Abstract
We develop biodegradable polymeric nanoparticles to facilitate non-viral gene transfer to human embryonic stem cells (hESC). Small (~200 nm) and positively charged (~10 mV) particles are formed by the self-assembly of cationic, hydrolytically-degradable, poly(beta-amino esters) and plasmid DNA. Varying the end-group of the polymer can tune the biophysical properties of the resulting nanoparticles and their gene delivery efficacy. An OCT4 driven GFP hES cell line was created to allow rapid identification of nanoparticles that facilitate gene transfer while maintaining an hESC undifferentiated state. Using this cell system, we synthesized nanoparticles that have gene delivery efficacy up to four times higher than the leading commercially available transfection agent, Lipofectamine 2000. Importantly, these materials have minimal toxicity and do not adversely affect hES colony morphology or cause non-specific differentiation.
Keywords: Gene Delivery, Nanoparticle, Stem Cell, Transfection, Polymer
The delivery of genes to stem cells can advance cell-based therapies and the field of tissue engineering. Given the right conditions, pluripotent stem cells can potentially differentiate into any cell type of the body, allowing wide therapeutic utility including treatments for autoimmune diseases, spinal chord injury, Parkinson’s disease, and cardiac tissue engineering, among many others.1 Gene delivery could allow for directed differentiation from a pluripotent stem cell into specific differentiated cell types of interest including hematopoietic cells, neurons, cardiomycytes, and osteoblasts, as well as reprogramming a differentiated cell back into a pluripotent state.2–4 Beyond controlled differentiation, ectopic expression of key growth and transcription factors could allow for elucidation of fundamental cell development pathways in vitro as well as regulation of growth in vivo once the cells are transferred to a patient. Gene delivery can also provide a mechanism for in vivo expression of secreted therapeutic proteins.
While there is promise, current approaches for gene transfer to hESCs are limited as there are safety concerns with viral approaches and non-viral methods have low efficacy. Forexample, testing in the H9 hESC line showed that commonly used lipid and polymer based transfection agents including FuGENE, LipofectAMINE Plus, and ExGen 500 transfected less than 10% of the hESCs5 Electroporation resulted in similarly low transfection and, unlike with murine ESC, significantly reduced viability. Nucleofection, a modified version of electroporation that includes the addition of a nucleofactor solution, can significantly improve gene transfer to achieve 20% transfection of hESCs whereas regular electroporation only achieves 5–6%.6 However, nucleofection, like regular electroporation, can cause high cell death. Many hESC gene transfer studies lack a reliable marker to separate transfected undifferentiated cells from differentiating, differentiated, and feeders.7 Studies from our group and others have shown that the differentiating cells on the periphery of cell colonies are the cells most likely to be transfected. Carry-over feeder cells may also be preferentially transfected over hard-to-transfect human stem cell colonies. For a quantitative analysis of transient transfection efficiency, a reliable undifferentiated hES marker is needed.
Viral gene transfer approaches for hESCs also have their limitations. Adenovirus serotype 5 has been shown to transduce just 11% of undifferentiated H9 hESCs and adeno associated virus (serotypes 2, 4, and 5) to transduce only 0.01% at best.8 The most effective viral strategy to date has been lentiviral vectors with about 40% transduction efficiency in H1 and H9 hESCs.9 Although the transgene expression can be further enhanced with concentrated virus, appropriate promoters, and drug selection after transduction, the safety concerns from insertional mutagenesis after viral integration are difficult to overcome. This issue limits their potential for human gene therapy. A safe and effective method for gene delivery to stem cells would be invaluable for the creation of new cell-based therapies.
We have developed a class of polymers, poly (β-amino esters), that are promising for non-viral gene delivery due to their ability to condense DNA into nanoparticles that facilitate cellular uptake and endosomal escape.10–12 Due to hydrolytic cleavage of the ester groups that compose these polymers, they are biodegradable and have low toxicity.10,13,14 These particles are also useful as they can be coated for ligand-specific delivery.15 Chemical modification of the ends of these polymers results in improved biomaterials that can deliver DNA to human umbilical vein endothelial cells (HUVECs) at levels comparable to adenovirus.16,17 Here we develop polymeric nanoparticles for non-viral gene transfer to undifferentiated hESCs. To ensure that the transfected hESC colonies remain in an undifferentiated state once transfected, we use hESCs targeted to stably express Oct4-driven GFP and monitor GFP levels as an indicator of the undifferentiated state of the hESCs. We demonstrate that these nanoparticles can be formulated to have high efficacy for gene delivery to hESCs while also maintaining a pluripotent, undifferentiated state.
