ERM-binding protein of 50 kDa (EBP50) and NHE3 kinase A regulatory protein (E3KARP) are closely related but show dramatically different dynamics in microvilli. The high dynamics of EBP50 is determined by a region in its tail and is inhibited by its PDZ domains, but is activated upon PDZ ligand binding. Proteomic analysis of the effects of EBP50 dynamics identifies a novel PDZ binding partner, IRSp53.
Abstract
The closely related apical scaffolding proteins ERM-binding phosphoprotein of 50 kDa (EBP50) and NHE3 kinase A regulatory protein (E3KARP) both consist of two postsynaptic density 95/disks large/zona occludens-1 (PDZ) domains and a tail ending in an ezrin-binding domain. Scaffolding proteins are thought to provide stable linkages between components of multiprotein complexes, yet in several types of epithelial cells, EBP50, but not E3KARP, shows rapid exchange from microvilli compared with its binding partners. The difference in dynamics is determined by the proteins’ tail regions. Exchange rates of EBP50 and E3KARP correlated strongly with their abilities to precipitate ezrin in vivo. The EBP50 tail alone is highly dynamic, but in the context of the full-length protein, the dynamics is lost when the PDZ domains are unable to bind ligand. Proteomic analysis of the effects of EBP50 dynamics on binding-partner preferences identified a novel PDZ1 binding partner, the I-BAR protein insulin receptor substrate p53 (IRSp53). Additionally, the tails promote different microvillar localizations for EBP50 and E3KARP, which localized along the full length and to the base of microvilli, respectively. Thus the tails define the localization and dynamics of these scaffolding proteins, and the high dynamics of EBP50 is regulated by the occupancy of its PDZ domains.
INTRODUCTION
Polarized epithelial cells are characterized by the presence of abundant microvilli on their apical domain. Microvilli consist of a core of bundled actin filaments tethered laterally to the plasma membrane. A critical component of these microvilli is the microfilament–membrane linking protein ezrin, a member of the ezrin/radixin/moesin (ERM) protein family. Ezrin is negatively regulated by an intramolecular association between its N-terminal 4.1 ERM (FERM) domain and its C-terminal tail domain. On activation by phosphorylation and phosphatidylinositol 4,5-bisphosphate binding, binding sites on the FERM domain for integral membrane proteins and the scaffolding protein ERM-binding protein of 50 kDa (EBP50) become accessible, as does the F-actin binding site in its C-terminus (Fehon et al., 2010). The conformational regulation of ezrin creates a regulated linkage between F-actin and the membrane proteins it associates with both directly and indirectly through EBP50 (Reczek and Bretscher, 1998).
EBP50, also known as Na+-H+ exchanger regulatory factor 1 (NHERF1), is a member of the NHERF family and is most closely related to NHE3 kinase A regulatory protein (E3KARP), which, like EBP50, has two postsynaptic density 95/disks large/zona occludens 1 (PDZ) domains and an ezrin-binding domain (EBD) at its C-terminal tail (Weinman et al., 1995; Reczek et al., 1997; Yun et al., 1997). EBP50 and E3KARP show distinct tissue distributions, yet both have an extremely high affinity for the FERM domain of ERM proteins and show some overlap in PDZ ligand specificity (Ingraffea et al., 2002; Shenolikar et al., 2004; Terawaki et al., 2006). We have shown that formation of microvilli on the choriocarcinoma-derived JEG-3 cell line requires EBP50, and specifically its ability to bind both to ezrin and PDZ1 ligands (Garbett et al., 2010; LaLonde et al., 2010). Additionally, mice lacking either ezrin or EBP50 show malformed microvilli, further suggesting the two proteins must function together for microvillar structure or regulation (Morales et al., 2004; Saotome et al., 2004).
Scaffolding proteins are generally believed to provide stable linkages between components of macromolecular complexes. However, EBP50 is a surprisingly dynamic component of microvilli when compared with two of its classical binding partners, ezrin and the PDZ ligand podocalyxin (Yu et al., 2007; Garbett and Bretscher, 2012). Interestingly, E3KARP, EBP50’s closest paralogue, shows dramatically slower exchange from microvilli (Garbett and Bretscher, 2012). Here we exploit the differences between EBP50 and E3KARP to reveal new mechanistic insights into the regulation of EBP50’s dynamics, localization, and binding partners.
RESULTS
Microvillar proteins show similar exchange rates in several polarized epithelial cell lines
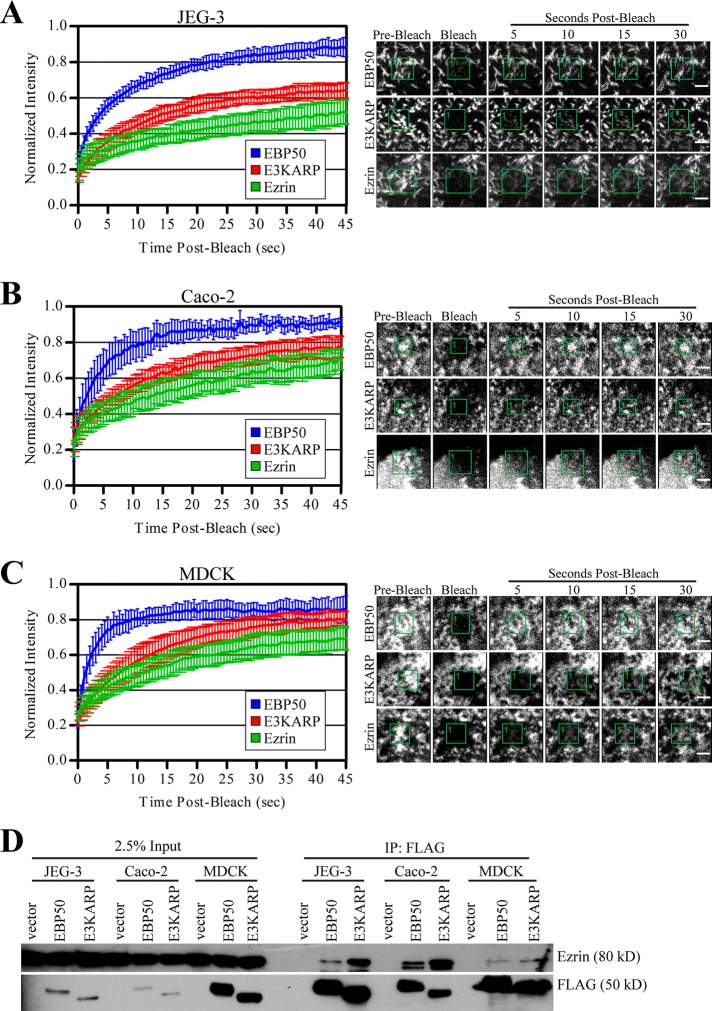
We have recently analyzed the dynamics of green fluorescent protein (GFP)-EBP50 and ezrin-GFP in the choriocarcinoma-derived JEG-3 cell line and have shown that EBP50 has remarkably rapid dynamics in microvilli (recovery in ∼5 s) compared with its binding partner ezrin (recovery in ∼30 s). Interestingly, EBP50’s most closely related family member, E3KARP, has slow dynamics (recovery in ∼20 s; Garbett and Bretscher, 2012). To see whether these effects extend to other epithelial cell lines, we used fluorescence recovery after photobleaching (FRAP) on Caco-2 and MDCK cells expressing low levels of GFP-EBP50, GFP-E3KARP, and ezrin-GFP (Figure 1, A–C). Despite polarized Caco-2 and MDCK cells having much shorter microvilli than JEG-3 cells, they show nearly identical exchange rates for EBP50, E3KARP, and ezrin Previously we have shown that mutations to EBP50 that reduce its dynamics also increase its ability to precipitate ezrin from JEG-3 cell lysate (Garbett and Bretscher, 2012). To see whether the slower dynamics of E3KARP was also representative of a stronger interaction with ezrin, we transiently expressed 3xFLAG-tagged EBP50 or E3KARP in JEG-3, Caco-2, and MDCK cells, performed FLAG immunoprecipitations, and probed for endogenous ezrin (Figure 1D). In all three cell lines, EBP50 precipitated less endogenous ezrin than seen with E3KARP, indicating the reduced dynamics of E3KARP was likely due to a more persistent interaction with ezrin. We continued our study using JEG-3 cells, which are easier to manipulate in culture and have microvilli that are more readily observed by confocal microscopy.
FIGURE 1:
EBP50, E3KARP, and ezrin show similar dynamic profiles in several polarized cell lines. Left, photobleaching recovery curves of GFP-tagged EBP50, E3KARP, and ezrin in JEG-3 (A), Caco-2 (B), and MDCK (C) cells. Error bars show SD; n ≥ 7 for all experiments. Right, representative time points for the recovery curves of the respective cells types. Scale bars: 2 μm. (D) 3xFLAG-tagged EBP50 and E3KARP expressed in JEG-3, Caco-2, and MDCK cells were immunoprecipitated (IP) and blotted for FLAG and endogenous ezrin.
