Abstract
Two BRCA2-like sequences are present in the Arabidopsis genome. Both genes are expressed in flower buds and encode nearly identical proteins, which contain four BRC motifs. In a yeast two-hybrid assay, the Arabidopsis Brca2 proteins interact with Rad51 and Dmc1. RNAi constructs aimed at silencing the BRCA2 genes at meiosis triggered a reproducible sterility phenotype, which was associated with dramatic meiosis alterations. We obtained the same phenotype upon introduction of RNAi constructs aimed at silencing the RAD51 gene at meiosis in dmc1 mutant plants. The meiotic figures we observed strongly suggest that homologous recombination is highly disturbed in these meiotic cells, leaving aberrant recombination events to repair the meiotic double-strand breaks. The ‘brca2' meiotic phenotype was eliminated in spo11 mutant plants. Our experiments point to an essential role of Brca2 at meiosis in Arabidopsis. We also propose a role for Rad51 in the dmc1 context.
Keywords: Arabidopsis, Brca2, Dmc1, meiosis, Rad51
Introduction
Germ-line mutations in the BRCA2 gene are associated with hereditary breast cancer in humans (Wooster et al, 1995; Scully and Livingston, 2000). Although accumulated data suggest that the Brca2 protein plays a role in the control of genome stability, its exact function remains unclear (Jasin, 2002; West, 2003). The demonstration of an interaction between Brca2 and Rad51, the eucaryotic RecA homolog, has suggested that Brca2 might be involved in homologous recombination (Sharan et al, 1997). Consistently, brca2 knockout mice show early embryo lethality associated with chromosomal rearrangements and breaks (Yu et al, 2000), a phenotype that is reminiscent of the one observed with rad51 knockout mice (Lim and Hasty, 1996; Tsuzuki et al, 1996). Furthermore, the formation of Rad51 foci following DNA damage is altered in brca2 mutant cells (Yu et al, 2000). Thus, Brca2 may be required to engage Rad51 in the recombinational repair of DNA damage.
Whereas Rad51 proteins are highly conserved, Brca2-related proteins are more divergent and sometimes absent from eucaryotic genomes. In humans, the BRCA2 gene encodes a protein of 3418 amino acids. Eight iterated ‘BRC repeats' in this protein are involved in its interaction with Rad51 (Bork et al, 1996; Wong et al, 1997; Chen et al, 1998). The BRC motifs are found only in Brca2-related proteins, but their number and location are subject to variation among species. For instance, only one BRC motif is present in the Ustilago maydis Brh2 (Brca2 homolog) protein (Kojic et al, 2002), while up to 15 appear in a Trypanosoma putative Brca2 (Lo et al, 2003). Except for the BRC motifs, overall conservation of the Brca2-related proteins is poor (Warren et al, 2002). A total of 800 residues of the C-terminal domain of the Brca2 protein (in a complex with Dss1), not comprising a BRC motif, can bind DNA (single-stranded (ss) or double-stranded (ds)) and stimulate Rad51-mediated recombination in vitro (Yang et al, 2002). Other work indicates that a single BRC repeat can interfere with the incorporation of Rad51 monomers into a nucleofilament (Pellegrini et al, 2002). These results illustrate the difficulty to understand the biological role of Brca2.
Correct segregation of chromosomes at meiosis relies upon their association into bivalents. In many organisms, bivalent formation relies upon homologous recombination (Roeder, 1997) and it was demonstrated in yeast to depend on the formation of recombination-initiating double-strand breaks (DSBs) by Spo11 (Bergerat et al, 1997; Keeney et al, 1997), and on the Rad51 and Dmc1 proteins, which are essential actors in the subsequent recombination (Masson and West, 2001). Consistently, spo11-1 and dmc1 Arabidopsis mutant plants are defective in bivalent formation, but they remain able to complete meiosis, in contrast to their yeast and mouse counterparts, which show meiotic delay, arrest or apoptosis (Couteau et al, 1999; Grelon et al, 2001).
Brca2 localizes upon human meiotic chromosomes during the formation of the synaptonemal complex (Chen et al, 1998). However, because of the recessive lethality of most brca2 mutant genes, the role of Brca2 in meiosis was difficult to investigate in mammals. Although mouse sterility was reported to accompany a mutation truncating the Brca2 protein at exon 11 (but not at exon 27) (Connor et al, 1997; McAllister et al, 2002), it is only recently that a meiotic phenotype was described in a partially rescued brca2−/− knockout mouse expressing the human BRCA2 gene (Sharan et al, 2003). Male meiocytes are found arrested in prophase I with normal Spo11 but disturbed Rad51 and Dmc1 foci. Aside mammalians, the phenotype of a brca2 mutant (brh2) isolated in Ustilago establishes that Brca2 contributes to DNA repair in this fungus. However, the mutant is unable to carry out meiosis, being arrested at a premeiotic stage (Kojic et al, 2002).
