Abstract
The coordination of chromatin remodeling with chromatin modification is a central topic in gene regulation. The yeast chromatin remodeling complex RSC bears multiple bromodomains, motifs for acetyl-lysine and histone tail interaction. Here, we identify and characterize Rsc4 and show that it bears tandem essential bromodomains. Conditional rsc4 bromodomain mutations were isolated, and were lethal in combination with gcn5Δ, whereas combinations with esa1 grew well. Replacements involving Lys14 of histone H3 (the main target of Gcn5), but not other H3 or H4 lysine residues, also conferred severe growth defects to rsc4 mutant strains. Importantly, wild-type Rsc4 bound an H3 tail peptide acetylated at Lys14, whereas a bromodomain mutant derivative did not. Loss of particular histone deacetylases suppressed rsc4 bromodomain mutations, suggesting that Rsc4 promotes gene activation. Furthermore, rsc4 mutants displayed defects in the activation of genes involved in nicotinic acid biosynthesis, cell wall integrity, and other pathways. Taken together, Rsc4 bears essential tandem bromodomains that rely on H3 Lys14 acetylation to assist RSC complex for gene activation.
Keywords: bromodomain, chromatin remodeling, histone acetylation, RSC
Introduction
Chromatin structural changes play central roles in controlling gene expression. Activation is often associated with the mobilization and modification of nucleosomes, the basic repeating unit of chromatin (Vignali et al, 2000). Nucleosomes are covalently modified by histone acetyltransferases (HATs), deacetylases (HDACs), and methyltransferases (HMTs), which target specific lysine residues in the histone subunits (Narlikar et al, 2002). Histone acetylation is generally associated with transcription activation, while deacetylation generally correlates with repression (Grunstein, 1997). Histone modifications help recruit additional factors including chromatin remodeling complexes, which utilize the energy of ATP to mobilize nucleosomes and render chromatin accessible to transcription factors (Havas et al, 2001). Two such remodeling complexes in Saccharomyces cerevisiae are the SWI/SNF complex and the highly related and essential RSC (Remodels the Structure of Chromatin) complex, both of which have close counterparts in higher eucaryotes.
One conserved motif found in all SWI/SNF family of remodelers is the bromodomain, a 110-amino-acid domain also found in HATs, TFIID components, and other remodeler complexes. In vitro, bromodomains bind to the amino-terminal tails of histones H3 and H4 (Ornaghi et al, 1999; Ladurner et al, 2003; Matangkasombut and Buratowski, 2003), and acetylation of lysine residues on these tails improves binding (Dhalluin et al, 1999; Hudson et al, 2000; Jacobson et al, 2000; Owen et al, 2000; Ladurner et al, 2003; Matangkasombut and Buratowski, 2003). These observations suggest that acetylated lysines on histone tails provide a platform for the recruitment of bromodomain-containing transcriptional regulators (Jenuwein and Allis, 2001; Hassan et al, 2002).
RSC subunits contain eight of the 15 bromodomains in yeast, suggesting that the recognition of chromatin modifications is an important aspect of RSC function. Characterized subunits include Sth1, the catalytic ATPase subunit of RSC (Du et al, 1998), and the double bromodomain proteins Rsc1 and Rsc2 (Cairns et al, 1999). Rsc1 and Rsc2 are similar in domain structure (multiple bromodomains, BAH domain, HMG domain) to the polybromo/BAF180 subunit of human hSWI/SNF-B (PBAF) complex, which functions as a cofactor for ligand-activated transcription on a chromatin template in vitro (Lemon et al, 2001). Taken together, bromodomain proteins are emerging as important regulators of chromatin remodeling and modifying complexes, but much remains to be learned about their binding determinants and their utilization in transcriptional regulation. Of particular interest is whether the tandem arrangement of bromodomains in certain proteins might enable combinatorial recognition of histone/factor modifications. Here, we identify and characterize the tandem double bromodomain protein Rsc4, and develop its connections to histone tail recognition, acetylation, and gene expression.
Results
Rsc4 identification and structure
RSC was purified to homogeneity from yeast cellular extracts as described previously (Cairns et al, 1996; Figure 1A), and peptides from Rsc4 were isolated and analyzed by MALDI-TOF mass spectrometry. Mass fingerprinting from Rsc4 uniquely identified the open reading frame (ORF) YKR008W. This result supports recent proteomic approaches for identifying protein complexes, which found Ykr008w in association with many other proteins, among them certain RSC components (Gavin et al, 2002; Sanders et al, 2002). Sequence comparisons using the CLUSTALW algorithm (Higgins et al, 1996) reveal two bromodomains (BD1 and BD2) separated by 36 amino acids (aa) (Figure 1B). This proximity raises the possibility that the two bromodomains pack against each other in a manner similar to the double bromodomains of TAF1 (Jacobson et al, 2000). RSC4 is an essential gene, as dissection of a sporulated RSC4/rsc4Δ heterozygous diploid yielded only two viable spores, and as a rsc4Δ strain was unable to lose a URA3-marked plasmid bearing wild-type (WT) RSC4 on medium containing 5-FOA (Figure 2B and data not shown), which prevents the growth of URA3+ strains.
Figure 1.
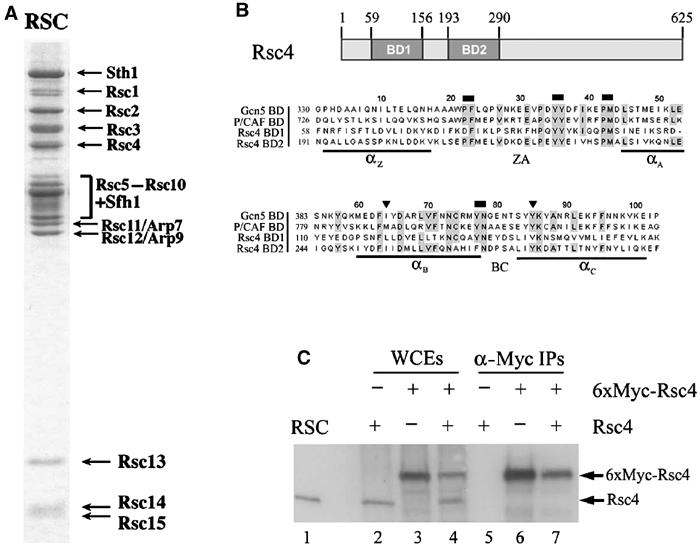
Identification of Rsc4. (A) Purified RSC complex stained with Coomassie dye (from Cairns et al, 1996). (B) Domain structure of Rsc4 and alignment of Rsc4 bromodomains with the bromodomains of yeast Gcn5 and P/CAF. Regions of identity are highlighted in gray. Triangles (▾) mark residues mutated in rsc4-2. Rectangles (⁃) mark paired residues mutated by site directed mutagenesis. (C) Stoichiometry of Rsc4 in RSC. Western analysis using anti-Rsc4 antiserum of whole-cell extracts (WCEs) and anti-Myc precipitations from three strains bearing different tagged RSC4 alleles. Extracts were prepared from YBC627 transformed with either 6xMyc-Rsc4 (p603) (lanes 3, 4, 6, and 7) or the empty vector (p415.MET25, p520) (lanes 2 and 5). Cells used in lanes 3 and 6 lost the untagged Rsc4 plasmid on SC+5-FOA.
Figure 2.
