Fission yeast MOZART1 homologue Mzt1 associates with γ-tubulin and is found at MTOCs. It plays an essential role in microtubule regulation, as well as in septum formation and cytokinesis. Recombinant Mzt1 interacts directly with GCP3Alp6. This interaction may assist MTOC activity of the γ-tubulin complex.
Abstract
In humans, MOZART1 plays an essential role in mitotic spindle formation as a component of the γ-tubulin ring complex. We report that the fission yeast homologue of MOZART1, Mzt1/Tam4, is located at microtubule-organizing centers (MTOCs) and coimmunoprecipitates with γ-tubulin Gtb1 from cell extracts. We show that mzt1/tam4 is an essential gene in fission yeast, encoding a 64–amino acid peptide, depletion of which leads to aberrant microtubule structure, including malformed mitotic spindles and impaired interphase microtubule array. Mzt1/Tam4 depletion also causes cytokinesis defects, suggesting a role of the γ-tubulin complex in the regulation of cytokinesis. Yeast two-hybrid analysis shows that Mzt1/Tam4 forms a complex with Alp6, a fission yeast homologue of γ-tubulin complex protein 3 (GCP3). Biophysical methods demonstrate that there is a direct interaction between recombinant Mzt1/Tam4 and the N-terminal region of GCP3Alp6. Together our results suggest that Mzt1/Tam4 contributes to the MTOC function through regulation of GCP3Alp6.
INTRODUCTION
Microtubule-organizing centers (MTOCs), as the name suggests, are involved in regulating microtubule (MT) architecture. They play essential roles in a wide range of fundamental biological activities, including cell proliferation and differentiation. In vitro studies have shown that two of the key components of microtubules, α- and β-tubulin, can self-assemble when a critical concentration is exceeded, indicating that MTOCs are not absolutely required for MT nucleation. However, MTOCs provide dominant MT nucleation sites at which MT assembly is initiated at lower, physiological concentrations, allowing a wide range of dynamic MT architecture to be generated in a cell (Mitchison and Kirschner, 1984). One of the key MTOC components is γ-tubulin. γ-Tubulin was originally identified in Aspergillus nidulans (Oakley and Oakley, 1989). It was purified from various higher eukaryotes, including Drosophila melanogaster and Xenopus laevis, as a component of a large multiprotein complex (Raff et al., 1993; Stearns and Kirschner, 1994) alongside the γ-tubulin complex proteins 2–6 (GCP2–6). This large complex was termed the γ-tubulin ring complex (γ-TuRC) based on its higher-order, three-dimensional, ring-like appearance (Zheng et al., 1995). The γ-TuRC caps the minus ends of MTs and facilitates directional MT nucleation (Zheng et al., 1995). It incorporates a smaller complex, the γ-tubulin small complex (γ-TuSC), consisting of GCP2, GCP3, and γ-tubulin. The γ-TuSC retains activity to facilitate MT nucleation, albeit at a substantially lower level than γ-TuRC (Oegema et al., 1999). It resembles the γ-tubulin complex (γ-TuC) of budding yeast (Knop and Schiebel, 1997). Reconstituted γ-TuSC employing recombinant budding yeast GCP2, GCP3, and γ-tubulin was found to assemble “ring-like” structures highly reminiscent of the γ-TuRC (Kollman et al., 2010). It has therefore been proposed that γ-TuSC contributes the core structure to γ-TuRC to generate the microtubule nucleation template (Kollman et al., 2010, 2011).
A protein called mitotic-spindle organizing protein associated with a ring of γ-tubulin 1 (MOZART1) was identified in a screen for novel human proteins that interact with γ-TuRC (Hutchins et al., 2010; Teixido-Travesa et al., 2010). Depletion of MOZART1 from HeLa cells reduced γ-tubulin recruitment to the centrosome, the major MTOC in higher eukaryotes, and generated spindle formation defects, highlighting MOZART1’s essential role in mitotic spindle formation (Hutchins et al., 2010). Of interest, although there is no obvious Mozart 1 homologue in budding yeast, a Mozart1 homologue, Tam4/Mzt1, was present in fission yeast (Hutchins et al., 2010; Bitton et al., 2011), making the organism highly attractive for the study of γ-TuC regulation and function.
The centrosome equivalent structure in yeast is the spindle pole body (SPB), a layered structure enriched in proteins that are homologous to the centrosome components (Wigge et al., 1998). Fission yeast homologues of GCP2–6, as well as the γ-tubulin Gtb1, localize to the SPB (Horio et al., 1991; Vardy and Toda, 2000; Heitz et al., 2001; Fujita et al., 2002; Venkatram et al., 2004; Anders et al., 2006). In fission yeast, there are also other MTOCs, namely interphase MTOC (iMTOC), which nucleates cytoplasmic MTs during interphase, and equatorial MTOC (eMTOC), which appears before cytokinesis and generates the postanaphase array (Hagan, 1998; Heitz et al., 2001; Sawin and Tran, 2006). All fission yeast γ-TuC components are found at the iMTOC as well as the eMTOC, showing a direct link between the γ-TuC and MTOC activity.
A mitotic spindle is composed of MTs and their accessory proteins. Therefore it is expected that MT misregulation directly causes spindle formation defects or spindle malfunction that leads to chromosome missegregation. Recent extensive studies also revealed that MTs play essential roles in “shaping the cell” by defining actin distribution during interphase and regulating cortical actin ring formation before cytokinesis (Hachet et al., 2012). Thus mutations in the components of the γ-TuC, the major MTOC constituent, are expected to induce a wide range of cellular defects in events including spindle formation and cytokinesis. Intriguingly, a mutant defective in the GCP2Alp4 function was reported to show septum formation defect as well as impaired MT regulation, indicating that γ-TuC is required for regulation of cytokinesis (Vardy et al., 2002).
Because γ-TuSC (GCP2, GCP3, and γ-tubulin) alone is not a potent MT nucleator, additional modulators are expected to enhance its activity (Oegema et al., 1999). GCP4–6 are attractive candidates. However, in fission yeast, whereas GCP2Alp4, GCP3Alp6, and γ-tubulinGtb1 are essential, cells devoid of GCP4Gfh1, GCP5Mod21, and GCP6Alp16 are viable, highlighting a distinction between the γ-TuSC and the other GCP proteins (Anders et al., 2006). Similarly, in Drosophila and Aspergillus, GCP4-6 are not required for cell survival (Verollet et al., 2006; Xiong and Oakley, 2009). Identification of Mzt1/Tam4 therefore prompted us to ask whether Mzt1/Tam4 plays a crucial role in activating the γ-TuSC. We found that Mzt1/Tam4 is a MTOC component and, unlike GCP4-6, is essential for microtubule regulation. It is also required for regulation of cytokinesis. Strikingly, it directly interacts with GCP3Alp6.
RESULTS
Identification of fission yeast MOZART1 homologue
Using an algorithm specifically designed to identify small open reading frames (ORFs) of <100 amino acids, we previously identified an ORF that was upregulated during meiosis, tam4+ (for “transcripts altered in meiosis”; Bitton et al., 2011). We predicted that the tam4+ gene encoded a protein of 64 amino acids. The tam4 gene locus was also identified through a homology search looking for a fission yeast homologues of human MOZART1 and was referred to as mzt1, reflecting the high level of homology to MOZART1 (Hutchins et al., 2010). Here we follow this nomenclature and refer to the gene as mzt1.
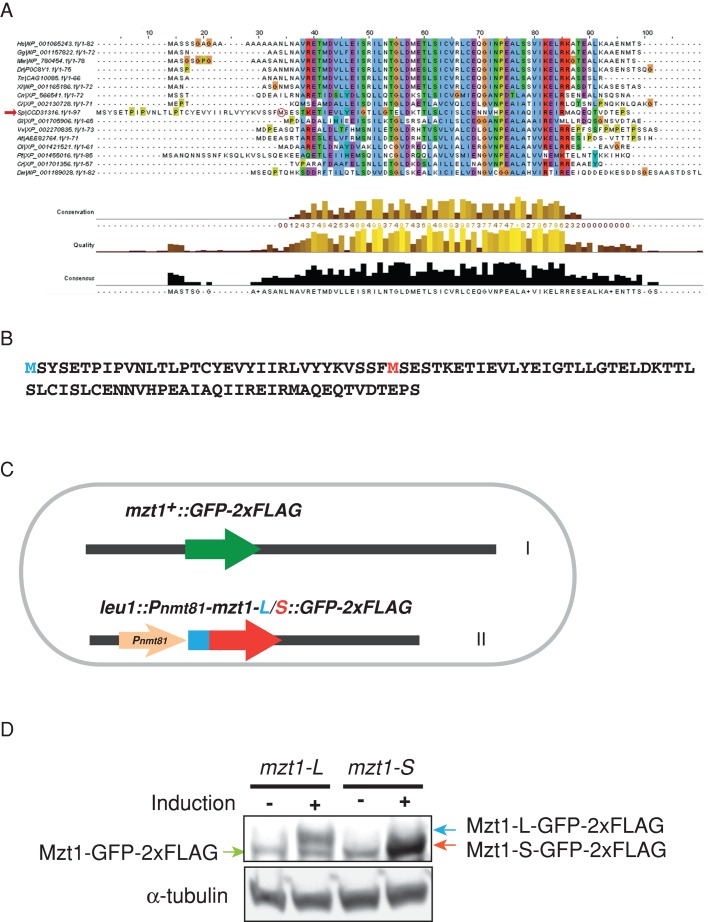
The previous study of Mzt1 protein predicted a longer, 97–amino acid, protein (Hutchins et al., 2010). The initiator methionine predicted by us corresponds to the methionine at position 34 of the Mzt1 protein predicted in their study, which we refer to as Mzt1-L. We noticed that the first 33 amino acids of Mzt1-L do not show substantial homology to MOZART1 homologues from a range of organisms (Figure 1A). This prompted us to examine which methionine is used as the start codon.
