes (Rink et al., 2005; Poteryaev et al., 2010). We found, in agreement with previous data (Kelly et al., 2009; Linford et al., 2012), a high degree of overlap between Rab11- and Rab14-positive membranes, suggesting that they are in close dynamic and functional continuity. In addition, myosin Va knockdown has a strong effect on the positioning of Rab11- and Rab14-positive membranes whereby they display striking perinuclear clustering in its absence. This suggests that myosin Va functions as a dynamic tether linking endosomal membranes to the actin cytoskeleton and thus maintaining their topological organization. This also suggests that membrane tethering to the actin cytoskeleton via myosin Va could be important for the formation of Rab domains on endosomal membranes, that is, myosin Va could participate in Rab-driven membrane remodeling and maturation. It remains to be determined whether myosin Va binds sequentially to Rab11 and Rab14 or can bind both proteins at the same time.
A myosin Va–driven Rab exchange mechanism is also conceivable for Rab11- and Rab8-positive membranes, which define subclasses of recycling endosomes (Roland et al., 2007). A variant of the “exchange” mechanism could be applied in the case of Rab3D, which is located on peripheral vesicles close to the plasma membrane and is likely to be involved in regulating the late steps of exocytosis, as is the case for other members of the Rab3 family. Rab11 membranes could be transported to the cell periphery by a kinesin-driven mechanism. The involvement of myosin Va in this long-range transport step seems to be excluded, as GFP-Rab11A membranes remain highly dynamic in its absence. Instead, it is likely that myosin Va would function to tether these vesicles to peripheral actin, and at this point, Rab11 may be exchanged for Rab3D, facilitating the formation of Rab3 vesicles. A role for Rab3 proteins, including Rab3D, in membrane remodeling and vesicle biogenesis was previously documented (Kogel and Gerdes, 2010; Kogel et al., 2010).
In the case of Rab6, it is noteworthy that myosin Va was not detected on Golgi/TGN membranes where the bulk of Rab6 is present. This suggests that Rab6 may only bind to myosin Va once it has interacted with Rab8 and/or Rab10. On the other hand, myosin Va depletion affects the morphology of Rab6-positive vesicles leading to the appearance of tubular structures. A tempting hypothesis is that upon recruitment to Rab8/Rab10-positive membranes, myosin Va serves as a docking platform for Rab6 vesicles exiting the Golgi complex. The tubules observed upon depletion of myosin Va could be the result of a defect in the docking/fusion of Rab6-positive vesicles with Rab8/Rab10-positive membranes due to loss of this docking platform. In this hypothesis, myosin Va could thus be an important player in the Rab6/Rab8 cascade known to operate in post-Golgi anterograde transport (Grigoriev et al., 2011). Indeed, Myo2 functions in a cascade with Ypt31/32 and Sec4 (yeast Rab8) in budding yeast (Jin et al., 2011; Santiago-Tirado et al., 2011).
In conclusion, our findings suggest that Rab GTPases are major regulators of myosin Va function in eukaryotes and suggest that myosin Va participates in Rab cascades involved in both remodeling and maturation of endocytic/recycling compartments and post-Golgi secretory pathways to coordinate cargo transport. The major challenge that remains is to fully delineate the cellular consequences of the no-less-than fifteen myosin Va/Rab GTPase interactions that are now known to exist.
MATERIALS AND METHODS
Antibodies and reagents
The following antibodies were used: rabbit anti–myosin Va (Sigma-Aldrich, St. Louis, MO); mouse anti-TfR (Sigma-Aldrich); mouse anti–β-tubulin (Sigma-Aldrich); mouse anti-Rab10 (clone 4E2; for immunofluorescence; Abcam, Cambridge, MA); rabbit anti-Rab10 (for Western blot; Sigma-Aldrich); sheep anti-TGN46 (AbD Serotec, Raleigh, NC); mouse anti-Rab3 (BD Biosciences, Franklin Lake, NJ); mouse anti-LAMP1 (BD Biosciences); mouse anti-GM130 (BD Biosciences); mouse anti–lysobisphosphatidic acid (LBPA; J. Gruenberg, University of Geneva, Geneva, Switzerland); rabbit anti-Rab3D (Sigma-Aldrich); rabbit anti-Rab8 (Sigma-Aldrich); rabbit anti-Rab14 (Sigma-Aldrich); rabbit anti-Rab6 (C-19; Santa Cruz Biotechnology, Santa Cruz, CA); human anti-Rab6 (AA2; Nizak et al., 2003); mouse anti-myc (Antibody Platform, Institut Curie, Paris, France); and rabbit anti-Rab39B (Proteintech, Chicago, IL). The anti-Rab11 antibody was raised in rabbits against GST-Rab11. See Supplemental Figure S7 for characterization of Rab antibody specificity.
