Expression of the adenovirus protein RIDα rescues the cholesterol storage phenotype in NPC1-deficient cells by inducing formation of lipid droplets. The function of RIDα is independent of NPC1 but dependent on NPC2 and the oxysterol-binding protein ORP1L. This study provides the first evidence that ORP1L plays a role in sterol transport and LD formation.
Abstract
Niemann–Pick disease type C (NPC) is caused by mutations in NPC1 or NPC2, which coordinate egress of low-density-lipoprotein (LDL)-cholesterol from late endosomes. We previously reported that the adenovirus-encoded protein RIDα rescues the cholesterol storage phenotype in NPC1-mutant fibroblasts. We show here that RIDα reconstitutes deficient endosome-to-endoplasmic reticulum (ER) transport, allowing excess LDL-cholesterol to be esterified by acyl-CoA:cholesterol acyltransferase and stored in lipid droplets (LDs) in NPC1-deficient cells. Furthermore, the RIDα pathway is regulated by the oxysterol-binding protein ORP1L. Studies have classified ORP1L as a sterol sensor involved in LE positioning downstream of GTP-Rab7. Our data, however, suggest that ORP1L may play a role in transport of LDL-cholesterol to a specific ER pool designated for LD formation. In contrast to NPC1, which is dispensable, the RIDα/ORP1L-dependent route requires functional NPC2. Although NPC1/NPC2 constitutes the major pathway, therapies that amplify minor egress routes for LDL-cholesterol could significantly improve clinical management of patients with loss-of-function NPC1 mutations. The molecular identity of putative alternative pathways, however, is poorly characterized. We propose RIDα as a model system for understanding physiological egress routes that use ORP1L to activate ER feedback responses involved in LD formation.
INTRODUCTION
Cholesterol plays an essential role in many aspects of eukaryotic membrane function. Excess unesterified cholesterol, which is toxic to cells, is tightly regulated by an elaborate network of feedback mechanisms (Simons and Ikonen, 2000; Maxfield and Tabas, 2005; Steck and Lange, 2010). Cholesterol levels are highest at the plasma membrane (PM) and lowest in the endoplasmic reticulum (ER), where many sterol regulatory proteins involved in homeostatic feedback responses reside (Lange et al., 2004). Therefore small changes in the cholesterol content of the ER allow for precise regulation of cholesterol homeostasis. The ER homeostatic machinery has two main components, which are likely regulated by different pools of active cholesterol (Kristiana et al., 2008): sterol regulatory element–binding protein (SREBP), which is converted to an active transcription factor regulating expression of multiple genes involved in cholesterol regulation when ER cholesterol levels are low (Goldstein et al., 2006); and acyl-CoA:cholesterol acyltransferase (ACAT), which esterifies excess free cholesterol into cholesterol esters that are stored in lipid droplets (LDs; Thiele and Spandl, 2008). LDs have been shown to be more than just storage compartments, as they have diverse functions, such as distributing lipids to endosomes and other organelles (Zehmer et al., 2009). Excess cholesterol also serves as a substrate for synthesis of regulatory side-chain oxysterols such as 27-hydroxycholesterol produced in mitochondria, which regulates SREBP processing and activates LXR nuclear transcription factors that regulate genes involved in reverse cholesterol transport (Olkkonen and Hynynen, 2009). Although much is known about cholesterol trafficking, questions remain about how endosomal cholesterol is transported to the ER and other organelles to regulate essential homeostatic responses.
A breakthrough in our understanding of intracellular cholesterol transport came from the study of Niemann–Pick disease type C (NPC). NPC is characterized by accumulation of low-density-lipoprotein (LDL)-cholesterol and other lipids in late endosomes/lysosomes (LE/Ly) termed lysosomal storage organelles (LSOs; Patterson et al., 2001; Chang et al., 2005). Although NPC is a relatively rare autosomal recessive disorder associated with progressive neurodegeneration and death, LSO-like compartments are found in a number of common human diseases, so studies with NPC have far-reaching clinical implications (Cianciola et al., 2011). NPC is caused by mutations in NPC1, a multispanning transmembrane protein localized to the limiting membrane of LE/Ly, or, less frequently in NPC2, a soluble protein found in the lumen of LE/Ly (Chang et al., 2005). LDL is taken up by cells through clathrin-mediated endocytosis of the LDL receptor (LDLR) and delivered to LE/Ly, where cholesterol esters are broken down by lysosomal acid lipase. NPC1 and NPC2 coordinate egress of LDL-cholesterol out of LE/Ly to other intracellular membranes through a direct interaction that depends on cholesterol and acidic pH (Deffieu and Pfeffer, 2011). Cholesterol egress out of LE/Ly also depends on the atypical LE phospholipid lysobisphosphatidic acid (LBPA; Chevallier et al., 2008). Although their ability to regulate cholesterol transport to ER homeostatic machinery is well established (Peake and Vance, 2010), whether NPC1 and NPC2 are also involved in transferring free cholesterol to mitochondria is controversial (Frolov et al., 2003; Charman et al., 2010). However, new research indicates that this transport step depends on NPC2 (Kennedy et al., 2012).
Host–pathogen interactions often illuminate previously unappreciated cell biological processes, and the study of host–pathogen interactions provides insight into the cell biology of both pathogen and host. We reported that the adenovirus (Ad) protein RIDα rescues the cholesterol storage phenotype of NPC1-mutant fibroblasts by a mechanism that appears to be silent in cells with functional NPC1 (Cianciola and Carlin, 2009). RIDα is a partial GTP-Rab7 mimic that interacts with two Rab7 effectors, Rab7-interacting lysosomal protein (RILP) and oxysterol-binding protein–related protein 1L (ORP1L; Shah et al., 2007). GTP-Rab7 forms a tripartite complex with RILP and ORP1L, which then interacts with the p150Glued dynactin subunit to recruit the dynein motor and regulate minus end–directed transport along microtubules (Johansson et al., 2007). Furthermore, in conjunction with Rab7, ORP1L senses LE cholesterol and regulates LE positioning through interactions with vesicle-associated membrane protein–associated ER protein (VAP) located on ER membranes (Rocha et al., 2009). It has been shown that members of the yeast oxysterol-binding protein (OSBP) family regulate nonvesicular sterol transport between closely apposed membranes (Schulz et al., 2009). Mammalian cells express 12 OSBP homologues, and many of these oxysterol-binding protein–related proteins reside in specific membrane compartments due to the presence of pleckstrin homology or transmembrane domains. Although two of the mammalian proteins lacking pleckstrin homology or transmembrane domains—ORP1S and ORP2—play a role in sterol transport from PM to ER and LDs, whether membrane-bound ORPs behave in a similar manner is not known (Jansen et al., 2011). Owing to the proximity of binding motifs in the short carboxy-terminal tail of RIDα (Shah et al., 2007), it is unlikely that RIDα binds RILP and ORP1L concomitantly, thus providing a unique model system to identify and study novel ORP1L functions. In this study, we report that RIDα rescues the NPC1 cholesterol storage phenotype by increasing LD formation via a mechanism that depends on ORP1L. These results provide the first evidence that ORP1L plays a role in sterol transport and LD formation independent of NPC1. Therefore RIDα provides a novel system to study a potentially redundant pathway for endosome-to-ER cholesterol transport.
RESULTS
NPC1 but not NPC2 cholesterol storage phenotype is rescued by RIDα
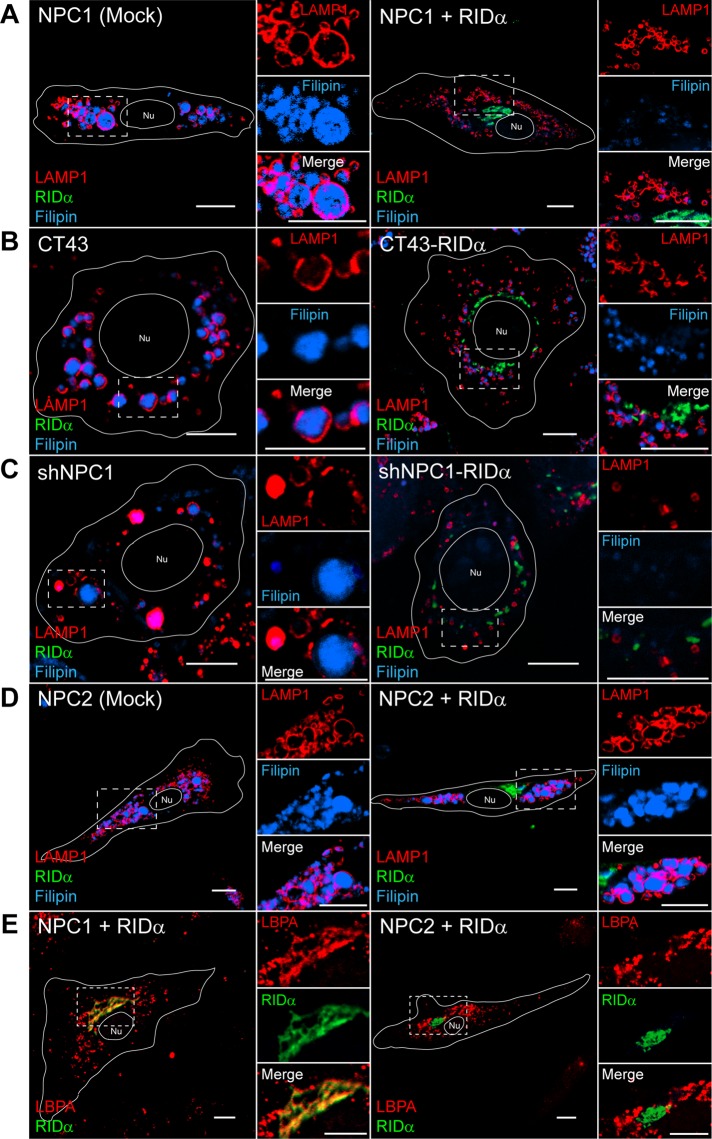
We previously showed that RIDα rescued the LSO cholesterol storage phenotype when transiently expressed in NPC1-mutant fibroblasts upon LDL loading (Figure 1A; Cianciola and Carlin, 2009). LSOs that accumulate in NPC disease cells can be visualized by costaining with LAMP1 to identify aberrant LEs and filipin, a fluorescent antibiotic that selectively binds free cholesterol. However, because of low transfection efficiency of patient fibroblasts, we are unable to perform biochemical assays in these cells. To further address the mechanism of how RIDα rescues the NPC1 cholesterol storage phenotype, we used two NPC1-deficient model systems with stable RIDα expression (Table 1): 1) CT43 cells, which are Chinese hamster ovary cells isolated after a chemical mutagenesis screen that harbor a mutation in NPC1 causing premature translational termination after 933 amino acids, producing a nonfunctional protein (Cruz et al., 2000), and 2) shNPC1 cells, which are immortalized human hepatocytes (IHHs) stably expressing shRNA to NPC1 (Ulatowski et al., 2011). CT43 cells and shNPC1 cells both displayed enlarged LAMP1/filipin-positive LEs indicative of LSOs upon LDL loading (Figure 1, B and C). Stable RIDα expression in either cell line rescued the NPC1-deficient cholesterol storage phenotype, as LSOs no longer persisted (Figure 1, B and C). We also tested whether RIDα expression could rescue the cholesterol storage phenotype in NPC2-mutant fibroblasts. NPC2-mutant fibroblasts displayed enlarged LAMP1/filipin-positive LEs upon LDL loading (Figure 1D); however, the LSO phenotype of NPC2-mutant fibroblasts was not affected by FLAG-RIDα expression (Figure 1D). Although we previously showed that RIDα colocalized with the atypical LE lipid LBPA in NPC1-mutant fibroblasts (Figure 1E; Cianciola and Carlin, 2009), in contrast, we found that RIDα did not localize with an LBPA-positive compartment in NPC2-mutant fibroblasts (Figure 1E). Collectively these data imply that the ability of RIDα to rescue the cholesterol storage phenotype in NPC1-deficient cells with different genetic defects depends on NPC2 and that RIDα function may involve an LBPA-positive compartment.
FIGURE 1:
RIDα rescues cholesterol storage phenotype in NPC1-deficient cells but not in NPC2-mutant fibroblasts. (A) Confocal images of NPC1-mutant fibroblasts mock transfected or transfected with FLAG-RIDα and stained with antibodies to LAMP1 and FLAG-RIDα and with filipin. (B) Confocal images of CT43 and CT43-RIDα cells stained with antibodies to LAMP1 and FLAG-RIDα and with filipin. (C) Confocal images of shNPC1 and shNPC1-RIDα cells stained with antibodies to LAMP1 and FLAG-RIDα and with filipin. (D) Confocal images of NPC2-mutant fibroblasts mock transfected or transfected with FLAG-RIDα and stained with antibodies to LAMP1 and FLAG-RIDα and with filipin. (E) Confocal images of NPC1- or NPC2-mutant fibroblasts transfected with FLAG-RIDα and stained with antibodies to LBPA and FLAG-RIDα. Boxed areas, regions of the image that were magnified. Bars, 10 μm. Nu, nucleus.