Two conserved BRCA2-like gene sequences are present in the Arabidopsis genome. In a yeast two-hybrid assay, the corresponding BRCA2 cDNAs interact with (i) AtRad51, confirming a typical Brca2 property, and (ii) a previously unrevealed partner, AtDmc1, linking AtBrca2 to meiosis. RNA interference (RNAi) allowed us to inactivate both Brca2 gene functions at meiosis (Waterhouse and Helliwell, 2003). To restrict potential somatic phenotypes, we placed RNAi constructs under the control of a meiotic promoter (pDMC1). The efficiency of this approach was ensured by our observation that Arabidopsis plants transformed by RNAi constructs targeted to the DMC1 transcripts at meiosis phenocopied dmc1 mutant plants. When RNAi/BRCA2 constructs under the control of the pDMC1 meiotic promoter were introduced into plants, 90% of the resulting transformants appeared partially sterile. This sterility was associated with meiotic defects that differed from those observed in dmc1 plants. Furthermore, introduction of a pDMC1∷RNAi/RAD51 construct into dmc1 plants resulted in a meiotic phenotype that exactly mimicked the pDMC1∷RNAi/BRCA2 phenotype. Collectively, these observations demonstrate an essential role of Brca2 in the control of homologous recombination at meiosis in Arabidopsis. We also provide clues to the role of Rad51 at meiosis in Arabidopsis.
Results
Two BRCA2 genomic sequences are recently duplicated in Arabidopsis
Two BRCA2-like sequences are present in the Arabidopsis thaliana genome. One of these sequences, BRCA2(IV), is located in a subtelomeric region of chromosome IV, close to the nucleolar organizer region (NOR). From its putative first codon to stop codon, it is 6258 nucleotides (nt) long and is the first open reading frame (ORF) (At4g00020/10) identified along this chromosome. The other BRCA2-like sequence (At5g01630) is also located near a chromosome extremity. From putative start to stop it is 5795 nt long and is the 62nd ORF of chromosome V (BRCA2(V)) (Figure 1A). Except for the presence of a 450 pb insertion in a putative intron, which prevented correct identification of the BRCA2(IV) genomic region by the Arabidopsis genome sequencing project, these two sequences are 96.8% identical and are probably the result of a recent duplication. The duplication is restricted to the BRCA2 locus—very little of the surrounding genomic sequences were involved. Close examination of their neighboring regions suggests that AtBRCA2(V) is the original copy—a nearby potential ORF (At5g01640), located 5′ to the BRCA2 region, on chromosome V, is split into two fragments around the AtBRCA2(IV) locus. Thus, the At5g01640 potential coding sequence from chromosome V is disrupted on chromosome IV; furthermore, it has accumulated numerous alterations leading to potential frameshift mutations (Figure 1A). The two BRCA2 sequences have conserved their potential ORFs, although one (BRCA2(IV)) is located close to the NOR, a region of high genomic instability, as manifested by rapid sequence divergence of the nearby fragment of At5g01640. Accession numbers for the isolated AtBRCA2 cDNAs are AJ488304 and AJ488305 for chromosome IV and chromosome V sequences, respectively.
Figure 1.
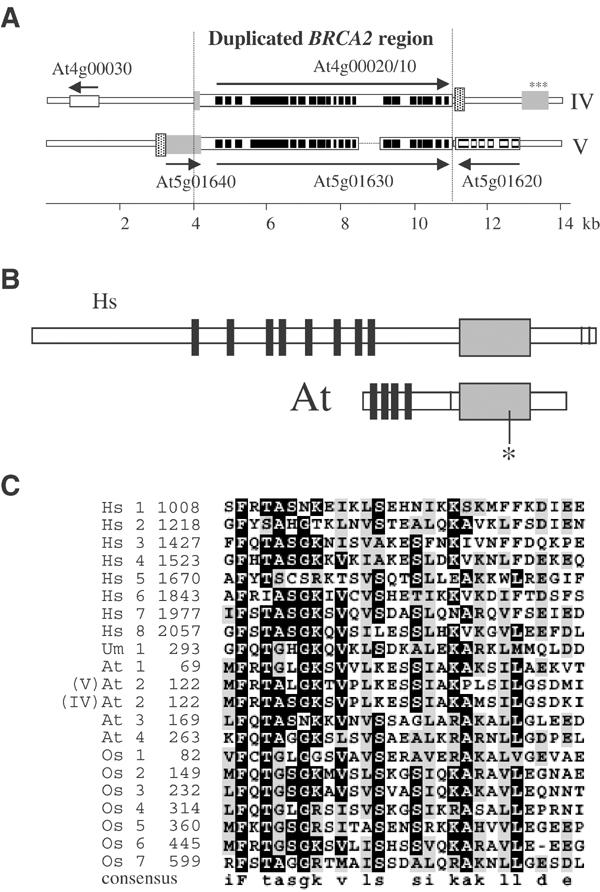
The Arabidopsis BRCA2 sequences. (A) Structure of the genomic BRCA2 loci on chromosomes IV and V. Arrows indicate potential ORFs. The 20 AtBRCA2 exons on each chromosome are shown as black boxes. The dotted line corresponds to a region in AtBRCA2(IV) that is not present in AtBRCA2(V). The gray boxes represent ORF At5g01640 on chromosome V that is disrupted into two fragments on chromosome IV. The dotted box is a sequence that is identical on both chromosomes IV and V. (B) Comparison of the Homo sapiens (Hs) and A. thaliana (At) Brca2 proteins. Thick black lines represent the BRC motifs, and thin black lines represent the NLS sequences. The gray box corresponds to the conserved C-terminal region; * indicates the position of the stop codon in the truncated Brca2(IV) protein. (C) Alignment of the BRC motifs from the human (Hs), Ustilago (Um), Arabidopsis (At) and rice (Os) Brca2 proteins. The numbers indicate their rank and amino-acid position respectively in the Brca2 proteins. These selected BRC motifs were chosen as most representative because they belonged to functionally defined Brca2 proteins (human and Ustilago) or plant putative orthologs (rice and Arabidopsis).