A region of the EBP50 tail absent from E3KARP confers a high exchange rate from microvilli
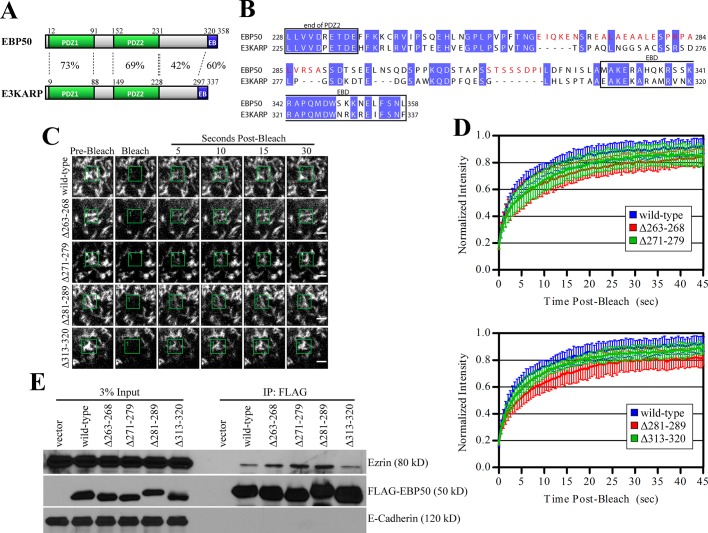
EBP50 and E3KARP are the most closely related members of the NHERF family, yet their dynamics in microvilli and ability to associate with ezrin, as seen by immunoprecipitation, are dramatically different (Figure 1; Garbett and Bretscher, 2012). They share the most identity in their PDZ domains (73% in PDZ1; 69% in PDZ2) and the EBD (60%) (Figure 2A). The region between their PDZ2 and EBD is more divergent, with only 42% shared identity. To see whether parts of this region contribute to the faster dynamics of EBP50, we deleted residues that were highly divergent from E3KARP (shown in red in Figure 2B) and examined their exchange from microvilli by FRAP (Figure 2, C and D).
FIGURE 2:
Unique regions in the tail of EBP50 have modest effects on its dynamics. (A) Schematic of EBP50 and E3KARP domains with percent identities indicated. (B) Sequence alignment of human EBP50 and E3KARP. Conserved residues are highlighted in blue, regions deleted in the EBP50 deletion mutants are in red. (C) Representative time points from photobleaching experiments of GFP-tagged EBP50 wild type and the indicated deletion mutants. Photobleached regions are indicated by green boxes. Scale bars: 2 μm. (D) Photobleaching recovery curves. Top: GFP-EBP50 wild type, Δ263–268, and Δ271–279; bottom: wild type, Δ281–289, and Δ313–320. Error bars show SD; n ≥ 9 for all experiments. (E) 3xFLAG-tagged EBP50 constructs expressed in JEG-3 cells were immunoprecipitated (IP) and blotted for FLAG, endogenous ezrin, and E-cadherin as a control.
We performed photobleaching experiments on JEG-3 cells transfected with GFP-tagged EBP50 wild-type or the various deletion mutants (Figure 2, B–D). Deletion of residues 263–268, 271–279, 281–289, or 313–320 all resulted in a slight reduction of EBP50 exchange from microvilli compared with wild-type. To see whether the modest effect on dynamics correlated with binding ezrin, we immunoprecipitated 3xFLAG-tagged wild-type EBP50 or the various tail deletions and blotted for endogenous ezrin (Figure 2E). Deletion of residues 263–268, 271–279, or 281–289 in the EBP50 tail resulted in a modest increase in the amount of coprecipitated ezrin compared with wild-type or a mutant with residues 313–320 deleted. Combined deletion of EBP50 residues 263–289 did not further enhance ezrin binding or dramatically reduce exchange from microvilli when compared with any of the smaller deletion mutants, suggesting there was no additive effect (unpublished data). This suggests that while these residues may contribute to the higher dynamic nature of EBP50 compared with E3KARP, they alone are not sufficient to account for their dramatic difference.
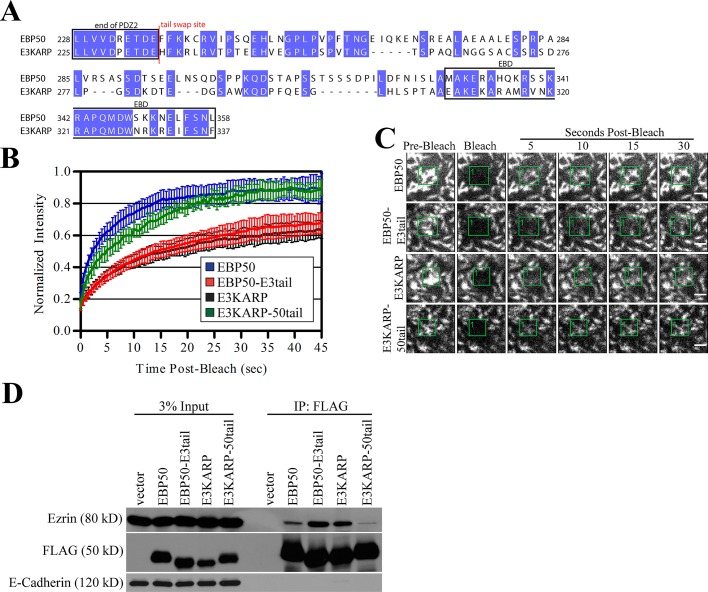
To determine whether other regions in the tail of EBP50 also contribute to its higher exchange from microvilli, we swapped the entire tail regions of EBP50 (residues 238–358) and E3KARP (residues 235–337) (Figure 3A) and again examined their exchange from microvilli by FRAP and ability to coprecipitate ezrin from JEG-3 lysates. Surprisingly, fusion of the tail of E3KARP to the EBP50 PDZ domains (EBP50-E3tail) showed greatly reduced dynamics compared with EBP50 and was indistinguishable from E3KARP (Figure 3, B and C). Additionally, fusion of the EBP50 tail to the E3KARP PDZ domains (E3KARP-50tail) greatly enhanced dynamics compared with E3KARP and was very similar to that of EBP50 (Figure 3, B and C). This suggests that the tail of EBP50 possesses a unique property that can fully account for EBP50’s highly dynamic nature, as it is able to transfer this ability to the E3KARP-50tail chimera.
FIGURE 3:
The tail of EBP50 contains a unique dynamic property that is absent in E3KARP. (A) Sequence alignment of human EBP50 and E3KARP. Conserved residues are highlighted in blue; the site where the tails of EBP50 and E3KARP were swapped is marked in red. (B) Photobleaching recovery curves of GFP-tagged EBP50, EBP50-E3tail, E3KARP, and E3KARP-50tail. Error bars show SD; n ≥ 9 for all experiments. (C) Representative time points from photobleaching experiments of GFP-tagged EBP50, E3KARP, EBP50-E3tail, and E3KARP-50tail. Photobleached areas are indicated by green boxes. Scale bars: 2 μm. (D) 3xFLAG-tagged EBP50 and E3KARP constructs expressed in JEG-3 cells were immunoprecipitated (IP) and blotted for FLAG, endogenous ezrin, and E-cadherin as a control.
To test the validity of this result, we again immunoprecipitated 3xFLAG-tagged versions of the tail chimeras and blotted for endogenous ezrin (Figure 3D). Both EBP50 and the E3KARP-50tail chimera precipitated very small amounts of ezrin, while E3KARP and the EBP50-E3tail chimera bound significantly more. This again indicates that the tail of EBP50 renders the protein highly dynamic, a property that is absent from E3KARP.