TABLE 1:
NPC1 genotypes in three NPC1-deficient model systems used in this study.
| Cell line | Genotype | RIDα expression |
|---|---|---|
| NPC1-mutant fibroblasts | Compound heterozygote for the I1061T and P237S alleles from NPC1 patient ( Pipalia et al., 2011) | Transient |
| NPC1 I1061T mutant fibroblasts | Homozygous for the I1061T mutation ( Gelsthorpe et al., 2008) | Transient |
| CT43 cells | Premature translational termination of NPC1 after 933 amino acids in Chinese hamster ovary cells ( Cruz et al., 2000) | Stable |
| shNPC1 cells | Stable shRNA knockdown of NPC1 in immortalized human hepatocytes ( Ulatowski et al., 2011) | Stable |
RIDα NPC1 rescue depends on LD formation
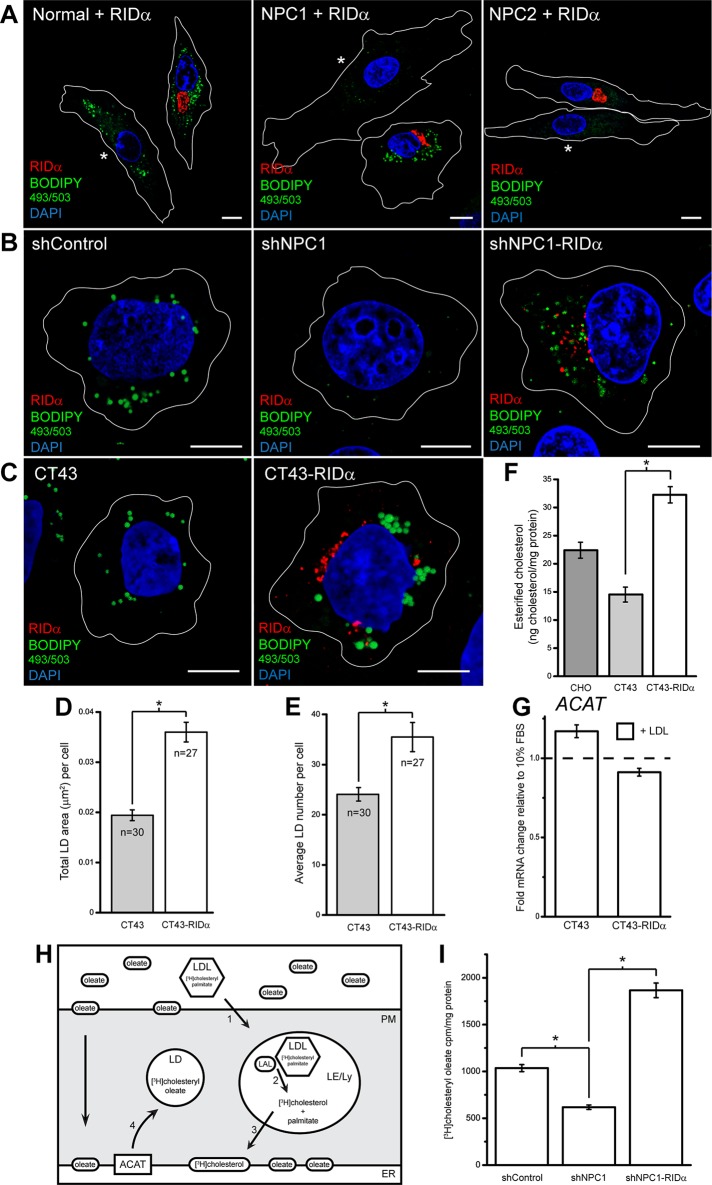
One mechanism for the regulation of cholesterol homeostasis is the esterification of free cholesterol for storage in LDs by the resident ER enzyme ACAT (Thiele and Spandl, 2008). To test whether RIDα uses this pathway to mediate NPC1 rescue, we examined LD formation in normal and NPC patient fibroblasts by staining with the neutral lipid stain BODIPY 493/503, which labels LDs. Normal fibroblasts exhibited abundant LDs in mock-transfected cells loaded with LDL (shown in the same field as FLAG-RIDα–transfected cells and designated with an asterisk), and RIDα expression had no discernible effect on LD formation in these cells (Figure 2A and Supplemental Figure S1A). In contrast, mock-transfected NPC1-mutant fibroblasts displayed an almost complete lack of BODIPY 493/503 staining (Figure 2A and Supplemental Figure S1B) due to a block in delivery of free cholesterol to the ER to act as a substrate for ACAT (Byers et al., 1989). Remarkably, FLAG-RIDα expression induced a dramatic increase in vesicular BODIPY 493/503 staining in NPC1-mutant fibroblasts upon LDL addition (Figure 2A and Supplemental Figure S1B). We also tested whether RIDα had a similar effect on LD formation in NPC2-mutant fibroblasts and found that, akin to LSO clearance, RIDα did not rescue LD formation when these cells were loaded with LDL (Figure 2A and Supplemental Figure S1C). Similar to NPC1-mutant fibroblasts, shNPC1 cells exhibited a dearth of BODIPY 493/503 staining compared with shControl cells, whereas shNPC1-RIDα cells displayed a striking increase in LD formation with LDL treatment (Figure 2B). We also assessed CT43 and CT43-RIDα cells for BODIPY 493/503 staining, and although LDs were present in CT43 cells, CT43-RIDα cells displayed an increase in LD formation compared with CT43 cells upon sterol addition (Figure 2C). Quantification of the microscopy (see Materials and Methods) showed that CT43-RIDα exhibited an increase in both the size (Figure 2D) and number (Figure 2E) of LDs compared with CT43 cells. Next we used the Amplex Red cholesterol assay to quantify cholesterol esters in CT43 and CT43-RIDα cells and discovered a statistically significant approximately twofold increase in esterified cholesterol in CT43-RIDα cells compared with CT43 cells when loaded with LDL (Figure 2F). As a control, we found that ACAT mRNA levels were not increased in CT43-RIDα cells compared with CT43 cells after LDL loading (Figure 2G), suggesting that changes in ACAT expression did not account for the increase in cholesterol esterification and LD formation seen in CT43-RIDα cells. The presence of LDs in NPC1-deficient CT43 cells compared with a lack of LDs in NPC1-mutant fibroblasts and shNPC1 cells may be attributed to the differences in the NPC1 genotype of these cells (Table 1). Alternatively, the CT43 cells may have acquired a gain-of-function mutation affecting LD formation during the chemical mutagenesis screen (Cruz et al., 2000). Nevertheless, RIDα was able to induce an increase in LD formation beyond what was present in CT43 cells.
FIGURE 2:
RIDα increases LD formation in NPC1-deficient cells. (A) Confocal images of normal and NPC1- and NPC2-mutant fibroblasts transfected with RIDα and stained with antibody to FLAG-RIDα and with BODIPY 493/503 and DAPI to visualize LDs and nuclei, respectively. Mock-transfected cells lacking FLAG-RIDα expression are shown in the same field and designated with an asterisk. (B) Confocal images of shControl, shNPC1, and shNPC1-RIDα cells stained with antibody to FLAG-RIDα and with BODIPY 493/503 and DAPI. (C) Confocal images of CT43 and CT43-RIDα cells stained with antibody to FLAG-RIDα and with BODIPY 493/503 and DAPI. (D, E) Quantification of average LD area (D) and average LD number (E) per cell in cells treated similarly to cells in C as described in Materials and Methods. Data are presented as mean ± SEM (*p < 0.001). (F) Quantification of esterified cholesterol in Chinese hamster ovary, CT43, and CT43-RIDα cells using the Amplex Red Cholesterol Assay kit as described in Materials and Methods. Values were normalized to total cellular protein and are displayed as mean ± SEM (*p < 0.001). (G) ACAT mRNA levels quantified by real-time PCR similarly to cells in Figure 4. Data are presented as mean ± SEM. (H) Experimental setup of cholesterol transport assay. Purified human LDL was labeled with [3H]cholesteryl palmitate, and cells were incubated with the labeled LDL and excess oleate. The labeled LDL was transported to Ly (step 1) and deesterified by lysosomal acid lipase (LAL; step 2). The liberated [3H]cholesterol can then be transported to the ER (step 3), where it can be reesterified by ACAT along with the excess oleate to form [3H]cholesteryl oleate and stored in LDs (step 4). (I) shControl, shNPC1, and shNPC1-RIDα cells were incubated with [3H]cholesteryl palmitate along with excess oleate as described in Materials and Methods. The [3H]cholesteryl oleate production was quantified, and values were normalized to total cellular protein and are displayed as mean ± SD (*p < 0.0001) from three independent experiments. Bars, 10 μm.
RIDα mediates transfer of LDL-cholesterol to ER for esterification
To determine whether RIDα mediates the transfer of LDL-cholesterol from LE/Ly to ER, we designed an experiment to follow the trafficking itinerary of exogenous cholesterol to the ER for esterification in NPC1-deficient cells. Our approach used LDL radiolabeled with [3H]cholesteryl palmitate, which was fed to cells in the presence of excess oleate (Figure 2H). Egress of radiolabeled cholesterol out of LE/Ly to the ER will favor reesterification with the fatty acid oleate to form [3H]cholesteryl oleate (Seo et al., 2001). shControl, shNPC1, and shNPC1-RIDα cells were treated with radiolabeled LDL along with excess oleate complexed to bovine serum albumin (BSA) as described in Materials and Methods, and lipids were extracted and analyzed by TLC along with appropriate standards. As expected, shNPC1 cells exhibited a significant decrease in [3H]cholesteryl oleate formation compared with shControl cells (Figure 2I). RIDα expression induced a statistically significant threefold increase in [3H]cholesteryl oleate formation in shNPC1-RIDα cells compared with shNPC1 cells (Figure 2I). This conclusively demonstrates that RIDα mediates the egress of LDL-cholesterol out of LE/Ly to the ER for reesterification and incorporation into LDs in NPC1-deficient cells.
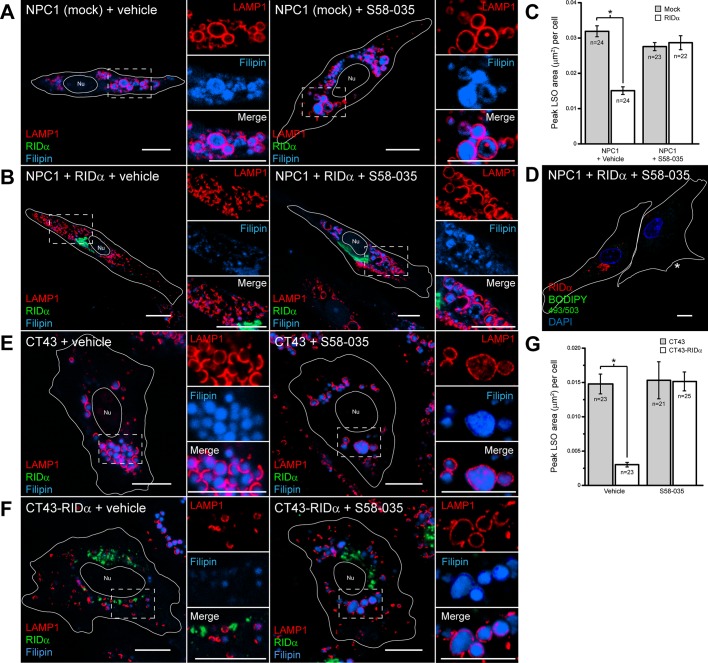
To further explore the effect of LD formation in RIDα LSO rescue, we treated NPC1-deficient cells with the prototypical ACAT inhibitor Sandoz 58-035 (S58-035; Ross et al., 1984). The LSO phenotype of mock-transfected NPC1-mutant fibroblasts was unaffected by vehicle treatment or 12-h treatment with S58-035 during LDL loading (Figure 3A); however, ACAT inhibitor treatment completely blocked the ability of RIDα to rescue the cholesterol storage phenotype in FLAG-RIDα–transfected NPC1-mutant fibroblasts (Figure 3B). We quantified the peak area of LAMP1/filipin-positive LSOs in mock-transfected and FLAG-RIDα–transfected NPC1-mutant fibroblasts upon LDL loading with vehicle and S58-035 treatment. RIDα expression induced a statistically significant decrease in peak LSO area in vehicle-treated cells compared with mock-transfected cells, whereas S58-035 treatment caused LSOs to persist in FLAG-RIDα–transfected cells (Figure 3C). As expected, S58-035 treatment also blocked LD formation in FLAG-RIDα–transfected NPC1-mutant fibroblasts stimulated with LDL (Figure 3D and Supplemental Figure S1D). To corroborate the results obtained in NPC1-mutant fibroblasts, we similarly loaded CT43 cells with LDL, treated them with vehicle or ACAT inhibitor for 12 h, and stained them for LAMP1 and filipin. The CT43 LSO phenotype was again unaffected by vehicle or S58-035 treatment (Figure 3E), and LSOs persisted in CT43-RIDα cells treated with S58-035 (Figure 3F). Similarly, quantification of confocal microscopy results showed a significant decrease in peak LSO area in vehicle treated CT43-RIDα cells compared with CT43 cells, and LSOs again persisted in CT43-RIDα cells treated with ACAT inhibitor (Figure 3G). Taken together these data indicate that RIDα promotes transport of free cholesterol to the ER, where it can be esterified by ACAT and stored in LDs in three different NPC1 mutant backgrounds.