The two BRCA2 cDNAs encode nearly identical putative proteins and are both expressed in flower buds
Using specific primers, we isolated two different BRCA2 cDNAs that correspond to either the chromosome IV (3456 nt) or the chromosome V (3468 nt) BRCA2 coding sequence, respectively, revealing that both BRCA2 sequences are transcriptionally active. Apparently, the 450 bp insertion located in intron 13 of the BRCA2 (IV) sequence does not impede its splicing. Both BRCA2 cDNA sequences are composed of 20 exons and 19 introns (Figure 1A). Divergence between the two spliced cDNAs is low (2.99% nucleotide substitutions), but only slightly lower than between their aligned introns (3.82%). These cDNAs encode, respectively, for 1151 amino acid (IV) and 1155 amino acid (V) long putative proteins, which are 94.5% identical to each other. The overall Arabidopsis proteins are shorter than the human Brca2 protein, which is comprised of 3418 amino acids. In the course of AtBRCA2 (IV) cDNA isolation, a sequence was obtained, which, due to erroneous splicing of intron 12, contained a 5 bp insertion, which truncated the putative Brca2 protein at amino acid 784 (see Figure 1B).
The presence of BRC motifs is the typical signature of Brca2 proteins (Bignell et al, 1997), and their presence in a wide range of eucaryotic Brca2 putative proteins was analyzed by Lo et al (2003). While eight BRC repeats were identified in the human Brca2 protein, only four such BRC motifs are present in the Arabidopsis putative Brca2 proteins (Figure 1B). The BRC motif number 2 is the only one that is different between the two proteins; the five amino-acid changes result, in particular, in loss of the consensus TASGK motif in the AtBrca2 (V) putative protein sequence (Figure 1C). The BRC motifs are found close to the N-terminal regions in Arabidopsis (between amino acids 67 and 288). Thus, the AtBrca2 proteins lack the region that is N-terminal to the BRC repeats in humans (Figure 1B). Apart from the BRC motifs, the Arabidopsis and human Brca2 proteins align well across 500 amino acids in their C-terminal regions (25% identities, 45% conservation). A nuclear localization sequence (NLS) can also be detected in the AtBrca2 putative protein sequences, at amino acid 457, a location that differs from that of the NLS motif in human Brca2.
RT–PCR experiments were conducted with oligonucleotides designed to allow specific examination of the relative expression levels of each BRCA2 gene. This approach indicated that, in contrast to RAD51, the BRCA2 genes are not induced following gamma irradiation (100 Gy); both BRCA2 genes are transcribed in flower buds to a similar level (see Figure 2A). This result is not surprising, as the duplication of the AtBRCA2 sequences includes a region that extends 5′ to their ATG start codon into the next ORF (Figure 1A). Thus, the two genes share the same promoter region and have conserved a similar transcription pattern.
Figure 2.