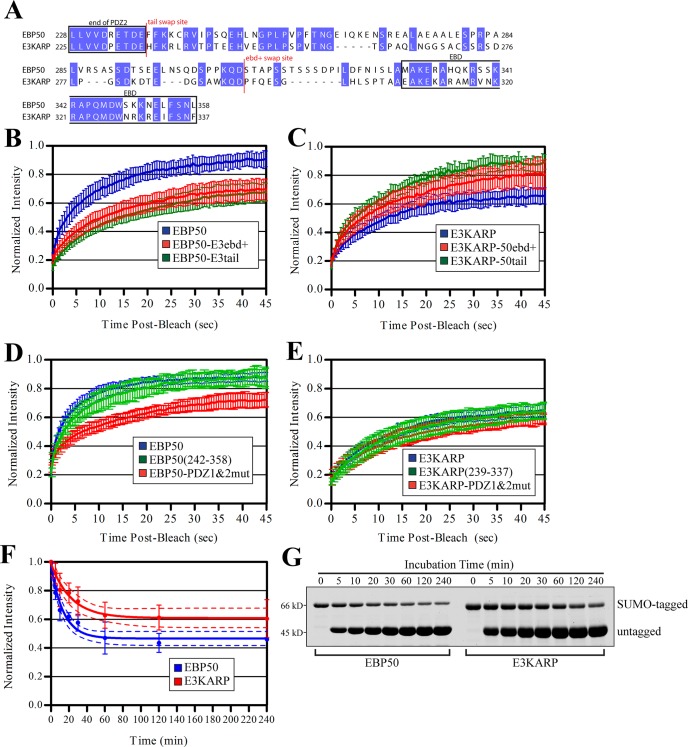
To determine whether a smaller region of the tail is capable of conferring this dynamic property, we next generated chimeras in which we swapped tail residues 294–337 of E3KARP onto residues 1–307 of EBP50 (EBP50-E3ebd+) and EBP50 tail residues 308–358 onto residues 1–293 of E3KARP (E3KARP-50ebd+) (Figure 4A). FRAP of GFP-tagged EBP50-E3ebd+ revealed that its exchange from microvilli was indistinguishable from EBP50-E3tail (Figure 4B). Importantly, the dynamics of GFP-E3KARP-50ebd+ was also similar to that of E3KARP-50tail (p value of 0.44) and different from E3KARP wild type (p value < 0.005) (Figure 4C). Taken together, this indicates that the last 50 residues of EBP50 contain this dynamic property, which is transferrable to E3KARP.
FIGURE 4:
The last ∼50 residues in tail of EBP50 are more dynamic than those in E3KARP both in vivo and in vitro. (A) Sequence alignment of human EBP50 and E3KARP. Conserved residues are highlighted in blue, the site where the tail and ebd+ regions of EBP50 and E3KARP were swapped is marked in red. (B) Photobleaching recovery curves of GFP-tagged EBP50, EBP50-E3tail, and EBP50-E3ebd+. Error bars show SD; n ≥ 10 for all experiments. (C) Photobleaching recovery curves of GFP-tagged E3KARP, E3KARP-50tail, and E3KARP-50ebd+. Error bars show SD. n ≥ 9 for all experiments. (D) Photobleaching recovery curves of GFP-tagged wild-type EBP50, EBP50(242-358), and EBP50-PDZ1&2mut. Error bars show SD; n ≥ 10 for all experiments. (E) Photobleaching recovery curves of GFP-tagged wild-type E3KARP, E3KARP(239-337), and E3KARP-PDZ1&2mut. Error bars show SD; n ≥ 11 for all experiments. (F) Graph of the normalized intensity of SUMO-tagged EBP50 or E3KARP that remained bound to ezrin FERM beads after incubation with untagged EBP50 or E3KARP for the times indicated. The data from three independent experiments were fitted to a single exponential decay curve shown as a solid line. Error bars indicate SD; dotted lines indicate the 95% confidence interval of the fit. (G) A representative experiment from (F). Limiting amounts of SUMO-EBP50 or E3KARP were prebound to ezrin FERM beads and competed off with untagged EBP50 and E3KARP for the times indicated. Gel was stained for total protein with IRDye.
Mutating the PDZ domains of EBP50 to abolish binding to PDZ ligands drastically reduces EBP50 exchange from microvilli (Garbett and Bretscher, 2012), yet the results presented above show that the tail renders EBP50 dynamic. We therefore wanted to explore whether the tail alone is dynamic, or whether it requires the presence of the PDZ domains. Strikingly, the tail alone (GFP-EBP50(242–358)) showed rapid turnover indistinguishable from that of full-length EBP50 (Figure 4D). We compared this result with the dynamics of GFP-tagged wild-type EBP50 and the PDZ1&2 point-inactivated mutant (PDZ1&2mut). Similar to what we have seen previously, EBP50-PDZ1&2mut showed a dramatic reduction in dynamics compared with wild-type. These results suggest that, in full-length EBP50, the PDZ domains negatively regulate the intrinsic dynamics of the tail, and this inhibition of dynamics is relieved upon PDZ ligand binding.
To determine whether the dynamics of E3KARP can be also be regulated by occupancy of its PDZ domains in a manner similar to EBP50, we performed photobleaching experiments of GFP-tagged wild-type E3KARP, PDZ1&2mut, and the tail of E3KARP alone (239–337). Interestingly, unlike EBP50, mutation of E3KARP's PDZ domains had no affect on its exchange from microvilli (Figure 4E). Additionally, the tail of E3KARP alone showed similar dynamics to that of both wild-type E3KARP and the PDZ1&2mut, altogether suggesting that E3KARP dynamics is not regulated by PDZ ligand binding.
The EBD of EBP50 has been shown to have a slightly higher affinity for the radixin FERM domain than that of E3KARP in vitro (Kd of 1.7 nM for EBP50, 9.5 nM for E3KARP; Terawaki et al., 2006). However, in the context of full-length proteins in JEG-3 cell lysate, E3KARP clearly precipitates more ezrin than is seen with EBP50 (Figure 3D). To examine this discrepancy in greater detail, we explored differences in the interactions of full-length EBP50 and E3KARP with ezrin FERM in vitro. To determine whether the off-rates of EBP50 and E3KARP from ezrin FERM differed, we first incubated 40 nM of purified SUMO-tagged EBP50 or E3KARP with 240 nM of FERM domain covalently coupled to beads. After a quick wash, we then added 1 μM of untagged EBP50 or E3KARP to the beads to saturate all FERM binding sites during the course of the experiment. After various amounts of time, the beads were quickly washed, and the remaining amount of bound SUMO-tagged EBP50 or E3KARP was quantified (Figure 4, F and G). Both EBP50 and E3KARP show dramatically slower exchange from ezrin in vitro than in vivo, similar to what we have already shown for EBP50 (Garbett and Bretscher, 2012). The intensities for each protein fitted well to a single exponential decay curve, and, interestingly, E3KARP had a slightly slower off-rate (half-life of ∼16 min) than EBP50 (half-life of ∼10 min). Further, a greater amount of SUMO-E3KARP remained bound to FERM after 4 h compared with SUMO-EBP50, again suggesting that its association with ezrin FERM in vitro persists longer than EBP50.
Identification of IRSp53-T as a novel EBP50 binding partner
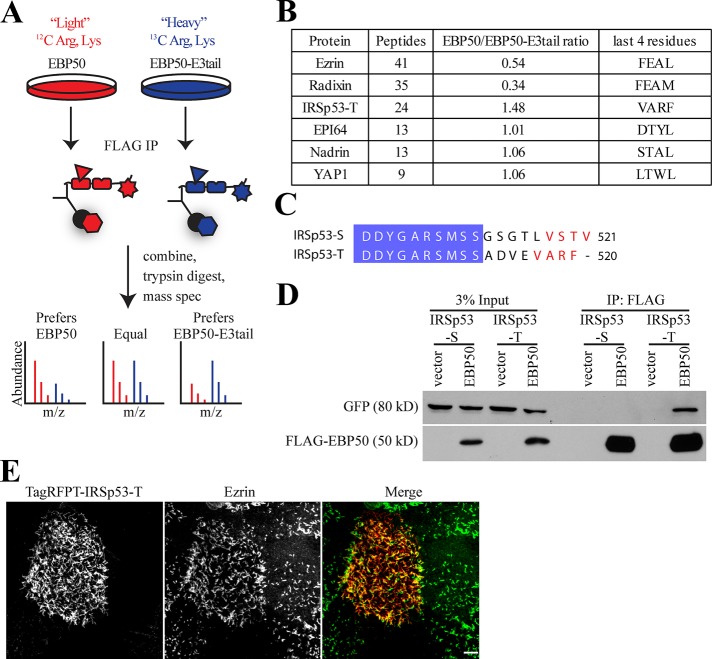
The difference in dynamics specified by the tail domains of EBP50 and E3KARP could be a direct result of a regulator interacting with the E3KARP tail to stabilize its interaction with ezrin, or a regulator binding to the EBP50 tail to enhance its release from ezrin. To search for such proteins, we used an unbiased proteomic approach. First, we generated stable JEG-3 cell lines expressing either 3xFLAG-tagged wild-type EBP50 or the chimeric fusion EBP50-E3tail. Next the cells were subjected to stable isotope labeling of amino acids in cell culture (SILAC), with the EBP50-expressing cells grown in “light” medium, and the EBP50-E3tail expressing cells grown in “heavy” medium to allow for uniform labeling. The 3xFLAG-EBP50 and 3xFLAG-EBP50-E3tail were separately immunoprecipitated, mixed, and subjected to trypsin digestion followed by mass spectrometry (Ong et al., 2002; Figure 5A). Consistent with our earlier results by Western blot (Figure 3D), the EBP50-E3tail chimera bound almost twice as much ezrin and three times as much radixin compared with EBP50 (Figure 5B). Interactions with known PDZ binding partners, such as EBP50 PDZ interactor of 64 kDa (EPI64), nadrin, and yes-associated protein-1, were unchanged in the EBP50-E3tail chimera (Mohler et al., 1999; Reczek and Bretscher, 2001). We also found a significant number of peptides for the N-terminal Inverse-Bin-Amphiphysin-RVS (I-BAR) domain containing protein insulin receptor substrate p53 (IRSp53) isoform–T, which preferentially bound ∼50% more to EBP50 than EBP50-E3tail (Figure 5B). Endogenous IRSp53-T runs at the same size as EBP50, so we were unable to confirm this interaction preference because its immunoblot signal was masked by the large quantity of 3xFLAG-EBP50 precipitated (unpublished data). However, due to the highly quantitative and sensitive nature of SILAC mass spectrometry, it is likely that IRSp53-T has a modest binding preference for EBP50 over EBP50-E3tail.