FIGURE 3:
Inhibition of ACAT blocks RIDα rescue of cholesterol storage phenotype in NPC1-deficient cells. (A, B) Confocal images of NPC1-mutant fibroblasts mock transfected (A) or transfected with RIDα (B) treated with dimethyl sulfoxide (DMSO) vehicle (left) or S58-035 (right) for 12 h and stained with antibodies to LAMP1 and FLAG-RIDα and with filipin. (C) Quantification of peak LSO area per cell in cells treated similarly to cells in A and B as described in Materials and Methods. Data are presented as mean ± SEM (*p < 0.001). (D) Confocal image of NPC1-mutant fibroblasts transfected with RIDα treated with S58-035 for 12 h and stained with antibody to FLAG-RIDα and with BODIPY 493/503 and DAPI. Mock-transfected cell is shown in the same field as designated by an asterisk. (E, F) CT43 (E) and CT43-RIDα cells (F) treated with DMSO vehicle (left) or S58–035 (right) for 12 h and stained with antibodies to LAMP1 and FLAG-RIDα and with filipin. (G) Quantification of peak LSO area per cell in cells treated similarly to cells in E and F as described in Materials and Methods. Data are presented as mean ± SEM (*p < 0.001). Boxed areas, regions of the image that were magnified. Bars, 10 μm. Nu, nucleus.
SREBP- and LXR-regulated gene expression is not affected by RIDα
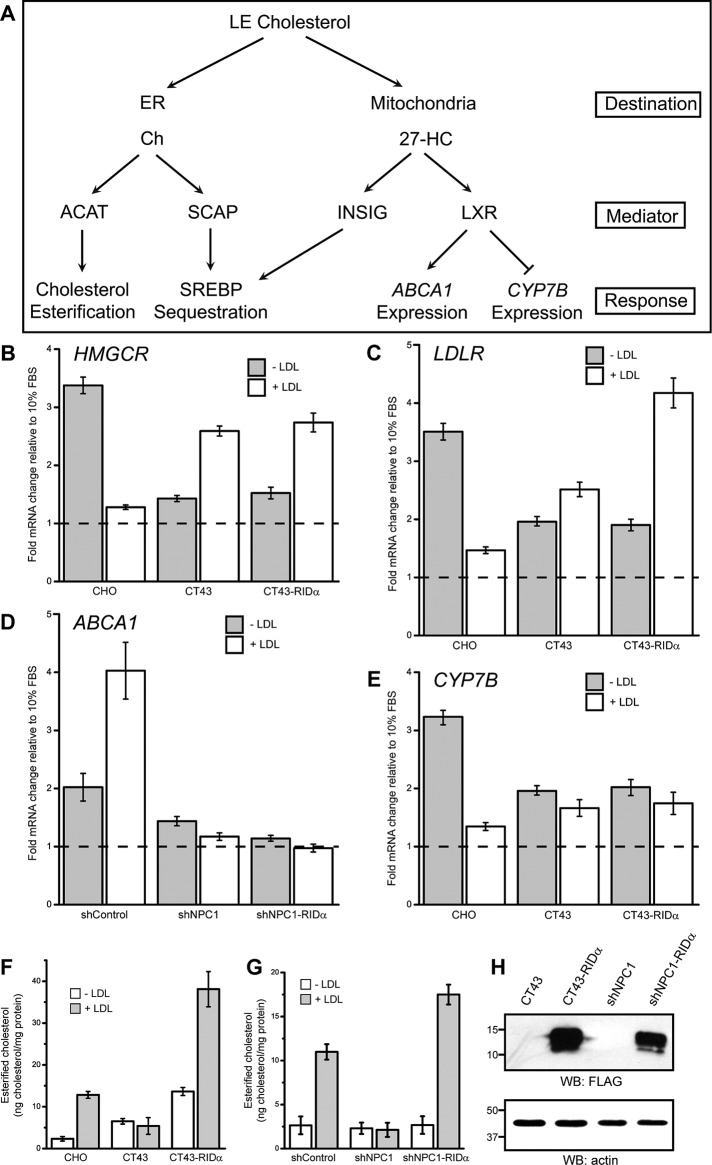
To further understand the role of RIDα in rescue of the NPC1 cholesterol storage phenotype, we tested the effect of RIDα on the regulation of cholesterol homeostatic gene expression. In normal cells cholesterol transport to the ER promotes sequestration of SREBP to inhibit expression of genes involved in increasing cholesterol levels, such as 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), the rate-limiting enzyme in cholesterol biosynthesis, and LDL receptor (LDLR), involved in internalization and endocytic transport of LDL-cholesterol (Figure 4A; Goldstein et al., 2006). This process is more complex in NPC1-deficient cells, as transport of cholesterol to the ER is blocked, causing SREBP-regulated gene expression to continue impervious to the rising free cholesterol levels in LSOs. We used real-time PCR to examine the effect of RIDα on expression of these SREBP-regulated genes. Homeostatic responses were assayed by comparing mRNA levels in lipid-starved Chinese hamster ovary and CT43 cell lines with and without supplemental LDL as described in Materials and Methods. CT43 cells were originally isolated from the parental cell line 25RA (Cruz et al., 2000), which contain a gain-of-function mutation in SREBP cleavage-activating protein (SCAP); however, this mutation has reverted back to wild type in the CT43 cells (Strauss et al., 2010). Therefore Chinese hamster ovary cells, with wild-type NPC1 and SCAP, were used to compare sterol-mediated gene regulation with CT43 and CT43-RIDα cells. Chinese hamster ovary cells displayed the expected increase in HMGCR and LDLR expression upon lipid starvation, which then decreased upon addition of LDL (Figure 4, B and C). CT43 cells with nonfunctional NPC1 showed lower levels of HMGCR and LDLR mRNA in lipid starved conditions compared with Chinese hamster ovary cells that actually increased upon LDL addition (Figure 4, B and C), consistent with a block in cholesterol transport to the ER and loss of homeostatic SREBP regulation. CT43-RIDα cells showed similar HMGCR mRNA levels compared with CT43 cells under lipid-starved and LDL-loaded conditions (Figure 4B), whereas LDLR mRNA levels were further increased upon LDL addition (Figure 4C). These results indicate that RIDα failed to restore proper SREBP homeostatic regulation for HMGCR and LDLR in NPC1-deficient cells.
FIGURE 4:
RIDα does not modulate sterol-regulated gene expression in NPC1-deficient cells with stable RIDα expression. (A) Schematic summarizing homeostatic responses to increased free cholesterol (Ch). High levels of ER Ch become esterified by ACAT and also inhibit the SREBP pathway by facilitating SCAP–SREBP interactions and ER retention. Increased mitochondrial Ch stimulates 27-hydroxycholesterol (27-HC) biosynthesis, which binds Insig, contributing to SREBP ER retention, and activates nuclear LXR transcription factors. (B–E) HMGCR (B), LDLR (C), ABCA1 (D), and CYP7B (E) mRNA levels quantified by real-time PCR. Values are expressed as relative units after internal normalization to glyceraldehyde 3-phosphate dehydrogenase mRNA levels and compared with control samples from the same cell lines cultured in 10% FBS, which was set to 1 (dashed line) from three independent experiments. Data are presented as mean ± SEM. (F, G) Quantification of esterified cholesterol in Chinese hamster ovary, CT43, and CT43-RIDα cells (F) or shControl, shNPC1, and shNPC1-RIDα cells (G) with and without 24 h treatment with LDL using the Amplex Red Cholesterol Assay kit as described in Materials and Methods. Values were normalized to total cellular protein and are displayed as mean ± SEM. (H) Equal aliquots of total cellular protein from CT43 and CT43-RIDα cells or shNPC1 and shNPC1-RIDα cells were immunoblotted with antibodies to FLAG or actin for loading control.
Another mechanism for cholesterol homeostasis is delivery of cholesterol to mitochondria, where it is metabolized to the oxysterol 27-hydroxycholesterol (Figure 4A; Russell, 2000). Oxysterols are present at much lower concentrations than cholesterol but are more potent regulators of cholesterol homeostasis (Wang et al., 1994). LXRs bind oxysterols and regulate expression of cholesterol homeostatic genes, including cholesterol efflux transporter ATP-binding cassette transporter A1 (ABCA1), and the P450 enzyme CYP7B (Ory, 2004). Although cholesterol trafficking to mitochondria functions independent of NPC1 (Charman et al., 2010), LXR-regulated genes like ABCA1 are misregulated in NPC1-deficient cells (Choi et al., 2003). Oxysterols also interact with Insigs, which promotes sequestration of SREBP in the ER to inhibit expression of cholesterol regulatory genes (Radhakrishnan et al., 2007; Sun et al., 2007). We used IHH cells expressing control or NPC1 shRNA to test the effect of RIDα on ABCA1 gene expression in response to sterol manipulation. shControl cells displayed the expected increase in ABCA1 mRNA expression under LDL-loaded conditions, whereas ABCA1 mRNA levels were unaffected upon sterol treatment in shNPC1 cells (Figure 4D), demonstrating that oxysterol production and LXR activation are perturbed with NPC1 knockdown. ABCA1 mRNA levels were unaffected after LDL loading in shNPC1-RIDα cells (Figure 4D), indicating that RIDα expression did not correct ABCA1 gene regulation in NPC1-deficient cells. In Chinese hamster ovary cells, CYP7B mRNA expression increased upon lipid starvation and decreased upon LDL addition, as expected (Figure 4E), since LXR downregulates CYP7B (Russell, 2000). CT43 cells showed no change in CYP7B mRNA levels upon LDL addition, and similar results were observed in CT43-RIDα cells (Figure 4E), demonstrating that RIDα does not regulate LXR gene regulation in NPC1-deficient cells. As a control, esterified cholesterol was measured under lipid-starved conditions with and without supplemental LDL in Chinese hamster ovary, CT43, and CT43-RIDα cells and shControl, shNPC1, and shNPC1-RIDα cells using the Amplex Red cholesterol assay to quantify cholesterol esters. Cholesterol ester formation was induced by LDL addition in Chinese hamster ovary cells but blocked in CT43 cells, as expected, whereas CT43-RIDα cells exhibited increased cholesterol ester formation upon LDL loading (Figure 4F). Similarly, cholesterol ester formation was induced by LDL addition in shControl cells but blocked in shNPC1 cells, as expected, whereas shNPC1-RIDα cells exhibited an increase in cholesterol ester formation upon LDL loading (Figure 4G). We also verified equivalent RIDα expression in CT43-RIDα and shNPC1-RIDα cells by Western blotting with antibody to FLAG (Figure 4H). Collectively these data indicate that RIDα does not rescue the NPC1 cholesterol storage phenotype by restoring SREBP-mediated gene regulation or promoting cholesterol transport to mitochondria for oxysterol production and subsequent LXR activity. They also suggest that RIDα directs free cholesterol to a specific pool in the ER, where it can be acted upon by ACAT independent of SREBP-mediated gene expression.