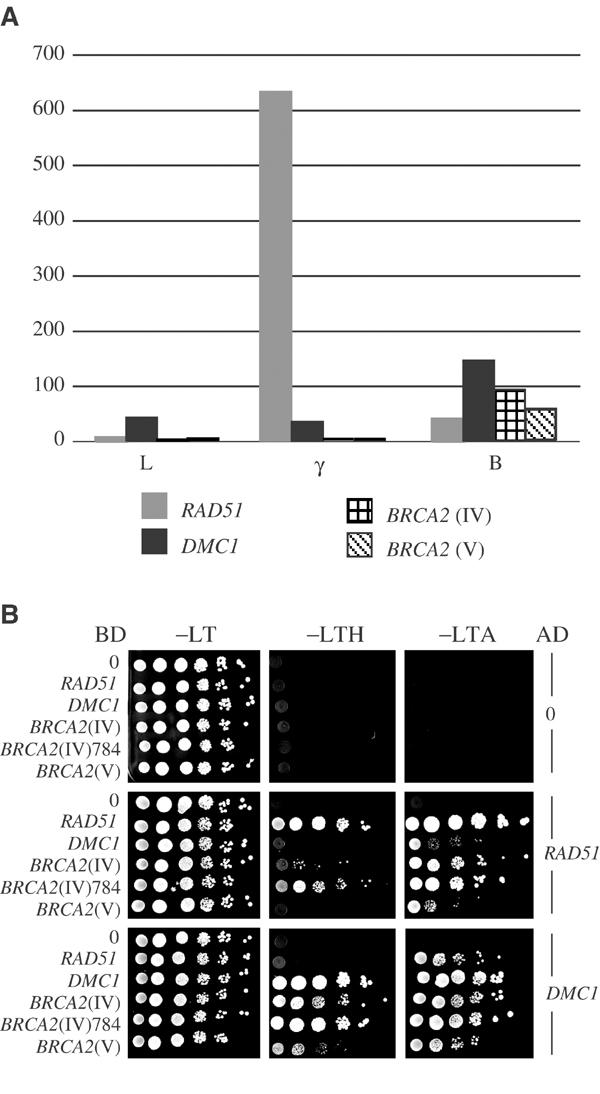
Expression of the AtBRCA2 genes and yeast two-hybrid interactions. (A) Real-time quantitative RT–PCR. Scale numbers are arbitrary. L, leaves; γ, gamma-irradiated leaves; B, flower buds. (B) Yeast two-hybrid interactions of Brca2 with Rad51 and Dmc1. Serial dilutions of yeast diploid strains containing combinations of the indicated activating domain (AD) and binding domain (BD) protein fusions. −LT, −LTH, −LTA correspond to dropout media without L, leucine; T, tryptophan; A, adenine. −LTH medium was supplemented with 3 mM 3-amino-triazole. BRCA2(IV)784 is the truncated Brca2 protein as described in the text.
In a yeast two-hybrid assay, AtBrca2 interacts with AtRad51 and AtDmc1
In addition to the presence of BRC motifs, a feature characteristic of the Brca2 proteins is their ability to interact with Rad51. This property was initially uncovered with the mammalian proteins, and was reported to be present also in the Ustilago Brh2 protein (Kojic et al, 2002). We examined this aspect of the AtBrca2 function, by the yeast two-hybrid procedure. The assay was positive when AtBRCA2 (IV) and AtRAD51 were introduced into the same yeast tester strain (Figure 2B). The results were more positive when the AtDMC1 and AtBRCA2 genes were introduced together. The AtBRCA2 (V) cDNA gave a less positive response when paired with either AtRAD51 or AtDMC1.
In the course of these experiments, we noted that the Rad51 and Dmc1 fusion proteins were active as homodimers and heterodimers to trigger reporter gene activation when coexpressed in the same yeast cells (Figure 2B). On the other hand, no interaction was revealed between the AtBrca2 proteins and any of the five other Rad51-related sequences found in Arabidopsis (data not shown).
Presence of a premature stop codon that results in a truncated N-terminal 784–amino-acid-long translation product did not prevent the interaction of AtBrca2 (IV) with AtRad51 or AtDmc1 (Figure 2B). The BRC motifs and the NLS are present in this truncated protein (see Figure 1B), consistent with the idea that, in Arabidopsis as in mammals, the BRC motifs are involved in the interaction of Brca2 with Rad51 (and Dmc1). The slight sequence divergence of BRC motif number 2 between the two Arabidopsis Brca2 proteins may be related to their difference in interacting efficiency—the sequence in AtBrca2 (IV) is closer to the consensus, particularly at the TASGK motif. Based on structural and mutational analysis, Shin et al (2003) defined sequence fingerprints along Rad51 that could identify the presence of Brca2-like activities in other organisms. Four amino acids were retained as typical of a functional interaction between HsRad51 and Brca2: Tyr205, Ser208, Ala209, Met251. At similar positions in the aligned protein sequences, we find Leu, Ala, Ser and Ser in AtRad51, and Leu, Ala, Ala and Arg in AtDmc1. These amino acids do not preclude that interactions exist. The human Dmc1 protein, which may also interact with Brca2, presents Asp, Ala, Ala and Arg residues at these same positions.
Setting up and assessing RNAi in meiotic cells
To characterize the function of the duplicated AtBRCA2 genes, we set up RNAi constructs to silence both genes at once (Figure 3). To ascertain the efficiency of that strategy, we introduced defined tester constructs. These RNAi constructs were placed under the control of the DMC1 promoter, which directs the expression of the meiotic DMC1 gene (Klimyuk and Jones, 1997; Doutriaux et al, 1998; Couteau et al, 1999). RNAi constructs were established in the pKannibal plasmid, which provides an intron sequence on either sides of which the target DNA sequence can be cloned in diverging orientations (see Figure 3; Helliwell and Waterhouse, 2003). The DNA sequence derived from the gene to be silenced was usually close to 400 bp in length. Our constructs contained respectively: (i) no target DNA (pDMC1∷RNAi/0), (ii) DNA aimed at silencing EMB506 (pDMC1∷RNAi/EMB506), a gene previously described as essential to embryo viability (Albert et al, 1999), in order to define whether the pDMC1 promoter could efficiently alleviate somatic phenotypes, (iii) DNA aimed at silencing the DMC1 gene (pDMC1∷RNAi/DMC1), in order to assess whether the RNAi procedure would create a phenocopy of the dmc1 mutant, and thus trigger efficient RNAi at meiosis, (iv) DNA aimed at silencing the BRCA2 genes (pDMC1∷RNAi/BRCA2) and (v) DNA aimed at silencing the RAD51 gene (pDMC1∷RNAi/RAD51).