Yeast two-hybrid assays
For large-scale screening of the wild-type (and DA and DN mutants) of the human Rab GTPase family for interactions with candidate Rab effector proteins, we developed a yeast two-hybrid “living chip” assay (Miserey-Lenkei, Lodeho, Jollivet, Mayeux, and Goud, unpublished data). Briefly, the yeast strain Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, gal4∆, gal80∆, met-, URA3::GAL1UAS-GAL1TATA-lacZ MEL1) was transformed with pLEXA-Rab constructs. Details of the primers used to generate the Rab (Q/L; DA) mutants are outlined in Supplemental Table S2. Transformants were selected on synthetic medium lacking tryptophan. Yeast stocks in 10% glycerol were distributed on 96-well plates and kept at −80°C. Before experiments, plates were replicated on yeast extract/peptone/dextrose–rich medium and then on selective medium at 30°C. The pGAD–myosin Va constructs were expressed in the yeast strain L40ΔGal4 (MATa, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, gal4∆, gal80∆, URA3::opLEXA-LacZ, LYS2:opLEXA-HIS3). Transformants were selected on synthetic medium lacking leucine. After 18 h of incubation on rich medium, Y187 and L40 strains mate and form diploids. Diploid cells containing the pLEX and pGAD plasmids were selected on synthetic medium lacking leucine and tryptophan and then plated for 3 or 6 d on synthetic medium lacking histidine. Histidine+ colonies were patched on selective plates and assayed for β-galactosidase activity.
Each positive interaction was retested individually by cotransforming pLEXA-Rab constructs and pGAD–myosin Va constructs into the L40 yeast strain (Janoueix-Lerosey et al., 1995).
Cell culture and transfection
A431, HeLa, and SH-SY5Y cells were cultured in DMEM (Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin/streptomycin and 2 mM glutamine. These cells were transfected with plasmid using XtremeGene 9 (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions. For silencing experiments HeLa cells were transfected with siRNA using HiPerFect (Qiagen, Valencia, CA), and SH-SY5Y cells were transfected with Lipofectamine RNAiMax (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. A431 cells stably expressing GFP-myosin Va FL fusion proteins were selected with 400 μg/ml G418 (Sigma-Aldrich) for 2 wks and colonies isolated with cloning rings.
Immunofluorescence microscopy
Cells were seeded on 10-mm glass coverslips and fixed with 4% paraformaldehyde and blocked/permeabilized with 0.05% saponin/0.2% bovine serum albumin. Cells were incubated with the indicated primary antibodies. The secondary antibodies used were Cy3–donkey anti-mouse, Cy3–donkey anti-rabbit, Cy5–donkey anti-rabbit, or Cy3–donkey anti-human from Jackson ImmunoResearch Laboratories (West Grove, PA) and Alexa488–donkey anti-mouse from Invitrogen. The cells were washed extensively with phosphate-buffered saline (PBS) between antibody incubations, and the coverslips were mounted in Mowiol. Images were recorded on a Zeiss LSM510 confocal microscope (Carl Zeiss, Jena, Germany) or a three-dimensional (3D) deconvolution microscope (Eclipse 80i; Nikon, Melville, NY) equipped with a piezo objective scanner (PIFOC) and a 100×/1.4 numerical aperture CFI Plan Apo objective lens for optical sectioning. The 3D multicolor image stacks were acquired using MetaMorph software (Molecular Devices, Sunnyvale, CA) through a cooled charge-coupled device camera (CoolSNAP HQ2; Photometrics, Tucson, AZ).