RIDα does not correct LE/Ly targeting of NPC1 I1061T mutant
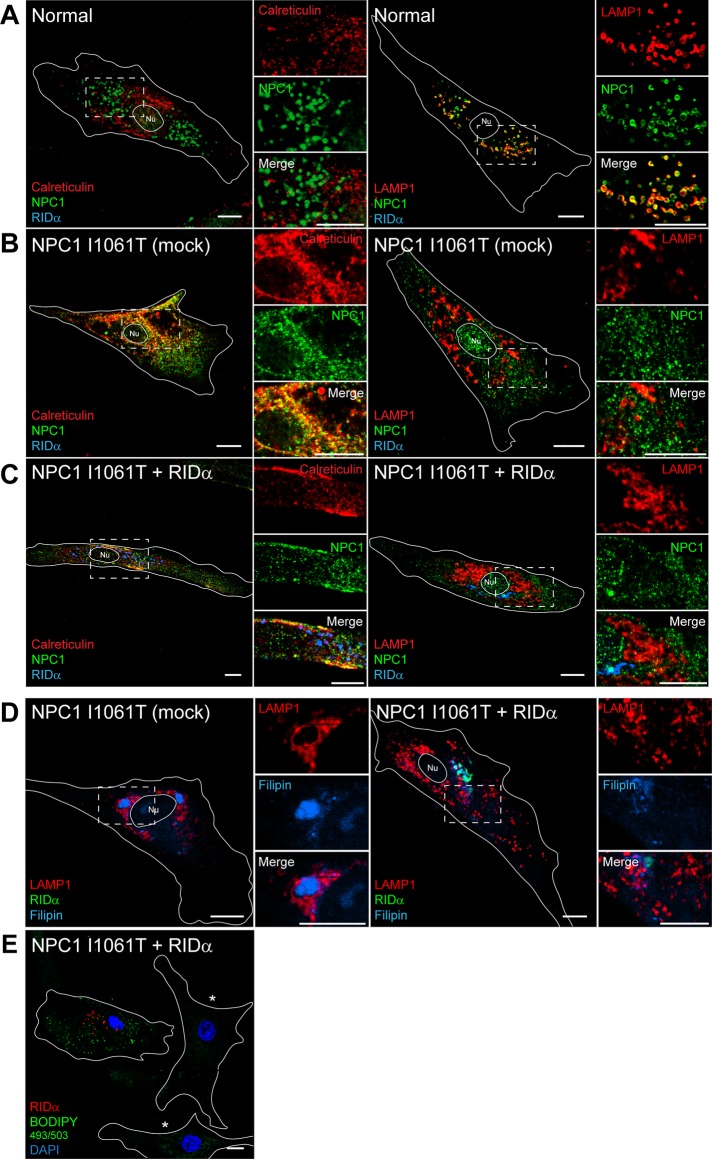
The NPC1 I1061T mutant can function in cholesterol egress but is retained in the ER and degraded (Gelsthorpe et al., 2008). Histone deacetylases (HDACs) are a diverse family of enzymes that are important mediators of chromatin remodeling and gene expression and also target nonhistone proteins for deacetylation (Glozak et al., 2005). Treatment of NPC1-mutant fibroblasts with HDAC inhibitor was shown to rescue the cholesterol storage phenotype (Munkacsi et al., 2011; Pipalia et al., 2011) by an unknown mechanism that involves increased expression of the NPC1 I1061T mutant protein and delivery to LE/Ly, where it can mediate cholesterol egress. Therefore one possible mechanism for RIDα NPC1 rescue is that RIDα promotes ER escape of NPC1 similar to HDAC inhibitor treatment. The NPC1 mutant fibroblasts used up to this point in this study are a compound heterozygote for the I1061T and P237S mutations in NPC1. To specifically test a role for RIDα in correcting the trafficking of the NPC1 I1061T mutant, we used homozygous NPC1 I1061T mutant fibroblasts and examined the colocalization of NPC1 with the ER marker calreticulin and the LE marker LAMP1 in cells transfected with FLAG-RIDα and loaded with LDL. Normal human fibroblasts exhibited colocalization of NPC1 with LAMP1 but not calreticulin (Figure 5A), demonstrating that wild-type NPC1 is efficiently trafficked out of the ER to its functional location on LAMP1-positive endosomes. In contrast, the NPC1 I1061T mutant protein exhibited significant overlap with calreticulin-positive ER and did not colocalize with LAMP1-positive LEs in mock-transfected homozygous NPC1 I1061T mutant fibroblasts (Figure 5B), confirming ER retention of the I1061T NPC1 mutant protein. However, RIDα did not correct the ER retention of the NPC1 I1061T mutant, as it remained colocalized with calreticulin but not LAMP1 (Figure 5C). FLAG-RIDα expression rescued the cholesterol storage phenotype in homozygous NPC1 I1061T mutant fibroblasts (Figure 5D) and induced formation of LDs upon LDL loading (Figure 5E). These data indicate that RIDα does not restore NPC1 function by promoting exit of the I1061T mutant protein from the ER.
FIGURE 5:
RIDα does not rescue ER retention of defective NPC1 I1061T mutant protein in patient fibroblasts. (A–C) Confocal images of normal (A), mock-transfected homozygous NPC1 I1061T mutant (B), or homozygous NPC1 I1061T mutant fibroblasts transfected with FLAG-RIDα (C) and stained with antibodies to NPC1 and FLAG-RIDα and costained with antibodies to calreticulin (left) or LAMP1 (right). (D, E) Confocal images of homozygous NPC1 I1061T mutant fibroblasts mock transfected (D, left) or transfected with FLAG-RIDα (D, right; E) and stained with antibodies to LAMP1 and FLAG-RIDα and with filipin (D) or with antibody to FLAG-RIDα and with BODIPY 493/503 and DAPI (E). Mock-transfected cells in E are shown in the same field and designated with an asterisk Boxed areas, regions of the image that were magnified. Bars, 10 μm. Nu, nucleus.
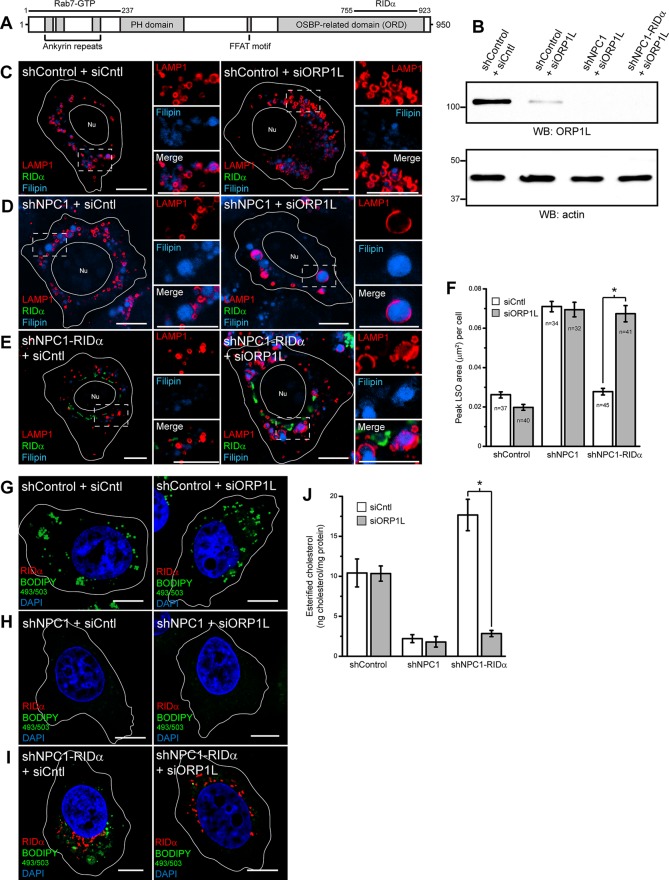
ORP1L is necessary for RIDα-induced formation of LDs
As previously noted, RIDα interacts with the Rab7 effector and cholesterol sensor ORP1L and colocalizes with ORP1L in a novel perinuclear compartment (Cianciola and Carlin, 2009). Whereas GTP-Rab7 interacts with the amino-terminal ankyrin repeats of ORP1L (Johansson et al., 2005), RIDα binds the carboxy-terminal oxysterol binding–related domain (ORD) involved in cholesterol and 25-hydroxycholesterol binding (Figure 6A; Shah et al., 2007; Suchanek et al., 2007). To test a role for ORP1L in RIDα-mediated increase in LD formation, shControl, shNPC1, and shNPC1-RIDα cells were treated with control small interfering RNA (siRNA) or ORP1L siRNA targeting the open reading frame (ORF), and equal aliquots of total cellular protein were immunoblotted for ORP1L and actin for loading control (Figure 6B). We routinely achieved >80% knockdown of ORP1L. Cells treated similarly to those in Figure 6B were loaded with LDL and then examined by confocal microscopy after cells were stained with antibodies to LAMP1 and FLAG-RIDα and with filipin. shControl cells treated with control siRNA displayed appropriate perinuclear LE staining with LAMP1 (Figure 6C). ORP1L knockdown induced LAMP1-positive LE scattering in shControl cells, as previously reported (Johansson et al., 2007). This was expected because ORP1L has an FFAT motif that directly binds the ER protein VAP, which regulates LE positioning (Rocha et al., 2009). However, ORP1L knockdown in shControl cells did not produce an LSO phenotype upon LDL addition (Figure 6C). shNPC1 cells displayed the NPC cholesterol storage phenotype in siCntl-treated cells, and knockdown of ORP1L did not exacerbate the phenotype (Figure 6D). shNPC1-RIDα cells showed the expected rescue of the LSO phenotype in siCntl-treated cells, and, in contrast, LSOs persisted after ORP1L knockdown (Figure 6E). Quantification of confocal images showed a statistically significant increase in peak LSO area in shNPC1-RIDα cells treated with siORP1L compared with control siRNA treatment (Figure 6F). Next we tested the role of ORP1L in LD formation in NPC1-deficient cells expressing FLAG-RIDα and loaded with LDL by BODIPY 493/503 staining of LDs. shControl cells treated with control siRNA displayed appropriate formation of LDs (Figure 6G and Supplemental Figure S2A) that were unaffected by ORP1L knockdown (Figure 6G and Supplemental Figure S2B), consistent with previous data demonstrating that ORP1L is not involved in LD formation (Jansen et al., 2011). shNPC1 cells exhibited a lack of LD staining with control siRNA treatment (Figure 6H and Supplemental Figure S2C), and ORP1L siRNA treatment had no effect on LD formation (Figure 6H and Supplemental Figure S2D). LD formation was induced in shNPC1-RIDα cells with control siRNA treatment (Figure 6I and Supplemental Figure S2E), but remarkably LD formation was severely limited in shNPC1-RIDα cells treated with siORP1L (Figure 6I and Supplemental Figure S2F). To confirm our results, we quantified the amount of esterified cholesterol in LDL-loaded shControl and shNPC1-RIDα cells treated with control and ORP1L siRNA using the Amplex Red cholesterol assay. ORP1L knockdown had no effect on cholesterol esterification in shControl cells, as expected, whereas siORP1L treatment significantly reduced cholesterol esterification in shNPC1-RIDα cells compared with siCntl treatment (Figure 6J).
FIGURE 6:
siRNA knockdown of ORP1L blocks RIDα rescue of cholesterol storage phenotype in NPC1-deficient cells. (A) Schematic representation of ORP1L. ORP1L has amino-terminal ankyrin repeats, pleckstrin homology domain (PH), FFAT motif, and carboxy-terminal oxysterol binding-related domain (ORD; Suchanek et al., 2007). GTP-Rab7 binding has been mapped to the ankyrin repeats (Johansson et al., 2005), and RIDα binds the ORD (Shah et al., 2007). (B) shControl cells were transfected with Cntl siRNA or shControl, shNPC1, or shNPC1-RIDα cells were transfected with ORP1L siRNA targeting the ORF, and equal aliquots of total cellular protein were immunoblotted with antibodies to ORP1L or actin for loading control. (C–E) Confocal images of shControl (C), shNPC1 (D), or shNPC1-RIDα cells (E) transfected with Cntl siRNA (left) or ORP1L siRNA targeting the ORF (right) and stained with antibodies to LAMP1 and FLAG-RIDα and with filipin. (F) Quantification of peak LSO area per cell in cells treated similarly to cells in C–E as described in Materials and Methods. Data are presented as mean ± SEM (*p < 0.001). (G–I) Confocal images of shControl (G), shNPC1 (H), or shNPC1-RIDα cells (I) transfected with Cntl siRNA (left) or ORP1L siRNA (right) and stained with antibody to FLAG-RIDα and with BODIPY 493/503 and DAPI. (J) Quantification of esterified cholesterol in shControl, shNPC1, and shNPC1-RIDα cells transfected with Cntl or ORP1L siRNA using the Amplex Red Cholesterol Assay kit as described in Materials and Methods. Values were normalized to total cellular protein and are displayed as mean ± SEM (*p < 0.001). Boxed areas, regions of the image that were magnified. Bars, 10 μm. Nu, nucleus.