FIGURE 1:
Identification of the Mzt1 initiation methionine. (A) Primary structure of fission yeast Mzt1. CLUSTALW multiple sequence alignment of predicted orthologues of human MOZART1. Fission yeast S. pombe Mzt1 is indicated with a red arrow. Species abbreviations are as follows: Hs, Homo sapiens; Gg, Gallus gallus; Mm, Mus musculus; Dr, Danio rerio; Tn, Tetraodon nigroviridis; Xt, Xenopus tropicalis; Cn, Cryptococcus neoformans; Ci, Ciona intestinalis; Sp, S. pombe; Gl, Giardia lamblia; At, Arabidopsis thaliana; Ol, Ostreococcus lucimarinus; Pt, Paramecium tetraurelia; Cr, Chlamydomonas reinhardtii; Dm, Drosophila melanogaster. (B) Two potential initiation methionines in the predicted Mzt1 ORF. Methionine in blue is the initiation methionine for the long Mzt1 sequence (mzt1-L) predicted by Hutchins et al. (2010). Methionine in red is the physiological initiation methionine for the short Mzt1 sequence (mzt1-S). (C) Schematic of the strain (KT3519 and KT3522) used to determine the initiation methionine of Mzt1. The mzt1 gene is tagged with GFP-2xFLAG at its endogenous locus, which is located on chromosome I. The mzt1-S or mzt1-L tagged with GFP-2×FLAG was integrated at the leu1+ locus (chromosome II) under the inducible nmt81 promoter. (D) The yeast strains KT3519 and KT3522, harboring mzt1-S or -L tagged with GFP-2xFLAG at the leu1+ gene locus, under the nmt81 promoter and mzt1-GFP-2xFLAG at its endogenous locus, were grown in the absence (–) or presence (+) of thiamine, which acts to repress the nmt81 promoter. Denatured whole-cell extracts were prepared from these cells, which were subjected to Western blotting. Monoclonal anti-FLAG antibody was used to probe for GFP-tagged Mzt1. α-Tubulin was used as a reference for loading. The blot shows that in the noninduced lanes (–), the bands present correspond to the Mzt1-S.
To identify the initiator methionine, we fused two constructs, mzt1-L (a longer ORF predicted using the methionine shown in blue in Figure 1B) and mzt1-S (a shorter ORF predicted using the methionine shown in red in Figure 1B), in frame, with green fluorescent protein (GFP)–2xFLAG at their 3′ ends and introduced at the leu1 locus under an inducible promoter nmt81 (Maundrell, 1993). The endogenous mzt1+ locus has also been fused, in frame, with GFP-2xFLAG at the 3′ end of its ORF immediately before the stop codon (Figure 1C). A comparison of the induced products and the endogenous Mzt1-GFP established that the endogenous molecule migrated with a molecular mass of 38 kDa, whereas induction of Mzt1-S-GFP-2xFLAG or Mzt1-L-GFP-2xFLAG by thiamine depletion to activate the nmt81 promoter produced protein that migrated at exactly the same mass as the endogenous Mzt1-GFP-2xFLAG or a higher–molecular weight form, respectively (Figure 1D). We therefore concluded that Mzt1-S represents the endogenous Mzt1 protein and the correct start methionine does indeed correspond to position 34 of the mzt1-L ORF, confirming Mzt1 as a 64–amino acid protein.
Mzt1 localizes to the MTOCs
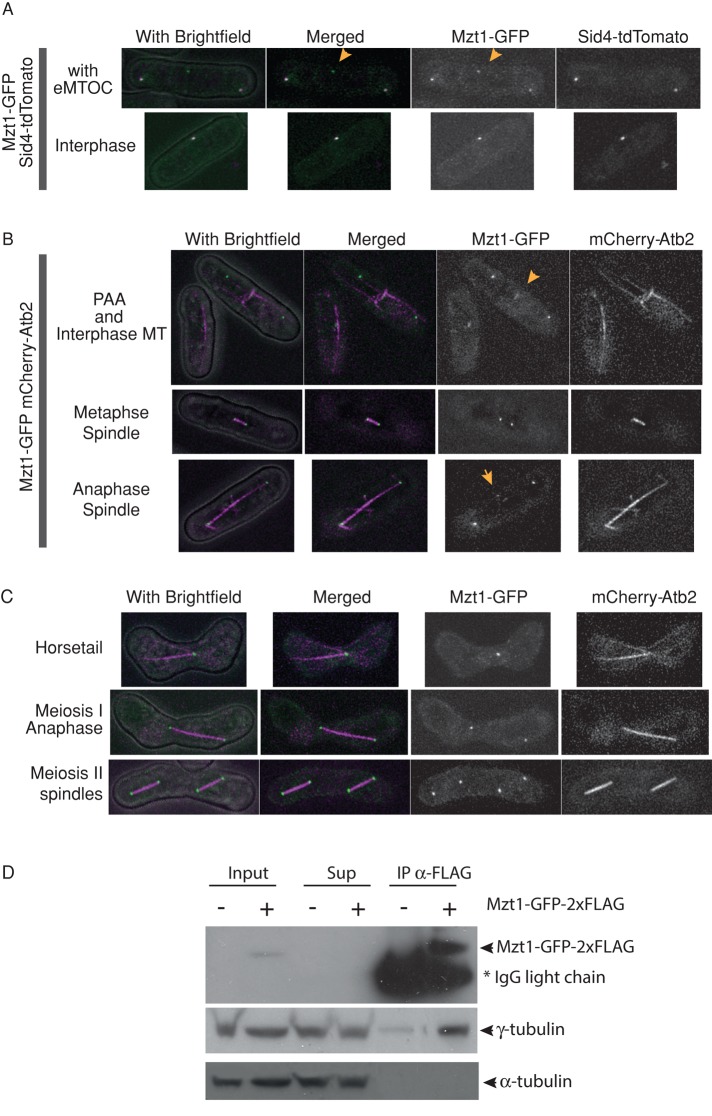
Human MOZART1 localizes at the centrosome and plays an essential role in spindle formation (Hutchins et al., 2010). To examine whether fission yeast Mzt1 plays a similar role, we observed its localization throughout the cell cycle and during meiotic differentiation. The endogenous mzt1 was tagged with GFP-2xFLAG at its 3′ end and introduced into a strain harboring either mCherry-tagged α-tubulin atb2 under the nda3 promoter (Masuda et al., 2006) or tdTomato-tagged sid4, a protein that localizes exclusively to the SPB at all stages of the cell cycle but not to the iMTOC or eMTOC (Tomlin et al., 2002). During the vegetative cell cycle, the Mzt1-GFP-2xFLAG signal was found not only at the SPB, but also at iMTOC and eMTOC (Figure 2, A and B), as observed for other γ-TuC components, Alp4 and Alp6 (Vardy and Toda, 2000). During meiotic differentiation, Mzt1 was found at the SPB (Figure 2C).
FIGURE 2:
Mzt1 localizes to the MTOCs and associates with γ-tubulin. (A) Localization of Mzt1 during the vegetative cell cycle in relation to the SPB component Sid4. Mzt1-GFP and Sid4-tdTomato signals were simultaneously observed using a confocal microscope. Mzt1 is tagged with GFP-2xFLAG in a cell that harbors tdTomato-tagged Sid4, an SPB component. Mzt1-GFP signal colocalizes with the Sid4-tdTomato signal that represents the SPB (top and bottom A cell that is forming septum (top) carries eMTOC, where Sid4-tdTomato signal is missing but Mzt1-GFP signal is found (yellow arrows). (B) Localization of Mzt1 during the vegetative cell cycle in relation to the MT structure. Fluorescence signals of Mzt1-GFP and mCherry-tagged α-tubulin, mCherry-Atb2, were simultaneously observed using a confocal microscope. Yellow arrows indicate Mzt1-GFP signals at the eMTOC. (C) Localization of Mzt1 during meiotic differentiation. Mzt1-GFP and mCherry-Atb2 signals were simultaneously captured using a confocal microscope. Mzt1-GFP signal was found at the MTOC. (D) Mzt1 associates with γ-tubulin. Cell extracts prepared from wild-type strains harboring Mzt1-GFP-2xFLAG (+) or nontagged Mzt1 (–) were subjected to immunoprecipitation (IP) assays with anti-FLAG antibody. The IP complexes were analyzed by Western blotting with antibodies against GFP and γ-tubulin. A high proportion of γ-tubulin is present in the anti-FLAG immunocomplex prepared from Mzt1-GFP-2xFLAG strain. The IgG light chain in the IP complexes detected by the secondary antibodies is indicated by asterisk.
Mzt1 is a part of the γ-TuC
Having seen the fission yeast Mzt1 localization, we next examined whether Mzt1 associates with γ-TuC. Cell extracts were prepared from asynchronous culture expressing Mzt1-GFP-2xFLAG, and using an anti-FLAG antibody, Mzt1-GFP-2xFLAG was immunoprecipitated. γ-Tubulin was found enriched in the immunocomplex (Figure 2D), a similar trend to human MOZART1 (Hutchins et al., 2010).