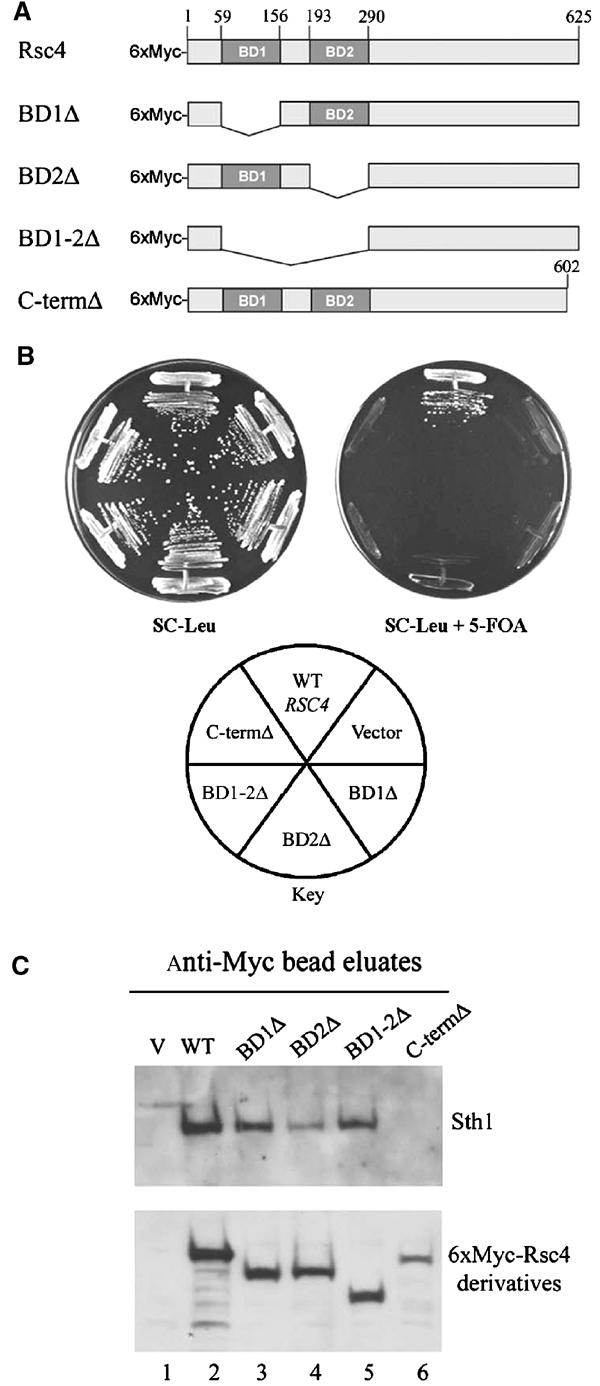
Deletion analysis of Rsc4 domains. (A) Diagram of Rsc4 bromodomain and C-terminal deletion constructs. (B) Rsc4 function requires both bromodomains and the C-terminus. Strains were incubated at 28°C for 2 days. Strains (YBC627) bearing pRS316.RSC4 and a LEU2-marked rsc4 derivative or control (pRS415MET25, 6xMyc.RSC4 (p603), 6xMyc.rsc4 BD1Δ (p804), 6xMyc.rsc4 BD2Δ (p1172), 6xMyc.rsc4 BD1-2Δ (p805), or 6xMyc.rsc4 C-termΔ (p809)) were grown on selective media with or without 5-FOA to enforce the loss of pRS316.RSC4. (C) Rsc4 mutant assembly into RSC. Extracts were prepared from strains used in (B) grown in the presence of pRS316.RSC4. Immune complexes were formed with anti-Myc antibody bound to beads, washed, eluted, immunoblotted, and probed with anti-Sth1 or anti-Myc antiserum.
Immunoprecipitation of 6xMyc-Rsc4 efficiently co-precipitates Sth1, the catalytic subunit of RSC, and this interaction is stable at high stringency (Figure 2C, lane 2, and data not shown), verifying the association of Rsc4 with RSC (and not just fortuitous co-purification). To determine the oligomeric state of Rsc4 in the RSC complex, we tested for co-immunoprecipitation of WT Rsc4 with 6xMyc-Rsc4 utilizing a polyclonal antibody that we raised against purified recombinant full-length Rsc4. This 6xMyc-Rsc4 derivative fully complements rsc4Δ (Figure 2B and data not shown). Immunoprecipitation of 6xMyc-Rsc4 with the anti-Myc antibody does not co-precipitate untagged Rsc4 (Figure 1C, lane 7) suggesting one copy of Rsc4 per RSC complex.
Both Rsc4 bromodomains are required for viability, and the C-terminus is essential for assembly into RSC
For structure–function analysis, we prepared plasmids encoding Rsc4 derivatives lacking either BD1, BD2, both BD1 and BD2, or a small portion (23 aa) of the C-terminus, each tagged with six copies of the myc epitope (Figure 2A). Derivatives were tested for complementation of a rsc4Δ mutation by assessing their ability to support growth following the loss of a WT RSC4-URA3 plasmid on medium containing 5-FOA. Whereas full-length 6xMyc-Rsc4 complements rsc4Δ, derivatives lacking BD1, BD2, or the C-terminus fail to complement, demonstrating that these domains are essential for Rsc4 function (Figure 2B). Derivative assembly into RSC complex was determined by co-precipitation of Sth1, using strains bearing a WT untagged copy of RSC4 to support viability. Whereas bromodomain deletion derivatives co-precipitated Sth1, the C-terminal truncation did not (Figure 2C). Taken together, the Rsc4 C-terminus mediates assembly into the RSC complex whereas the bromodomains perform an alternative essential function.
Site-directed mutagenesis of Rsc4 bromodomains
To better understand the role of the two bromodomains in Rsc4 function, site-directed mutations (SDMs) were isolated based on mutagenesis and structural studies of the Gcn5 and P/CAF bromodomains (Dhalluin et al, 1999; Owen et al, 2000; Mujtaba et al, 2002). The bromodomain utilizes a left-handed four-helix bundle (with helices termed Z, A, B, and C) to form an architectural platform for two helix-connecting loop regions (ZA and BC). These loops form a significant portion of the binding pocket for acetyl-lysine recognition, and also recognize residues flanking the acetyl-lysine, which make important contributions to binding specificity. Residues in the ZA loop are predicted to make essential acetyl-lysine contacts (Y364 of Gcn5 and Y760 of P/CAF) (Dhalluin et al, 1999; Owen et al, 2000; Mujtaba et al, 2002). The BC loop residue N407 of Gcn5 (N134 in BD1 and N268 in BD2 of Rsc4) coordinates a network of water-mediated hydrogen bonds, or ‘water ring,' with a carbonyl group on the acetyl and is also critical for acetyl-lysine binding (Owen et al, 2000). In addition, the conserved adjacent aromatic residue (Y406 of Gcn5 and Y802 of P/CAF) interacts directly with residues flanking the acetyl-lysine, and is important for tail recognition (Dhalluin et al, 1999; Owen et al, 2000; Mujtaba et al, 2002).
Converting the conserved Y92 and Y93 residues (ZA loop) to alanines in BD1 did not affect Rsc4 function, nor did independently changing the analogous Y225 and Y226 residues in BD2 (Table I, SDM1 and SDM2; and see Figure 1B). However, combining these mutations in BD1 and BD2 conferred lethality (Table I, SDM3) without significantly affecting protein stability or assembly into RSC (see Supplementary Figure 1). Thus, the bromodomains appear partially redundant; a reduction in the function of one bromodomain can make viability reliant on full function of the other tandem bromodomain partner. Conversion of the conserved Y133 and N134 residues to alanines in BD1 conferred slow growth at 38°C (Table I, SDM4; and Figure 1B), whereas the analogous F267 N268 alanine substitutions (in BD2) conferred no phenotype (Table I, SDM5). Thus, the two bromodomains do not strictly rely on identical residue positions for substrate recognition. However, we again observe greater than additive defects in combination (Table I, SDM6). Similar relationships are observed with replacements at other positions (Table I, SDM7-11). Taken together, our targeted amino-acid replacements identified residues important for Rsc4 bromodomain function and provide evidence for partial redundancy, although the complete loss of either bromodomain confers lethality.
Table 1.