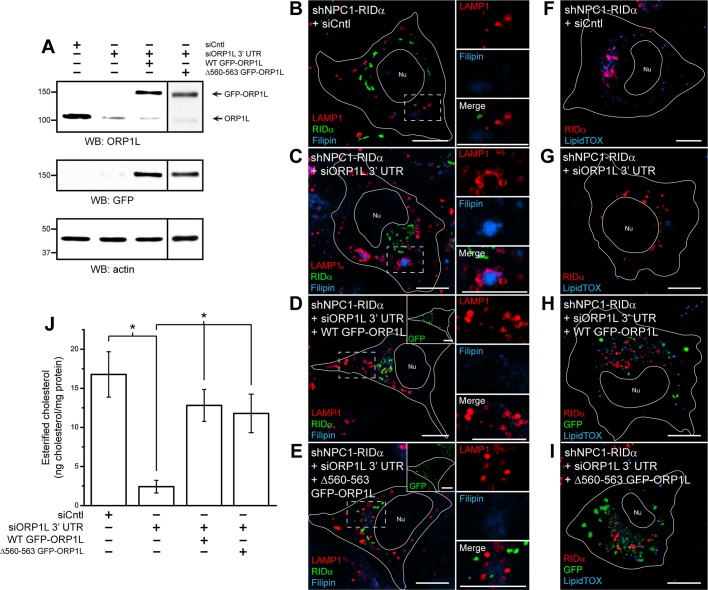
To further our understanding of the involvement of ORP1L in the ability of RIDα to rescue the NPC1 cholesterol storage phenotype, we developed a knockdown/rescue system in which ORP1L expression was silenced with siRNA directed against the 3′ untranslated region (UTR) and then rescued with green fluorescent protein (GFP)–ORP1L overexpression, which was resistant to knockdown. shNPC1-RIDα cells were transfected with Cntl siRNA or ORP1L siRNA targeting the 3′ UTR or cotransfected with ORP1L siRNA targeting the 3′ UTR along with wild-type GFP-ORP1L or a sterol binding mutant (Δ560–563 GFP-ORP1L; Vihervaara et al., 2011). Equal aliquots of total cellular protein were then immunoblotted for ORP1L and GFP, with actin for loading control (Figure 7A). We routinely achieved ∼90% knockdown of ORP1L, whereas GFP-ORP1L expression was unaffected. shNPC1-RIDα cells treated similarly to those in Figure 7A were loaded with LDL and then examined by confocal microscopy after cells were stained with antibodies to LAMP1 and FLAG-RIDα and with filipin. shNPC1-RIDα cells treated with siCntl showed the expected rescue of the LSO phenotype (Figure 7B), and knockdown of ORP1L again caused LSOs to persist (Figure 7C). However, shNPC1-RIDα cells transfected with siORP1L targeting the 3′ UTR and overexpressing wild-type GFP-ORP1L displayed a lack of enlarged LAMP1/filipin-positive LEs indicative of LSOs upon LDL loading (Figure 7D). Of interest, rescue of ORP1L expression with the sterol binding mutant Δ560–563 GFP-ORP1L in shNPC1-RIDα cells caused a reduction in the size of LAMP1/filipin-positive LEs consistent with RIDα NPC1 rescue (Figure 7E), as well as with the hypothesis that RIDα binding the ORP1L ORD transforms its ability to sense cholesterol (Shah et al., 2007). Next we used the ORP1L knockdown/rescue technique to study LD formation in shNPC1-RIDα and loaded with LDL by LipidTOX deep red staining of LDs. LipidTOX deep red neutral lipid stain was used because the BODIPY 493/503 emission overlapped with that of GFP, and LipidTOX deep red colocalizes exclusively with BODIPY 493/503 (Klapper et al., 2011). LD formation was induced in shNPC1-RIDα cells treated with control siRNA (Figure 7F), and LD formation was again inhibited when cells were treated with ORP1L siRNA targeting the 3′ UTR (Figure 7G). LD formation was induced in shNPC1-RIDα cells cotransfected with siORP1L targeting the 3′ UTR and wild-type GFP-ORP1L (Figure 7H), as well as in Δ560–563 GFP-ORP1L–transfected cells (Figure 7I). We then used the Amplex Red cholesterol assay to quantify cholesterol esters in shNPC1-RIDα cells with knockdown/rescue of ORP1L expression. Knockdown of ORP1L targeting the 3′ UTR caused a statistically significant decrease in the formation of esterified cholesterol compared with control siRNA treatment, and rescue of ORP1L expression with wild-type or Δ560–563 GFP-ORP1L restored the ability of RIDα to induce an increase in esterified cholesterol (Figure 7J). Collectively these data reveal that RIDα NPC1 rescue depends on ORP1L but is independent of its ability to bind cholesterol and suggest for the first time a possible role for ORP1L in transport of cholesterol to the ER and subsequent LD formation in the absence of functional NPC1.
FIGURE 7:
Overexpression of ORP1L after siRNA knockdown restores the ability of RIDα to rescue the cholesterol storage phenotype in NPC1-deficient cells. (A) shNPC1-RIDα cells were transfected with Cntl siRNA or ORP1L siRNA targeting the 3′ UTR or cotransfected with ORP1L siRNA targeting the 3′ UTR along with wild-type or Δ560–563 GFP-ORP1L, and equal aliquots of total cellular protein were immunoblotted with antibodies to ORP1L, GFP, or actin for loading control. (B–E) Confocal images of shNPC1-RIDα cells transfected with Cntl siRNA (B), ORP1L siRNA targeting the 3′ UTR (C), or cotransfected with ORP1L siRNA targeting the 3′ UTR and wild-type (D) or Δ560–563 GFP-ORP1L (E) and stained with antibodies to LAMP1 and FLAG-RIDα and with filipin. (F–I) Confocal images of shNPC1-RIDα cells transfected with Cntl siRNA (F), ORP1L siRNA targeting the 3′ UTR (G), or cotransfected with ORP1L siRNA targeting the 3′ UTR and wild-type (H) or Δ560–563 GFP-ORP1L (I) and stained with antibody to FLAG-RIDα and with LipidTOX deep red. (J) Quantification of esterified cholesterol in shNPC1-RIDα cells transfected with Cntl siRNA or ORP1L siRNA targeting the 3′ UTR or cotransfected with ORP1L siRNA targeting the 3′ UTR along with wild-type or Δ560–563 GFP-ORP1L using the Amplex Red Cholesterol Assay kit as described in Materials and Methods. Values were normalized to total cellular protein and are displayed as mean ± SEM (*p < 0.001). Boxed areas, regions of the image that were magnified. Bars, 10 μm. Nu, nucleus.
Ad-expressed RIDα induces formation of LDs independent of NPC1
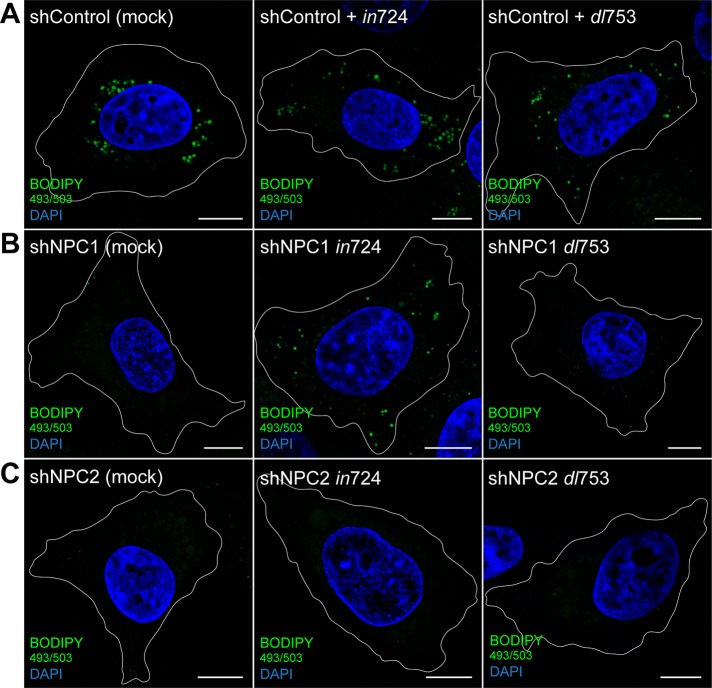
Our studies reveal a role for RIDα in the formation of LDs independent of NPC1. To address physiological relevance, we next sought to determine whether RIDα plays a similar role in LD formation during infection with group C Ad serotype 2. We first verified that NPC1 and NPC2 are not required for Ad infection. This was determined by showing that shControl, shNPC1, and shNPC2 cells infected with an Ad that overexpresses RIDα (in724) all produced similar levels of early region 1A (E1A), the first virally encoded protein synthesized in acute infections (Supplemental Figure S3; Nevins et al., 1979). Infected cells were then stained for LDs with BODIPY 493/503. Mock-infected shControl cells displayed appropriate formation of LDs when loaded with LDL (Figure 8A), and infection of shControl cells with RIDα-overexpressing in724 Ad had no effect on LD formation (Figure 8A). Mock-infected shNPC1 cells displayed a lack of LDs when loaded with LDL (Figure 8B); however, shNPC1 cells infected with the RIDα-overexpressing in724 virus displayed a marked increase in LD formation with LDL load (Figure 8B). Mock-infected shNPC2 cells also displayed a lack of LDs upon LDL loading (Figure 8C). However, similar to ectopic expression of tagged RIDα in NPC2-mutant fibroblasts, infection of shNPC2 cells with RIDα-overexpressing in724 Ad did not induce formation of LDs with LDL load (Figure 8C). Next we infected cells with a RIDα-null Ad (dl753) to show that RIDα is necessary for LD formation. Similar to infection with in724, shControl cells infected with RIDα-null dl753 Ad displayed appropriate formation of LDs upon LDL loading (Figure 8A). As expected, infection of shNPC1 cells with the RIDα-null dl753 Ad did not induce formation of LDs (Figure 8B), demonstrating that RIDα is the viral protein mediating alterations to the LD phenotype. Finally, infection of shNPC2 cells with the RIDα-null dl753 Ad had no effect on LD formation (Figure 8C). Together these data demonstrate that RIDα induces LD formation by an NPC1-independent mechanism during an acute Ad infection as well when expressed ectopically as a tagged transgene.
FIGURE 8:
RIDα induces formation of LDs during Ad infection of NPC1-deficient cells. (A– C) Confocal images of shControl (A), shNPC1 (B), or shNPC2 cells (C) mock infected or infected with RIDα-overexpressing in724 (middle) or RIDα-null dl753 (right) Ad2. Cells were stained with BODIPY 493/503 and DAPI. Bars, 10 μm.
DISCUSSION
Cholesterol homeostasis is precisely regulated by free cholesterol levels in the ER, although relatively little is known about the mechanism of cholesterol transport to the ER. In this study, we demonstrate that RIDα regulates delivery of LDL-cholesterol to the ER by a mechanism independent of NPC1 but dependent on NPC2 and ORP1L. RIDα induces formation of LDs in NPC1-deficient cells, and inhibition of ACAT blocks the ability of RIDα to rescue the NPC1 cholesterol storage phenotype. Of interest, RIDα had no effect on the expression of the SREBP- or LXR-regulated genes that we tested. Therefore our data indicate that RIDα mediates transport of LDL-cholesterol specifically to an ER pool of cholesterol that is accessible to ACAT for esterification and storage in LDs. This strongly supports the concept of distinct ER cholesterol pools capable of regulating different aspects of cholesterol homeostasis (Kristiana et al., 2008). Despite the failure to restore SREBP feedback control, the storage of cholesterol esters in LDs of RIDα-expressing NPC1-deficient cells may be less toxic than the accumulation of free cholesterol in LSOs (Abi-Mosleh et al., 2009). Therefore up-regulation of a physiological pathway that has been coopted by RIDα and induces LD formation may constitute a significant therapeutic advance for NPC1 treatment.
We showed that RIDα rescues the cholesterol storage phenotype in three different NPC1-deficient cell models, further supporting that RIDα function is independent of NPC1 (Cianciola and Carlin, 2009). However, RIDα function depends on NPC2, as RIDα is unable to rescue the cholesterol storage phenotype of NPC2-mutant fibroblasts. Of interest, RIDα colocalized with LBPA in NPC1-mutant fibroblasts but not in NPC2-mutant fibroblasts, indicating that LBPA plays an important role in RIDα function. LBPA is a critical regulator of cholesterol trafficking (Chevallier et al., 2008) and enhances the transfer of cholesterol between membranes in the presence of NPC2 (Xu et al., 2008). NPC1 and NPC2 cooperate to transport cholesterol out of LEs in vivo, although in vitro data indicate that NPC2 can mediate incorporation of cholesterol into donor membranes independent of NPC1 (Cheruku et al., 2006; Xu et al., 2008). Recently it was shown that cholesterol transport to mitochondria depends on NPC2 but not NPC1, suggesting that NPC2 alone can transfer cholesterol from the lumen into the LE-limiting membrane in vivo (Kennedy et al., 2012). Our results support this model by which NPC2 delivers cholesterol to the endosome-limiting membrane, from which RIDα and ORP1L can mediate its transport to the ER. This also implies that NPC1 has functions independent of NPC2, potentially in the generation of LE–ER membrane contact sites mediated by interaction with the ER-resident protein ORP5 (Du et al., 2011; Du and Yang, 2013).