FIGURE 5:
IRSp53-T binds to EBP50 and localizes to microvilli. (A) Experimental schematic of SILAC mass spectrometry to compare binding-partner differences of EBP50 and EBP50-E3tail. Briefly, JEG-3 cells stably expressing 3xFLAG-tagged versions of EBP50 or EBP50-E3tail were labeled with either light or heavy arginine and lysine, respectively. After being immunoprecipitated, samples were trypsin digested and analyzed by mass spectrometry. The ratio of light:heavy peptide can be compared to yield highly sensitive and quantitative differences in protein abundance. (B) Table of SILAC mass spectrometry results. The number of peptides and overall ratio of binding to EBP50/EBP50-E3tail is indicated. The last four C-terminal amino acids are also listed for reference. (C) Sequence of alignment of human IRSp53-S and IRSp53-T isoforms. Conserved regions are highlighted in blue; the PDZ binding motif of IRSp53-S and potential PDZ binding site in IRSp53-T are in red. (D) 3xFLAG-tagged vector alone or EBP50 were immunoprecipitated from transiently transfected cells also expressing either GFP-IRSp53-S or IRSp53-T and blotted for FLAG and GFP. (E) Maximum projection image of JEG-3 cells expressing TagRFPT-IRSp53-T (red) and ezrin (green). Scale bar: 5 μm.
Interestingly, another isoform of IRSp53, IRSp53-S, is an effector of Cdc42 that is involved in filopodia formation and apical lumen polarity (Krugmann et al., 2001; Cohen et al., 2011). IRSp53-T and IRSp53-S only differ in their last 8–9 C-terminal residues (Figure 5C). IRSp53-S has a C-terminal PDZ binding motif (-VSTV) that interacts with the PDZ domain–containing proteins nematode Lin7 and mammalian Lin7 homologue (Hori et al., 2003). Interestingly, IRSp53-T also ends in a potential PDZ binding motif (-VARF; Millard et al., 2007). To examine whether both IRSp53-S and IRSp53-T are capable of binding to EBP50, we performed immunoprecipitations from JEG-3 cells transiently cotransfected with either GFP-tagged IRSp53-S or IRSp53-T and 3xFLAG-tagged EBP50 or vector control (Figure 5D). Only GFP-IRSp53-T coprecipitated efficiently with EBP50, while IRSp53-S was not detectable. Because IRSp53-T interacts with EBP50, we next determined whether it localized to microvilli. Expression of TagRFPT-tagged IRSp53-T in JEG-3 cells revealed that it is highly enriched in microvilli where it colocalizes with ezrin (Figure 5E).
IRSp53-T binds to the PDZ1 domain of EBP50 and has slow exchange from microvilli
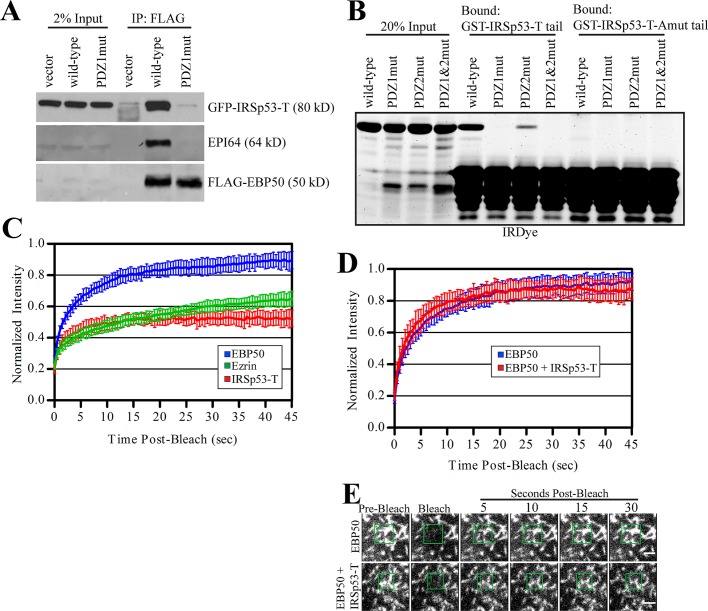
To further investigate how IRSp53-T interacts with EBP50, we performed immunoprecipitations from JEG-3 cells transiently transfected with GFP-IRSp53-T and 3xFLAG-tagged versions of either EBP50 wild-type, a PDZ1 mutant, or vector control (Figure 6A). Both GFP-IRSp53-T and endogenous EPI64, a known PDZ1 binding partner, bound very strongly to wild-type EBP50 but not to the PDZ1 mutant or vector control. To determine whether the interaction between IRSp53-T and EBP50 occurs directly through the potential PDZ binding motif in the tail of IRSP53-T and the PDZ1 domain of EBP50, we created glutathione S-transferase (GST) fusions of the IRSp53-T wild-type tail and a mutant with an alanine added (IRSp53-T-Amut), which blocks PDZ interactions, and performed GST pull downs against purified recombinant untagged full-length EBP50 (Figure 6B). The GST-IRSp53-T-Amut tail did not bind to any of the EBP50 constructs used. GST-IRSp53-T wild-type tail efficiently precipitated wild-type EBP50 and the PDZ2 mutant to a lesser degree, but did not bind to the PDZ1 or PDZ1&2 mutants. Altogether, this demonstrates that IRSp53-T directly interacts with the PDZ1 domain of EBP50 via its C-terminal PDZ binding motif.
FIGURE 6:
IRSp53-T binds to the PDZ1 domain of EBP50 and shows slow exchange from microvilli. (A) 3xFLAG-tagged vector alone, EBP50 wild-type, or a PDZ1 mutant were immunoprecipitated from transiently transfected cells also expressing GFP-IRSp53-T and blotted for FLAG, GFP, and endogenous EPI64. (B) GST-IRSp53-T tail or a mutant with an additional alanine added to the C-terminus (Amut) were used to precipitate purified full-length EBP50 wild-type or PDZ1, PDZ2, and PDZ1&2 mutants. Gel was stained for total protein with IRDye. (C) Photobleaching recovery curves of GFP-tagged EBP50, ezrin, and IRSp53-T. Error bars show SD; n ≥ 10 for all experiments. (D) Photobleaching recovery curves of GFP-tagged EBP50 alone or in cells also expressing TagRFPT-IRSp53-T. Error bars show SD; n ≥ 10 for all experiments. (E) Representative time points from photobleaching experiments of cells expressing GFP-EBP50 alone or with TagRFPT-IRSp53-T. Photobleached regions are indicated by green boxes. Scale bars: 2 μm.
Previously we have shown that EBP50 has considerably higher dynamics than one of its PDZ binding partners, podocalyxin (PCX), likely due to the fact that PCX is a transmembrane protein (Garbett and Bretscher, 2012). IRSp53-T, which has an I-BAR domain, Src homology 3 (SH3) domain, and a C-terminal PDZ binding motif, could be tightly associated with the membrane via its I-BAR domain. To determine whether IRSp53-T is stably associated with microvilli or exchanges more rapidly in a manner similar to EBP50, we performed photobleaching experiments on JEG-3 cells transiently expressing GFP-tagged EBP50, ezrin, and IRSp53-T (Figure 6C). While EBP50 showed very fast dynamics, with a large portion recovered after only 5 s, IRSp53-T recovered very slowly, in a manner almost indistinguishable from ezrin. Further, a large fraction of IRSp53-T was immobile, suggesting that it was strongly associated with the membrane or underlying cytoskeleton (Figure 6C).