Impact of site-directed mutations on RSC4 function
| Phenotype | |||||
|---|---|---|---|---|---|
| Mutant | BD1 mutation | BD2 mutation | 30°C | 35°C | 38°C |
| SDM1 | Y92A, Y93A | – | +++ | +++ | +++ |
| SDM2 | – | Y225A, Y226A | +++ | +++ | +++ |
| SDM3 | Y92A, Y93A | Y225A, Y226A | Inviable | ||
| SDM4 | Y133A, N134A | – | ++ | ++ | +/− |
| SDM5 | – | F267A, N268A | +++ | +++ | +++ |
| SDM6 | Y133A, N134A | F267A, N268A | + | + | − |
| SDM7 | P99A, M100H | – | +++ | +++ | ++ |
| SDM8 | – | P232A, M233H | +++ | +++ | +++ |
| SDM9 | – | P212A, F213A | +++ | +++ | +++ |
| SDM10 | P99A, M100H | P232A, M233H | +++ | +++ | +++ |
| SDM11 | P99A, M100H | P212A, F213A | Inviable | ||
| Growth was assessed from 10-fold dilutions spotted to plates after 3 days at the indicated temperature and compared to WT (defined as +++). ‘++' indicates slightly reduced viability (fewer colonies) but near WT colony size. ‘+' indicates moderately reduced viability and smaller colony size. ‘+/−' indicates severely reduced viability and extremely small colony size. ‘−' indicates no growth. | |||||
Isolation and characterization of conditional mutations in Rsc4 bromodomains
For functional and suppression analysis, we sought temperature-sensitive (Ts−) rsc4 bromodomain alleles (rsc4 BDTs−) with ideal properties: alleles that display WT growth at 28°C and inviability at 38°C, encode stable proteins that assemble well into RSC at the nonpermissive temperature, and encode single amino-acid substitutions in each of the bromodomains. To acquire such alleles, we targeted random mutations to the region of RSC4 encoding both bromodomains using a plasmid shuffle strategy (see Materials and methods). The screen yielded 10 Ts− mutants (Figure 3A).
Figure 3.
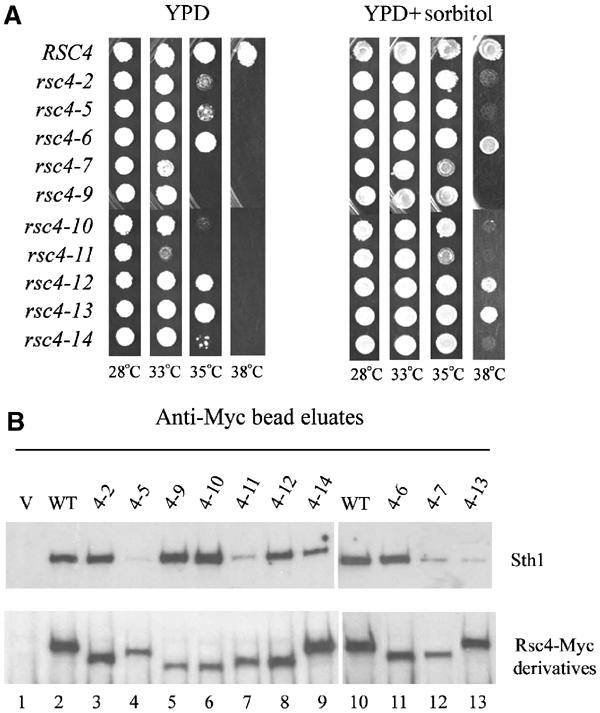
Conditional mutations in RSC4 bromodomains. (A) Growth of rsc4 BDTs− mutants. Strains are transformants of YBC627 where pRS316.RSC4 is exchanged for rsc4 BDTs− plasmids: pRS314.RSC4 (p1060), pRS314.rsc4-2 (p1083), pRS314.rsc4-5 (p1084), pRS314.rsc4-6 (p1085), pRS314.rsc4-7 (p1086), pRS314.rsc4-9 (p1087), pRS314.rsc4-10 (p1088), pRS314.rsc4-11 (p1089), pRS314.rsc4-12 (p1090), pRS314.rsc4-13 (p1091), pRS314.rsc4-14 (p1092). (B) Assembly of rsc4 BDTs− mutants into RSC. Extracts were prepared from strains transformed with plasmids bearing rsc4 BDTs− alleles fused to Myc epitopes grown in the presence of pRS316.RSC4 at 35°C. Immune complexes were formed with anti-Myc antibody bound to beads, washed, eluted, and immunoblotted with anti-Sth1 or anti-Myc antiserum. The migration of the Myc-Rsc4 derivatives deviates from WT due to deletion of Myc epitopes by homologous recombination.
The majority of the alleles obtained bore mutations in both bromodomains, again supporting the notion of partial redundancy, whereas the few alleles obtained bearing a mutation in a single bromodomain typically resulted in lower protein stability or assembly (see below and Figure 3B). However, as alleles bearing multiple mutations were not separated, their individual contributions have not been determined. We proceeded with three alleles: rsc4-2, rsc4-7, and rsc4-11. rsc4-2 has two mutations, one in each bromodomain (L120P, Y275H; Figure 1B), and satisfies all the criteria listed above. Therefore, rsc4-2 represents the most informative and useful allele identified in our random screen, and all further studies focused on this allele; rsc4-7 and rsc4-11 were chosen for comparative purposes as they are stronger Ts− alleles (see Materials and methods for all alleles/replacements). Most rsc4 BDTs− alleles encoded stable Rsc4 derivatives, although their mobility varied due to deletion of one or more of the six Myc epitopes by spurious homologous recombination (Figure 3B and data not shown). The rsc4 BDTs− derivatives varied in their expression levels and competency for assembly, as determined by co-precipitation of Sth1 (Figure 3B). We note that reducing the abundance of Rsc4 protein 10-fold below WT levels (through repression using a MET25 promoter) has no phenotypic consequence (data not shown).
Certain RSC mutants show a cell wall defect at elevated temperatures that can be suppressed by osmotic stabilizers (Angus-Hill et al, 2001). Similarly, we found that growth on 1 M sorbitol, 200 mM CaCl2, or 100 mM MgCl2 partially suppressed the temperature-sensitive phenotype of the rsc4 BDTs− mutants (Figure 3A and data not shown), supporting a role for Rsc4 in assisting RSC in maintaining cell wall integrity. Certain rsc mutants (such as rsc3 or sth1) accumulate in G2/M at the nonpermissive temperature (Tsuchiya et al, 1998; Angus-Hill et al, 2001) and may be linked to defects in centromeric chromatin (Hsu et al, 2003), whereas other rsc mutants lack this property. We observed little or no cell cycle bias with rsc4 mutants (data not shown), suggesting that Rsc4 function is not linked to centromere function.
Genetic cooperativity between Rsc4 bromodomains and the Gcn5 histone H3 acetyltransferase
As bromodomains recognize acetylated histone tails, we tested for genetic interactions between rsc4 bromodomain mutations and mutations in the HAT complexes SAGA/ADA and NuA4. Gcn5 is the catalytic subunit of the SAGA and ADA complexes and primarily acetylates histone H3 at lysine 14 (Grant et al, 1998). Esa1 is the catalytic subunit of the NuA4 complex and primarily acetylates histone H4 at lysines 5, 8, 12, and 16 (Allard et al, 1999). ESA1 is an essential gene, and the esa1-L327S allele is Ts− and displays a modest reduction in H4 acetylation levels genome wide (Clarke et al, 1999), but shows significant reductions at certain genes (Suka et al, 2002). esa1-L254P is a stronger Ts− allele and displays a greater reduction in H4 acetylation (Clarke et al, 1999).
Interestingly, rsc4 BDTs− mutations were lethal in combination with a gcn5 deletion at 28°C (Figure 4A), suggesting that Rsc4 function is highly sensitive to the acetylation status of histone H3. In striking contrast, combinations involving the esa1-L327S and esa1-L254P alleles showed no growth defects at 28°C (Figure 4B and data not shown). A slight lowering (by 2°C) of the nonpermissive temperature was observed with rsc4-2 esa1-L327S combinations (Figure 4C); however, this only occurs over a very narrow temperature range (32–34°C). Taken together, rsc4 BDTs− mutants are entirely reliant on proper H3 tail acetylation, whereas they display only partial and conditional reliance on proper H4 acetylation.
Figure 4.