Rescue of the cholesterol storage phenotype and induction of LD formation in NCP1-deficient cells by RIDα was also found to depend on ORP1L. As previously stated, ORP1L is an effector protein of GTP-Rab7 and a member of the family of oxysterol-binding proteins, which have been linked to many aspects of lipid homeostasis (Olkkonen et al., 2012). Of interest, two members of that family that lack membrane localization motifs, ORP1S and ORP2, were directly implicated in the transport of cholesterol to the ER and into LDs (Jansen et al., 2011). We previously showed that RIDα interacts with ORP1L in vitro (Shah et al., 2007) and colocalizes with ORP1L in FLAG-RIDα–expressing cells (Cianciola and Carlin, 2009). Therefore it became an attractive candidate to test its role in RIDα function. siRNA-mediated knockdown of ORP1L in NPC1-deficient cells completely blocked the ability of RIDα to rescue the LSO phenotype and induce formation of LDs. To counteract any potential off-target effects of a single siRNA construct directed against the ORF of ORP1L, we developed a knockdown/rescue technique using siRNA targeting the 3′ UTR of ORP1L along with expression of wild-type GFP-ORP1L, which was resistant to gene silencing. Using this technique, we were able to demonstrate that restoration of ORP1L expression supported the ability of RIDα to rescue the LSO phenotype and induce formation of LDs in NPC1-deficient cells. Of interest, expression of an ORP1L mutant defective in sterol binding (Δ560–563) also supported RIDα NPC1 rescue, demonstrating that RIDα function is independent of cholesterol sensing/binding by ORP1L. Collectively these data suggest for the first time that ORP1L is capable of playing a role in the transport of cholesterol from endosomes to the ER and LDs in cells lacking functional NPC1 in the presence of RIDα.
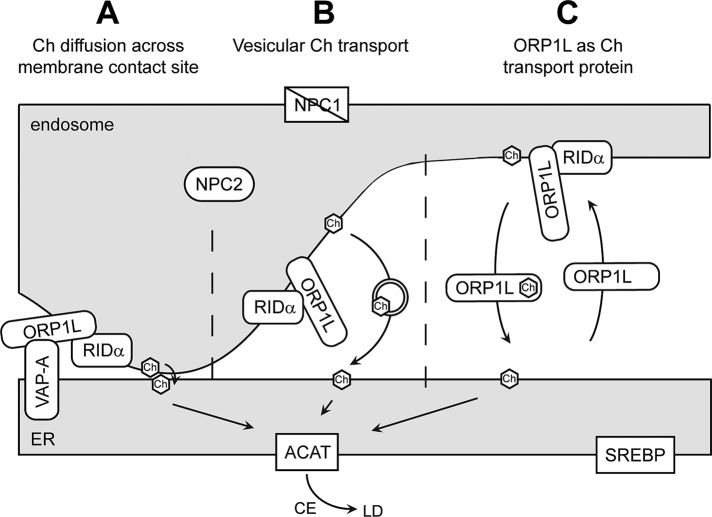
ORP1L binds cholesterol, and this interaction has a profound effect on its conformation downstream of GTP-Rab7 (Rocha et al., 2009). High LE cholesterol levels occlude the ORP1L FFAT motif involved in VAP-A binding, whereas this motif is exposed when LE cholesterol levels are low. In this way, cholesterol levels and ORP1L in association with Rab7 regulate the positioning of LEs. Because RIDα interacts with ORP1L in its cholesterol binding ORD (Shah et al., 2007), we hypothesize that RIDα may modulate the effect of cholesterol on ORP1L conformation. Thus, ORP1L may be able to promote VAP-A interactions in the presence of high cholesterol while bound to RIDα. We previously showed that RIDα does not localize to LAMP1-positive LEs but promotes formation of a hybrid intermediate endosome compartment positive for ORP1L via an unconventional autophagy-based pathway (Cianciola and Carlin, 2009). We hypothesize that RIDα binding to ORP1L allows for exposure of the FFAT motif responsible for binding to VAP-A on the ER surface independent of cholesterol levels. The ORP1L/VAP-A interaction would create endosome–ER contact sites that allow for cholesterol to be transported to the ER by way of passive diffusion down a concentration gradient between closely apposed membranes (Figure 9A). Although we favor this mechanism, the possibility exists that ORP1L may facilitate ER cholesterol trafficking through vesicular transport (Figure 9B) or by acting as a cholesterol transport protein (Figure 9C). However, the notion that ORP1L acts as a cholesterol transport protein is not supported in the literature, and our data indicate that cholesterol binding by ORP1L is unessential for RIDα NPC1 rescue. The yeast oxysterol-binding protein Kes1/Osh4p binds dehydroergosterol and phosphatidylinositol 4-phosphate in a hydrophobic pocket for transport of these lipids between membranes along opposite routes in vitro (de Saint-Jean et al., 2011). However, reports have also demonstrated that Osh proteins do not play a role in the transport of sterol from PM to ER and LDs in living cells (Georgiev et al., 2011) and that a Kes1/Osh4p sterol-binding mutant is a gain-of-function mutation that inhibits cell growth (Alfaro et al., 2011; Mousley et al., 2012). We propose that RIDα reengineers ORP1L to act as a bridge linking cholesterol-loaded LEs with VAP-A–positive ER by transforming its cholesterol-sensing capacity in NPC1-deficient cells. Alternatively, ORP1L may have additional functions aside from mediating endosome–ER membrane contact sites with VAP-A that have yet to be identified. Further experiments are underway to investigate the role of ORP1L in ER cholesterol transport during RIDα NPC1 rescue.
FIGURE 9:
Model depicting hypothetical mechanisms of RIDα/ORP1L-mediated transport of cholesterol from endosome to ER in NPC1-deficient cells. Cholesterol may be trafficked from endosomes to the ER by vesicular or nonvesicular mechanisms, including membrane contact sites, where close apposition of donor and acceptor membranes supports transport down a concentration gradient (A), vesicular transport, where cholesterol is packaged in transport vesicles to be delivered to the ER (B), or by transport proteins that bind cholesterol in a hydrophobic pocket protected from the cytosol and transport it to the ER (C). RIDα mediates the transport of cholesterol to the ER independent of NPC1 but dependent on NPC2 and ORP1L. Once delivered, cholesterol is acted upon by ACAT for incorporation into LDs and does not affect SREBP-regulated gene expression. CE, cholesterol ester; Ch, cholesterol.
The host–pathogen interaction is a powerful model system that provides insight into fundamental cell biological processes. Ad has already provided such insight, as the Ad type I transmembrane protein E3/19K was used to identify a consensus amino acid motif responsible for ER retention (Jackson et al., 1990). In this study we focused on the function of RIDα from the group C Ad serotype 2. Ad2 is a nonenveloped, double-stranded DNA virus taken up by clathrin-mediated endocytosis after its binding to CAR and integrin coreceptors (Meier and Greber, 2003). Ad2 escapes from endosomes undergoing early-to-late endosome maturation by a process known as Rab conversion, by which the small GTPase Rab5 associated with early endosomes is replaced by Rab7 necessary for LE function (Rink et al., 2005; Zeng and Carlin, 2013). Ad2 then attaches to dynein molecular motors and undergoes microtubule-dependent transport to the nucleus, where the virus replicates (Kellar et al., 2004). We showed previously that cells infected with a RIDα-null virus are essentially devoid of normal early and late endosomes but accumulate markers for these organelles in aberrant compartments that are also enriched for free cholesterol and LBPA (Cianciola and Carlin, 2009). Escape of Ad2 from endosomes may disrupt feedback control circuits that couple endocytic flux with Rab conversion. It is also possible Ad2 has a direct role in inhibiting Rab conversion similar to polyoma virus, which sequesters the Vps39 component of the Rab5–Rab7 exchange machinery in the nucleus (Feng et al., 2011). RIDα expression appears to restore endocytic function by increasing free cholesterol egress capacity to the ER, where it is converted to cholesterol esters and stored as LDs. One possibility is that this activity may preserve endosome integrity and function by preventing accumulation of free cholesterol, which impairs endocytic flux by inhibiting Rab7 and other small GTPases (Lebrand et al., 2002; Ganley and Pfeffer, 2006). A functional endocytic apparatus is necessary to protect infected cells from a number of apoptosis-inducing cytokines (TRAIL, TNF, and Fas ligand) by clearing their receptors from the cell surface (Shisler et al., 1997; Shah et al., 2007). Alternatively, LDs are emerging as important targets in a number of diverse host–pathogen interactions. Thus LDs may be a source of energy during Ad replication, provide a lipid metabolite required for assembly of infectious Ad particles, or enable an anti-immunity strategy (Saka and Valdivia, 2012). Finally Ad36, which has been linked to morbid obesity, triggers formation of LDs by a mechanism involving the Ad36 early region 4 open reading frame (E4orf; Esposito et al., 2012). In contrast to Ad2 E4orf, Ad36 E4orf transcriptionally up-regulates a number of genes involved in LD biogenesis (Wang et al., 2010). E3 genes often entail serotype-specific sequence variations implicated in Ad-induced pathology (Fessler et al., 2004). It will be interesting to determine whether RIDα encoded by Ad36 contributes to the lipid storage phenotype by disrupting LD biogenesis or turnover.
To summarize, these studies describe the first virally encoded protein directly involved in cholesterol trafficking and LD formation in mammalian cells. Because intracellular pathogens often take on features of their hosts to gain evolutionary advantage, these data indicate an unexpected role for LD in the Ad life cycle. Similar to diverse intracellular pathogens, RIDα undoubtedly subverts a previously unrecognized host cholesterol homeostasis regulatory process involving ORP1L. Understanding how RIDα regulates intracellular cholesterol trafficking and metabolism will identify new molecular targets and improve treatment options for patients with NPC and other cholesterol storage diseases.
MATERIALS AND METHODS
Antibodies and reagents
The following antibodies were used: actin mouse monoclonal antibody (Sigma, St. Louis, MO); calreticulin chicken polyclonal antibody (Abcam, Cambridge, MA); E1A mouse monoclonal antibody (BD Biosciences, San Jose, CA); FLAG-M2 mouse monoclonal antibody and FLAG rat monoclonal antibody (Sigma); GFP rabbit polyclonal antibody (Abcam); hamster- and human-specific LAMP1 mouse monoclonal antibodies (Developmental Studies Hybridoma Bank, Iowa City, IA); LBPA mouse monoclonal antibody (Echelon Biosciences, Salt Lake City, UT); NPC1 rabbit polyclonal antibody (Novus Biologicals, Littleton, CO); ORP1L rabbit monoclonal antibody (Abcam); and RIDα rabbit polyclonal antibody produced as described in Hoffman et al. (1992). Fluorescent- and horseradish peroxidase–tagged secondary antibodies were from Jackson ImmunoResearch (West Grove, PA); filipin and S58–035 were from Sigma; 4′,6-diamidino-2-phenylindole (DAPI), BODIPY 493/503, and LipidTOX deep red neutral lipid stain were from Invitrogen (Carlsbad, CA).
Cell culture and Ad stocks
To create a stable population of CT43 or shNPC1 cells expressing FLAG-RIDα (referred to as CT43-RIDα, and shNPC1-RIDα, respectively), we transfected GP2-293 retrovirus packaging cells stably expressing FLAG-RIDα with pVSV-G plasmid using Trans-IT 293 transfection reagent (Mirus Bio, Madison, WI). Pantropic retrovirus-containing medium was collected 48 h later and added to CT43 or shNPC1 cells, and cells were selected for drug resistance in media supplemented with 200 μg/ml G418 and maintained in media supplemented with 50 μg/ml G418. Stable expression of RIDα was verified by immunoblotting and immunostaining with FLAG and RIDα antibodies.
Chinese hamster ovary cells were grown in MEMα. CT43 (kindly provided by T. Y. Chang, Dartmouth, Hanover, NH) and CT43-RIDα cells were grown in Ham's F12 medium; human normal (GM05659), NPC1 (GM03123), homozygous NPC1 I1061T mutant (kindly provided by D. Ory, Washington University in St. Louis, St. Louis, MO), and NPC2 (GM18455) patient fibroblasts were grown in MEM; IHH cells stably expressing control, NPC1, or NPC2 shRNA (shControl, shNPC1, and shNPC2 respectively; kindly provided by D. Manor, Case Western Reserve University, Cleveland, OH; Ulatowski et al., 2011) and shNPC1-RIDα cells were grown in DMEM supplemented with 10 μg/ml puromycin; and GP2–293 cells were grown in DMEM. Delipidated fetal bovine serum (FBS) was prepared using Cab-O-Sil (Sigma) as described (Lala et al., 1997; Haas et al., 2007). Unless otherwise noted, cells were incubated with appropriate media plus 5% delipidated FBS for 24 h, followed by delipidated FBS media supplemented with 20 μg/ml (S58-035 treated cells) or 50 μg/ml LDL for 24 h. CT43 cells were lipid starved in Ham's F12 medium plus 5% delipidated FBS, 1.5 mM CaCl2, and 35 μM oleic acid.
Ad stocks were grown in HEK293 cells, and titers were determined by plaque assay using standard techniques. Cells were acutely infected with ∼100 plaque-forming units/cell and analyzed after 24 h (Hoffman and Carlin, 1994). Two adenovirus mutants were used in this study: in724 is a splice-donor insertion mutant that overexpresses the RIDα protein (Brady and Wold, 1988), and dl753 has an internal deletion of 207 nucleotides in the RIDα ORF and does not produce RIDα (Brady and Wold, 1987).
LDL preparation
LDL was isolated from fresh human plasma (obtained from the Cleveland Clinic Blood Bank, Cleveland, OH) by sequential ultracentrifugation (Havel et al., 1955) and extensively dialyzed against 0.9% NaCl and 0.02% EDTA, pH 7.4.