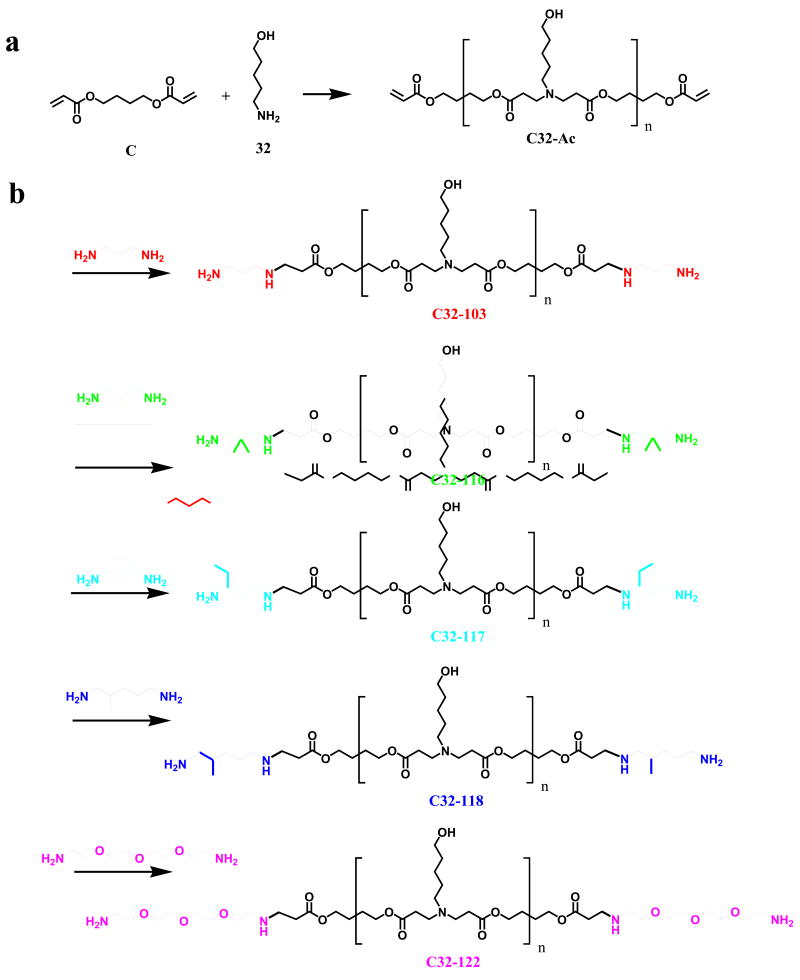
End-modified poly(β-amino esters) were synthesized in a two step procedure. First, acrylate terminated C32 (C32-Ac) was synthesized via the conjugate addition of 5-amino-1-pentanol to 1,4-butanediol diacrylate at a 1.2:1 molar ratio of diacrylate monomer to amine monomer (Supporting Information). Next, this polymer was end-modified by reacting C32-Ac with an excess of various diamine monomers (Figure 1) in THF and then purified by precipitation. Polymer/DNA nanoparticles were formed by vortexing polymer and DNA solutions together in 25 mM sodium acetate and waiting ten minutes for particle self-assembly (Supporting Information). All particles were formulated at a 50:1 weight ratio of poly(beta-amino ester) to DNA. These particles were found to generally have a small size (~200 nm) and positive zeta potential (~10 mV) as measured by dynamic light scattering and phase analysis light scattering. However, slight changes to the end-group of the polymer can change theses properties (Figure 2A). For example, particles formed with polymer C32-116 are much larger (400 nm) and less positively charged (4 mV). Structurally, C32-116 is almost identical to C32-103 and C32-117 (Figure 1). The only difference is that C32-116 has two methyl groups near the terminal amine, whereas C32-103 does not have these groups and C32-117 has an ethyl group instead. C32-118, which has a slightly longer hydrocarbon spacer between the terminal amine and the base C32 polymer, forms nanoparticles that are the smallest in size (160 nm) and the most positively charged (16 mV). C32-122, which has a slightly longer hydrophilic spacer between the terminal amine and the base C32 polymer forms nanoparticles of average size, but higher than average zeta potential(15 mV).
Figure 1.
Synthesis of end-modified poly(beta-amino esters). Each end of C32-117 and C32-118 may be in either of the configurations shown.
Figure 2.