Because IRSp53-T bound preferentially to EBP50 over the EBP50-E3tail chimera by SILAC mass spectrometry (Figure 5B), we next examined whether overexpression of IRSp53-T alters EBP50 dynamics in microvilli. JEG-3 cells were transiently cotransfected with both TagRFPT-IRSp53-T and GFP-EBP50, and cells expressing both proteins were subjected to GFP photobleaching (Figure 6, D and E). Coexpression of TagRFPT-IRSp53-T had no effect on EBP50 dynamics when compared with cells expressing only GFP-EBP50. The implications of this result are discussed in the following section.
The tails of E3KARP and EBP50 promote different localizations within microvilli
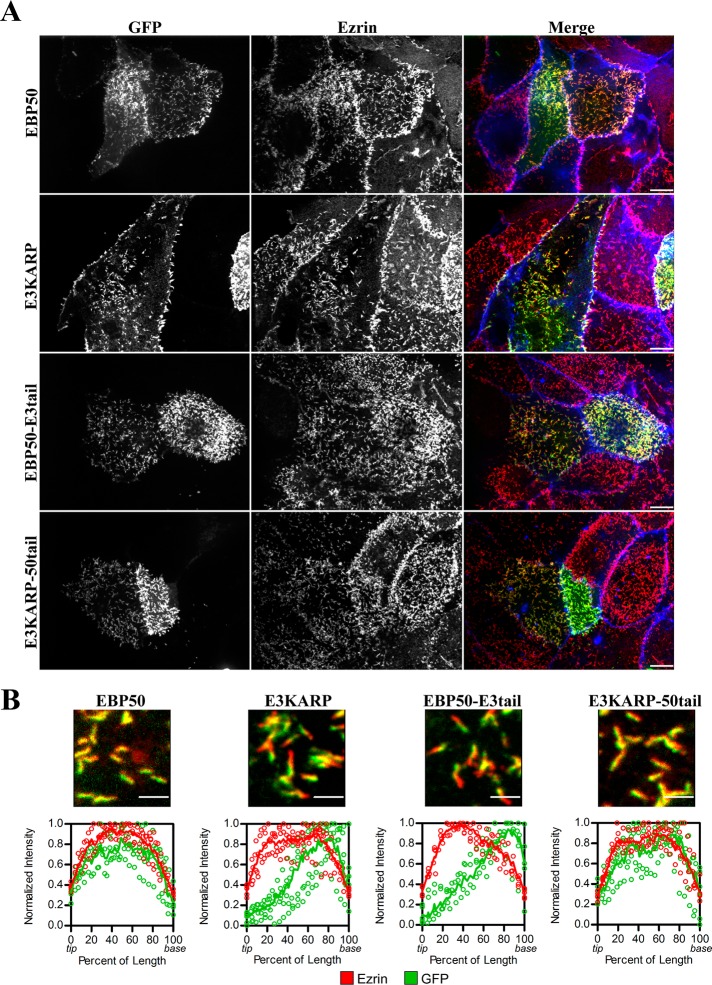
It has been suggested that E3KARP localizes toward the terminal web region below microvilli in renal proximal tubules (Wade et al., 2003). Using high-resolution confocal microscopy of JEG-3 cells, we have shown that we are able to localize microvillar proteins to different subdomains within a single microvillus (Hanono et al., 2006; Viswanatha et al., 2012). To further explore the localization differences between EBP50 and E3KARP on the submicrovillar scale, we expressed GFP-EBP50, GFP-E3KARP, or the chimeras GFP-EBP50-E3tail and GFP-E3KARP-50tail in JEG-3 cells and visualized the GFP signal in relation to endogenous ezrin (Figure 7A). The fluorescence intensity of GFP and ezrin was then measured within multiple microvilli and fitted to a locally weighted scatterplot-smoothing (LOWESS) function. In cells with both high and moderate levels of GFP fluorescence, EBP50 and E3KARP-50tail both localized along the full-length of microvilli as determined by ezrin staining (Figure 7B). In contrast, in cells with moderate levels of GFP fluorescence, E3KARP and EBP50-E3tail both showed a strong localization toward the base of microvilli that also persisted slightly into the terminal web area, where ezrin staining disappears (Figure 7B). Deletion of EBP50 residues 263–268, 271–279, 281–289, or 313–320 did not have any effect on its localization within microvilli (Supplemental Figure S1). Additionally, the tails of EBP50 (residues 242–358) and E3KARP (residues 239–337) alone show similar localizations to full-length EBP50 and E3KARP, respectively (Figure S2), although the base localization of the E3KARP tail alone was less prevalent than with full-length E3KARP. Therefore E3KARP localizes toward the base of microvilli, and this is a characteristic of its tail, which can shift EBP50 to the microvillar base, as seen with the EBP50-E3tail chimera.
FIGURE 7:
The tail of E3KARP causes preferential localization to the base of microvilli. (A) Representative maximum projection images of JEG-3 cells expressing GFP-tagged EBP50, E3KARP, or the chimeras EBP50-E3tail and E3KARP-50tail. GFP signal is shown in green; ezrin staining in red; actin in blue. Scale bars: 10 μm. (B) Top, highly magnified regions of microvilli containing GFP-tagged EBP50, E3KARP, or the tail chimeras, and stained for ezrin (red). Scale bars: 2 μm. Bottom, graphs of normalized intensity of ezrin (red) and GFP (green) of multiple microvilli (n ≥ 5) were plotted (open circles) as a function of the percent of the total microvillar length with 0 and 100% representing the tip and base respectively. The data were fitted to a LOWESS function (shown as a solid line).
DISCUSSION
Microvilli are a hallmark of the apical aspect of polarized epithelial cells. Yet, despite being discovered more than 50 years ago (Granger and Baker, 1950), their regulation and contribution to cell physiology is still poorly understood. A recently discovered role for the importance of brush border–derived vesicles in preventing pathogenic infection is perhaps just one of many functions for microvilli (Shifrin et al., 2012). Central components of epithelial cell microvilli are the membrane cytoskeletal linker ezrin and its binding partner, the scaffolding protein EBP50. In this paper, we report how the tail of EBP50, in comparison with its closely related paralogue E3KARP, contributes to its intrinsic dynamics, binding partner selection, and localization. The findings presented here are of general significance, as we have found nearly identical differences in dynamics and ezrin binding between EBP50 and E3KARP in epithelial cells derived from three different tissues—JEG-3 cells from placenta, Caco-2 cells from the colon, and MDCK cells from kidney. During final preparation of this manuscript, another study examining rabbit EBP50 and E3KARP reported similar results over longer timescales in opossum kidney cells and found that the slower turnover of E3KARP is important for its regulation of the N+/H+ exchanger NHE3 (Yang et al., 2013).
We first show that the high rate of dynamics exhibited by EBP50 is determined by its tail domain, and further localize the region contributing the dynamics to the C-terminal 50 residues. This area contains the most highly conserved region of the tail, so the observed dynamics is most likely important and therefore preserved by evolution. Remarkably, we find that the tail alone shows dynamics indistinguishable from the full-length wild-type protein. In earlier studies, and reproduced here, we showed that the high dynamics of wild-type EBP50 in vivo is dependent upon functional PDZ domains—a mutant unable to bind ligands shows slow dynamics similar to that of E3KARP. Thus the two PDZ domains, when unoccupied negatively, regulate the dynamics conferred by the tail, and this negative regulation is alleviated by ligand binding to its PDZ domains. This finding strongly supports our previous suggestion that EBP50 functions as a biological governor—when ligands are present at a low concentration it has dampened dynamics, but as ligand concentration increases, it becomes more dynamic and presumably less efficient at restraining ligands in the microvillus. In support of this model, we have shown that overexpression of an artificial PDZ ligand, trafficked to microvilli in an EBP50-independent manner (dystroglycan fused to the EPI64 tail), is able to cause a modest increase in EBP50 dynamics (Garbett and Bretscher, 2012).
Because EBP50 has numerous binding partners and diverse roles in receptor surface retention, trafficking, and signaling pathways (Weinman et al., 2006; Ardura and Friedman, 2011), we tried to identify PDZ ligands that are sensitive to the dynamics of EBP50. We performed SILAC-based mass spectrometry comparing ligands of wild-type EBP50 to those of a chimera with its tail replaced with that of E3KARP (EBP50-E3tail). We identified a novel EBP50 PDZ1 ligand, IRSp53-T, which shows a modest binding preference for the more dynamic wild-type EBP50 and localizes in vivo to microvilli. However, overexpression of IRSp53-T has no detectable affect on EBP50 exchange from microvilli. This may not be surprising, given the large number of expected PDZ ligands inside a microvillus, where EBP50 may require multiple short-lived interactions with numerous PDZ ligands to regulate its dynamics. Another important component of microvilli, brush border myosin I, also provides a linkage between the plasma membrane and actin cytoskeleton, which allows for distribution of membrane tension forces across microvilli and the apical domain (Nambiar et al., 2009; Tyska and Nambiar, 2010). The function of EBP50’s dynamics could also create a rapid-feedback system to govern the amount of membrane–cytoskeletal linkage in an effort to quickly adapt to membrane surface tension forces, which have been shown to be important for endocytosis from the apical domain of polarized epithelial cells (Boulant et al., 2011).