LDL labeling and cholesterol transport assay
Starting with fresh human plasma, LDL was labeled with [1,2-3H]cholesteryl palmitate (50 Ci/mmol; American Radiolabeled Chemicals, St. Louis, MO) by a lipid dispersion technique (Morton and Zilversmit, 1981) to yield 14,750 dpm 3H/μg protein. shControl, shNPC1, and shNPC1-RIDα cells were incubated with media plus 5% delipidated FBS for 24 h, followed by incubation with 50 μg/ml [3H]cholesteryl palmitate–labeled LDL in the same media along with 250 μM oleate-BSA for 24 h. Cells were collected and suspended in 1 ml of phosphate-buffered saline (PBS), lipids were extracted using standard techniques (Thompson et al., 1971), and the lipid-containing hexane phase was dried under N2. Lipids were fractionated on 5% silver nitrate–impregnated TLC plates in a developing system of hexanes:diethyl ether (97:3 vol/vol), and bands were identified by comigration with authentic standards (cholesterol, cholesteryl oleate, and cholesteryl palmitate). Bands were scraped, and 3H content was determined by scintillation counting. Protein concentration was determined by the method of Peterson (1977).
Confocal microscopy
Normal and NPC1- and NPC2-mutant fibroblasts were mock treated or transfected with 3 μg of FLAG-RIDα plasmid DNA (Cianciola et al., 2007) using the Normal Human Dermal Fibroblast Nucleofector kit and Nucleofector device (Lonza, Walkersville, MD). For ACAT inhibitor studies, cells were incubated with 20 μg/ml LDL, as well as 10 μg/ml S58-035, for 12 h before staining. Cells were seeded on poly-l-lysine–treated coverslips, perforated with 0.5% β-escin, fixed with 3% paraformaldehyde, incubated with primary antibodies for 1 h at room temperature and fluorescent-tagged secondary antibodies for 16 h at 4°C, and mounted on glass slides using SlowFade (Invitrogen). Fixed cells were incubated with 50 μg/ml filipin to detect free cholesterol, 10 μg/ml BODIPY 493/503 to detect LDs, and DAPI to detect nuclei for 16 h at 4°C. Confocal images were acquired with a laser-scanning microscope (LSM 510 Meta; Carl Zeiss, Jena, Germany) and accompanying software using diode (excitation 405 nm), argon (excitation 488 nm), and HeNe (excitation 543 and 633) lasers and 63× or 100× Plan-Apochromat numerical aperture 1.4 objectives. Filipin was excited with the diode laser, and emissions were collected between 411 and 486 nm. When necessary, brightfield images were collected, and nuclei and cell boundaries were drawn using Illustrator and overlaid onto respective confocal images. All images were processed with Photoshop CS5 and Illustrator CS5 software (Adobe).
Cell harvesting and immunoblotting
Cells were washed with PBS and lysed in radioimmunoprecipitation assay buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM ethylene glycol tetraacetic acid, 5 mM EDTA, 1% NP-40, 0.5% Na deoxycholate, and 0.1% SDS). Equal protein (determined by Bradford assay [Bio-Rad, Hercules, CA]) was resolved by SDS–PAGE and transferred to nitrocellulose for immunoblotting using standard methods. Signals were quantified using ImageJ processing software (National Institutes of Health, Bethesda, MD).
LD quantification
Confocal stacks of BODIPY 493/503–labeled cells were created by collecting images every 0.2 μm through the entire cell, and projections were created using the accompanying software. LDs were counted for each cell and data presented as mean LD number per cell ± SEM from three independent experiments. From the same projections, total LD area was quantified using MetaMorph software. Total cell area was also quantified from brightfield images using MetaMorph software. Total LD area was calculated by dividing total LD area by the total cell area to normalize the data. Data are presented as mean total LD area per cell ± SEM from three independent experiments.
Peak LSO area quantification
Confocal images were collected, and the peak area of LAMP1/filipin-positive LSOs was quantified using MetaMorph software. Brightfield images were also collected, and total cell area was quantified using MetaMorph software. Peak LSO area per cell was calculated by dividing the peak LSO area by the total cell area to normalize the data. Data are presented as mean peak LSO area per cell ± SEM from three independent experiments.
Quantification of esterified cholesterol
Chinese hamster ovary, CT43, and CT43-RIDα cells, or shControl, shNPC1, and shNPC1-RIDα cells transfected with control or ORP1L siRNA duplexes, were treated with 50 μg/ml LDL as described, washed twice with PBS, scraped in 600 μl of PBS, and homogenized by 10 passages through a 22-gauge needle. Aliquots were analyzed for total (plus cholesterol esterase) and free (minus cholesterol esterase) cholesterol using the Amplex Red Cholesterol Assay kit (Invitrogen) according to the manufacturer's instructions, and fluorescence was read at 590 nm using a Tecan GENios Pro plate reader and xFluor4 GENios Pro software. Esterified cholesterol was derived by subtracting free cholesterol from total cholesterol. Esterified cholesterol content was normalized to total protein as determined by Bradford assay after addition of 10× radioimmunoprecipitation assay buffer to 1× the remaining cell homogenate. Data are represented as mean nanograms esterified cholesterol per milligram of protein ± SEM from three independent experiments.
Gene expression experiments
Chinese hamster ovary, CT43, and CT43-RIDα cells, or shControl, shNPC1, and shNPC1-RIDα cells, were cultured in media supplemented in 10% FBS for 48 h, media supplemented with 5% delipidated FBS for 48 h, or media supplemented with 5% delipidated FBS for 24 h followed by the same media plus 50 μg/ml LDL for 24 h. RNA extracted using the ToTALLY RNA kit (Applied Biosystems, Carlsbad, CA) was reverse transcribed using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). Target-gene mRNA levels were analyzed by real-time PCR using standard hydrolysis probe (TaqMan) techniques and a glyceraldehyde 3-phosphate dehydrogenase internal control. mRNA expression levels were plotted as fold change relative to mRNA levels for control cells cultured in standard media containing 10% FBS for each individual cell line. PCR was performed on a real-time PCR system (model 7500; Applied Biosystems), and products were analyzed using 7500 System SDS software, version 1.3 (Applied Biosystems). Probe and primer pairs listed in Supplemental Table S1 were designed using the Universal Probe Library Assay Design Center (Roche, Indianapolis, IN). Probes and PCR master mix were from Roche, and primers were from Operon.
ORP1L knockdown and rescue
For ORP1L-knockdown experiments, siRNA duplexes were transfected using Oligofectamine transfection reagent (Invitrogen) and used for experiments 2 d later. siRNA for ORP1L targeting the ORF (UGCCAGUGCCGGAUUCUGAdTdT; Johansson et al., 2007) and AllStars Negative Control siRNA were from Qiagen (Valencia, CA). For ORP1L knockdown and rescue experiments, siRNA duplexes were cotransfected with wild-type or Δ560–563 GFP-ORP1L (kindly provided by V. M. Olkkonen, Minerva Foundation Institute for Medical Research, Helsinki, Finland) using Oligofectamine reagent and used for experiments 2 d later. siRNA for ORP1L targeting the 3′ UTR (UAGUUCCAUAAAGCUAAAGdTdT) was from Sigma.
Statistical analysis
Statistical analyses for Amplex Red cholesterol quantification, LD quantification, and LSO quantification were performed using the Student's t test (SigmaStat). Data are represented as mean ± SEM from three independent experiments. p < 0.001 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Maryanne Pendergast and the Neurosciences Imaging Center for assistance with confocal microscopy, T. Y. Chang for CT43 cells, Lynn Ulatowski and Danny Manor for IHH cells and critical review of the manuscript, D. Ory for homozygous NPC1 I1061T mutant fibroblasts, V. M. Olkkonen for GFP-ORP1L expression plasmids, and all laboratory members for their support and discussions. This work was supported by Public Health Service RO1 Grant GM081498 to C.R.C. N.L.C. was supported in part by a Peter G. Pentchev Research Fellowship from the National Niemann–Pick Disease Foundation.
Abbreviations used:
- ABCA1
ATP-binding cassette transporter A1
- ACAT
acyl-CoA:cholesterol acyltransferase
- Ad
adenovirus
- HDAC
histone deacetylase
- HMGCR
3-hydroxy-3-methylglutaryl-CoA reductase
- IHH
immortalized human hepatocyte
- LBPA
lysobisphosphatidic acid
- LD
lipid droplet
- LDLR
low-density-lipoprotein receptor
- LE
late endosome
- LSO
lysosomal storage organelle
- Ly
lysosome
- NPC
Niemann–Pick disease type C
- ORD
oxysterol binding–related domain
- ORF
open reading frame
- ORP1L
oxysterol-binding protein–related protein 1L
- RILP
Rab7-interacting lysosomal protein
- S58-035
Sandoz 58-035
- SREBP
sterol regulatory element–binding protein
- UTR
untranslated region
- VAP
vesicle-associated membrane protein–associated endoplasmic reticulum protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-10-0760) on September 11, 2013.
REFERENCES
- Abi-Mosleh L, Infante RE, Radhakrishnan A, Goldstein JL, Brown MS. Cyclodextrin overcomes deficient lysosome-to-endoplasmic reticulum transport of cholesterol in Niemann-Pick type C cells. Proc Natl Acad Sci USA. 2009;106:19316–19321. doi: 10.1073/pnas.0910916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro G, Johansen J, Dighe SA, Duamel G, Kozminski KG, Beh CT. The sterol-binding protein Kes1/Osh4p is a regulator of polarized exocytosis. Traffic. 2011;12:1521–1536. doi: 10.1111/j.1600-0854.2011.01265.x. [DOI] [PubMed] [Google Scholar]
- Brady HA, Wold WSM. Identification of a novel sequence that governs both polyadenylation and alternative splicing in region E3 of adenovirus. Nucleic Acids Res. 1987;15:9397–9416. doi: 10.1093/nar/15.22.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady HA, Wold WS. Competition between splicing and polyadenylation reactions determines which adenovirus region E3 mRNAs are synthesized. Mol Cell Biol. 1988;8:3291–3297. doi: 10.1128/mcb.8.8.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers DM, Rastogi SR, Cook HW, Palmer FB, Spence MW. Defective activity of acyl-CoA:cholesterol O-acyltransferase in Niemann-Pick type C and type D fibroblasts. Biochem J. 1989;262:713–719. doi: 10.1042/bj2620713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TY, Reid PC, Sugii S, Ohgami N, Cruz JC, Chang CC. Niemann-Pick type C disease and intracellular cholesterol trafficking. J Biol Chem. 2005;280:20917–20920. doi: 10.1074/jbc.R400040200. [DOI] [PubMed] [Google Scholar]
- Charman M, Kennedy BE, Osborne N, Karten B. MLN64 mediates egress of cholesterol from endosomes to mitochondria in the absence of functional Niemann-Pick Type C1 protein. J Lipid Res. 2010;51:1023–1034. doi: 10.1194/jlr.M002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheruku SR, Xu Z, Dutia R, Lobel P, Storch J. Mechanism of cholesterol transfer from the Niemann-Pick Type C2 protein to model membranes supports a role in lysosomal cholesterol transport. J Biol Chem. 2006;281:31594–31604. doi: 10.1074/jbc.M602765200. [DOI] [PubMed] [Google Scholar]
- Chevallier J, Chamoun Z, Jiang G, Prestwich G, Sakai N, Matile S, Parton RG, Gruenberg J. Lysobisphosphatidic acid controls endosomal cholesterol levels. J Biol Chem. 2008;283:27871–27880. doi: 10.1074/jbc.M801463200. [DOI] [PubMed] [Google Scholar]
- Choi HY, Karten B, Chan T, Vance JE, Greer WL, Heidenreich RA, Garver WS, Francis GA. Impaired ABCA1-dependent lipid efflux and hypoalphalipoproteinemia in human Niemann-Pick type C disease. J Biol Chem. 2003;278:32569–32577. doi: 10.1074/jbc.M304553200. [DOI] [PubMed] [Google Scholar]
- Cianciola NL, Carlin CR. Adenovirus RID-α activates an autonomous cholesterol regulatory mechanism that rescues defects linked to Niemann-Pick disease type C J. Cell Biol. 2009;187:537–552. doi: 10.1083/jcb.200903039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciola NL, Carlin CR, Kelley TJ. Molecular pathways for intracellular cholesterol accumulation: common pathogenic mechanisms in Niemann–Pick disease Type C and cystic fibrosis. Arch Biochem Biophys. 2011;515:54–63. doi: 10.1016/j.abb.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciola NL, Crooks D, Shah AH, Carlin C. A tyrosine-based signal plays a critical role in the targeting and function of adenovirus RIDa protein. J Virol. 2007;81:10437–10450. doi: 10.1128/JVI.00399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JC, Sugii S, Yu C, Chang TY. Role of Niemann-Pick type C1 protein in intracellular trafficking of low density lipoprotein-derived cholesterol. J Biol Chem. 2000;275:4013–4021. doi: 10.1074/jbc.275.6.4013. [DOI] [PubMed] [Google Scholar]