(a) Particle size (left axis, black bars) and zeta potential (right axis, white bars) of end-modified poly(beta-amino esters)/DNA particles (mean + SD). (b) hES viability 24 hrs after transfection with end-modified poly(beta-amino esters) or Lipofectamine 2000 (mean + SD). Polymeric nanoparticles composed of C32-103, C32-116, C32-118, and C32-122 do not show cytotoxicity. C32-117 shows some cytotoxicity (p<0.01) compared to untreated cells, but is not statistically more cytotoxic than Lipofectamine 2000.
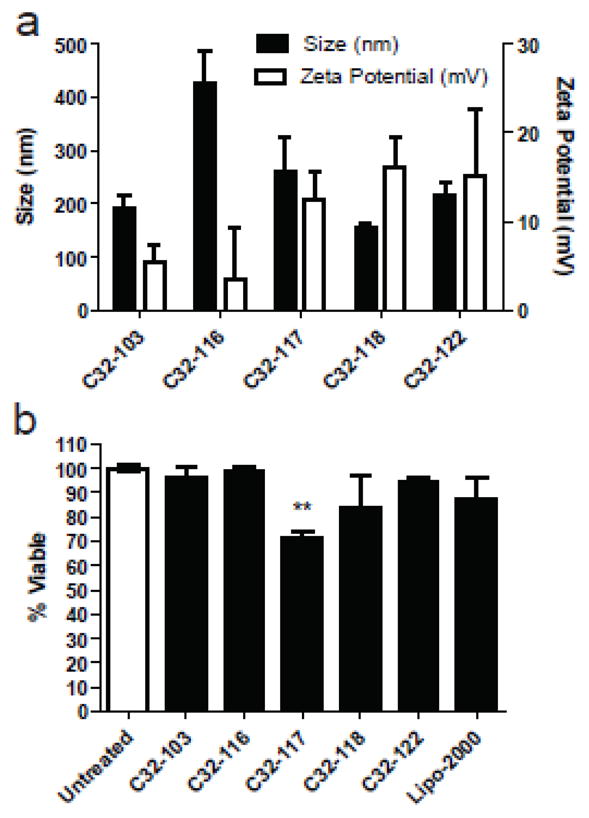
To measure cell viability, cellular metabolic activity was measured using the Cell Titer 96 Aqueous One Solution assay kit (Promega, Madison, WI, USA) 24 hrs post-transfection (Supporting Information). When these particles are added to hES colonies, generally there is minimal cytotoxicity. C32-103, C32-116, C32-118, and C32-122 nanoparticles each show the same viability as untreated controls (Figure 2B). However, C32-117 nanoparticles show some loss of viability compared to untreated controls (70% viable), although this is not statistically different from the small loss of viability with Lipofectamine 2000. When the levels of gene transfer are quantified by flow cytometry, the poly(beta amino ester) nanoparticles enable transfection of up to 50% of human embryonic stem cell colonies. Dramatically, C32-118 nanopartcles (Figure 3A) are able to significantly transfect hES colonies whereas Lipofectamine 2000, the leading commercially available non-viral vector is only able to transfect cells on the periphery. Representative fluorescent micrographs of this GFP transfection can be seen in Figure 3. For this particular transfection, it is important to note that some of these cells may be differentiated and others may be feeder cells. To avoid this, hES BG02-Oct4-GFP cells were made by introducing hOct4-GFP-puro construct into hES cells (Supporting Information). In this construct, the GFP reporter gene is expressed from the Oct4 promoter that is active when cells are in an undifferentiated state. Upon differentiation, the Oct4 promoter is gradually inactivated and therefore the GFP reporter is down-regulated. This line expresses all pluripotent stem cell markers and forms teratomas after being grafted into severe combined immunodeficient mice (SCIS). With these cells, undifferentiated hESCs can be clearly distinguished from differentiated stem cells and/or feeder cells.
Figure 3.
Poly(beta-amino esters) form nanoparticles with high gene delivery efficacy to hESC colonies. (a) Poly(beta-amino ester) C32-118/DNA particles have a small, spherical shape (scale bar is 100 nm). Fluorescence micrographs are shown following treatment with (b) Lipofectamine 2000 and (c) C32-118. Two-dimensional FACS gating of GFP expression (FL1) of hESCs: (d) untreated (e) Lipofectamine 2000 (f) C32-118.