Mzt1 is essential and required for microtubule regulation
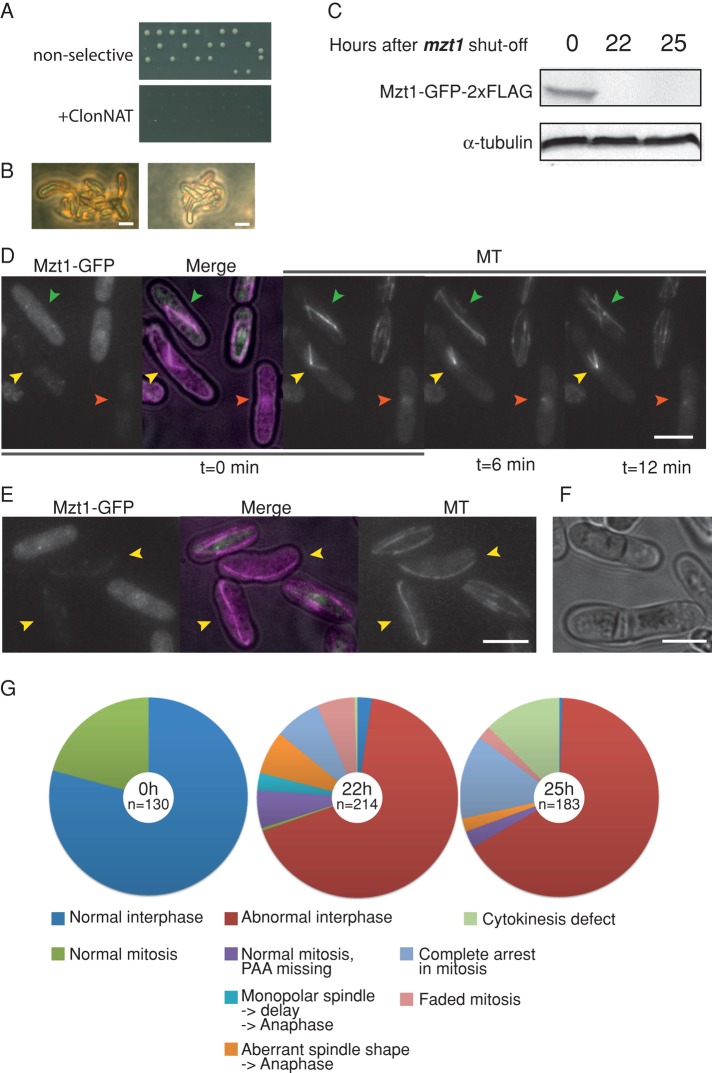
To gain further insight into Mzt1 function, we generated a strain from which the mzt1 ORF was deleted. A diploid strain harboring one copy of mzt1+ and one copy of a mzt1 deletion allele marked with ClonNAT was analyzed by tetrad dissection (Figure 3A). Only two of the four spores in each tetrad set were viable, and each of these was sensitive to ClonNAT, indicating that mzt1+ is an essential gene. Cells in the microcolonies that did not grow showed bent morphology (Figure 3B) reminiscent of the alp mutants that affect microtubule regulation (Radcliffe et al., 1998).
FIGURE 3:
Mzt1 is essential, and its depletion causes abnormal MT organization and cytokinesis defect. (A) Tetrad analysis of KT3449 (h90/h90 mzt1+/mzt1::ClonNAT). Only two viable colonies out of four tetrad progenies emerged. These viable colonies are ClonNAT sensitive. (B) Those that did not form viable colonies in the tetrad analysis formed microcolonies of ∼10–30 cells that show bent-cell morphology. Scale bar, 10 μm. (C) A Mzt1 shut-off strain KT3711 that also harbors mCherry-atb2 was first cultured in the MM media in the absence of thiamine to allow expression of Mzt1-GFP. The culture was then supplemented with thiamine (2 μM), and Mzt1-GFP expression was repressed for 22 and 25 h. Whole-cell extracts were prepared, and the level of Mzt1-GFP was examined by Western blotting with anti-GFP antibody. (D, E) Cells from time 0 and 25 h after Mzt1 depletion were mixed, and mCherry-Atb2 signals were observed. Cells with Mzt1-GFP signals (time-zero sample) did not show MT defects, whereas cells without Mzt1-GFP signal (indicated by yellow and orange arrowheads) showed mitotic spindle formation defects (D) or interphase MT defect (E). In D, an anaphase spindle in a nondepleted cell (green arrowhead) elongated without a delay, whereas a monopolar spindle (yellow arrowhead) did not proceed to anaphase. Cells carrying a mitotic MTOC with very low MTOC activity (orange arrowhead) were also observed. In E, interphase cells devoid of Mzt1-GFP (yellow arrowheads) have fewer MTs, indicating that the iMTOC activity was low in these cells. Scale bar, 10 μm. (F) At 25 h after depletion of Mzt1, cells with aberrant septum started to accumulate. Scale bar, 10 μm. (G) Summary of the Mzt1 depletion phenotypes. Cells depleted of Mzt1 for 0, 22, and 25 h were filmed for 20 min at 1.5-min intervals (representative images in Supplemental Figure S1) and classified into categories as graphically illustrated in Supplemental Figure S2. Longer depletion increased the population of cells with cytokinesis defect.
To further examine the terminal phenotype, we generated a strain in which the endogenous mzt1 is deleted but the cells were kept alive by mzt1, tagged with GFP-2xFLAG, and integrated at the leu1 locus under the regulation of the nmt81 promoter, allowing it to be repressed by addition of thiamine. This strain also expresses mCherry-tagged α-tubulin atb2, making it possible to monitor the status of microtubule organization.
At 22 h after repression of the nmt81 promoter through the addition of thiamine, the Mzt1-GFP-2xFLAG levels declined below the detection level in Western blotting using anti-GFP antibody (Figure 3C). The GFP signal from the cells also dropped, and aberrant microtubule structures accumulated (Figure 3, D and E, Supplemental Figure S1, and Supplemental Movies S1– S3). Both interphase and spindle microtubules were affected (Figure 3, D and E; summarized in Figure 3G). Some cells had mitotic spindles of aberrant morphology that proceeded into anaphase, indicating that the spindle assembly checkpoint is compromised in Mzt1-depleted cells. After 25 h of Mzt1 depletion, 14% of cells were arrested during mitosis and could not proceed to anaphase (Figure 3, D, yellow arrowhead, and G). Of interest, after a longer depletion period (25 h), more cells were found to have defects in cytokinesis (Figure 3, F and G).
Mzt1 is not required for assembly of Alp6-Alp4-Gtb1
The phenotype of the mzt1 shut-off strain is very similar to the phenotypes observed in mutants defective in GCP2Alp4 and GCP3Alp6 (Vardy and Toda, 2000). Because GCP2Alp4 and GCP3Alp6 form γ-TuC together with γ-tubulin Gtb1 (Vardy and Toda, 2000), we examined whether Mzt1 influences γ-TuSC formation. FLAG-tagged GCP3Alp6, nontagged GCP2Alp4, nontagged γ-tubulinGtb1, and nontagged Mzt1 were generated by in vitro translation, and the efficiency of γ-TuSC formation was assessed in the presence and absence of Mzt1. All components were translated together, and an anti-FLAG antibody was used to isolate GCP3Alp6 immunocomplex (Figure 4). In the supernatant fraction, only a trace of GCP3Alp6 was found, indicating that GCP3Alp6 was efficiently captured by anti-FLAG antibody. A substantial proportion of the remaining components were found in the supernatant, indicating that they were present in excess in the reaction to reconstitute γ-TuSC. Although Mzt1 was incorporated into the GCP3Alp6 immunocomplex, it did not affect Alp6-Alp4-Gtb1 complex formation, as seen by comparable protein levels of each component in the complex in the absence or presence of Mzt1. Our result indicates that Mzt1 is unlikely to regulate γ-TuSC formation.
FIGURE 4:
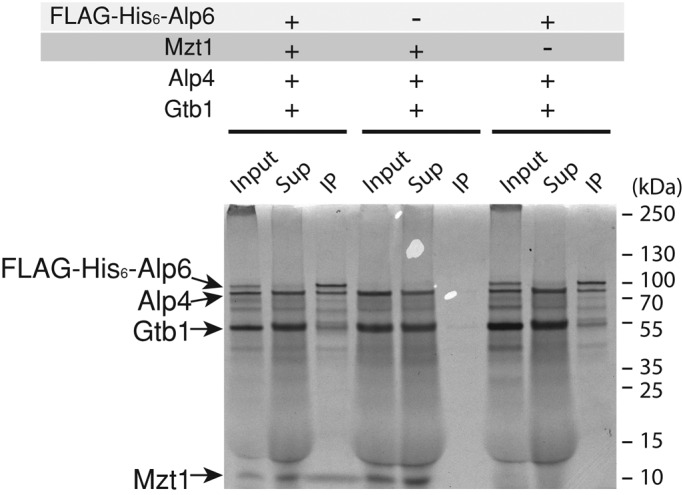
Mzt1 is not required for the assembly of the γ-TuSC. The ability of Mzt1 to enhance γ-TuSC complex formation was assessed using proteins produced by an in vitro translation system (IVT). The Alp6 was N-terminally tagged with FLAG-His6, and the rest of the components (Alp4, Gtb1, and Mzt1) were untagged. Samples were prepared as a single IVT reaction in order to produce proteins together in the presence of [S35]methionine. These were subjected to immunoprecipitation using an anti-FLAG antibody. The IP complexes were separated using SDS–PAGE and visualized by autoradiography. Thirty-five percent equivalent of cell extracts used for IP samples were loaded as input and supernatant samples. The ratio of Alp6:Alp4:γ-tubulin in the IP complex did not change in the presence or absence of Mzt1.