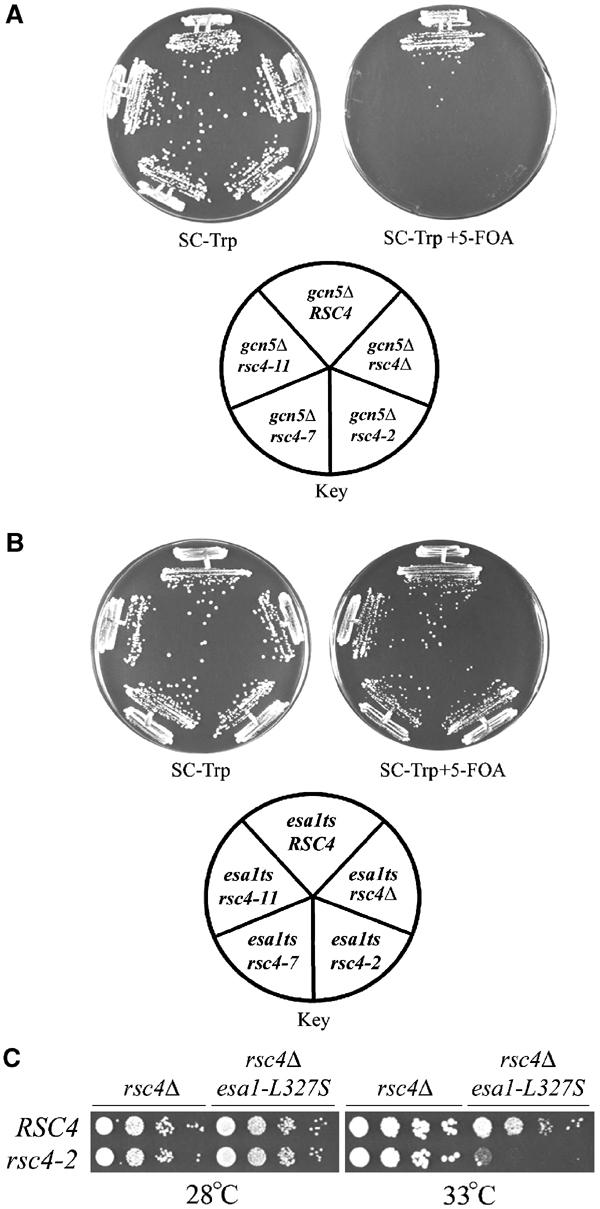
Genetic interactions between Rsc4 bromodomains and HATs. (A) rsc4 BDTs− mutants are lethal in combination with gcn5Δ. Key indicates genotype of strains. YBC1622 was transformed with rsc4 BDTs− plasmids or controls: pRS314.RSC4 (p1060), pRS314, pRS314.rsc4-2 (p1083), pRS314.rsc4-7 (p1086), or pRS314.rsc4-11 (p1089). Strains were grown at 28°C on selective media with or without 5-FOA to enforce the loss of pRS316.RSC4. (B) rsc4 BDTs− mutants are viable in combination with esa1-L327S at 28°C. YBC1796 was transformed with the rsc4 BDTs− plasmids listed above and plated as above. (C) rsc4-2 is lethal in combination with esa1-L327S only at elevated temperatures. YBC627 and YBC1796 were transformed with pRS314.RSC4 or pRS314.rsc4-2, and plated to 5-FOA to enforce loss of pRS316.RSC4. The 10-fold dilutions were spotted to YPD and grown for 2–3 days at the indicated temperatures.
Genetic interactions between Rsc4 and histone N-terminal tails
To test whether rsc4-2 gcn5Δ lethality was due to defects in acetylation of particular residues of histone tails, the rsc4-2 allele was combined with specific histone H3 and H4 tail mutations and tested for phenotypes (see Materials and methods). Individual replacements involving Lys5, 8, 12, or 16 of the H4 tail had, at most, a slight impact on viability and no impact on growth (Zhang et al, 1998), and combinations with rsc4-2 conferred little or no additional growth defect (Table II). Certain replacements involving two H4 residues slightly reduced viability, but combinations with rsc4-2 typically had little additional impact. In contrast, strong genetic interactions were observed between rsc4-2 and replacements involving Lys14 of H3. Substitution of glutamine, arginine, or glycine for lysine at position 14 in isolation conferred almost no growth defect, whereas combinations with rsc4-2 conferred extremely slow growth. We note that the acetyl-lysine binding pocket (for bromodomains characterized structurally) utilizes conserved hydrophobic residues (and a coordinated water ring) that contact the acetyl group itself. Although glutamine (like acetyl-lysine) is uncharged, several lines of evidence suggest that it should not fully mimic acetyl-lysine: the acetyl moiety is absent in glutamine, and glutamine is significantly shorter and contains different terminal functional groups. In accordance with this reasoning, the K14Q substitution enhanced rsc4-2 phenotypes rather than suppressing them. In addition, enhancement of rsc4-2 phenotypes was specific for combinations involving Lys14 of H3, as replacements involving Lys9 had no effect (Table II). Since Lys14 of histone H3 is the preferred site of acetylation by Gcn5 (Kuo et al, 1996), these results support the specificity observed in our combinations with HAT mutations and point to H3 Lys14 as a critical residue for Rsc4 bromodomain function.
Table 2.
Genetic interactions between rsc4-2 and histone N-terminal tail mutations
| Growth on 5-FOA at 33°C | ||||
|---|---|---|---|---|
| Plasmids | H3 | H4 | RSC4 | rsc4-2 |
| pWZ414-F13 | WT | WT | ++++ | ++++ |
| pWZ414-F30 | K9Q | WT | ++++ | ++++ |
| pWZ414-F53 | K9R | WT | ++++ | ++++ |
| pWZ414-F31 | K14Q | WT | ++++ | −/+ |
| pWZ414-F36 | K14G | WT | ++++ | −/+ |
| pWZ414-F43 | K14R | WT | ++++ | −/+ |
| pWZ414-F23 | WT | K5Q | ++++ | ++++ |
| pWZ414-F22 | WT | K5R | ++++ | +++ |
| pWZ414-F25 | WT | K16Q | ++++ | ++++ |
| pWZ414-F26 | WT | K16G | ++++ | ++++ |
| pWZ414-F24 | WT | K16R | +++ | ++ |
| pWZ414-F51 | WT | K5,12Q | +++ | +++ |
| pWZ414-F52 | WT | K5,12R | ++++ | ++ |
| pWZ414-F47 | WT | K8,16Q | ++++ | +++ |
| pWZ414-F49 | WT | K8,16R | +++ | ++ |
| pWZ414-F48 | K14Q | K8,16Q | ++++ | −/+ |
| pWZ414-F50 | K14R | K8,16R | +++ | − |
| Strains WZY42 (RSC4) and YBC1931 (rsc4-2) carrying Ycp50-HHT2-HHF2 were transformed with plasmids bearing the indicated mutations (Zhang et al, 1998) and then tested on medium containing 5-FOA for the ability to grow at 33°C. ‘+++' and ‘++' indicate reduced viability (fewer colonies), but near WT colony size. ‘−/+' indicates severely reduced viability and extremely small colony size. ‘−' indicates no growth. Growth was assessed after 3–4 days and compared to WT (defined as ++++). | ||||
Suppression of rsc4 bromodomain mutations by deletion of HDACs and components of repression complexes
As loss of histone H3 acetylation exacerbates rsc4 BDTs− mutations, we tested whether increased acetylation levels might suppress rsc4 BDTs− mutations through combinations involving HDAC mutations. Sir2 is the founding member of the NAD-dependent family of HDACs, and it deacetylates lysine residues on histone H3 and H4 (Imai et al, 2000). In addition, there are five different NAD-independent HDACs in yeast: Hda1, Hos1, Hos2 (part of the Set3C complex), Hos3, and Rpd3 (part of the Sin3 complex). These HDACs vary widely in their substrate specificity (histone tail and residue) and also in their impact on both genome-wide and locus-specific acetylation levels.
To test for suppression of rsc4 BDTs− alleles, we crossed a rsc4Δ strain bearing WT RSC4 on a URA3-marked plasmid to strains containing deletions of the above-mentioned HDACs, transformed in plasmids containing the rsc4 BDTs− mutations, and selected for loss of the WT RSC4 on 5-FOA media. Cells were then spotted to YPD plates, incubated at 28, 33, 35, or 38°C and compared for growth. Interestingly, deletion of RPD3 did not suppress rsc4 BDTs− mutations, whereas deletion of HDA1, HOS1, HOS2, HOS3, or SIR2 conferred partial suppression (Figure 5A and B and data not shown). Deletion of any of these latter five HDACs suppressed the rsc4-2 allele partially at 35°C and weakly at 38°C. Partial suppression of the stronger rsc4-7 and rsc4-11 alleles was also detected. Hos2 is part of the Set3C complex, and set3 null also partially suppressed the rsc4 BDTs− alleles (Figure 5B and data not shown), suggesting that suppression is a property of the Set3C complex.
Figure 5.