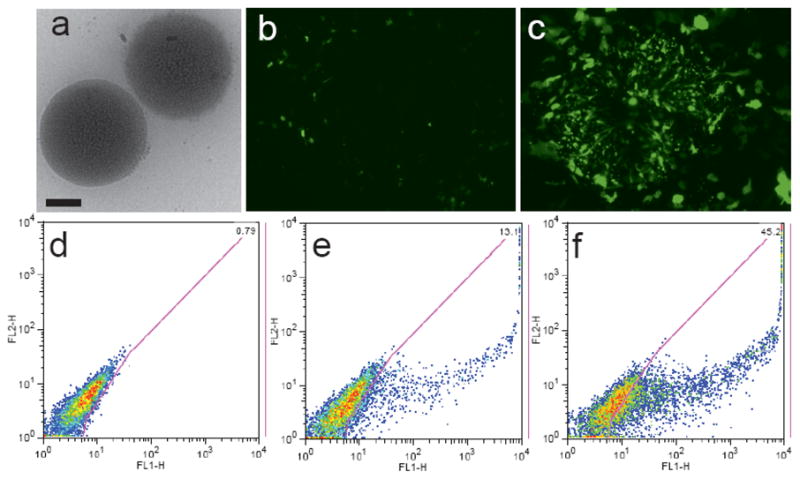
With this new cell system, FURW was delivered to encode red fluorescent protein (RFP) as a reporter rather than GFP. Positive transfection of undifferentiated hES cells was determined by counting cells positive for both RFP and GFP. The results for polymeric gene delivery to undifferentiated hES cells that remain undifferentiated following transfection are shown in Figure 4. Poly(beta-amino esters) showed superior gene delivery efficacy compared to Lipofectamine 2000. Cell viability was high and colony morphology was normal as shown in Figure 4D.
Figure 4.
FACSgating of Oct4 GFP+ undifferentiated hESCs and RFP+ transfected hESCs for (a) free DNA and (b) C32-122/DNA nanoparticles. (c) Transfection of hESC colonies for different poly(beta-amino esters) (mean + SD). C32-117 (p<0.01) and C32-118 (p<0.05) are significantly better at gene delivery than Lipofectamine 2000. Micrograph of representative hESC colony, one week post-transfection with C32-122/DNA nanoparticles by (d) light microscopy, (e) GFP fluorescence (showing undifferentiation) and (f) RFP fluorescence (showing persistence of transfection).
Interestingly, small structural changes to just the ends of the gene delivery polymer, including single carbon changes, can significantly increase transfection efficacy to hESC colonies. These differences may be due in part to the biophysical properties of the nanoparticles, with smaller particles (~200 nm) being more favorable than larger particles (~400 nm) and positively charged particles (~15 mV) being more favorable than weakly charged particles (~5 mV). Nanoparticles formed with polymers such as C32-118 may also potentially target hESCs compared to other cell types. For example, here we show that C32-118 nanoparticles transfect hESCs more effectively than C32-103 nanoparticles. Yet, we have previously shown that in the case of differentiated human primary cells, human umbilical vein endothelial cells (HUVECs), C32-118 nanoparticles transfect 70% fewer cells than C32-103 nanoparticles.17 Thus, end-modification of polymers used to form gene delivery nanoparticles may be a useful strategy to promote cell-specific delivery when multiple cell types or cells of different differentiation state are present. In contrast, alternate polymer end-modification may create polymeric gene delivery nanoparticles with high delivery to a range of human cell types. For example, here we show that C32-117 nanoparticles enabled the highest transfection of hESC colonies as is also true with HUVECs.17 Importantly, the poly(beta-amino esters) used here had up to four-fold the efficacy at gene transfer to undifferentiated human embryonic stem cell colonies than the leading non-viral vector, Lipofectamine 2000 (22% positive with C32-117 vs. 5.8% positive with Lipofectamine 2000). This facile method for gene transfer works in the presence or absence of serum and without the need for physical or electrical methods that can enhance gene transfer, but can also significantly lower cell viability.
In comparison to viral vectors, the biodegradable polymers used here may be safer for certain therapeutic applications. Whereas retroviruses, including lentivirus vectors, can cause non-specific integration that may lead to mutation and cancer, the non-viral gene delivery approach used here transfers an episomal plasmid to the cells that is expressed transiently. We confirmed that gene expression is transient by measuring the delivery of both fluorescent protein reporter encoding genes and antibiotic resistance encoding genes over time. Transient expression may also be useful for temporal control of the expression of growth factors. As a biological tool to study the role that varying genes play in development, viral vectors are slow to prepare and screen as each gene needs to be separately cloned into each vector. In contrast, a non-viral approach using nanoparticles that spontaneously self-assemble when mixed with DNA, as used here, facilitates high-throughput screening of gene combinations to study development, transdifferentiation, and/or reprogramming.