Mzt1 interacts with Alp6, the GCP3 homologue
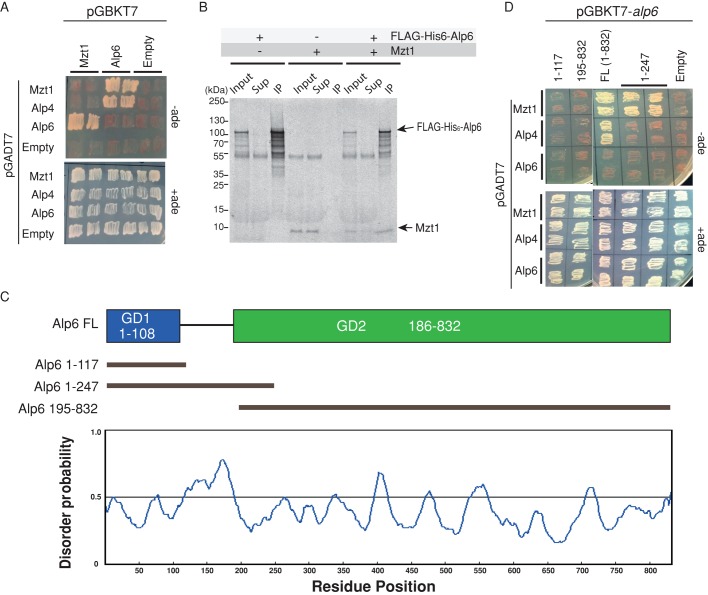
To obtain more clues about Mzt1’s mode of action in regulation of microtubules, we set up a yeast two-hybrid assay (Y2H) to evaluate Mzt1 interaction with Alp6 and Alp4. Although a positive interaction in the Y2H does not guarantee direct interaction of two proteins, because the budding yeast proteins may act as a bridging complex, it serves as a convenient tool to probe potential protein–protein interactions. Full-length Alp6, Alp4, and Mzt1 were put into yeast two-hybrid vectors pGADT7 (activation domain) and pGBKT7 (DNA-binding domain; Clontech) and tested for their interactions. Among them, pGBKT7-alp4 construct was dropped from the analysis, as it self-activated the system. In addition, gtb1, which encodes γ-tubulin, could not be included in the analysis because neither pGADT7-gtb1 nor pGBKT7-gtb1 showed interaction with Alp6 constructs, a known interactor of γ-tubulin. The pGADT7-alp4 showed interaction with pGBKT7-alp6, as expected. Of interest, however, pGADT7-alp4 showed negative interaction with pGBKT7-mzt1, suggesting that Alp4 and Mzt1 are unlikely to interact directly (Figure 5A). On the other hand, pGBKT7-mzt1 did interact with pGADT7-alp6. This interaction between Mzt1 and Alp6 was further verified when the bait and prey vectors were switched (combination of pGADT7-mzt1 and pGBKT7-alp6; Figure 5A). In line with this observation, full-length nontagged Mzt1 and full-length FLAG-hexahistidine (His6)-Alp6, generated by in vitro translation, interacted directly when FLAG-His6-Alp6 was pulled down by anti-FLAG antibody (Figure 5B).
FIGURE 5:
Mzt1 interacts with the N-terminal end of Alp6. (A) γ-TuC components that directly interact with Mzt1 were explored using yeast 2-hybrid system. Interactions between Mzt1, GCP3Alp6, GCP2Alp4 fused to Gal4 activation domain (pGADT7), and Mzt1 and GCP3Alp6 fused to Gal4 DNA-binding domain (pGBKT7) were assessed using the adenine auxotroph. Cell growth in the absence of adenine indicated positive interaction between the proteins tested. Mzt1 interacted with GCP3Alp6, and GCP3Alp6 showed interaction with Mzt1. When constructs were tested against empty vectors, none showed interaction. Thus all constructs were valid for this assay. (B) Direct interaction between full-length Mzt1 and FLAG-His6-Alp6. The proteins were prepared by the in vitro translation system in the presence of [35S]methionine, followed by immunoprecipitation using an anti-FLAG antibody. The IP complexes were separated using SDS–PAGE and visualized by autoradiography. Twelve percent equivalent of cell extracts used for IP samples were loaded as input and supernatant samples. (C) The Alp6 truncation constructs were generated based on the disorder probability prediction of Alp6 using RONN. Alp6 is highly disordered between residues 120 and 180. Three constructs were designed: 1) 1–117, which consists of the highly ordered, first globular domain (GD1) of Alp6; 2) 1–247, which consists of the GD1 followed by the disordered region; and 3) 196–832, which consists of the larger structured domain (GD2). (D) Mapping of the Mzt1-interacting region of Alp6 with the yeast two-hybrid system. Interactions between the Alp6 regions 1–117, 195–832, full length (1–832), and 1–249 fused to Gal4-binding domain (pGBKT7) and Mzt1 fused to Gal4 activation domain (pGADT7) were assessed using the adenine auxotroph. Cell growth in the absence of Ade indicated a positive interaction between proteins tested. Mzt1 interacted with Alp6 in its region 1–247.
Secondary structure prediction suggests that Alp6 is comprised of a short, globular domain at its N-terminus (1–108, here termed GD1) followed by an unstructured disordered stretch that links to a long, globular structured region (186–832, termed GD2; Figure 5C). To explore which region of Alp6 was responsible for its interaction with Mzt1, we generated three constructs, pGBKT7-alp6(1-117), pGBKT7-alp6(1-247), and pGBKT7-alp6(195-832), corresponding to GD1, GD1 plus flexible region, and GD2, respectively. No interaction was observed with pGBKT7-alp6(1-117) or pGBKT7-alp6(195-832); however, the construct containing the flexible region, pGBKT7-alp6(1-247), showed a positive interaction with pGADT7-mzt1 (Figure 5D).
Mzt1 directly interacts with the N-terminal end of Alp6
To determine whether there is direct interaction between Mzt1 and Alp6, we generated a full-length, recombinant, C-terminally His6-tagged Mzt1 protein (Mzt1-His6) and an Alp6 1–186 fragment tagged with FLAG-His6 at its N-terminus (FLAG-His6-Alp61-186). Each protein was expressed in Escherichia coli and purified separately as described in Materials and Methods.
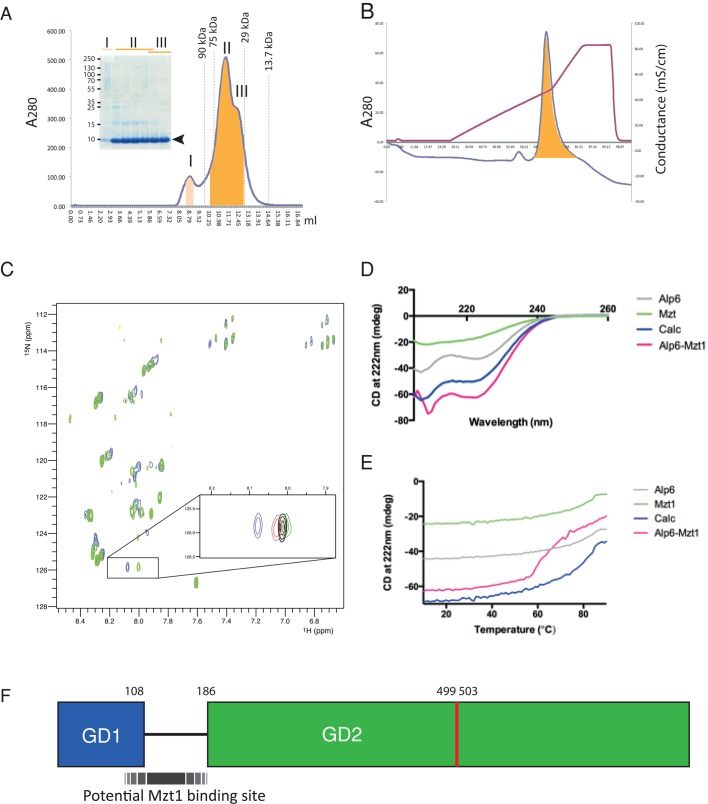
Purified Mzt1 migrated as a single band on SDS–PAGE with a molecular weight of ∼9 kDa (Figure 6A). However, size exclusion chromatography of purified Mzt1-His6 suggested that Mzt1 exists as several oligomeric species. The major species has an apparent molecular weight of 50 kDa (peak II). Two other species have apparent molecular weights of 130 (peak I) and 35 kDa (peak III), respectively (Figure 6A). In contrast to Mzt1, Alp61-186 purified as a single species (Figure 6B).