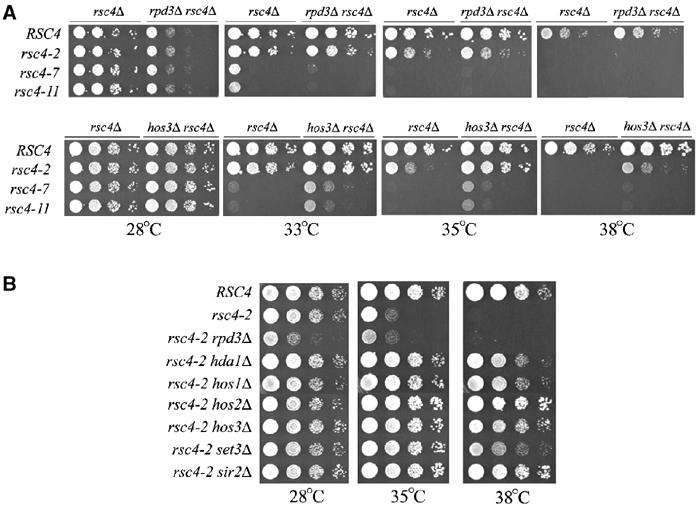
Genetic interactions between Rsc4 bromodomains and HDACs. (A) rsc4 BDTs− mutations are suppressed by specific HDAC deletions. rsc4Δ (YBC627), rpd3Δ rsc4Δ (YBC1698), and hos3Δ rsc4Δ (YBC1789) were transformed with plasmids bearing WT RSC4 or rsc4 BDTs− alleles and grown on 5-FOA media to enforce loss of pRS316.RSC4. The 10-fold dilutions were spotted to YPD and grown for 2–3 days at the indicated temperatures. (B) rsc4-2 is suppressed at 35 and 38°C by deletion of several HDACs, but not by others. rsc4Δ (YBC627), rpd3Δ rsc4Δ (YBC1698), hda1Δ rsc4Δ (YBC1821), hos1Δ rsc4Δ (YBC1702), hos2Δ rsc4Δ (YBC1727), hos3Δ rsc4Δ (YBC1789), set3Δ rsc4Δ (YBC1882), and sir2Δ rsc4Δ (YBC1892) were transformed with pRS314.rsc4-2 (p1083), and grown on 5-FOA media to enforce loss of pRS316.RSC4. The 10-fold dilutions were spotted to YPD and grown for 2–3 days at the indicated temperatures.
To compare their relative strength as rsc4 BDTs− suppressors, we tested the entire panel of suppressor candidates discussed above with the rsc4-2 allele on a single plate at three temperatures. We found that deletion of HDA1, HOS1, HOS2, HOS3, SIR2, or SET3 suppressed the rsc4-2 allele to a similar extent at 35 and 38°C (Figure 5B). Since we observed only partial suppression by these HDAC deletions, and since HDAC deletions cannot suppress bromodomain deletion mutations (data not shown), HDAC deletions do not simply bypass Rsc4 function. Rather, the presence of the acetyl moiety on the histone tail may be important for a partially crippled bromodomain to interact, and the loss of this modification may drop binding below a crucial threshold (see Discussion).
Rsc4 bromodomains bind to histone H3 peptides acetylated at Lys14 in vitro
To determine the histone tail specificity of Rsc4, we tested for interaction with a panel of biotinylated histone tail peptides that either bear or lack acetylation at specific lysine residues. Two Rsc4 derivatives were produced by in vitro transcription/translation, WT and SDM3. SDM3 bears replacements predicted to impair acetyl-lysine recognition (see Table I) but still produces a stable protein that assembles into RSC in vivo (see Supplementary Figure 1). Equimolar amounts of each biotinylated peptide (in vast excess over Rsc4 protein) were bound separately to streptavidin agarose beads. Peptide binding to the beads was estimated as 95–100% efficient (see Supplementary Figure 2). The peptide beads were then incubated with equal levels of Rsc4 protein. The bound material was washed extensively with buffers of moderate stringency, eluted in sample buffer, and tested for retention by immunoblot analysis (see Materials and methods). We find that WT Rsc4 protein, but not the SDM3 derivative, shows preferential binding to the histone H3 peptide acetylated at Lys14 compared to the unmodified peptide (Figure 6A, compare lanes 3 and 4 with lanes 9 and 10). Furthermore, WT Rsc4 protein shows little binding to the unacetylated or acetylated H4 tail peptides tested (Figure 6A, lanes 5 and 6).
Figure 6.
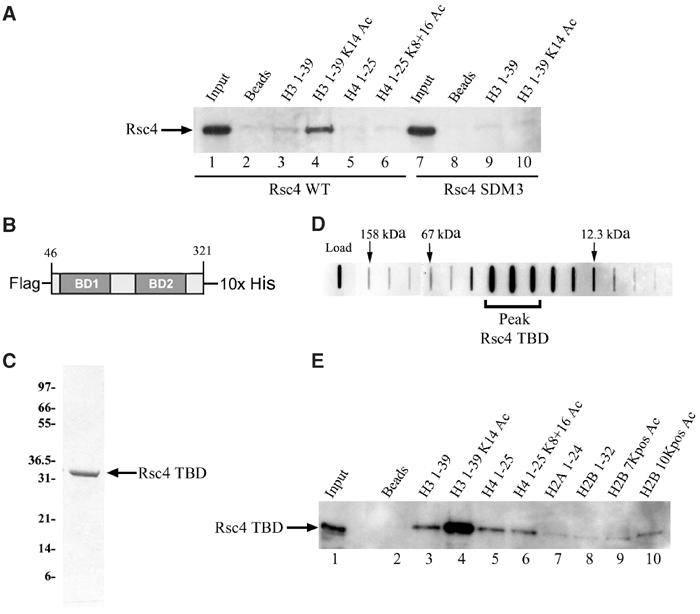
Binding of Rsc4 to histone tail peptides. (A) Binding of WT and Rsc4 SDM3 (each bearing 2xMyc epitopes) to biotinylated N-terminal histone tail peptides on streptavidin beads, assessed by Western blot. Peptides are labeled based on histone, amino-acid sequence, and modification state. In all, 50% of the eluates and 5% of the input were resolved by SDS–PAGE, blotted, and analyzed using anti-Myc antiserum. (B) Diagram of recombinant Rsc4 TBD construct. (C) Coomassie-stained gel of purified full-length recombinant Rsc4 TBD. (D) Slot blot analysis of purified Rsc4 TBD following size exclusion chromatography. Blot was probed with anti-Rsc4 antiserum. Fraction peaks for three molecular weight standards are indicated. (E) Western blot analysis of purified Rsc4 TBD bound to histone tail peptides. The H2B 7Kpos Ac peptide is acetylated at seven lysines (Lys 6, 7, 11, 16, 17, 21, and 22) and H2B 10Kpos Ac is acetylated at all 10 lysines (Lys 3, 6, 7, 11, 16, 17, 21, 22, 30, and 31). In all, 50% of the eluates and 2% of the input were resolved by SDS–PAGE, blotted, and analyzed using anti-Rsc4 antiserum.
To determine whether the binding specificity was due solely to the tandem bromodomain region of Rsc4, we expressed a recombinant version of the Rsc4 tandem bromodomain region (Rsc4 TBD, residues 46–321) that bore a Flag epitope at the amino terminus and a 10X-histidine tag at the carboxyl terminus (Figure 6B). Full-length Rsc4 TBD was purified to homogeneity by tandem affinity chromatography using nickel agarose beads followed by anti-Flag agarose beads (Figure 6C). Size exclusion chromatography of the purified protein revealed a monomeric derivative of approximately 33 kDa (Figure 6D). Rsc4 TBD was then tested for peptide binding specificity in the format described above (with additional peptide substrates), and showed clear preference for H3 acetylated at Lys14 (Figure 6E, lane 4). Rsc4 TBD bound poorly to unmodified and acetylated H4 peptides (Figure 6E, lanes 5 and 6) and at essentially background levels to unmodified or acetylated H2A and H2B peptides (Figure 6E, lanes 7–10). Taken together, these data demonstrate that the tandem bromodomain region of Rsc4 preferentially recognizes histone H3 acetylated at Lys14.