- Deffieu MS, Pfeffer SR. Niemann–Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc Natl Acad Sci USA. 2011;108:18932–18936. doi: 10.1073/pnas.1110439108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol. 2011;195:965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Kumar J, Ferguson C, Schulz TA, Ong YS, Hong W, Prinz WA, Parton RG, Brown AJ, Yang H. A role for oxysterol-binding protein–related protein 5 in endosomal cholesterol trafficking. J Cell Biol. 2011;192:121–135. doi: 10.1083/jcb.201004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Yang H. Endosomal cholesterol trafficking: protein factors at a glance. Acta Biochim Biophys Sin. 2013;45:11–17. doi: 10.1093/abbs/gms095. [DOI] [PubMed] [Google Scholar]
- Esposito S, Preti V, Consolo S, Nazzari E, Principi N. Adenovirus 36 infection and obesity. J Clin Virol. 2012;55:95–100. doi: 10.1016/j.jcv.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Feng H, Kwun HJ, Liu X, Gjoerup O, Stolz DB, Chang Y, Moore PS. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One. 2011;6:e22468. doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessler S, Delgado-Lopez F, Horwitz M. Mechanisms of E3 modulation of immune and inflammatory responses. Curr Top Microbiol Immunol. 2004;273:113–135. doi: 10.1007/978-3-662-05599-1_4. [DOI] [PubMed] [Google Scholar]
- Frolov A, Zielinski SE, Crowley JR, Dudley-Rucker N, Schaffer JE, Ory DS. NPC1 and NPC2 regulate cellular cholesterol homeostasis through generation of low density lipoprotein cholesterol-derived oxysterols. J Biol Chem. 2003;278:25517–25525. doi: 10.1074/jbc.M302588200. [DOI] [PubMed] [Google Scholar]
- Ganley IG, Pfeffer SR. Cholesterol accumulation sequesters Rab9 and disrupts late endosome function in NPC1-deficient cells. J Biol Chem. 2006;281:17890–17899. doi: 10.1074/jbc.M601679200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsthorpe ME, Baumann N, Millard E, Gale SE, Langmade SJ, Schaffer JE, Ory DS. Niemann-Pick Type C1 I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J Biol Chem. 2008;283:8229–8236. doi: 10.1074/jbc.M708735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev AG, Sullivan DP, Kersting MC, Dittman JS, Beh CT, Menon AK. Osh proteins regulate membrane sterol organization but are not required for sterol movement between the ER and PM. Traffic. 2011;12:1341–1355. doi: 10.1111/j.1600-0854.2011.01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Haas D, et al. Abnormal serol metabolism in holoprosencephaly: studies in cultured lymphoblasts. J Med Genet. 2007;44:298–305. doi: 10.1136/jmg.2006.047258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havel RJ, Eder HA, Bragdon JH. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J Clin Invest. 1955;34:1345–1353. doi: 10.1172/JCI103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P, Carlin C. Adenovirus E3 protein causes constitutively internalized EGF receptors to accumulate in a prelysosomal compartment, resulting in enhanced degradation. Mol Cell Biol. 1994;14:3695–3706. doi: 10.1128/mcb.14.6.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P, Yaffe MB, Hoffman BL, Yei S, Wold WSM, Carlin C. Characterization of the adenovirus E3 protein that down-regulates the epidermal growth factor receptor. J Biol Chem. 1992;267:13480–13487. [PubMed] [Google Scholar]
- Jackson MR, Nilsson T, Peterson PA. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Ohsaki Y, Rita Rega L, Bittman R, Olkkonen VM, Ikonen E. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic. 2011;12:218–231. doi: 10.1111/j.1600-0854.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- Johansson M, Lehto M, Tanhuanpaa K, Cover TL, Olkkonen VM. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol Biol Cell. 2005;16:5480–5492. doi: 10.1091/mbc.E05-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuiji C, Olkkonen VM, Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7–RILP–p150Glued, ORP1L, and the receptor βlll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellar SA, Pfister KK, Crystal RG, Leopold PL. Cytoplasmic dynein mediates adenovirus binding to microtubules. J Virol. 2004;78:10122–10132. doi: 10.1128/JVI.78.18.10122-10132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BE, Charman M, Karten B. Niemann-Pick type C2 protein contributes to the transport of endosomal cholesterol to mitochondria without interacting with NPC1. J Lipid Res. 2012;53:2632–2642. doi: 10.1194/jlr.M029942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapper M, Ehmke M, Palgunow D, Böhme M, Matthäus C, Bergner G, Dietzek B, Popp J, Döring F. Fluorescence-based fixative and vital staining of lipid droplets in Caenorhabditis elegans reveal fat stores using microscopy and flow cytometry approaches. J Lipid Res. 2011;52:1281–1293. doi: 10.1194/jlr.D011940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiana I, Yang H, Brown AJ. Different kinetics of cholesterol delivery to components of the cholesterol homeostatic machinery: implications for cholesterol trafficking to the endoplasmic reticulum. Biochim Biophys Acta. 2008;1781:724–730. doi: 10.1016/j.bbalip.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Lala D, Syke P, Lazarchik S, Mangelsdorf D, Parker K, Heyman R. Activation of the orphan nuclear receptor steroidogenic factor 1 by oxysterols. Proc Natl Acad Sci USA. 1997;94:4895–4900. doi: 10.1073/pnas.94.10.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange Y, Ye J, Steck TL. How cholesterol homeostasis is regulated by plasma membrane cholesterol in excess of phospholipids. Proc Natl Acad Sci USA. 2004;101:11664–11667. doi: 10.1073/pnas.0404766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrand C, Corti M, Goodson H, Cosson P, Cavalli V, Mayran N, Faure J, Gruenberg J. Late endosome motility depends on lipids via the small GTPase Rab7. EMBO J. 2002;21:1289–1300. doi: 10.1093/emboj/21.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- Meier O, Greber UF. Adenovirus endocytosis. J Gene Med. 2003;5:451–462. doi: 10.1002/jgm.409. [DOI] [PubMed] [Google Scholar]
- Morton RE, Zilversmit DB. A plasma inhibitor of triglyceride and cholesteryl ester transfer activities. J Biol Chem. 1981;256:11992–11995. [PubMed] [Google Scholar]
- Mousley CJ, et al. A sterol-binding protein integrates endosomal lipid metabolism with TOR signaling and nitrogen sensing. Cell. 2012;148:702–715. doi: 10.1016/j.cell.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkacsi AB, et al. An “exacerbate-reverse” strategy in yeast identifies histone deacetylase inhibition as a correction for cholesterol and sphingolipid transport defects in human Niemann-Pick type C disease. J Biol Chem. 2011;286:23842–23851. doi: 10.1074/jbc.M111.227645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR, Ginsberg HS, Blanchard J-M, Wilson MC, Darnell JE. Regulation of the primary expression of the early adenovirus transcription units. J Virol. 1979;32:727–733. doi: 10.1128/jvi.32.3.727-733.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkonen VM, Hynynen R. Interactions of oxysterols with membrane proteins. Mol Aspects Med. 2009;30:1233–1233. doi: 10.1016/j.mam.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Olkkonen VM, Zhou Y, Yan D, Vihervaara T. Oxysterol-binding proteins—emerging roles in cell regulation. Eur J Lipid Sci Technol. 2012;114:634–643. [Google Scholar]
- Ory DS. Nuclear receptor signaling in the control of cholesterol homeostasis: have the orphans found a home. Circ Res. 2004;95:660–670. doi: 10.1161/01.RES.0000143422.83209.be. [DOI] [PubMed] [Google Scholar]
- Patterson MC, Vanier MT, Suzuki K, Morris JA, Carstea E, Neufeld EB, Blanchette-Mackie JE, Pentchev PG. Niemann Pick disease type C: a lipid trafficking disorder. In: Scriver CR, editor. In: The Online Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. pp. 3611–3633. [Google Scholar]
- Peake KB, Vance JE. Defective cholesterol trafficking in Niemann-Pick C-deficient cells. FEBS Lett. 2010;584:2731–2739. doi: 10.1016/j.febslet.2010.04.047. [DOI] [PubMed] [Google Scholar]
- Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Pipalia NH, Cosner CC, Huang A, Chatterjee A, Bourbon P, Farley N, Helquist P, Wiest O, Maxfield FR. Histone deacetylase inhibitor treatment dramatically reduces cholesterol accumulation in Niemann-Pick type C1 mutant human fibroblasts. Proc Natl Acad Sci USA. 2011;108:5620–5625. doi: 10.1073/pnas.1014890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc Natl Acad Sci USA. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7–RILP–p150Glued and late endosome positioning. J Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AC, Go KJ, Heider JG, Rothblat GH. Selective inhibition of acyl coenzyme A:cholesterol acyltransferase by compound 58–035. J Biol Chem. 1984;259:815–819. [PubMed] [Google Scholar]
- Russell DW. Oxysterol biosynthetic enzymes. Biochim Biophys Acta. 2000;1529:126–135. doi: 10.1016/s1388-1981(00)00142-6. [DOI] [PubMed] [Google Scholar]
- Saka HA, Valdivia R. Emerging roles for lipid droplets in immunity and host-pathogen interactions. Annu Rev Cell Dev Biol. 2012;28:411–437. doi: 10.1146/annurev-cellbio-092910-153958. [DOI] [PubMed] [Google Scholar]
- Schulz TA, Choi M-G, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, Prinz WA. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol. 2009;187:889–903. doi: 10.1083/jcb.200905007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo T, Oelkers PM, Giattina MR, Worgall TS, Sturley SL, Deckelbaum RJ. Differential modulation of ACAT1 and ACAT2 transcription and activity by long chain free fatty acids in cultured cells. Biochemistry. 2001;40:4756–4762. doi: 10.1021/bi0022947. [DOI] [PubMed] [Google Scholar]
- Shah AH, Cianciola NL, Mills JL, Sonnichsen FD, Carlin C. Adenovirus RIDa regulates endosome maturation by mimicking GTP-Rab7. J Cell Biol. 2007;179:965–980. doi: 10.1083/jcb.200702187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisler J, Yang C, Walter B, Ware CF, Gooding LR. The adenovirus E3-10.4/14.5 complex mediates loss of cell surface Fas (CD95) and resistance to Fas-induced apoptosis. J Virol. 1997;71:8299–8306. doi: 10.1128/jvi.71.11.8299-8306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Ikonen E. How cells handle cholesterol. Science. 2000;290:1721–1726. doi: 10.1126/science.290.5497.1721. [DOI] [PubMed] [Google Scholar]
- Steck TL, Lange Y. Cell cholesterol homeostasis: mediation by active cholesterol. Trends Cell Biol. 2010;20:680–687. doi: 10.1016/j.tcb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss K, Goebel C, Runz H, Möbius W, Weiss S, Feussner I, Simons M, Schneider A. Exosome secretion ameliorates lysosomal storage of cholesterol in Niemann-Pick Type C Disease. J Biol Chem. 2010;285:26279–26288. doi: 10.1074/jbc.M110.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchanek M, Hynynen R, Wohlfahrt G, Lehto M, Johansson M, Saarinen H, Radzikowska A, Thiele C, Olkkonen VM. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem J. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LP, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci USA. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele C, Spandl J. Cell biology of lipid droplets. Curr Opin Cell Biol. 2008;20:378–385. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Thompson JN, Erdody P, Brien R, Murray TK. Fluorometric determination of vitamin A in human blood and liver. Biochem Med. 1971;5:67–89. doi: 10.1016/0006-2944(71)90076-7. [DOI] [PubMed] [Google Scholar]
- Ulatowski L, et al. Altered vitamin E status in Niemann-Pick type C disease. J Lipid Res. 2011;52:1400–1410. doi: 10.1194/jlr.M015560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara T, Uronen R-L, Wohlfahrt G, Bjorkhem I, Ikonen E, Olkkonen VM. Sterol binding by OSBP-related protein 1L regulates late endosome motility and function. Cell Mol Life Sci. 2011;68:537–551. doi: 10.1007/s00018-010-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Yu Y, Zhang XH, Floyd EZ, Cefalu WT. Human adenovirus 36 decreases fatty acid oxidation and increases de novo lipogenesis in primary cultured human skeletal muscle cells by promoting Cidec/FSP27 expression. Int J Obes. 2010;34:1355–1364. doi: 10.1038/ijo.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Farver W, Kodukula S, Storch J. Regulation of sterol transport between membranes and NPC2. Biochemistry. 2008;47:11134–11143. doi: 10.1021/bi801328u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehmer JK, Huang Y, Peng G, Pu J, Anderson RGW, Liu P. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9:914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Carlin CR. Host cell autophagy modulates early stages of adenovirus infections in airway epithelial cells. J Virol. 2013;87:2307–2319. doi: 10.1128/JVI.02014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.