Figure 3.
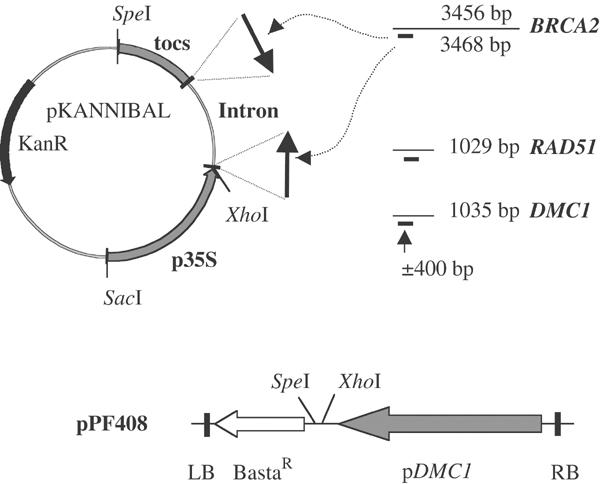
RNAi constructs. Fragments of the genes to be targeted by RNAi were cloned in diverging orientation in pKannibal. The RNAi constructs were then transferred into pPF408 so as to be under the control of the pDMC1 promoter. pPF408 also contains the T-DNA left and right borders (LB and RB), and the BastaR cassette to allow transformation and selection of transformed plants.
Plants transformed by the T-DNA construct that contained only the intron sequence from pKannibal under the control of the DMC1 promoter (pDMC1∷RNAi/0) had wild-type phenotype. This indicated that the DMC1 promoter did not trigger a dmc1 phenotype per se, as could have resulted from some other silencing mechanism (trans-inactivation). Anthers from two of these plants were isolated and their meiocytes (pollen mother cells, PMC) dissected prior to DAPI staining. In these plants, meiosis was identical to that in wild-type plants (see Figures 4A–G). We could identify all typical prophase I through telophase II stages, showing (i) the regular organization of five pairs of chromosomes into five bivalents (five pachytene and two diakinesis) among 17 prophase I stages, and one metaphase I stage (Figures 4A–D), before (ii) their successive segregation into two pools of five distinct univalents (eight anaphase I stages were counted as in Figure 4E) and then (iii) four sets of five chromosomes (one anaphase II and one telophase II stages were found, as in Figures 4F and G).
Figure 4.
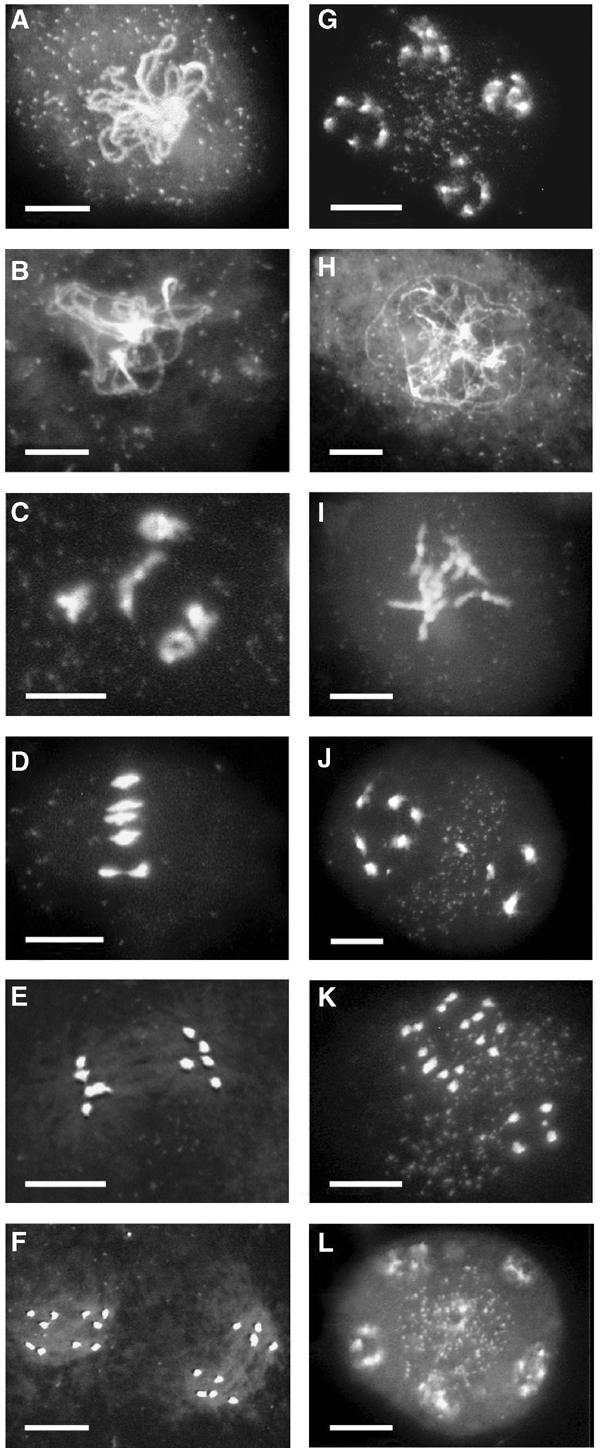
Meiosis in wild-type pDMC1∷RNAi/0 and pDMC1∷RNAi/DMC1-transformed plants. (A–G) DAPI staining of chromosomes in meiocytes from pDMC1∷RNAi/0-transformed plants: (A) pachytene stage showing chromosome synapsis; (B) diplotene stage, the synapsed chromosomes start separating; (C) diakinesis stage, the five bivalents are attached at the chiasma; (D) metaphase I with five aligned bivalents; (E) anaphase I, bivalents segregate into two sets of five univalents; (F) anaphase II with four groups of five chromosomes; (G) telophase II, the chromosomes start decondensing. (H–L) DAPI staining of chromosomes in meiocytes from pDMC1∷RNAi/DMC1-transformed plants: (H) prophase I; (I) diakinesis stage with independent univalents; (J) anaphase I with 10 randomly segregating univalents; (K) anaphase II with 20 chromosomes separating; (L) telophase II, with five or six groups of decondensed chromosomes. Bar, 10 μm.