Effect of Rsc4 bromodomain mutations on transcription
We then assessed the impact of rsc4 bromodomain mutations on gene expression by performing transcriptional profiling on rsc4-2, rsc4-7, and rsc4-11 strains at their permissive (30°C) and nonpermissive (35 or 37°C) temperatures. Different alleles and temperatures were employed to help attribute changes to rsc4 as opposed to temperature shift alone. Only a few genes were affected in the rsc4 BDTs− mutants at the permissive temperature: 6–11 genes were downregulated more than two-fold, and 10–15 genes were upregulated more than two-fold (see Supplementary Tables S4, S5, S8, S9, S12, and S13). At nonpermissive temperatures, between 47 and 111 genes were downregulated more than two-fold and 123–194 genes were upregulated more than two-fold. The transcription profiles of the mutants grown at nonpermissive temperatures were remarkably similar, with rsc4-7 and rsc4-11 affecting nearly identical sets of genes (compare Supplementary Tables S6 with S10 and S7 with S11). Common classes of affected genes included the upregulation of genes that regulate cell wall integrity and the response to cell stress, as well as genes involved in iron homeostasis (see Supplementary Table S1). Misregulation of genes involved in cell wall integrity has been observed with other rsc mutants (Angus-Hill et al, 2001). Interestingly, transcript levels of several genes involved in nicotinic acid biosynthesis and transport were decreased in all three rsc4 mutants (see Supplementary Table S1). Additionally, genes involved in mRNA splicing and mating were also decreased (see Supplementary Table S1). To ensure that our microarray results reported true differences in mRNA abundance, we performed S1 nuclease analysis on several transcripts and observed similar changes (see Supplementary Figure 3 and data not shown). Expression profiles of rsc4-2 mutants are similar to those of rsc4-7 and rsc4-11 mutants, but also include additional gene classes. Most striking is the large number (39) of ribosomal proteins and histone gene loci (known targets of RSC) that are downregulated in rsc4-2 at 37°C (see Supplementary Table S2).
Recently, a connection between RSC and Pol III transcription has been demonstrated, as genome-wide localization reveals RSC at virtually all Pol III targets (Ng et al, 2002). Furthermore, protein–protein interactions occur between the Pol III machinery and Rsc4, but this interaction does not involve the tandem bromodomain region of Rsc4 (M Werner, in preparation). Indeed, rsc4 BDTs− mutations do not significantly affect the transcript levels of the Pol III targets tested, including SCR1 or the unspliced version of either RPR1 or the tRNA tF(GAA)P2 (data not shown). Thus, Rsc4 appears to bear three distinct regions: one for interaction with Pol III components, one for assembly into RSC, and tandem bromodomains for histone tail interaction and regulation of Pol II transcription.
Discussion
Bromodomains are found in chromatin regulatory complexes and have been characterized as histone tail and acetyl-lysine recognition motifs in vitro, but how they are utilized in vivo is poorly understood. RSC is an essential and abundant chromatin remodeling complex bearing eight of the 15 bromodomains present in S. cerevisiae, making RSC an attractive complex to study bromodomain function. Here, we identify Rsc4 as an integral and essential member of RSC and show that its tandem bromodomains are essential for viability, cooperate with H3 acetylation in vivo, are utilized to recognize H3 Lys14 acetylation in vitro, and are important for the expression of certain genes.
Rsc4 bears essential tandem bromodomains
RSC appears to have two types of double bromodomain proteins: Rsc4 with two essential tandem bromodomains, and the paralogs Rsc1/Rsc2, which have a nonessential N-terminal bromodomain separated from a second essential bromodomain by a large intervening region (Cairns et al, 1999). Thus, the tight bromodomain spacing in Rsc4 better resembles the tandem bromodomain protein TAF1 (a member of the human TFIID complex) (Jacobson et al, 2000) and its yeast counterparts, Bdf1 and Bdf2, alternative members of yeast TFIID. Certain results with Rsc4 are similar conceptually to those obtained with Bdf1 (Matangkasombut and Buratowski, 2003); strong phenotypes were observed only when the proposed acetyl-lysine recognition sites are impaired in both bromodomains. This relationship suggests a similar function and partial redundancy between the two tandem bromodomains in these proteins. However, as emphasized below, Rsc4 and Bdf1 show essentially opposite tail specificity and genetic interactions.
rsc4 bromodomain mutants require H3 Lys14 and proper H3 acetylation for viability
Our results show a strong link between Rsc4 bromodomains and histone H3 acetylation. First, deletion of the H3 HAT Gcn5 is lethal in combination with a rsc4 BDTs− mutation. Also compelling is the strength and specificity of the genetic interactions between H3 Lys14 and rsc4 BDTs− mutations. As all three amino-acid replacements (glutamine, glycine, or arginine) at position 14 conferred (with rsc4-2) the same strong phenotype, a lysine appears to be required. Rsc4 function depended only weakly on histone H4 acetylation as a rsc4 BDTs− mutation only lowers the nonpermissive temperature for esa1-L327S by 2°C, does not appreciably affect the growth of the stronger esa1-L254P allele, and only very modestly affects the growth of several histone H4 lysine mutants.
Suppression of rsc4 bromodomain mutations by HDAC deletions
Here we provided the first evidence that an HDAC deletion can partially suppress a bromodomain mutation. However, the breadth of hdac suppression was surprising: only rpd3Δ failed to show partial suppression. These results may reflect HDAC tail/residue specificity. For example, although Rpd3 can deacetylate all core histones, it prefers H4 tails in vivo (Rundlett et al, 1998; Suka et al, 2001). As Rsc4 function appears largely independent of H4 acetylation status, the lack of suppression by rpd3Δ would appear consistent with our results. Although we cannot provide a definitive explanation for the breadth of hdac suppression, it seems reasonable to propose that rsc4 BDTs− mutations may cause defects in the activation at many genes, which together confer temperature sensitivity. These gene targets may be repressed by different HDACs, and, therefore, removing any single HDAC may enable partial expression of only a subset of Rsc4 targets, and thus confer only partial suppression of rsc4 BDTs− temperature sensitivity.
Rsc4 binds histone H3 tails acetylated at lysine 14
Our biochemical experiments involving histone tail peptides reveal an interaction between the Rsc4 tandem bromodomains and the histone H3 tail acetylated at lysine 14. Binding by Rsc4 to H3 Lys14Ac utilizes a pair of conserved tyrosine residues in the ZA loop, residues that are utilized by other bromodomains for interaction with the acetyl moiety on the lysine (Ornaghi et al, 1999; Owen et al, 2000). However, our data present an apparent paradox: Rsc4 bromodomains are essential whereas the acetylation of H3 Lys14 is not essential, as yeast strains bearing substitutions at H3 Lys14 are viable. Analysis of other bromodomain structures shows that the bromodomain (and the ZA loop in particular) also interacts with residues flanking the modified lysine residue; thus both the acetyl-lysine and flanking residues likely contribute to the binding energy. In keeping with this idea, the Rsc4 TBD derivative interacted weakly with the unmodified H3 tail. One explanation for the paradox is that Rsc4 bromodomain mutants have moderately reduced interactions with both the acetyl-lysine and the flanking residues. These impaired bromodomains may now require acetylation at H3 Lys14 to bind nucleosomes above a critical threshold required for remodeling RSC targets. An alternative (but related) explanation is that Rsc4 bromodomains bind two different determinants on the nucleosome, only one of which is acetylated H3 Lys14. By this model, interaction with H3 Lys14Ac and the second determinant together provide sufficient binding energy for interaction and target remodeling. Here again, the impaired bromodomains may rely on the presence of the acetylation at H3 Lys14 to avoid falling below a critical binding threshold.
Tandem bromodomain utilization and specialization
Interestingly, rsc4 mutations and bdf1 mutations show precisely opposite genetic interactions: bdf1 alleles display strong genetic interactions with H4 lysine mutations, lethality in combination with esa1-L327S (the identical allele used in our studies), and little effect in combination with gcn5 (Matangkasombut and Buratowski, 2003). These results strongly suggest that the tandem bromodomains of Bdf1 are functionally distinct from those in Rsc4. Indeed, we find that a Rsc4 chimera bearing a precise replacement of its double bromodomain region with that of Bdf1 fails to complement rsc4Δ, even though the chimeric protein is expressed and assembles well into the RSC complex (data not shown). Taken together, we suggest that yeast cells have developed two types of tandem bromodomain proteins, each displaying very different tail-modification specificity, in order to broaden their repertoire of histone tail recognition.