EBP50 undergoes allosteric interdomain motions on submicrosecond timescales that are involved in the regulation of its formation of multiprotein complexes and PDZ domain specificity. Interestingly, binding to the FERM domain of ezrin has been shown to affect allosteric distances between the PDZ domains, and binding to PDZ ligands has been shown to affect conformation and backbone dynamics (Farago et al., 2010; Bhattacharya et al., 2013). This, combined with our current and previous results, strongly suggests that binding to either ezrin FERM domain or to PDZ ligands induces conformational changes in EBP50 over very short timescales that can regulate both the specificity and duration of its interactions in microvilli.
In addition to the differences in dynamics between EBP50 and E3KARP, we have shown that they have distinct localizations in microvilli, again with their tails being the defining region. Thus EBP50 localizes along the length of a microvillus, whereas E3KARP localizes to the base. Localization to the base of microvilli is not a consequence of slower dynamics, because a PDZ1&2 mutant of EBP50 that shows slow turnover is still able to localize along the full length of a microvillus (Garbett et al., 2010). Ezrin is activated, at least in part, by phosphorylation of Thr-567 (Fehon et al., 2010). Phosphorylated ezrin, and its predominant kinase in JEG-3 cells, lymphocyte-oriented kinase, both localize toward the distal tip region of microvilli (Hanono et al., 2006; Viswanatha et al., 2012). This suggests, surprisingly, that E3KARP seems to have a preferential association with a region of microvilli reduced in Thr-567 phosphorylation. Further, the Thr-567 phosphorylated species of ezrin has a half-life of ∼1 min, which suggests that turnover of ezrin phosphorylation, and therefore its active/closed conformation, may also play a role in the regulation of EBP50 and E3KARP dynamics (Viswanatha et al., 2012).
The significance of the interaction between EBP50 and IRSp53-T in microvilli remains to be determined. IRSp53 contains an I-BAR domain that is involved in remodeling membrane by inducing membrane curvature and an SH3 domain responsible for its regulation of actin dynamics via interactions with Wiskott-Aldrich syndrome protein family proteins, mammalian enabled, the formin mDia, and EGFR kinase substrate 8 (Scita et al., 2008; Ahmed et al., 2010). Given the previously identified role for IRSp53-S in filopodia formation (Krugmann et al., 2001; Millard et al., 2005), it will be interesting to see whether IRSp53-T has a similar function in microvilli, although this may be difficult to separate from its role in apical lumen formation (Cohen et al., 2011).
In summary, we have uncovered a novel mechanism showing that EBP50 dynamics is defined by its tail domain and regulated by the occupancy of its PDZ domains and that the tail domains of EBP50 and E3KARP target them to distinct regions of the microvillus. Our studies illuminate that microvilli are much more dynamic and complex structures than generally envisioned, and our current efforts are to understand their regulation and how they contribute to the function of epithelial cells.
MATERIALS AND METHODS
Antibodies and reagents
The mouse anti-FLAG and mouse anti–E-cadherin antibody were purchased from Sigma-Aldrich (St. Louis, MO) and BD Biosciences (Franklin Lakes, NJ), respectively. The mouse anti-GFP antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The rabbit antisera and affinity-purified antibodies against full-length human ezrin and EPI64 were described previously (Bretscher, 1989; Hokanson and Bretscher, 2012). Goat anti-mouse and anti-rabbit secondary antibodies conjugated to horseradish peroxidase were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA) and MP Biomedicals (Solon, OH), respectively. Alexa Fluor 568 donkey anti-rabbit antibody and Alexa Fluor 660–conjugated phalloidin were purchased from Invitrogen (Carlsbad, CA).
DNA constructs and sequence alignment
N-terminal GFP and SUMO-tagged EBP50 constructs were created in pEGFP-C2 (Takara Bio, Shiga, Japan) and pE-SUMO (LifeSensors, Malvern, PA), respectively, and were previously described (Garbett et al., 2010). GFP-tagged ezrin was cloned into pEGFP-N2 (Takara Bio, Shiga, Japan). N-terminal GFP and SUMO-tagged E3KARP constructs were created in pEGFP-C2 and pE-SUMO, respectively. GFP-tagged IRSp53-T and IRSp53-S were created by tailored PCR from IMAGE clone #4125501 (Open Biosystems, Huntsville, AL) after deletion of a three-nucleotide insertion (nucleotides 1163–1165) and insertion into pEGFP-C1. TagRFPT-IRSp53-T was created by PCR from a vector expressing TagRFPT that was provided by R. Tsien (University of California, San Diego, La Jolla, CA [ Shaner et al., 2008]). GST-tagging of the last 86 residues of the IRSp53-T tail was created by PCR and insertion into pGEX-6P1 (GE Healthcare, Waukesha, WI). The addition of an alanine (Amut) to the GST-IRSp53-T tail was generated by PCR. The 3xFLAG-tagged EBP50 and E3KARP constructs were created using PCR and inserted into pQCXIP (BD Biosciences). Untagged ezrin FERM (residues 1–298) in pQE16 (Qiagen, Valencia, CA) was previously described (Reczek et al., 1997). The internal deletions of EBP50 (Δ263–268, Δ271–279, Δ281–289, Δ313–320, and Δ263–289) were generated by two steps of overlapping PCR and inserted into pEGFCP-C2 or pQCXIP. All chimeric fusions of EBP50 and E3KARP were created by two steps of overlapping PCR and inserted into pEGFP-C2 and pQCXIP. EBP50-E3tail corresponds to EBP50(1–237) and E3KARP(235–337) residues. E3KARP-50tail corresponds to E3KARP(1–234) and EBP50(238–358) residues. EBP50-E3ebd+ corresponds to EBP50(1–307) and E3KARP(294–337) residues. E3KARP-50ebd+ corresponds to E3KARP(1–293) and EBP50(308–358) residues.
Human protein sequences for EBP50, E3KARP, IRSp53-S, and IRSp53-T were aligned using the ClustalW method in MegAlign (Lasergene; DNAStar, Madison, WI) and processed for publication using Jalview (Waterhouse et al., 2009).
Cell culture and transfection
JEG-3, Caco-2, and MDCK, and Phoenix-AMPHO cells (American Type Culture Collection, Manassas, VA) were maintained in a 5% CO2 humidified atmosphere at 37°C. JEG-3 cells were cultured in MEM with 10% fetal bovine serum (FBS), MDCK in DMEM with 5% FBS, and Caco-2 and Phoenix-AMPHO in DMEM with 10% FBS. JEG-3 cells were transfected with polyethylenimine (Polysciences, Warrington, PA) and 1–2 μg plasmid DNA, as previously described (Hanono et al., 2006). Caco-2 and MDCK cells were transfected with XtremeGENE HP DNA transfection reagent (Roche, Indianapolis, IN) and Lipofectamine 2000 (Invitrogen), respectively, according to the manufacturers’ instructions.
For generation of stable JEG-3 cell lines expressing either 3xFLAG-EBP50 or 3xFLAG-EBP50-E3tail, Phoenix-AMPHO cells were cotransfected with the above constructs in pQCXIP in addition to a plasmid encoding VSV-G using polyethylenimine. The resulting retroviruses were then used to infect JEG-3 cells, which were then selected and maintained with 2 μg/ml puromycin (Sigma-Aldrich).