Most plants transformed by the pDMC1∷RNAi/EMB506 construct were alive (95%), to the contrary of plants transformed by the same RNAi construct under the control of a plant constitutive promoter (p35S). In this latter case, 95% of the transformed plants bleached and died prematurely following selection (data not shown). This allowed us to consider that the pDMC1 promoter was adequate to avoid most somatic expression of the RNAi constructs.
pDMC1∷RNAi/DMC1-transformed plants are phenocopies of dmc1 mutant plants
When the construct aimed at silencing DMC1 at meiosis (pDMC1∷RNAi/DMC1) was introduced into Arabidopsis, we found that 90% (18/20) of the transformed plants were partially sterile. This result allowed us to conclude that RNAi was efficient and dominant, since T1 plants were affected. Although seed set was low in the pDMC1∷RNAi/DMC1 plants, enough seeds were collected to grow T2 and then T3 plants. The partial sterility phenotype remained consistent through three successive generations. In the absence of selection, some fertile plants segregated that did not contain the RNAi construct, indicating that RNAi is reversible and correlates with the presence of the RNAi transgene. To further ascertain that the pDMC1∷RNAi/DMC1 plants phenocopied the dmc1 mutation, we examined the meiotic products and meiosis in these plants. The presence of polyads in the anthers of the pDMC1∷RNAi/DMC1 plants (data not shown) indicated that meiosis was affected in these transformed plants.
DAPI staining of chromosomes in PMCs confirmed that meiosis was profoundly disturbed in the pDMC1∷RNAi/DMC1 plants. Two independently transformed plants, randomly chosen from among the T1 partially sterile transformants, were found to reproduce the Arabidopsis dmc1 mutant phenotype (Figures 4H–L). The lack of bivalents and the segregation defects were as previously described for dmc1 mutants (Couteau et al, 1999). We saw only 10 univalents (18 images corresponded to the diakinesis stage, as in Figure 4I) that segregated randomly following the first (20 anaphase I images as in Figure 4J) and the second meiotic division (seven anaphase II images as in Figure 4K) respectively. Sometimes, one or two bivalents could be observed, which may reflect intermediate RNAi efficiency in these cells. In no case did we find that sister chromatid segregation, which takes place during the second meiotic division, was disturbed, either 10 or 20 chromosomes could always be distinguished. So, as in the case of the original dmc1 mutant plants, bivalent formation at meiosis was defective when plants were transformed with RNAi constructs aimed at silencing the DMC1 gene at meiosis.