Image analysis
Pearson's colocalization coefficient was calculated using Zeiss ZEN 2009 software. To calculate the “compaction factor” (Delevoye et al., 2009), fixed cells were colabeled with endosome and Golgi markers, and images were acquired on an epifluorescence microscope. A region of interest (ROI) was drawn around the Golgi in ImageJ (National Institutes of Health, Bethesda, MD), and after background subtraction the fluorescence intensity of the endosome marker in the ROI was divided by the total cellular fluorescence.
Time-lapse fluorescence microscopy
Transfected cells were grown on glass-bottomed 35-mm dishes (Iwaki; Asahi Techno Glass, Tokyo, Japan). Time-lapse imaging was performed at 37°C on a spinning-disk confocal microscope mounted on an inverted motorized microscope (DMIRE2; Leica, Wetzlar, Germany) equipped with a CSUXI spinning disk head (Yokogawa, Tokyo, Japan) and temperature and CO2 controller (Life Imaging Services, Reinach, Switzerland). GFP was excited with a 491-nm laser line (MAG Biosystems, Santa Fe, NM). Images were captured on a QuantEM 512SC camera (Photometrics). A z-stack of seven planes was acquired every 2 s for a total duration of 2 min. Movies were generated by compiling three-dimensional maximum-intensity projections in MetaMorph. A Gaussian filter was applied to the time-lapse movies for better visual analysis.
Electron microscopy
Cells were fixed with a mixture of 2% paraformaldehyde and 0.125% glutaraldehyde (both from Electron Microscopy Sciences, Hatfield, PA) in phosphate buffer for 2 h. Cells were processed for ultracryomicrotomy and double immunogold labeled using PAG10 and PAG15 (Cell Microscopy Center, Utrecht University, Utrecht, Netherlands) as reported (Miserey-Lenkei et al. 2007). All samples were analyzed using an FEI CM120 electron microscope (FEI, Hillsboro, OR), and digital acquisitions were made with a numeric camera (Keen View; Soft Imaging System, Münster, Germany).
Coimmunoprecipitation assays and subcellular fractionation
HeLa cells growing in six-well plates were transfected with plasmids using XtremeGene 9 transfection reagent (Roche) according to the manufacturer's instructions. At 24 h posttransfection the cells were washed with ice-cold PBS and lysed in the well in NP-40 buffer (20 mM Tris, pH 7.4, 50 mM KCl, 0.5% NP-40) supplemented with a protease inhibitor cocktail. Lysates were cleared by centrifugation at 10,000 × g at 4°C for 10 min. Mouse anti-GFP antibody (Roche) prebound to protein G–Sepharose (GE Healthcare Life Sciences, Piscataway, NJ) was added to the cleared lysates and rotated in the cold room for 4 h. Complexes were pelleted by centrifugation at 500 × g for 3 min and washed with 10 volumes of NP-40 buffer. This step was repeated for a total of three times, and after the final wash the complexes were resuspended in protein sample buffer and subjected to Western blot analysis. Secondary horseradish peroxidase–coupled antibodies were purchased from Jackson ImmunoResearch Laboratories.
For the subcellular fractionation cells were resuspended in fractionation buffer (250 mM sucrose, 3 mM imidazole) supplemented with a protease inhibitor cocktail (Roche) and 1 mM CaCl2 or 5 mM EGTA. Cells were lysed by freeze–thaw and passage through a 26-gauge needle. The membrane fraction was isolated from a postnuclear supernatant by centrifugation at 100,000 × g for 30 min in a TLA120.1 rotor (Beckman Coulter, Brea, CA). The high-speed supernatant (cytosolic fraction) was removed, and the pellet (membrane fraction) was resuspended in an equal volume of fractionation buffer. Fractions were analyzed by SDS–PAGE.
RNA interference
All siRNA oligonucleotides were purchased from Sigma-Aldrich. See Supplemental Table S3 for sequences. In all cases an siRNA oligonucleotide targeting firefly luciferase (siLuc) was used as a control.
Supplementary Material
Acknowledgments
We thank E. Coudrier for critical reading of the manuscript, A. Miyawaki for reagents, and S. Miserey-Lenkei and A. Nolan for technical assistance. We thank V. Fraisier and L. Sengmanivong for their microscopy expertise and the Nikon Imaging Center at the Institut Curie-CNRS. This work was supported by the Institut Curie, the Centre National de la Recherche Scientifique, the Agence Nationale pour la Recherche (ANR Grant 2010 BLAN 122902 to B.G.), the Science Foundation Ireland (SFI09/IN1/B2629 to M.M.C.), and the Health Research Board (PD/2005/25) and a joint HRB/Marie Curie Mobility Fellowship (MCPD/2009/6) to A.J.L. C.P.H. is supported by an Irish Cancer Society Research Fellowship Award (CRF11HOR).