FIGURE 6:
Confirmation of a direct interaction between Mzt1 and Alp61-186. (A) Purification of recombinant 15N-labeled Mzt1 from bacteria cell extracts. Gel filtration chromatography of 15N-Mzt1-His6 on a Superdex 75 10/300 GL column. Molecular weight markers are indicated by dashed lines. Molecular weights of peaks I, II, and III are estimated to be ∼130, 50, and 35 kDa, respectively. Given that the predicted molecular weight of Mzt1-His6 is expected to be 9.1 kDa, peak I may represent dodecamer, peak II heptamer or hexamer, and peak III tetramer or trimer. SDS–PAGE analysis (4–12% gradient gel) confirmed that all eluted peaks were Mzt1-His6 (arrowhead, molecular weight ∼9 kDa). Peaks II and III were combined, buffer exchanged into NMR buffer (20 mM phosphate, 50 mM NaCl, 2 mM DTT, 10% D2O, pH 6.5), and concentrated for NMR. (B) Purification of recombinant nonlabeled Alp61-186 from bacteria cell extracts. Anion exchange chromatography of Alp61-186 (HiTrap Q HP) using a 10 mM to 1 M sodium chloride gradient (purple line). (C) 1H,15N HSQC spectra of 80 μM 15N-labeled Mzt1 in the absence (blue) and presence (green) of 160 μM Alp61-186. Inset, zoomed-in region showing the progressive chemical shift changes upon addition of 40 (red), 80 (black), and 160 μM (green) Alp61-186. The progressive spectral changes on addition of Alp61-186 are indicative of a direct interaction. (D) CD spectra of Mzt1 alone (green), Alp6 alone (gray), and 1:1 complex of Mzt1:Alp6. The complex trace (pink) shows markedly more secondary structure than the summation of the two isolated proteins (blue). (E) Denaturation profiles for Mzt1 alone (green), Alp6 alone (gray), and a 1:1 complex of Mzt1:Alp6 (pink) were measured by monitoring the change in CD at 222 nm with increasing temperature. Neither Mzt1 nor Alp6 alone showed any cooperative unfolding. However, Mzt1 and Alp6 together showed cooperative unfolding, suggesting that they form a complex. This denaturation profile of the complex was significantly different from the calculated profile of the sum of the two individual components (blue). (F) Schematic diagram of Alp6, highlighting the predicted Mzt1-binding region. Whereas recombinant Mzt1 interacts with Alp61-186, yeast two-hybrid analysis shows that Mzt1 fails to interact with Alp61-117. The Mzt1-binding region of GCP3Alp6 is predicted to include the flexible region linking GD1 and GD2 but not the region including the residues (499–503, indicated in red) corresponding to the proposed “hinge” region of human GCP3 (Guillet et al., 2011).
Nuclear magnetic resonance (NMR) spectroscopy was used to explore whether there is a direct interaction between Mzt1-His6 and FLAG-His6-Alp61-186. The 1H,15N heteronuclear single quantum coherence (HSQC) spectrum of 15N-labeled Mzt1-His6 shows a number of sharp signals, but many of the signals are broadened, consistent with an oligomeric form of Mzt1 rather than a monomeric species. Addition of increasing amounts of unlabeled FLAG-His6-Alp61-186 to 15N-labeled Mzt1-His6 induced progressive spectral changes in the Mzt1 spectrum, strongly supporting that there is a direct interaction between Mzt1 and Alp61-186 (Figure 6C).
To further explore the nature of the complex between Mzt1 and Alp6, we compared the thermal denaturation circular dichroism (CD) spectra of the individual proteins and the complex. The CD spectra of Alp61-186 suggest that it is predominantly α-helical (Figure 6D). Mzt1 is at least partly folded and also has partial helical character (Figure 6D). Of interest, denaturation of either Mzt1 or Alp61-186 showed a progressive rather than a strongly cooperative unfolding transition (Figure 6E). However, denaturation of the 1:1 mixture of Alp6 and Mzt1 revealed a much more cooperative unfolding transition (Figure 6E), supporting the conclusion that there is a cooperatively folded interface between the two proteins. This is consistent with the CD spectrum of the complex, which suggests that the complex may be somewhat more helical in character than the sum of the two isolated proteins.
DISCUSSION
In this study, we show that the fission yeast MOZART1 homologue Mzt1 is a MTOC component that plays an essential role in microtubule regulation. We show that, strikingly, Mzt1 directly interacts with the N-terminal region of GCP3Alp6.
MOZART1 is highly conserved
MOZART1 is a highly conserved protein identified through interaction screenings of the γ-TuRC (Hutchins et al., 2010; Teixido-Travesa et al., 2010). It plays essential roles in MT regulation, including mitotic spindle formation (Hutchins et al., 2010; Masuda et al., 2013; this study). The previous study predicted that the fission yeast MOZART1 homologue Mzt1 was a 97–amino acid protein (Hutchins et al., 2010). Here we show that the initiator methionine corresponds to the methionine 34 of the previously annotated mzt1 ORF. Consequently, Mzt1 is a protein of 64 amino acids, similar to MOZART1 homologues from other species (Figure 1A). This information allowed us to carry out structural analysis of Mzt1 using recombinant Mzt1 protein in this study.
Recently Arabidopsis MOZART1 homologues GIP1/GIP1a and GIP2/GIP1b were reported to be components of γ-TuC and to play a role in MT regulation (Janski et al., 2012; Nakamura et al., 2012). Of interest, GIP1/GIP1a and GIP2/GIP1b were originally identified as proteins that interact with GCP3 (Janski et al., 2008). Furthermore, as we show here in fission yeast (Figures 5 and 6), the N-terminal end of GCP3 was identified as an interacting region (Janski et al., 2012). Collectively these studies suggest that MOZART1 is likely to play a crucial conserved role in regulating the γ-TuSC. Intriguingly, no MOZART1 homologue has been identified in the budding yeast genome, so the exact mechanism of how γ-TuSC regulation occurs in this yeast without MOZART1 is unclear. It could be that an unidentified small protein similar to Mzt1 plays a similar role or that an interacting molecule (such as Spc110; Knop et al., 1997) contains a domain that plays the role of MOZART1. One major difference between the budding yeast and fission yeast is that the budding yeast uses the SPB as the sole MTOC, whereas the fission yeast has iMTOC and eMTOC, which define interphase cytoplasmic MT architecture. The MOZART1 homologue may help to generate and regulate various types of MTOCs.
Mzt1 mode of action
It has been proposed that the poor MT nucleation activity observed in the γ-TuSC may stem from the GCP3 conformation, which prevents even placement of the 13 γ-tubulin molecules from which 13 MT protofilaments are nucleated to form a single microtubule fiber of 13 protofilaments (Kollman et al., 2010). They proposed that if the hinge region of GCP3 in the reconstituted γ-TuSC is “straightened,” 13 γ-tubulin molecules will be evenly spaced, allowing lateral interactions of the γ-tubulin molecules as well as of the 13 MT protofilaments. Consequently, the efficiency of MT nucleation is expected to be enhanced (Kollman et al., 2010). This model is based on the assumption that one γ-TuRC consists of 6.5 units of γ-TuSC, which holds 13 γ-tubulin molecules. Although these numbers are not exactly the same as in a recent in vivo study, which estimated seven γ-TuSC, approximately two additional GCP3, and three additional γ-tubulin molecules at a single MT minus end (Erlemann et al., 2012), they are in reasonably good agreement with each other, and the model is likely to represent the overall situation. The question is what is the GCP3 modulator that “straightens” GCP3. It has been proposed that GCP4 is an attractive candidate based on its crystal structure, which shows an ideal angle to serve as a solid splint for GCP3 (Guillet et al., 2011), although GCP4 is not essential in fission yeast, Aspergillus, or Drosophila. Based on our findings, we propose that Mzt1 is another good candidate. First, Mzt1 is essential, as is the case for GCP2, GCP3, and γ-tubulin (Figure 3). Second, it directly interacts with GCP3 but not with GCP2, implying direct regulation of GCP3 (Figure 5A). Third, binding of Mzt1 to Alp61-186 brings better protein folding, indicating that the interaction is likely to affect GCP3Alp6 core structure (Figure 6, D and E).
If Mzt1 acts as the GCP3 modulator to “straighten up” the GCP3, our data suggest that Mzt1 may not be a mere splint. The predicted “hinge” region of GCP3Alp6, residues 499–503 (based on the predicted human GCP3 hinge region, 551–555; Guillet et al., 2011), is distal to the GCP3Alp6 N-terminal region (1–186), which interacts with Mzt1 (Figure 6F and Supplementary Figure S3). The N-terminal end of budding yeast GCP3Spc98 has been mapped to the base of the γ-TuSC (Choy et al., 2009). Considering the fact that purified recombinant Mzt1 forms oligomers, one possibility is that Mzt1 forms a multimer complex that fits deep inside the funnel-shaped γ-tubulin ring-like complex and adjusts the angle of the GCP3 (Figure 7). Narrowing down the Mzt1 interaction region in GCP3Alp6 and performing structural analysis of the Mzt1-Alp6 complex are essential to finding out the precise molecular mechanism by which Mzt1 regulates the γ-TuC.