Live-cell imaging and FRAP
Transfected cells grown in 35-mm glass bottom dishes (MatTek, Ashland, MA) were washed in phosphate-buffered saline (PBS) and then maintained in low-sodium bicarbonate phenol red-free MEM (Sigma-Aldrich) supplemented with 25 mM HEPES (pH 7.4), with 10% FBS and GlutaMAX (Invitrogen). Live cells were imaged by time-lapse microscopy on a spinning disk (CSU-X; Yokogawa, Tokyo, Japan) with a spherical aberration correction device, a 100×/1.46 numerical aperture (NA) objective (Leica, Wetzlar, Germany) on an inverted microscope (DMI6000B; Leica) and an electron-multiplied charge-coupled device camera (QuantEM; Photometrics, Tucson, AZ) at 37°C in an environmental chamber (Okolab, Ottaviano, Italy) controlled by SlideBook version 5.5 (Intelligent Imaging Innovations, Denver, CO). Regions selected for FRAP were illuminated either with a digital mirror illumination system (Mosaic; Andor Technology, Belfast, Northern Ireland) coupled to a 488-nm 400-W argon laser or with a point-scanner galvanometer-based system (Vector; Intelligent Imaging Innovations) coupled to a 473-nm, 50-mW diode-pumped solid-state laser. For all FRAP experiments, an independent region was also monitored to control for photobleaching during the observation period. Movies were processed using SlideBook and analyzed using Excel (Microsoft, Redmond, WA) and Prism (GraphPad, La Jolla, CA). The expression levels of each GFP-tagged construct were determined by their fluorescence intensity, and FRAP recoveries of cells with varying expression levels were compared to ensure that differences in recoveries were not due to variations in expression. The normalized data from multiple biological replicates were fitted to a curve, and samples with overlapping SD were subjected to a two-way analysis of variance using Prism.
Immunofluorescence
JEG-3 cells grown on glass coverslips were fixed in 3.7% formaldehyde/PBS for 10 min at room temperature and then permeabilized in 0.2% Triton X-100/PBS for 5 min. Cells were then washed in PBS and incubated in primary antibody in 3% FBS in PBS. After cells were washed in PBS, secondary antibody and phalloidin stain were added in 3% FBS in PBS. After further washing in PBS, coverslips were mounted onto glass slides using Vectashield (H-1000; Vector Laboratories, Burlingame, CA), and images were then acquired on a CSU-X spinning-disk microscope with spherical aberration correction device, 100×/1.46 NA objective on an inverted microscope (DMI6000B) and either an HQ2 CCD camera (Photometrics) or QuantEM EMCCD camera equipped with 2× magnifying lens controlled by SlideBook software. Images were then processed using SlideBook and Photoshop (Adobe, San Jose, CA).
Measurements of protein localization within microvilli were performed in SlideBook. Lines were drawn from tip to base for multiple microvilli (their orientation was determined by scanning through multiple z-slices), and intensity values for GFP and ezrin were exported into Excel. Variations in length between microvilli were accounted for by converting intensities for each pixel into a percentage of the total length of the microvillus, with 0 and 100% representing the tip and base, respectively. The normalized intensity values for each microvillus were then plotted as a function of the percentage of microvillar length and fitted to a LOWESS function in Prism.
Purification of recombinant proteins
Ezrin FERM was purified as previously described (Reczek et al., 1997). Briefly, induced cells were lysed in 180 mM KH2PO4 (pH 7.0) with complete protease inhibitor (Roche), lysed by sonication (Branson Ultrasonics, Danbury, CT), centrifuged and run over a hydroxyapatite column (Pall Corporation, Port Washington, NY), and eluted by linear gradient of 180–800 mM KH2PO4. Ezrin FERM–containing fractions were pooled and dialyzed against 20 mM 2-(N-morpholino)ethanesulfonic acid (MES), 150 mM NaCl (pH 6.7); applied to an S-Sepharose column (GE Life Sciences); and eluted by linear gradient of 0.15–1.0 M NaCl. Purified FERM was conjugated to cyanogen bromide (CNBr)-activated Sepharose (Sigma-Aldrich) by first dialyzing against C Buffer (0.1 M NaHCO3, 0.5 M NaCl, pH 8.3) and then adding to hydrated CNBr beads for a final concentration of 36.4 μM on the resin. Beads were then washed and blocked in 0.25 M glycine (pH 8.0) overnight. Beads were washed again and then stored as a 50% slurry in H buffer (50 mM Tris, 150 mM NaCl, pH 7.4).
SUMO-tagged EBP50 and E3KARP constructs were purified as described previously (LaLonde et al., 2010). Briefly, induced bacterial pellets were lysed in binding buffer (20 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, pH 7.4, 1% Triton X-100) by sonication, centrifuged and run over a His gravitrap column (GE Life Sciences), washed, and eluted in elution buffer (20 mM sodium phosphate, 500 mM NaCl, 500 mM imidazole, pH 7.4). Eluate was dialyzed into binding buffer, cleaved with 6xHis-Ulp1 for 40 min at 30°C to remove the SUMO-tag, and then run over another His gravitrap column; and flow-through containing untagged protein was collected and dialyzed into 150 mM NaCl, 10 mM Tris (pH 7.4) with 1 mM dithiothreitol.
Bacterial pellets induced to express GST-IRSp53-T tail or GST-IRSp53-T-Amut tail were lysed in 150 mM NaCl, 20 mM Tris (pH 7.4), 0.1% β-mercaptoethanol, and 0.1% Triton X-100 with complete protease inhibitor by sonication; centrifuged and added to hydrated glutathione agarose (Sigma-Aldrich); washed; and left as a 50% slurry for binding assays.
In vitro binding assays
The in vitro competition assay was described previously (Garbett and Bretscher, 2012). Briefly, excess ezrin FERM domain coupled to CNBr beads at a final concentration of 240 nM was mixed with 40 nM of soluble SUMO-tagged EBP50 or E3KARP in 300 mM NaCl, 0.1% Triton X-100, 5% glycerol, and 20 mM Tris (pH 7.4), and nutated for 1 h at 4°C. Beads were spun and quickly washed once, and then buffer containing excess untagged EBP50 or E3KARP at 1 μM was added for the indicated times to compete with prebound SUMO-tagged EBP50 or E3KARP, respectively. Beads were spun, quickly washed once, and then boiled in sample buffer and separated by SDS–PAGE. The gel was stained with IRDye (LI-COR Biosciences, Lincoln, NE) and then imaged using an infrared imaging system (Odyssey; LI-COR Biosciences). The normalized data were fitted to a curve using Prism.
For binding assays between the IRSp53-T tail and EBP50, GST-IRSp53-T and GST-IRSp53-T-Amut tail coupled to glutathione agarose were used to precipitate 10 μg of the soluble untagged EBP50 constructs in binding buffer (300 mM NaCl, 0.1% Triton X-100, 5% glycerol, 20 mM Tris, pH 7.4) at 4°C. Beads were then washed in binding buffer, boiled in sample buffer, and separated by SDS–PAGE. The gel was stained with IRDye and then imaged using an Odyssey infrared imaging system.
Immunoprecipitation and Western blotting
JEG-3 cells transiently expressing 3xFLAG-EBP50 and E3KARP constructs were lysed in lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1% IGEPAL-630, 50 mM NaF, 0.1 mM Na3VO4, 10 mM β-GP, and 2.5% glycerol) and incubated with anti-FLAG M2 affinity resin (Sigma-Aldrich) while nutating at 4°C for 2 h. The resin was then washed three times in lysis buffer with 0.1% IGEPAL-630, and the remaining bound sample was eluted using 3xFLAG-peptide (Sigma-Aldrich) for 45 min at room temperature. Protein samples were boiled in sample buffer and separated by SDS–PAGE, transferred to Immobilon-P (EMD Millipore, Billerica, MA) for Western blotting, and then visualized using ECL (GE Healthcare).
SILAC and mass spectrometry
For SILAC, JEG-3 stable cell lines expressing 3xFLAG-EBP50 or 3xFLAG-EBP50-E3tail were grown for ∼3 wk in MEM (Thermo Fisher Scientific, Lafayette, CO) containing dialyzed FBS (Invitrogen) and either [12C]arginine and lysine or [13C]arginine and lysine (Sigma-Aldrich), respectively.
FLAG immunoprecipitations were as described above with slight modifications for mass spectrometry processing, as described previously (Smolka et al., 2007; Viswanatha et al., 2012). Briefly, after immunoprecipitation, bound protein was instead eluted in 50 mM Tris (pH 8.0) and 1% SDS and then precipitated with 50% ethanol, 49.9% acetone, and 0.1% acetic acid. Protein samples were then mixed, trypsin digested (Promega, Madison, WI), and desalted in a C18 column (Waters, Milford, MA). The tryptic peptides were dehydrated in a speed vacuum and dissolved in 80% acetonitrile and 1% formic acid for fractionation by hydrophilic interaction chromatography. The resulting fractions were dried, dissolved in 0.1% trifluoroacetic acid, and injected into a mass spectrometer (Qexactive LC-MS/MS; Thermo Fisher Scientific). The data were analyzed using Proteome Discoverer (Thermo Fisher Scientific).
Supplementary Material
Acknowledgments
We are indebted to Marcus Smolka and Francisco Bastos de Oliveira for assistance with mass spectrometry. We are grateful to Roger Tsien for generously providing the plasmid encoding TagRFPT. We also thank members of the Bretscher lab for helpful comments and suggestions. This work was supported by National Institutes of Health (NIH) grant GM-036652 to A.B. D.G. and R.V. were partially supported by NIH training grant 5T32GM007273.