Abbreviations used:
- CC
coiled-coil
- DA
dominant-active
- DN
dominant-negative
- EEA1
early endosomal antigen 1
- GTD
globular tail domain
- MD
motor domain
- RNAi
RNA interference
- siRNA
small interfering RNA
- TfR
transferrin receptor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0236) on September 4, 2013.
The authors declare that they have no conflict of interest.
REFERENCES
- Chen W, Feng Y, Chen D, Wandinger-Ness A. Rab11 is required for trans-Golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol Biol Cell. 1998;9:3241–3257. doi: 10.1091/mbc.9.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darchen F, Goud B. Multiple aspects of Rab protein action in the secretory pathway: focus on Rab3 and Rab6. Biochimie. 2000;82:375–384. doi: 10.1016/s0300-9084(00)00219-4. [DOI] [PubMed] [Google Scholar]
- Delevoye C, et al. AP-1 and KIF13A coordinate endosomal sorting and positioning during melanosome biogenesis. J Cell Biol. 2009;187:247–264. doi: 10.1083/jcb.200907122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos C, et al. Rab27A and its effector MyRIP link secretory granules to F-actin and control their motion towards release sites. J Cell Biol. 2003;163:559–570. doi: 10.1083/jcb.200302157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AR, Voeltz GK. Rab10 GTPase regulates ER dynamics and morphology. Nat Cell Biol. 2013;15:169–178. doi: 10.1038/ncb2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada P, Kim J, Coleman J, Walker L, Dunn B, Takizawa P, Novick P, Ferro-Novick S. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2003;163:1255–1266. doi: 10.1083/jcb.200304030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Kuroda TS, Mikoshiba K. Slac2-a/melanophilin, the missing link between Rab27 and myosin Va: implications of a tripartite protein complex for melanosome transport. J Biol Chem. 2002;277:12432–12436. doi: 10.1074/jbc.C200005200. [DOI] [PubMed] [Google Scholar]
- Giannandrea M, et al. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. Am J Hum Genet. 2010;86:185–195. doi: 10.1016/j.ajhg.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I, et al. Rab6, Rab8, and MICAL3 cooperate in controlling docking and fusion of exocytotic carriers Rab6 regulates transport and targeting of exocytotic carriers. Curr Biol. 2011;21:967–974. doi: 10.1016/j.cub.2011.04.030. [DOI] [PubMed] [Google Scholar]
- Hammer JA, 3rd, Sellers JR. Walking to work: roles for class V myosins as cargo transporters. Nat Rev Mol Cell Biol. 2011;13:13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- Huang JD, Mermall V, Strobel MC, Russell LB, Mooseker MS, Copeland NG, Jenkins NA. Molecular genetic dissection of mouse unconventional myosin-VA: tail region mutations. Genetics. 1998;148:1963–1972. doi: 10.1093/genetics/148.4.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume AN, Collinson LM, Hopkins CR, Strom M, Barral DC, Bossi G, Griffiths GM, Seabra MC. The leaden gene product is required with Rab27a to recruit myosin Va to melanosomes in melanocytes. Traffic. 2002;3:193–202. doi: 10.1034/j.1600-0854.2002.030305.x. [DOI] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Jollivet F, Camonis J, Marche PN, Goud B. Two-hybrid system screen with the small GTP-binding protein Rab6. Identification of a novel mouse GDP dissociation inhibitor isoform and two other potential partners of Rab6. J Biol Chem. 1995;270:14801–14808. doi: 10.1074/jbc.270.24.14801. [DOI] [PubMed] [Google Scholar]