We have demonstrated the use of self-assembled, biodegradable polymeric nanoparticles as a tool for non-viral gene transfer to human embryonic stem cell colonies. Small modifications to the ends of the polymers were found to significantly improve the transfection efficacy of the polymer/DNA particles. Lead polymers enabled high gene expression while maintaining high viability, normal cell morphology, and an undifferentiated state. With further studies, these biodegradable, non-integrating vectors may be a more useful alternative for high-throughput biological screening and a safer alternative to viruses for use in regenerative medicine.
STATISTICS
Statistical calculations were performed using GraphPad Prism 5 for Windows. All graphs show mean ± standard deviation. To analyze transfection efficacy, one-way analysis of variance was used to calculate statistical significance (p<0.01). The Bonferroni post-test was used to measure differences between each of the polymeric nanoparticles and Lipofectamine 2000, a leading non-viral commercially available transfection agent. To analyze cell viability, one-way analysis of variance and the Bonferroni post-test was also used to calculate statistical significance (p<0.01). Each batch of nanoparticles was compared to untreated cells as well as Lipofectamine 2000.
Supplementary Material
Acknowledgments
We thank Yong Zhang for assistance with TEM and the CMSEExperimental Facilities supported in part by the MRSEC Program of the National Science Foundation under Award Number DMR 02-13282. This work was supported by NIH Grant EB 00244 and R01-DE016516-03.
Footnotes
SUPPORTING INFORMATION
Detailed description of the materials and methods is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Yates F, Daley GQ. Gene Therapy. 2006;13:1431–1439. doi: 10.1038/sj.gt.3302854. [DOI] [PubMed] [Google Scholar]
- 2.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Eiges R, Schuldiner M, Drukker M, Yanuka O, Itskovitz-Eldor J, Benvenisty N. Curr Biol. 2001;11:514–518. doi: 10.1016/s0960-9822(01)00144-0. [DOI] [PubMed] [Google Scholar]
- 6.Lakshmipathy U, Pelacho B, Sudo K, Linehan JL, Coucouvanis E, Kaufman DS, Verfaillie CM. Stem Cells. 2004;22:531–543. doi: 10.1634/stemcells.22-4-531. [DOI] [PubMed] [Google Scholar]
- 7.Liew CG, Draper JS, Walsh J, Moore H, Andrews PW. Stem cells. Vol. 25. Dayton, Ohio: 2007. pp. 1521–1528. [DOI] [PubMed] [Google Scholar]
- 8.Smith-Arica JR, Thomson AJ, Ansell R, Chiorini J, Davidson B, McWhir J. Cloning Stem Cells. 2003;5:51–62. doi: 10.1089/153623003321512166. [DOI] [PubMed] [Google Scholar]
- 9.Xia X, Zhang Y, Zieth CR, Zhang SC. Stem Cells Dev. 2007;16:167–176. doi: 10.1089/scd.2006.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynn DM, Langer R. J Am Chem Soc. 2000;122:10761–10768. [Google Scholar]
- 11.Anderson DG, Lynn DM, Langer R. Angew Chem Int Ed Engl. 2003;42:3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 12.Akinc A, Lynn DM, Anderson DG, Langer R. J Am Chem Soc. 2003;125:5316–5323. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- 13.Anderson DG, Peng W, Akinc A, Hossain N, Kohn A, Padera R, Langer R, Sawicki JA. Proc Natl Acad Sci U S A. 2004;101:16028–16033. doi: 10.1073/pnas.0407218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green JJ, Shi J, Chiu E, Leshchiner ES, Langer R, Anderson DG. Bioconjug Chem. 2006;17:1162–1169. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- 15.Green JJ, Chiu E, Leshchiner ES, Shi J, Langer R, Anderson DG. Nano Lett. 2007;7:874–879. doi: 10.1021/nl062395b. [DOI] [PubMed] [Google Scholar]
- 16.Zugates GT, Peng W, Zumbuehl A, Jhunjhunwala S, Huang YH, Langer R, Sawicki JA, Anderson DG. Mol Ther. 2007 doi: 10.1038/sj.mt.6300132. [DOI] [PubMed] [Google Scholar]
- 17.Green JJ, Zugates GT, Tedford NC, Huang Y, Griffith LG, Lauffenburger DA, Sawicki JA, Langer R, Anderson DG. Adv Mat. 2007;19:2836–2842. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.