Abbreviations used:
- E3KARP
NHE3 kinase A regulatory protein
- EBP50
ERM-binding protein of 50 kDa
- EPI64
EBP50 PDZ interactor of 64 kDa
- ERM
ezrin/radixin/moesin
- FERM
4.1 ERM
- I-BAR
Inverse-Bin-Amphiphysin-Rvs
- IRSp53
insulin receptor substrate p53
- PDZ
postsynaptic density 95/disks large/zona occludens-1
- SH3
Src homology 3
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-06-0330) on August 28, 2013.
REFERENCES
- Ahmed S, Goh WI, Bu W. I-BAR domains, IRSp53 and filopodium formation. Semin Cell Dev Biol. 2010;21:350–356. doi: 10.1016/j.semcdb.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Ardura JA, Friedman PA. Regulation of g protein-coupled receptor function by Na+/H+ exchange regulatory factors. Pharmacol Rev. 2011;63:882–900. doi: 10.1124/pr.110.004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Ju JH, Orlova N, Khajeh JA, Cowburn D, Bu Z. Ligand-induced dynamic changes in extended PDZ domains from NHERF1. J Mol Biol. 2013;425:2509–2528. doi: 10.1016/j.jmb.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S, Kural C, Zeeh J-C, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol. 2011;13:1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J Cell Biol. 1989;108:921–930. doi: 10.1083/jcb.108.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Fernandez D, Lázaro-Diéguez F, Müsch A. The serine/threonine kinase Par1b regulates epithelial lumen polarity via IRSp53-mediated cell-ECM signaling. J Cell Biol. 2011;192:525–540. doi: 10.1083/jcb.201007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farago B, Li J, Cornilescu G, Callaway DJE, Bu Z. Activation of nanoscale allosteric protein domain motion revealed by neutron spin echo spectroscopy. Biophys J. 2010;99:3473–3482. doi: 10.1016/j.bpj.2010.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, Bretscher A. PDZ interactions regulate rapid turnover of the scaffolding protein EBP50 in microvilli. J Cell Biol. 2012;198:195–203. doi: 10.1083/jcb.201204008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, LaLonde DP, Bretscher A. The scaffolding protein EBP50 regulates microvillar assembly in a phosphorylation-dependent manner. J Cell Biol. 2010;191:397–413. doi: 10.1083/jcb.201004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger B, Baker RF. Electron microscope investigation of the striated border of intestinal epithelium. Anat Rec. 1950;107:423–441. doi: 10.1002/ar.1091070409. [DOI] [PubMed] [Google Scholar]
- Hanono A, Garbett D, Reczek D, Chambers DN, Bretscher A. EPI64 regulates microvillar subdomains and structure. J Cell Biol. 2006;175:803–813. doi: 10.1083/jcb.200604046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson DE, Bretscher AP. EPI64 interacts with Slp1/JFC1 to coordinate Rab8a and Arf6 membrane trafficking. Mol Biol Cell. 2012;23:701–715. doi: 10.1091/mbc.E11-06-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Konno D, Maruoka H, Sobue K. MALS is a binding partner of IRSp53 at cell-cell contacts. FEBS Lett. 2003;554:30–34. doi: 10.1016/s0014-5793(03)01074-3. [DOI] [PubMed] [Google Scholar]
- Ingraffea J, Reczek D, Bretscher A. Distinct cell type-specific expression of scaffolding proteins EBP50 and E3KARP: EBP50 is generally expressed with ezrin in specific epithelia, whereas E3KARP is not. Eur J Cell Biol. 2002;81:61–68. doi: 10.1078/0171-9335-00218. [DOI] [PubMed] [Google Scholar]
- Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A. Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol. 2001;11:1645–1655. doi: 10.1016/s0960-9822(01)00506-1. [DOI] [PubMed] [Google Scholar]
- LaLonde DP, Garbett D, Bretscher A. A regulated complex of the scaffolding proteins PDZK1 and EBP50 with ezrin contribute to microvillar organization. Mol Biol Cell. 2010;21:1519–1529. doi: 10.1091/mbc.E10-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Fütterer K. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 2005;24:240–250. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard TH, Dawson J, Machesky LM. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J Cell Sci. 2007;120:1663–1672. doi: 10.1242/jcs.001776. [DOI] [PubMed] [Google Scholar]
- Mohler PJ, Kreda SM, Boucher RC, Sudol M, Stutts MJ, Milgram SL. Yes-associated protein 65 localizes p62(c-Yes) to the apical compartment of airway epithelia by association with EBP50. J Cell Biol. 1999;147:879–890. doi: 10.1083/jcb.147.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales FC, Takahashi Y, Kreimann EL, Georgescu MM. Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia. Proc Natl Acad Sci USA. 2004;101:17705–17710. doi: 10.1073/pnas.0407974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar R, McConnell RE, Tyska MJ. Control of cell membrane tension by myosin-I. Proc Natl Acad Sci USA. 2009;106:11972–11977. doi: 10.1073/pnas.0901641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteom. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Reczek D, Berryman M, Bretscher A. Identification of EBP50: a PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D, Bretscher A. The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J Biol Chem. 1998;273:18452–18458. doi: 10.1074/jbc.273.29.18452. [DOI] [PubMed] [Google Scholar]
- Reczek D, Bretscher A. Identification of EPI64, a TBC/rabGAP domain-containing microvillar protein that binds to the first PDZ domain of EBP50 and E3KARP. J Cell Biol. 2001;153:191–206. doi: 10.1083/jcb.153.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome I, Curto M, McClatchey AI. Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev Cell. 2004;6:855–864. doi: 10.1016/j.devcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Scita G, Confalonieri S, Lappalainen P, Suetsugu S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008;18:52–60. doi: 10.1016/j.tcb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenolikar S, Voltz JW, Cunningham R, Weinman EJ. Regulation of ion transport by the NHERF family of PDZ proteins. Physiology. 2004;19:362–369. doi: 10.1152/physiol.00020.2004. [DOI] [PubMed] [Google Scholar]
- Shifrin DA, McConnell RE, Nambiar R, Higginbotham JN, Coffey RJ, Tyska MJ. Enterocyte microvillus-derived vesicles detoxify bacterial products and regulate epithelial-microbial interactions. Curr Biol. 2012;22:627–631. doi: 10.1016/j.cub.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen S, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terawaki S, Maesaki R, Hakoshima T. Structural basis for NHERF recognition by ERM proteins. Structure. 2006;14:777–789. doi: 10.1016/j.str.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Tyska MJ, Nambiar R. Myosin-1a: a motor for microvillar membrane movement and mechanics. Commun Integr Biol. 2010;3:64–66. doi: 10.4161/cib.3.1.10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanatha R, Ohouo PY, Smolka MB, Bretscher A. Local phosphocycling mediated by LOK/SLK restricts ezrin function to the apical aspect of epithelial cells. J Cell Biol. 2012;199:969–984. doi: 10.1083/jcb.201207047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JB, Liu J, Coleman RA, Cunningham R, Steplock DA, Lee-Kwon W, Pallone TL, Shenolikar S, Weinman EJ. Localization and interaction of NHERF isoforms in the renal proximal tubule of the mouse. Am J Physiol Cell Physiol. 2003;285:1494–1503. doi: 10.1152/ajpcell.00092.2003. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinman EJ, Hall RA, Friedman PA, Liu-Chen L-Y, Shenolikar S. The association of NHERF adaptor proteins with G protein-coupled receptors and receptor tyrosine kinases. Annu Rev Physiol. 2006;68:491–505. doi: 10.1146/annurev.physiol.68.040104.131050. [DOI] [PubMed] [Google Scholar]
- Weinman EJ, Steplock D, Wang Y, Shenolikar S. Characterization of a protein cofactor that mediates protein kinase A regulation of the renal brush border membrane Na+-H+ exchanger. J Clin Invest. 1995;95:2143–2149. doi: 10.1172/JCI117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, et al. NHERF2 protein mobility rate is determined by a unique c-terminal domain that is also necessary for its regulation of NHE3 protein in OK cells. J Biol Chem. 2013;288:16960–16974. doi: 10.1074/jbc.M113.470799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Chen JY, Lin YY, Shen KF, Lin WL, Chien CL, ter Beest MB, Jou TS. A bipartite signal regulates the faithful delivery of apical domain marker podocalyxin/Gp135. Mol Biol Cell. 2007;18:1710–1722. doi: 10.1091/mbc.E06-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CH, Oh S, Zizak M, Steplock D, Tsao S, Tse CM, Weinman EJ, Donowitz M. cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc Natl Acad Sci USA. 1997;94:3010–3015. doi: 10.1073/pnas.94.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.