FIGURE 7:
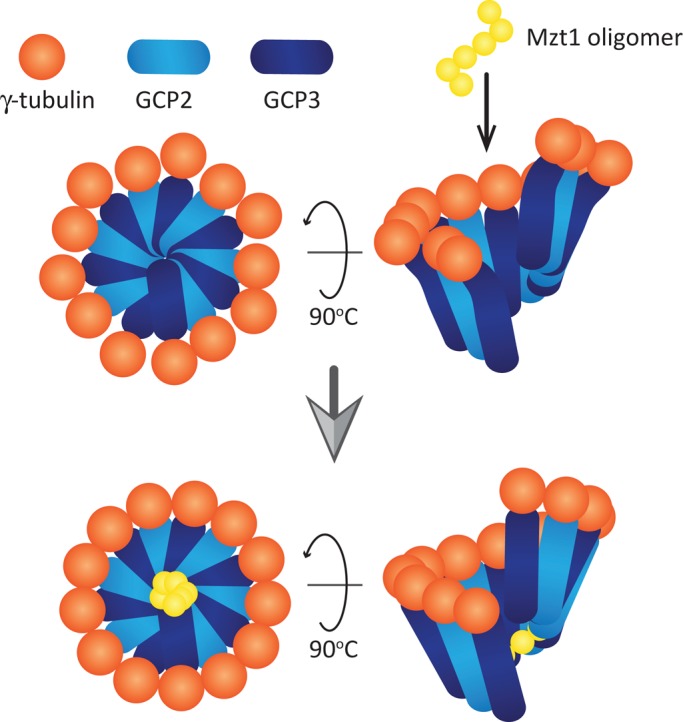
A speculative model for Mzt1 function to activate γ-TuC. Model adapted from the one proposed by Kollman et al. (2011). A ring-like γ-TuSC made of GCP2, GCP3, and γ-tubulin carries 13 γ-tubulins, which are not evenly distributed. Mzt1 oligomer incorporated into the γ-TuC may be recruited to the bottom of the ring-like structure via its interaction with GCP3. The Mzt1 oligomer stabilizes the whole complex, resulting in the 13 γ-tubulin molecules being more evenly distributed and leading to more efficient MT nucleation.
Mzt1 may also regulate the γ-TuC in other ways, although it is reasonable to assume that its prime effector is GCP3, to which Mzt1 directly binds. An alternative mechanism could be that Mzt1 acts as an adaptor to recruit other essential MTOC regulators, such as Spc110, to GCP3.
Spc110 and MOZART1 in γ-TuC activation
In the budding yeast γ-TuSC reconstitution experiment, addition of an amino-terminal Spc110 fragment (Spc1101-220) increased stability of the γ-TuSC and its MT nucleation activity (Kollman et al., 2010). Spc110 helped to recruit the γ-TuSC to the desired nucleation site, the SPB, by binding to both GCP2Spc97 and GCP3Spc98 (Knop and Schiebel, 1997). By doing so, it also may help γ-TuSC to obtain rigorous MTOC activity. Spc110 is related to centrosomin in higher eukaryotes, and it is highly interesting to know whether a similar event occurs in other organisms. Fission yeast has three Spc110-related proteins, Mto1, Mto2, and Pcp1 (Flory et al., 2002; Sawin et al., 2004; Venkatram et al., 2004, 2005; Janson et al., 2005; Samejima et al., 2005; Zimmerman and Chang, 2005). It is important to examine whether Mzt1 affects Alp6-Mto1, Alp6-Mto2, or Alp6-Pcp1 complex formation.
Role of Mzt1 in recruiting the γ-TuC to the MTOC
In HeLa cells, MOZART1 depletion removed the γ-tubulin signal from the centrosome (Hutchins et al., 2010). In Arabidopsis, GIP1/2 depletion resulted in a decline in the signal intensity of γ-tubulin, GCP3, and GCP2 at MT nucleation sites, although the signal was not completely abolished (Janski et al., 2012). A similar trend was observed in fission yeast, in which decrease of the γ-TuC signal was seen at the SPB (Masuda et al., 2013). These observations indicate that Mzt1 affects γ-TuC recruitment to the MTOC site. This may be a direct consequence of Mzt1 depletion if Mzt1 acts as a bridge between γ-TuC and the MTOC site. Alternatively, it may be an indirect effect in which a conformational change of GCP3 brought about by Mzt1 helps to anchor the γ-TuC to the MT nucleation site. Although we prefer the latter model due to the low molecular weight of Mzt1, further analysis is required to determine the exact molecular mechanism.
Involvement of γ-TuC in the regulation of cytokinesis
Although the immediate effect of Mzt1 depletion was mitotic spindle formation defect and impaired interphase MT arrangement, when Mzt1 was depleted for a longer time, cells with the cytokinesis defect started to accumulate, indicating a role of γ-TuC in the regulation of cytokinesis (Figure 3, F and G). This phenotype is consistent with previous studies, in which impairment of γ-TuC by an alp4-1891 mutant led to septum formation defects with a misregulated septum initiation network (Vardy et al., 2002). In addition, loss of Mto1, a γ-TuC–associating protein, also leads to cytokinesis defect (Venkatram et al., 2004, 2005; Samejima et al., 2005). Furthermore, when human γ-tubulin mutants were introduced into fission yeast to evaluate the mutants’ functionality, some of the mutants showed a cytokinesis defect (Hendrickson et al., 2001). Collectively these observations strongly suggest that γ-TuC plays a key role in the regulation of cytokinesis.
In summary, we identified the fission yeast MOZART1 homologue, Mzt1, as an essential MTOC protein of 64 amino acids that regulates MT organization. Of importance, we showed that it binds directly to the N-terminal of GCP3Alp6. We propose that this binding may help to alter the arrangement of Alp6 and γ-TuC, facilitating microtubule growth. Further analysis of Mzt1 using fission yeast, a highly tractable model organism, is likely to reveal the essential mechanism of how the γ-TuC regulates MT organization.
MATERIALS AND METHODS
Yeast strains and media
Schizosaccharomyces pombe strains used in this study are listed in Table 1. General genetic methods and media for S. pombe were described previously (Tanaka et al., 2005). A chromosome-integration vector to generate the Pnda3-GFP-atb2<<aur1r and Pnda3-mCherry-atb2<<aur1r alleles was a generous gift from Hisahiro Masuda and Takashi Toda (Cancer Research UK, London Research Institute, UK). Sexual differentiation of yeast cells was induced on solid SPA plates as previously described (Tanaka et al., 2005).
TABLE 1:
Fission yeast strains used in this study.
| Figure 1D | |
| KT3519 | h90 ade6-M216 leu1+::Pnmt81-mzt1-S-GFP-2xFLAG mzt1-GFP-2xFLAG::KanR |
| KT3522 | h90 ade6-M216 leu1+::Pnmt81-mzt1-L-GFP-2xFLAG mzt1-GFP-2xFLAG::KanR |
| Figure 2A | |
| KT4164 | h90 ade6-M216 leu1.32 sid4-tdTomato::HygR mzt1-GFP-2xFLAG::KanR |
| Figure 2, B and C | |
| KT3228 | h90 ade6-M216 leu1.32 Pnda3-mCherry-atb2-aur1R mzt1-GFP-2xFLAG::KanR |
| Figure 2D | |
| KT301 | h90 ade6-M216 leu1.32 |
| KT3224 | h90 ade6-M216 leu1.32 mzt1-GFP-2×FLAG::KanR |
| Figure 3, A and B | |
| KT3449 | h90/h90 ade6-M216/ade6-M210 leu1.32/leu1.32 mzt1+/mzt1::ClonNATR |
| Figure 3, C–G | |
| KT3711 | h90 ade6-M216 leu1+::Pnmt81-mzt1-S-GFP-2xFLAG Pnda3-mCherry-atb2-aur1R mzt1::HygR |
Gene deletion and GFP-2xFLAG and mCherry tagging of genes were performed using the direct chromosomal integration method (Wach, 1996; Bahler et al., 1998; Funaya et al., 2012). For tdTomato tagging, pFA6a-tdTomato-kanMX6 and pFA6a-tdTomato-hphMX6 were generated by replacing GFP(S65T) of pFA6a-GFP(S65T)-kanMX6 plasmid (Bahler et al., 1998) or pFA6a-GFP(S65T)-hphMX6 plasmid (Sato et al., 2005) with a tdTomato cassette (Shaner et al., 2004), which was a generous gift from Roger Tsien (University of California, San Diego).
Introduced mutations or deletions were confirmed by DNA sequencing (Protein Nucleic Acid Chemistry Laboratory, University of Leicester). Chromosome integration of mzt1-S and mzt1-L under the nmt81 promoter at the leu1 gene locus was carried out using the pDUALGFH81 vector (Matsuyama et al., 2004) kindly provided by Akihisa Matsuyama and Minoru Yoshida (RIKEN, Japan). Briefly, DNA fragments corresponding to the mzt1-S or mzt1-L were amplified from the wild-type yeast genome using a pair of primers nmt-tam4S-F (5′-ACTTTCTGACTTATAGTCGCTTTGTTAAAATGTCTGAATCTACCAAAGAGACAATAGAAG-3′) or nmt-tam4L-F (5′-TTTCTGACTTATAGTCGCTTTGTTAAAATGAGCTACTCCGAGACGCCTATTCCAGTAAAC-3′) and Du-tamT-R (5′-GTGGCGCGCCGGATCCTCTAGAGTCGACTACTTGTCATCGTCGTCCTTGTAGTCCTTGTCA-3′). In addition, three parts of the pDUALGFH81 were also amplified. One of them was amplified using a pair of primers leu1-nmt-F (5′- CGTGACTGGGAAAACCCTGGCGTTAGATCAGAAAATTATCGCCATAAAAGACAGAATAAG-3′) and nmt-tam4S-R (5′-CTTCTATTGTCTCTTTGGTAGATTCAGACATTTTAACAAAGCGACTATAAGTCAGAAAG-3′) or leu1-nmt-F and nmt-tam4L-R (5′-TTTACTGGAATAGGCGTCTCGGAGTAGCTCATTTTAACAAAGCGACTATAAGTCAGAAAG-3′). The second part was amplified using a pair of primers Du-tamT-F (5′-CAAGGACTACAAGGACGACGATGACAAGTAGTCGACTCTAGAGGATCCGGCGCGCCAC-3′) and leu1-5′-R (5′-CAAA CGA GCA ATA CGA GAA ACT TCT TCC AAA C-3′). The third part was amplified using a pair of primers leu1-3′-F (5′-TTA GCT GAAA CTT CCAA CCC TCC TGC TCC-3′) and leu1-nmt-R (5′-TATTCTGTCTTTTATGGCGATAATTTTCTGATCTAACGCCAGGGTTTTCCCAGTCACGAC-3′). The resultant three PCR fragments from the pDUALGFH81 vector were “sewn” with the mzt1-S or mzt1-L PCR fragment by a PCR using the primers leu1-3′-F and leu1-5′-R. The generated fragment was transformed into yeast strains that carry leu1.32 mutation, and cells were selected for recovery from leucine auxotroph. Correct chromosome integration was examined with genomic PCR, and the leu1 gene locus was sequenced to confirm that no additional mutation was introduced into mzt1-S or mzt1-L.