Meiosis is profoundly disturbed in the pDMC1∷RNAi/BRCA2-transformed plants
The pDMC1∷RNAi/BRCA2 construct was introduced into Arabidopsis, 20 transformed plants were grown to maturity, and all were confirmed to contain the transgene. No somatic phenotype was observed in the pDMC1∷RNAi/BRCA2-transformed plants. However, when flowers emerged, it was obvious that most (18/20) were affected in their fertility. As previously observed, the partial sterility phenotype was stable through three successive generations and reversible. The observation of polyads in the cleared anthers indicated that meiosis was affected in the sterile pDMC1∷RNAi/BRCA2 plants. PMCs were extracted from the anthers of the pDMC1∷RNAi/BRCA2-transformed plants and their meiotic behavior was examined under fluorescent microscopy following DAPI staining. Examination of three T1 plants revealed that meiosis was profoundly disturbed (see Figure 5). The meiotic defects obviously differed from the dmc1 mutant phenotype. Because each of three different constructs (whether aimed at silencing no target, or DMC1 or BRCA2) produced differing phenotypes, we conclude that the RNAi strategy has the requisite specificity.
Figure 5.
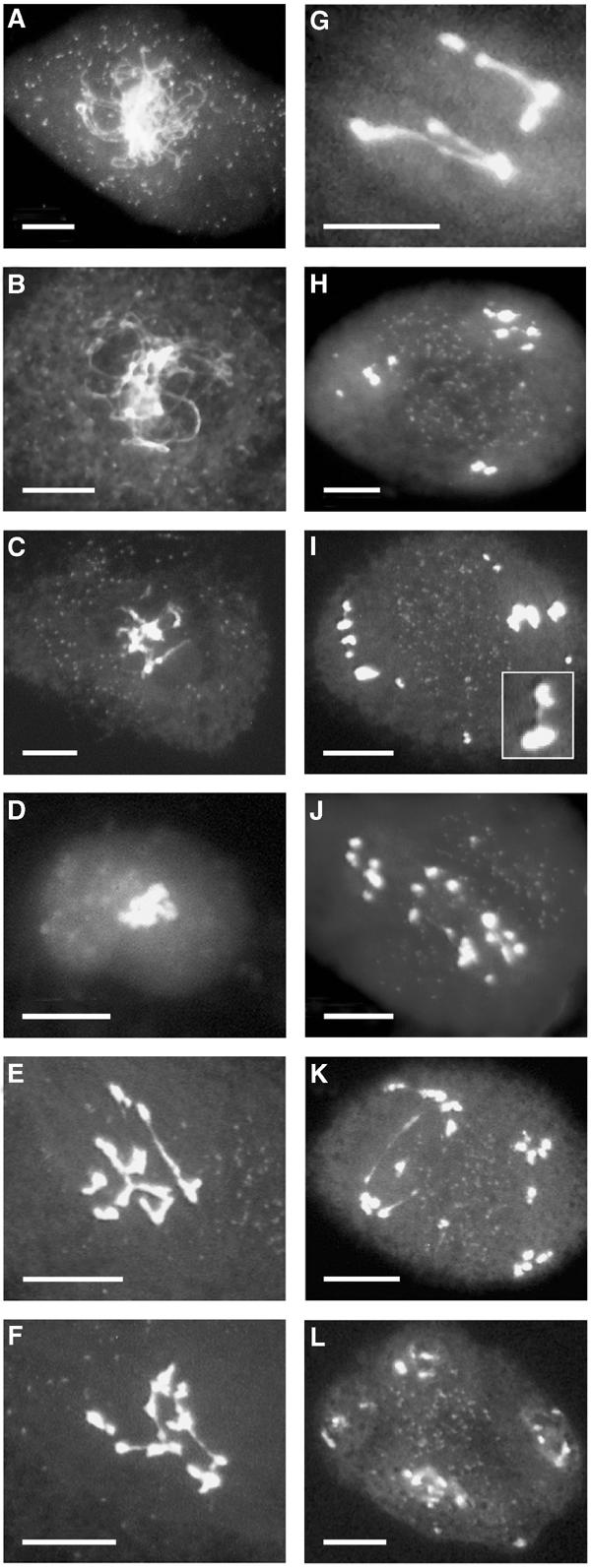
Meiosis in wild-type pDMC1∷RNAi/BRCA2-transformed plants. DAPI staining of chromosomes in meiocytes from pDMC1∷RNAi/ BRCA2-transformed plants: (A, B) prophase I stages; (C) diakinesis stage with entangled chromosomes; (D) metaphase I, showing highly condensed and entangled chromosomes; (E–G) ‘failed anaphase I' with evidences of polyvalent structures and bridges extending between the chromosomes; (H–J) anaphase I through anaphase II stages with varying chromosome numbers of heterogeneous sizes (H: 10 chromosomes; I: 13 independent structures; J: 17 chromosome structures), bridges are often visible extending from one structure to another (see detail in I); (K) ‘failed anaphase II' showing ‘laggards' extending along a spindle axis; (L) telophase II with four heterogeneous groups of decondensed chromosomes, and elements being left aside. Bar, 10 μm.