One interesting proposal for tandem bromodomain function is their use in cooperative recognition of two modifications. Indeed, the presence of multiple tandem bromodomains in proteins such as polybromo/BAF180 (and other orthologs) presents the opportunity for the simultaneous recognition of multiple modifications. In TAF1 (TAFII250), the tandem bromodomains pack together and utilize their ZA loops as part of the interface (Jacobson et al, 2000), raising the possibility that the binding of a ligand at one bromodomain may influence the binding properties of the other bromodomain. Studies on TAF1 clearly show preferred interaction with diacetylated H4 tails as compared to nonacetylated H4 tails; however, cooperativity per se was not tested (Jacobson et al, 2000). Thus, whether Rsc4, TAF1, or other tandem bromodomains display cooperative binding remains an important unresolved question. For Rsc4, approaching this issue first requires determining whether the bromodomains in Rsc4 both recognize H3 Lys14Ac (and whether this involves two H3 tails on the same nucleosome), or whether H3 Lys14Ac recognition involves only one of the two bromodomains, with the other bromodomain recognizing a second uncharacterized determinant.
Impact of Rsc4 bromodomain mutations on gene expression
RSC functions in both activation and repression of target genes (Moreira and Holmberg, 1999; Angus-Hill et al, 2001; Damelin et al, 2002; Ng et al, 2002). However, we find that combinations of rsc4-2 with hat or hdac mutations show opposite relationships, lethality versus suppression, strongly suggesting that Rsc4 promotes activation. In keeping with this notion, we observe downregulation of several classes of genes at nonpermissive temperatures including several in the nicotinic acid biosynthesis pathway. Although microarray cannot determine whether an effect is direct, we note that several genes downregulated in the rsc4 BDTs− microarray profiles are occupied by RSC (Damelin et al, 2002; Ng et al, 2002). This set includes genes that demonstrate some of the strongest downregulation, such as BNA6, TNA1, SMX3, and YKR049C, suggesting that these may be direct targets of Rsc4. Nicotinic acid pathway genes are unlikely to underlie the temperature sensitivity of rsc4 BDTs− strains, however, as nicotinic acid supplementation provides no suppression. Although we also observe genes that are upregulated in the mutants, few of these show significant occupancy by RSC. Recently, bdf1Δ gene expression profiles have shown a subtelomeric bias of downregulated genes, suggesting that Bdf1 forms an antisilencing barrier at telomeres and other heterochromatin–euchromatin boundaries (Ladurner et al, 2003). However, genes regulated in rsc4 BDTs− mutants do not cluster to telomeric or subtelomeric regions, further highlighting the differences between Rsc4 and Bdf1 function.
In conclusion, we show that Rsc4 bromodomains are essential for viability, directly recognize acetylated H3 Lys14, cooperate with H3 Lys14 acetylation for their function, show suppression relationships with certain HDACs, and are important for the expression of certain genes. These results highlight the important role bromodomains play in the coordination of chromatin remodeling with histone acetylation in transcriptional regulation, and reveal new properties of tandem bromodomain function.
Materials and methods
Media, genetic methods, and strains
Standard procedures were used for media preparation, transformations, integrations, sporulation, and tetrad analysis. All strains are derivatives of S288C. Strain genotypes are listed in Table III.
Table 3.
Yeast strains
| Strains | Mating type | Genotype | Source |
|---|---|---|---|
| YBC627 | MAT α | rsc4Δ::HIS3 [p164; RSC4; CEN URA3] leu2Δ1 ura3-52 trp1Δ63 his3Δ200 lys2-12δ | This work |
| YBC1622 | MAT α | gcn5Δ::LEU2 rsc4Δ::HIS3 [p164; RSC4; CEN URA3] leu2Δ1 ura3-52 trp1Δ63 his3Δ200 lys2-12δ | This work |
| YBC1796 | MAT a | esa1Δ::HIS3 esa1-L327S::LEU2 rsc4Δ::HIS3 [p164; RSC4; CEN URA3] leu2 ura3-52 trp1 his3Δ200 lys2-12δ | This work |
| YBC1947 | MAT a | esa1Δ::HIS3 esa1-L254P::LEU2 rsc4Δ::HIS3 [p164; RSC4; CEN URA3] leu2 ura3-52 trp1 his3Δ200 lys2-12δ | This work |
| WZY63 | MAT a | hht1-hhf1::pWZ405-F2F9-LEU2 hht2-hhf2::pWZ403-F4F10-HIS3 [pWZ414-F13; HHT2-HHF2; CEN TRP1] ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 | Zhang et al (1998) |
| YBC1924 | MAT a | Same as WZY63 except rsc4-2 integrated into RSC4 locus | This work |
| YBC1931 | MAT a | Same as YBC1924 except Ycp50-copyII (HHT2-HHF2) instead of pWZ414-F13 | This work |
| WZY42 | MAT a | Same as WZY63 except Ycp50-copyII (HHT2-HHF2) instead of pWZ414-F13 | Zhang et al (1998) |
| YBC1698 | MAT a | rpd3Δ::KanMX rsc4Δ::HIS3 [p164; RSC4; CEN URA3] leu2 ura3 trp1Δ63 his3 lys2 | This work |
| YBC1821 | MAT α | hda1Δ::KanMX rsc4Δ::HIS3 [p164; RSC4; CEN URA3] leu2 ura3 trp1Δ63 his3 lys2 | This work |
| YBC1702 | MAT α | hos1Δ::KanMX rsc4Δ::HIS3 [p164; RSC4; CEN URA3] leu2 ura3 trp1Δ63 his3 lys2 | This work |
| YBC1727 | MAT a | hos2Δ::KanMX rsc4Δ::HIS3 [p164; RSC4; CEN URA3] leu2 ura3 trp1Δ63 his3 lys2 | This work |
| YBC1789 | MAT α | hos3Δ::KanMX rsc4Δ::HIS3 [p164; RSC4; CEN URA3] leu2 ura3 trp1Δ63 his3 lys2 | This work |
| YBC1882 | MAT α | set3Δ::KanMX rsc4Δ::HIS3 [p164; RSC4; CEN URA3] leu2 ura3 trp1Δ63 his3 lys2 | This work |
| YBC1892 | MAT α | sir2Δ::KanMX rsc4Δ::HIS3 [p164; RSC4; CEN URA3] leu2Δ1 ura3-52 trp1Δ63 his3Δ200 lys2-12δ | This work |
Plasmids
Details of plasmid constructions are available upon request. All plasmid constructs generated through PCR were sequence verified. The RSC4 gene was isolated from a genomic cosmid as a 4.2 kb BamHI fragment and subcloned into pRS316 (URA3, CEN). PCR-generated ORF of 6xMyc.RSC4 was subcloned into the methionine-repressible p415.MET25 (Mumberg et al, 1994) to create p603. 6xMyc.rsc4 BD1Δ (p804) lacks aa 59–156, 6xMyc.rsc4 BD2Δ (p1172) lacks aa 193–290, 6xMyc.rsc4 BD1-2 Δ (p805) lacks aa 59–290, and 6xMyc.rsc4 C-termΔ (p809) inserts a stop codon after aa 602. The rsc4 BDTs− mutants were isolated as N-terminal Myc fusions in p415MET25. The pRS314 series of rsc4 BDTs− mutant plasmids were created by subcloning the indicated RSC4 derivative without Myc tags into pRS314 (TRP1, CEN). DNA fragments encoding WT Rsc4 (p1513) or Rsc4 SDM3 (p1516) tagged with 10X HIS and two Myc epitopes at the N-terminus were subcloned into pCITE-4b (Novagen) for in vitro transcription and translation.