Fluorescence microscopy
Live imaging of the cells was performed at 30°C using Leica SP5 (objective, HCX PL APO Lbd. BI, 63×/numerical aperture [NA] 1.4; Figure 2, A and B) and Olympus FV1000 (objective, UPlanSAPO, 60×/NA 1.35; Figure 2C) laser scanning confocal microscopes and a Nikon Eclipse Ti-E microscope (objective, Plan Apo VC, 100×/NA 1.4) with an Andor iXon EM-DU897 camera and CoolLED pE-1 LED excitation system (Figure 3, D–F, and Supplemental Figure S1) as previously described (Funaya et al., 2012). Cells were immobilized on glass-bottom dishes (MatTek Corporation, Ashland, MA) coated with lectin from Bandeiraea simplicifolia (Sigma, Dorset, UK) and incubated with minimal media. For each time point, images along the z-axis were taken every 0.4 μm to fully cover the thickness of the cell. Obtained images were processed by Huygens Essential (Scientific Volume Imaging, Hilversum, Netherlands) deconvolution software, and maximum z-projection images were generated by ImageJ (National Institutes of Health, Bethesda, MD).
Two-hybrid assays
Two-hybrid analysis of interacting proteins was performed as previously described (Tanaka et al., 2005) using yeast strain AH109. ORFs of alp4+, alp6+, and mzt1+ were introduced into the pGADT7 and pGBKT7 vectors (Clontech, Saint Germain en laye, France), and adenine auxotroph was used as a read-out for the interaction.
Western blotting
Fission yeast whole-cell extracts were prepared by trichloroacetic acid (TCA) extraction to avoid protein degradation. A total of 2.5 × 108 cells was collected by filtration and denatured by 5 ml of 20% TCA. Cell pellets were rinsed once with 1 ml of 20% TCA and resuspended in 250 μl of 20% TCA. Equal volume of acid-washed glass beads (Sigma) was added, and the cells were disrupted by vortexing using FastPrep 24 homogenizer (MP Biomedicals, Illkirch, France) at speed 6.5 for 1 min for three times. Cell extracts were collected, and 1 ml of 5% TCA was added. The samples were then centrifuged for 10 min at 4°C at 14,000. Supernatant was removed, and the pellet was washed with 750 μl of 100% ethanol. The pellet was finally resuspended in 100 μl of 1 M Tris (pH 8.0) and 100 μl of Laemmli 3× loading buffer. The samples were denatured at 100°C for 5 min and spun down, and 20 μl of the supernatant was analyzed by SDS–PAGE.
To detect α-tubulin, monoclonal antibody TAT-1, a generous gift from Keith Gull (University of Oxford, UK), was used at 1:3000 dilution. To detect Mzt1-GFP-2xFLAG, either anti-GFP monoclonal antibody (final concentration 0.2 μg/ml; 11814460001; Roche, Basel, Switzerland) or anti-FLAG monoclonal antibody (final concentration 0.5 μg/ml; F1804; Sigma) was used. To detect γ-tubulin in immunoprecipitation complexes, monoclonal anti-γ-tubulin–specific primary antibody (1:5000 dilution; T6557; Sigma) was used, followed by anti-mouse immunoglobulin G (IgG) light-chain–specific secondary antibody (Millipore, Billerica, MA) to avoid overlap of the molecular weights of γ-tubulin and IgG heavy chain.
Immunoprecipitation
To prepare Mzt1 immunocomplex from the strains KT301 and KT3224, cells were grown in YE+ade to a cell density of 1 × 107 cells/ml. One minute before harvesting the cells, phenylmethylsulfonyl fluoride (PMSF) was added to a final concentration of 1 mM. A total of 4 × 108 cells was collected in a 1.5-ml screw-cap tube, rinsed with CDC2 STOP buffer (Moreno et al., 1991), snap-frozen in liquid nitrogen, and stored at −80°C. Native soluble cell extracts were prepared by adding 400 μl of KS buffer (7.5% glycerol, 50 mM Tris, pH 7.4, 100 mM NaCl, 5 mM ethylene glycol tetraacetic acid [EGTA], 1 mM EDTA, 1% Triton X-100, 1 mM PMSF, Complete protease inhibitors [Roche]) and 500 μl of glass beads. Cells were disrupted using Fastprep24 (speed 6.5, 20 s×2; MP Biomedicals). The cell extracts were collected and centrifuged for 10 min at 13,000 rpm, and the supernatant was added to 25 μl of Dynabeads Protein G (Invitrogen, Paisley, UK), loaded, and cross-linked with 5 μg of anti-FLAG antibody (T6557; Sigma). Immunoprecipitation was performed on ice. The bound protein was washed three times in KS buffer to remove any non–specifically bound protein, and the final pellet was resuspended in 40 μl of KS buffer and 40 μl of 3× Laemmli loading buffer (LB; Laemmli, 1970) and boiled for 5 min, and the supernatant was run on the gel.
Protein production using in vitro translation system
Gene fragments encoding FLAG-His6-Alp6, nontagged Alp4, nontagged Gtb1, and nontagged Mzt1 were cloned in the pLEICS12 vector (Protex [Protein Expression Laboratory], University of Leicester), which has a T7 promoter. Proteins were produced using the TNT T7 Coupled Reticulocyte Lysate System (Promega, Madison, WI) in the presence of [35S]methionine.
To assess γ-TuC formation, pull-downs were performed using anti-FLAG antibody–conjugated Dynabeads Protein G, as described earlier, in CDK5RAP2 buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.2, 150 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1 mM dithiothreitol [DTT], 0.5% NP-40; Choi et al., 2010). The bound complex on the beads was washed twice with CDK5RAP2 buffer, and the pellet was resuspended with 10 μl of CDK5RAP2 buffer and 10 μl of 3× loading dye and loaded onto the gel.
Expression and purification of 15N-Mzt1 and Alp61-186
Constructs were expressed in E. coli Rosetta cells cultured in LB or, for preparation of 15N-labeled samples, in 2M9 minimal media containing 1 g of [15N]ammonium chloride/l. The C-terminally His-tagged Mzt1 was purified using HisTrap resin (GE Healthcare, Buckinghamshire, UK), followed by gel filtration on a Superdex 75 10/300 GL column (GE Healthcare). His-tagged Alp61-186 was purified on nickel-nitriloacetic acid, followed by anion exchange chromatography using a HiTrap Q HP 5-ml column (GE Healthcare). The proteins were dialyzed into 20 mM phosphate, 50 mM NaCl, and 2 mM DTT, pH 6.5. Protein purity was analyzed by SDS–PAGE and protein concentrations determined using Qubit.
CD spectroscopy
CD spectra were recorded using a Chirascan-plus CD spectrometer. Far-ultraviolet CD spectra were recorded at 20°C over the wavelength range 200–250 nm in a quartz cell of 0.1-cm path length (time per points of 0.5 s). Proteins were dissolved in 20 mM sodium phosphate (pH 6.5) and 50 mM NaCl at a concentration of 20 μM. For denaturation studies, the unfolding of α-helices was followed at 222 nm.
NMR of Mzt1 and Alp6
The purified Mzt1 and Alp6 were dialyzed into 20 mM phosphate, 50 mM NaCl, and 2 mM DTT, pH 6.5, overnight at 4°C. Proteins concentrations were determined using Qubit. All 1H,15N HSQC experiments were performed at 25°C using a Bruker AVANCE AVII 800 spectrometer equipped with CryoProbe. A series of HSQC spectra was measured for 15N-labeled protein alone (80 μM) and then in the presence of increasing amounts of unlabeled protein.