- Jin Y, Sultana A, Gandhi P, Franklin E, Hamamoto S, Khan AR, Munson M, Schekman R, Weisman LS. Myosin V transports secretory vesicles via a Rab GTPase cascade and interaction with the exocyst complex. Dev Cell. 2011;21:1156–1170. doi: 10.1016/j.devcel.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junutula JR, De Maziere AM, Peden AA, Ervin KE, Advani RJ, van Dijk SM, Klumperman J, Scheller RH. Rab14 is involved in membrane trafficking between the Golgi complex and endosomes. Mol Biol Cell. 2004;15:2218–2229. doi: 10.1091/mbc.E03-10-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly EE, Horgan CP, Adams C, Patzer TM, Ní Shúilleabháin DM, Norman JC, McCaffrey MW. Class I Rab11-family interacting proteins are binding targets for the Rab14 GTPase. Biol Cell. 2009;102:51–62. doi: 10.1042/BC20090068. [DOI] [PubMed] [Google Scholar]
- Kelly EE, Horgan CP, Goud B, McCaffrey MW. The Rab family of proteins: 25 years on. Biochem Soc Trans. 2012;40:1337–1347. doi: 10.1042/BST20120203. [DOI] [PubMed] [Google Scholar]
- Kogel T, Gerdes HH. Roles of myosin Va and Rab3D in membrane remodeling of immature secretory granules. Cell Mol Neurobiol. 2010;30:1303–1308. doi: 10.1007/s10571-010-9597-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogel T, Rudolf R, Hodneland E, Hellwig A, Kuznetsov SA, Seiler F, Sollner TH, Barroso J, Gerdes HH. Distinct roles of myosin Va in membrane remodeling and exocytosis of secretory granules. Traffic. 2010;11:637–650. doi: 10.1111/j.1600-0854.2010.01048.x. [DOI] [PubMed] [Google Scholar]
- Lambert J, Naeyaert JM, Callens T, De Paepe A, Messiaen L. Human myosin V gene produces different transcripts in a cell type-specific manner. Biochem Biophys Res Commun. 1998;252:329–333. doi: 10.1006/bbrc.1998.9644. [DOI] [PubMed] [Google Scholar]
- Larance M, et al. Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J Biol Chem. 2005;280:37803–37813. doi: 10.1074/jbc.M503897200. [DOI] [PubMed] [Google Scholar]
- Li XD, Jung HS, Mabuchi K, Craig R, Ikebe M. The globular tail domain of myosin Va functions as an inhibitor of the myosin Va motor. J Biol Chem. 2006;281:21789–21798. doi: 10.1074/jbc.M602957200. [DOI] [PubMed] [Google Scholar]
- Li XD, Jung HS, Wang Q, Ikebe R, Craig R, Ikebe M. The globular tail domain puts on the brake to stop the ATPase cycle of myosin Va. Proc Natl Acad Sci USA. 2008;105:1140–1145. doi: 10.1073/pnas.0709741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linford A, Yoshimura S, Nunes Bastos R, Langemeyer L, Gerondopoulos A, Rigden DJ, Barr FA. Rab14 and its exchange factor FAM116 link endocytic recycling and adherens junction stability in migrating cells. Dev Cell. 2012;22:952–966. doi: 10.1016/j.devcel.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Tokarev AA, Jin Y, Mulholland J, Weisman LS, Segev N. Direct interaction between a myosin V motor and the Rab GTPases Ypt31/32 is required for polarized secretion. Mol Biol Cell. 2008;19:4177–4187. doi: 10.1091/mbc.E08-02-0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- Mercer JA, Seperack PK, Strobel MC, Copeland NG, Jenkins NA. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 1991;349:709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- Miserey-Lenkei S, et al. Rab6-interacting protein 1 links Rab6 and Rab11 function. Traffic. 2007;8:1385–1403. doi: 10.1111/j.1600-0854.2007.00612.x. [DOI] [PubMed] [Google Scholar]
- Miserey-Lenkei S, Chalancon G, Bardin S, Formstecher E, Goud B, Echard A. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat Cell Biol. 2010;12:645–654. doi: 10.1038/ncb2067. [DOI] [PubMed] [Google Scholar]
- Morel E, Parton RG, Gruenberg J. Annexin A2-dependent polymerization of actin mediates endosome biogenesis. Dev Cell. 2009;16:445–457. doi: 10.1016/j.devcel.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Nagashima K, Torii S, Yi Z, Igarashi M, Okamoto K, Takeuchi T, Izumi T. Melanophilin directly links Rab27a and myosin Va through its distinct coiled-coil regions. FEBS Lett. 2002;517:233–238. doi: 10.1016/s0014-5793(02)02634-0. [DOI] [PubMed] [Google Scholar]
- Ng EL, Wang Y, Tang BL. Rab22B's role in trans-Golgi network membrane dynamics. Biochem Biophys Res Commun. 2007;361:751–757. doi: 10.1016/j.bbrc.2007.07.076. [DOI] [PubMed] [Google Scholar]