Isolation of rsc4 BDTs− mutants and site-directed mutants
The region encoding the double bromodomain (bp 92–982) of RSC4 was mutagenized utilizing the inherent error rate of Taq in PCR using oligonucleotides BC796 (5′ GGAAAACATCCTAAAAACCAAG 3′) and BC797 (5′ GATATAGCTAACGCACCTGCTG 3′). The PCR product was co-transformed into YBC627 with 6xMyc.rsc4 BD1-2Δ (p805) linearized with NotI. The PCR product was inserted into the gapped plasmid by homologous recombination and cells bearing the repaired plasmid were selected on medium lacking leucine and pooled together to create a library (31 000 colonies). Cells were plated and 2500 colonies lacking pRS316.RSC4 were selected with 5-FOA and tested for conditional growth at 38°C via replica plating. Plasmids were recovered, subcloned, and fully sequenced. The rsc4-7 allele has a single mutation in BD2 (L241P), whereas rsc4-11 bears a mutation in BD1 and in the linker between the two bromodomains (C130S, L182P). Additional Ts− alleles used were as follows: rsc4-5 D122N, N286S; rsc4-6 D66V, Y133H; rsc4-9 Y38C, V147I, D277V, L297S; rsc4-10 N59Y, V260A, I274V; rsc4-12 Q131R, L189E; rsc4-13 L120P; rsc4-14 M145V, M149V, T279P.
Plasmids bearing site-directed mutations in RSC4 were prepared using the Quick Change Method (Stratagene), subcloned, and fully sequenced.
Analysis of rsc4-2 with histone tail mutants
The rsc4-2 allele was exchanged for the WT allele at the chromosomal RSC4 locus in WZY63 (gift from S Dent) by targeted replacement to create strain YBC1931. YBC1931 was transformed with a series of TRP1-marked plasmids containing mutant H3 alleles (with WT H4), mutant H4 alleles (with WT H3), or mutations in both histone genes (gifts from S Dent). Loss of the URA3-marked plasmid bearing WT H3 and H4 was enforced through growth on 5-FOA. Phenotypes of the rsc4-2 histone double mutations were assessed by spot dilution analysis at 33°C, the highest temperature at which the rsc4-2 allele displayed WT growth.
Anti-Rsc4 antibody
Recombinant full-length Rsc4 with a 10X-histidine tag was expressed in Escherichia coli BL21 Codon (+) cells (Stratagene) and purified by binding to nickel resin (Qiagen) under denaturing conditions. A measure of 2 mg of purified protein was used for injection into rabbits (Covance Inc.).
Extract preparation and immunoprecipitations
Whole-cell extracts were prepared as described previously (Cairns et al, 1999). For RSC assembly assays, all Rsc4 derivatives were tagged with Myc epitopes and analyzed in the presence of untagged WT RSC4 (p164) for complementation. For the rsc4 BDTs− mutants, cultures were grown at 35°C in SC media lacking leucine and uracil. Anti-Myc antibody (clone 9E10) was conjugated to protein G beads and incubated with 400 μg extract for 3 h. Precipitates were recovered, washed twice with IP wash buffer (50 mM Tris–Cl (pH 7.5), 250 mM NaCl, 1 mM EDTA, 10% glycerol, 0.05% Tween-20), and eluted in 4 × SDS sample buffer. SDS–PAGE gels were immunoblotted to PVDF membrane and developed as indicated.
Rsc4 TBD protein purification
Recombinant Rsc4 bearing tandem bromodomains (Rsc4 TBD, residues 46–321) and both a Flag and 10X-histidine tag (N- and C-terminal, respectively) was expressed in BL21 DE3 E. coli and purified using standard methods. The purified protein was analyzed by size exclusion chromatography on a Superdex 200 column (Amersham Pharmacia Biotech) followed by slot blot analysis of odd fractions utilizing anti-Rsc4 antiserum.
Histone tail binding assay
Biotinylated histone tail peptides were bound to streptavidin beads (Invitrogen) in a ratio of 20 nmol of peptide to 100 μl bed volume of beads in peptide binding buffer (PBB) (20 mM Tris (pH 7.5), 150 mM NaCl, 5% glycerol, 0.05% Tween-20, 1 mM EDTA, 1 mM β-mercaptoethanol, protease inhibitors) with 0.5 mg/ml protease-free BSA. Beads were washed five times with PBB, blocked with protease-free BSA, and resuspended in a 50% slurry with PBB and 0.5 mg/ml protease-free BSA. WT and SDM3 mutant 10xHIS-2xMyc-Rsc4 were in vitro transcribed/translated using a reticulocyte lysate system as described by the manufacturer (Promega) with the addition of 10 mM KCl. The lysate (20 μl) was incubated for 1.5 h with 20 μl of peptide-bound beads, washed twice with PBB and twice with PBB containing 200 mM NaCl. The Rsc4 protein was eluted by boiling in 4 × SDS sample buffer prior to Western blot analysis. For binding studies involving the recombinant Rsc4 TBD, 15 μl of the bead slurry was rotated at 4°C for 3 h with 500 ng of purified Rsc4 TBD. The beads were washed twice with PBB and twice with PBB containing 250 mM NaCl. The Rsc4 TBD protein was eluted as above and analyzed by Western blot.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplemental Microarray Analysis Materials and Methods
Acknowledgments
We thank Sharon Dent and Lorraine Pillus for generous gifts of strains and/or plasmids. We are grateful to Douglas Roberts, Daniel Richardson, and Matthew Gordon for assistance with microarray analysis. We also thank Alisha Schlichter and Pierre Thuriaux for comments on the manuscript. This work was supported by the National Institutes of Health (GM60415 to BRC; CA24014 for core facilities) the Human Frontier Science Program and the Howard Hughes Medical Institute. Margaret Kasten is a Research Associate and Brad Cairns is an Assistant Investigator with the Howard Hughes Medical Institute.
References
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR (2001) A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell 7: 741–751 [DOI] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD (1996) RSC, an essential, abundant chromatin-remodeling complex. Cell 87: 1249–1260 [DOI] [PubMed] [Google Scholar]
- Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F (1999) Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol Cell 4: 715–723 [DOI] [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L (1999) Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol 19: 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin M, Simon I, Moy TI, Wilson B, Komili S, Tempst P, Roth FP, Young RA, Cairns BR, Silver PA (2002) The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol Cell 9: 563–573 [DOI] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491–496 [DOI] [PubMed] [Google Scholar]
- Du J, Nasir I, Benton BK, Kladde MP, Laurent BC (1998) Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics 150: 987–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR III, Workman JL (1998) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94: 45–53 [DOI] [PubMed] [Google Scholar]
- Grunstein M (1997) Histone acetylation in chromatin structure and transcription. Nature 389: 349–352 [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL (2002) Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379 [DOI] [PubMed] [Google Scholar]
- Havas K, Whitehouse I, Owen-Hughes T (2001) ATP-dependent chromatin remodeling activities. Cell Mol Life Sci 58: 673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266: 383–402 [DOI] [PubMed] [Google Scholar]
- Hsu JM, Huang J, Meluh PB, Laurent BC (2003) The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol Cell Biol 23: 3202–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE (2000) Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J Mol Biol 304: 355–370 [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R (2000) Structure and function of a human TAFII250 double bromodomain module. Science 288: 1422–1425 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383: 269–272 [DOI] [PubMed] [Google Scholar]
- Ladurner AG, Inouye C, Jain R, Tjian R (2003) Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol Cell 11: 365–376 [DOI] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R (2001) Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414: 924–928 [DOI] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski S (2003) Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell 11: 353–363 [DOI] [PubMed] [Google Scholar]
- Moreira JM, Holmberg S (1999) Transcriptional repression of the yeast CHA1 gene requires the chromatin-remodeling complex RSC. EMBO J 18: 2836–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba S, He Y, Zeng L, Farooq A, Carlson JE, Ott M, Verdin E, Zhou MM (2002) Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol Cell 9: 575–586 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2002) Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev 16: 806–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornaghi P, Ballario P, Lena AM, Gonzalez A, Filetici P (1999) The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J Mol Biol 287: 1–7 [DOI] [PubMed] [Google Scholar]
- Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA (2000) The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J 19: 6141–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392: 831–835 [DOI] [PubMed] [Google Scholar]
- Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA (2002) Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol 22: 4723–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N, Luo K, Grunstein M (2002) Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet 32: 378–383 [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8: 473–479 [DOI] [PubMed] [Google Scholar]
- Tsuchiya E, Hosotani T, Miyakawa T (1998) A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res 26: 3286–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL (2000) ATP-dependent chromatin-remodeling complexes. Mol Cell Biol 20: 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY (1998) Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J 17: 3155–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplemental Microarray Analysis Materials and Methods