Supplementary Material
Acknowledgments
We are grateful to Iain Hagan (Paterson Institute for Cancer Research, UK) for critical reading of the manuscript and stimulating discussions. We are thankful to Hirohisa Masuda and Takashi Toda for stimulating discussions and communication of unpublished results. We thank Louise Fairall and Neil Bate (University of Leicester, UK) for helpful suggestions and Xiaowen Yang (Protex [Protein Expression Laboratory], University of Leicester) and Kees Straatman (Leicester Advanced Imaging Facility, University of Leicester, UK) for support, technical assistance, and helpful advice. We thank all the lab members for their valuable support and discussion. We are indebted to Keith Gull, Akihisa Matsuyama, Minoru Yoshida, Hirohisa Masuda, Takashi Toda, Roger Tsien, and Masa Sato for providing antibodies and reagents. This work was supported by the Wellcome Trust Institutional Strategic Support Fund (097828/Z/11/Z).
Abbreviations used:
- CD
circular dichroism
- MT
microtubule
- MTOC
microtubule organizing center
- NMR
nuclear magnetic resonance
- γ-TuC
γ-tubulin complex
- γ-TuRC
γ-tubulin ring complex
- γ-TuSC
γ-tubulin small complex
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0253) on September 4, 2013.
Present addresses:*MRC Laboratory of Molecular Biology, Cambridge Biomedical Campus, Cambridge CB2 0QH, United Kingdom;
†Department of Genetics, Evolution and Environment, University College London, London WC1E 6BT, United Kingdom.
REFERENCES
- Anders A, Lourenco PC, Sawin KE. Noncore components of the fission yeast gamma-tubulin complex. Mol Biol Cell. 2006;17:5075–5093. doi: 10.1091/mbc.E05-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd., Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bitton DA, Wood V, Scutt PJ, Grallert A, Yates T, Smith DL, Hagan IM, Miller CJ. Augmented annotation of the Schizosaccharomyces pombe genome reveals additional genes required for growth and viability. Genetics. 2011;187:1207–1217. doi: 10.1534/genetics.110.123497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YK, Liu P, Sze SK, Dai C, Qi RZ. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol. 2010;191:1089–1095. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy RM, Kollman JM, Zelter A, Davis TN, Agard DA. Localization and orientation of the gamma-tubulin small complex components using protein tags as labels for single particle EM. J Struct Biol. 2009;168:571–574. doi: 10.1016/j.jsb.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlemann S, Neuner A, Gombos L, Gibeaux R, Antony C, Schiebel E. An extended gamma-tubulin ring functions as a stable platform in microtubule nucleation. J Cell Biol. 2012;197:59–74. doi: 10.1083/jcb.201111123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory MR, Morphew M, Joseph JD, Means AR, Davis TN. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 2002;13:47–58. [PubMed] [Google Scholar]
- Fujita A, Vardy L, Garcia MA, Toda T. A fourth component of the fission yeast gamma-tubulin complex, Alp16, is required for cytoplasmic microtubule integrity and becomes indispensable when gamma-tubulin function is compromised. Mol Biol Cell. 2002;13:2360–2373. doi: 10.1091/mbc.02-01-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funaya C, et al. Transient structure associated with the spindle pole body directs meiotic microtubule reorganization in S. pombe. Curr Biol. 2012;22:562–574. doi: 10.1016/j.cub.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet V, et al. Crystal structure of gamma-tubulin complex protein GCP4 provides insight into microtubule nucleation. Nat Struct Mol Biol. 2011;18:915–919. doi: 10.1038/nsmb.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Bendezu FO, Martin SG. Fission yeast: in shape to divide. Curr Opin Cell Biol. 2012;24:858–864. doi: 10.1016/j.ceb.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Hagan IM. The fission yeast microtubule cytoskeleton. J Cell Sci. 1998;111:1603–1612. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- Heitz MJ, Petersen J, Valovin S, Hagan IM. MTOC formation during mitotic exit in fission yeast. J Cell Sci. 2001;114:4521–4532. doi: 10.1242/jcs.114.24.4521. [DOI] [PubMed] [Google Scholar]
- Hendrickson TW, Yao J, Bhadury S, Corbett AH, Joshi HC. Conditional mutations in gamma-tubulin reveal its involvement in chromosome segregation and cytokinesis. Mol Biol Cell. 2001;12:2469–2481. doi: 10.1091/mbc.12.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M. The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- Hutchins JR, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janski N, Herzog E, Schmit AC. Identification of a novel small Arabidopsis protein interacting with gamma-tubulin complex protein 3. Cell Biol Int. 2008;32:546–548. doi: 10.1016/j.cellbi.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Janski N, Masoud K, Batzenschlager M, Herzog E, Evrard JL, Houlne G, Bourge M, Chaboute ME, Schmit AC. The GCP3-interacting proteins GIP1 and GIP2 are required for gamma-tubulin complex protein localization, spindle integrity, and chromosomal stability. Plant Cell. 2012;24:1171–1187. doi: 10.1105/tpc.111.094904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson ME, Setty TG, Paoletti A, Tran PT. Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J Cell Biol. 2005;169:297–308. doi: 10.1083/jcb.200410119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast gamma-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol. 2011;12:709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Masuda H, Mori R, Yukawa M, Toda T. Fission yeast MOZART1/Mzt1 is an essential γ-tubulin complex component required for complex recruitment to the microtubule organizing center, but not its assembly. Mol Biol Cell. 2013;24:2894–2906. doi: 10.1091/mbc.E13-05-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Toda T, Miyamoto R, Haraguchi T, Hiraoka Y. Modulation of Alp4 function in Schizosaccharomyces pombe induces novel phenotypes that imply distinct functions for nuclear and cytoplasmic gamma-tubulin complexes. Genes Cells. 2006;11:319–336. doi: 10.1111/j.1365-2443.2006.00946.x. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Shirai A, Yashiroda Y, Kamata A, Horinouchi S, Yoshida M. pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast. 2004;21:1289–1305. doi: 10.1002/yea.1181. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Microtubule assembly nucleated by isolated centrosomes. Nature. 1984;312:232–237. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Yagi N, Kato T, Fujita S, Kawashima N, Ehrhardt DW, Hashimoto T. Arabidopsis GCP3-interacting protein 1/MOZART 1 is an integral component of the gamma-tubulin-containing microtubule nucleating complex. Plant J. 2012;71:216–225. doi: 10.1111/j.1365-313X.2012.04988.x. [DOI] [PubMed] [Google Scholar]
- Oakley CE, Oakley BR. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, Zheng Y. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe P, Hirata D, Childs D, Vardy L, Toda T. Identification of novel temperature-sensitive lethal alleles in essential beta-tubulin and nonessential alpha 2-tubulin genes as fission yeast polarity mutants. Mol Biol Cell. 1998;9:1757–1771. doi: 10.1091/mbc.9.7.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff JW, Kellogg DR, Alberts BM. Drosophila gamma-tubulin is part of a complex containing two previously identified centrosomal MAPs. J Cell Biol. 1993;121:823–835. doi: 10.1083/jcb.121.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima I, Lourenco PC, Snaith HA, Sawin KE. Fission yeast mto2p regulates microtubule nucleation by the centrosomin-related protein mto1p. Mol Biol Cell. 2005;16:3040–3051. doi: 10.1091/mbc.E04-11-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Lourenco PC, Snaith HA. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr Biol. 2004;14:763–775. doi: 10.1016/j.cub.2004.03.042. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Tran PT. Cytoplasmic microtubule organization in fission yeast. Yeast. 2006;23:1001–1014. doi: 10.1002/yea.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of gamma-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kohda T, Yamashita A, Nonaka N, Yamamoto M. Hrs1p/Mcp6p on the meiotic SPB organizes astral microtubule arrays for oscillatory nuclear movement. Curr Biol. 2005;15:1479–1486. doi: 10.1016/j.cub.2005.07.058. [DOI] [PubMed] [Google Scholar]
- Teixido-Travesa N, Villen J, Lacasa C, Bertran MT, Archinti M, Gygi SP, Caelles C, Roig J, Luders J. The gammaTuRC revisited: a comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol Biol Cell. 2010;21:3963–3972. doi: 10.1091/mbc.E10-05-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlin GC, Morrell JL, Gould KL. The spindle pole body protein Cdc11p links Sid4p to the fission yeast septation initiation network. Mol Biol Cell. 2002;13:1203–1214. doi: 10.1091/mbc.01-09-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L, Fujita A, Toda T. The gamma-tubulin complex protein Alp4 provides a link between the metaphase checkpoint and cytokinesis in fission yeast. Genes Cells. 2002;7:365–373. doi: 10.1046/j.1365-2443.2002.00530.x. [DOI] [PubMed] [Google Scholar]
- Vardy L, Toda T. The fission yeast gamma-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 2000;19:6098–6111. doi: 10.1093/emboj/19.22.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram S, Jennings JL, Link A, Gould KL. Mto2p, a novel fission yeast protein required for cytoplasmic microtubule organization and anchoring of the cytokinetic actin ring. Mol Biol Cell. 2005;16:3052–3063. doi: 10.1091/mbc.E04-12-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram S, Tasto JJ, Feoktistova A, Jennings JL, Link AJ, Gould KL. Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol Biol Cell. 2004;15:2287–2301. doi: 10.1091/mbc.E03-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verollet C, Colombie N, Daubon T, Bourbon HM, Wright M, Raynaud-Messina B. Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J Cell Biol. 2006;172:517–528. doi: 10.1083/jcb.200511071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Oakley BR. In vivo analysis of the functions of gamma-tubulin-complex proteins. J Cell Sci. 2009;122:4218–4227. doi: 10.1242/jcs.059196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman S, Chang F. Effects of γ-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol Biol Cell. 2005;16:2719–2733. doi: 10.1091/mbc.E04-08-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.