- Nizak C, Monier S, del Nery E, Moutel S, Goud B, Perez F. Recombinant antibodies to the small GTPase Rab6 as conformation sensors. Science. 2003;300:984–987. doi: 10.1126/science.1083911. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Rahajeng J, Panapakkam Giridharan SS, Cai B, Naslavsky N, Caplan S. MICAL-L1 is a tubular endosomal membrane hub that connects Rab35 and Arf6 with Rab8a. Traffic. 2012;13:82–93. doi: 10.1111/j.1600-0854.2011.01294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell. 2007;18:2828–2837. doi: 10.1091/mbc.E07-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland JT, Lapierre LA, Goldenring JR. Alternative splicing in class v myosins determines association with rab10. J Biol Chem. 2009;284:1213–1223. doi: 10.1074/jbc.M805957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G, Brennwald P. Yeast homologues of lethal giant larvae and type V myosin cooperate in the regulation of Rab-dependent vesicle clustering and polarized exocytosis. Mol Biol Cell. 2011;22:842–857. doi: 10.1091/mbc.E10-07-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Tirado FH, Legesse-Miller A, Schott D, Bretscher A. PI4P and Rab inputs collaborate in myosin-V-dependent transport of secretory compartments in yeast. Dev Cell. 2011;20:47–59. doi: 10.1016/j.devcel.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seperack PK, Mercer JA, Strobel MC, Copeland NG, Jenkins NA. Retroviral sequences located within an intron of the dilute gene alter dilute expression in a tissue-specific manner. EMBO J. 1995;14:2326–2332. doi: 10.1002/j.1460-2075.1995.tb07227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M, Hume AN, Tarafder AK, Barkagianni E, Seabra MC. A family of Rab27-binding proteins. Melanophilin links Rab27a and myosin Va function in melanosome transport. J Biol Chem. 2002;277:25423–25430. doi: 10.1074/jbc.M202574200. [DOI] [PubMed] [Google Scholar]
- Thirumurugan K, Sakamoto T, Hammer JA, 3rd, Sellers JR, Knight PJ. The cargo-binding domain regulates structure and activity of myosin 5. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus KM, Gushchin MI, Lui H, Hazelwood L, Krementsova EB, Volkmann N, Hanein D. Effect of calcium on calmodulin bound to the IQ motifs of myosin V. J Biol Chem. 2007;282:23316–23325. doi: 10.1074/jbc.M701636200. [DOI] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Wagner W, Brenowitz SD, Hammer JA., 3rd Myosin-Va transports the endoplasmic reticulum into the dendritic spines of Purkinje neurons. Nat Cell Biol. 2011;13:40–48. doi: 10.1038/ncb2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Patel A, Lee YL, Provance DW, Jr, Vought VE, Cosgrove MS, Mercer JA, Langford GM. Myosin5a tail associates directly with Rab3A-containing compartments in neurons. J Biol Chem. 2011;286:14352–14361. doi: 10.1074/jbc.M110.187286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolner S, Bement WM. Unconventional myosins acting unconventionally. Trends Cell Biol. 2009;19:245–252. doi: 10.1016/j.tcb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bowers B, Rao K, Wei Q, Hammer JA., 3rd Visualization of melanosome dynamics within wild-type and dilute melanocytes suggests a paradigm for myosin V function In vivo. J Cell Biol. 1998;143:1899–1918. doi: 10.1083/jcb.143.7.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wang F, Rao K, Sellers JR, Hammer JA., 3rd Rab27a is an essential component of melanosome receptor for myosin Va. Mol Biol Cell. 2002a;13:1735–1749. doi: 10.1091/mbc.01-12-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XS, Rao K, Zhang H, Wang F, Sellers JR, Matesic LE, Copeland NG, Jenkins NA, Hammer JA., 3rd Identification of an organelle receptor for myosin-Va. Nat Cell Biol. 2002b;4:271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.