In the pDMC1∷RNAi/BRCA2-transformed plants, we saw no chromosome pairing, no synapsis and no bivalent formation among 74 meiosis I figures (Figures 5A–D). Most of the observed figures suggested chromosome entangling that was too severe to estimate the number of independent chromosome structures. Second, in 27 anaphase I figures, with less than 10 distinct staining bodies, the chromosomes seemed to be pulled apart with extreme difficulty and in an uncoordinated fashion. These figures were interpreted as ‘failed anaphase I' (Figures 5E–G). In 39 other meiocytes, we could distinguish from 10 to 20 (or more) independent chromosomal structures, which were heterogeneous in size and shape (Figures 5H–J). Finally, we counted 17 meiocytes, which we classified as ‘failed anaphase II' because they presented more than 10 chromosomal elements, improperly separating along independent spindles, with elements sometimes being left aside (Figure 5K). These observation indicate that (i) homologous recombination did not take place correctly (otherwise one should find stages with defined bivalents), (ii) chromosomes were more tightly connected or entangled than in wild-type meiosis, (iii) chromosome movements were poorly coordinated (otherwise only 5, 10 or 20 chromosome structures should be seen, as in wild-type or dmc1 plants), (iv) some chromosome fragmentation had occurred (otherwise the number of defined structures should never be greater than 20) and (v) sister chromatid separation was affected (as anaphase II was often disturbed).
Transformation by pDMC1∷RNAi/RAD51 alters dmc1 mutant phenotype
Constructs aimed at silencing the RAD51 transcripts at meiosis did not affect the fertility of transformed plants (30 transformed plants examined), and meiotic events of the T1 were indistinguishable from those of wild type (data not shown).
We introduced the pDMC1∷RNAi/RAD51 construct into heterozygous (T0) dmc1+/− plants. Following selection and genotyping, we were able to define T1 transformed progeny plants that were either heterozygous or homozygous for the dmc1 mutation (as a result of T0 plants segregation). As for wild-type plants, transformed heterozygous T1 dmc1+/− plants were fully fertile, although they contained the pDMC1∷RNAi/RAD51 construct, and DAPI staining revealed no meiosis defect in these plants (data not shown). Results were strikingly different when one T1 dmc1−/− mutant plant was found to be transformed—presence of the pDMC1∷RNAi/RAD51 construct completely altered the dmc1 phenotype (see Figure 6). The same phenotype was observed in T2 dmc1−/− plants segregating from four independently transformed and fertile heterozygous T1 dmc1+/− plants. We may thus consider that the absence of phenotype in the wild-type or heterozygous dmc1+/− context was not due to inefficacy of the RNAi construct at silencing RAD51, but rather its undetectability (at least at the cytological scale), which was uncovered in dmc1 mutant plants.
Figure 6.
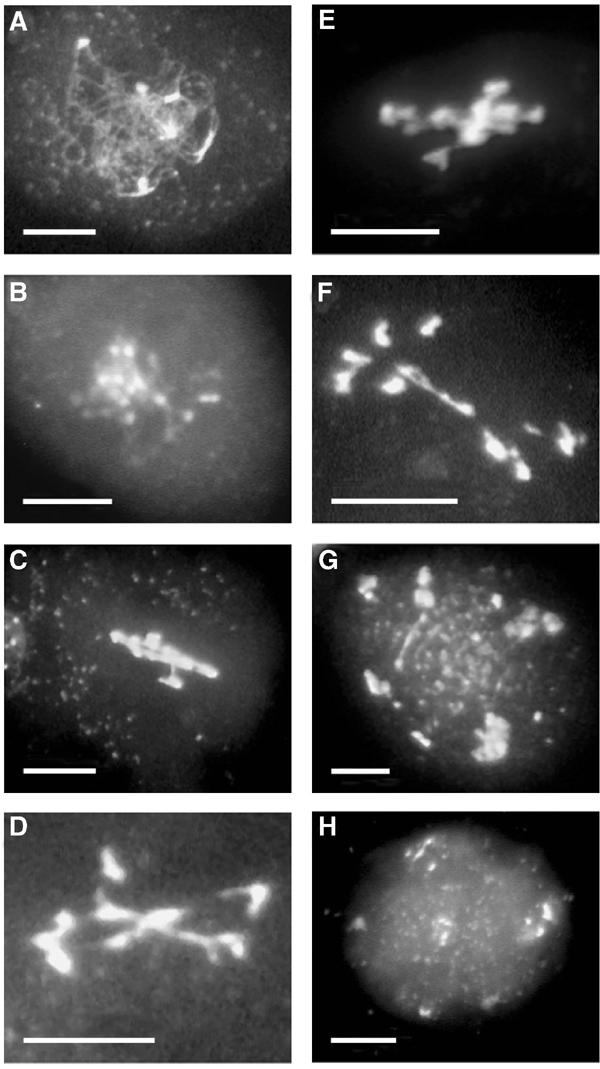
Meiosis in dmc1 pDMC1∷RNAi/RAD51-transformed plants. (A) prophase I stage; (B) diakinesis stage showing chromosome entangling; (C) metaphase I stage with highly condensed chromosomes; (D–F) anaphase I stages with evidence of polyvalent structures and varying chromosome numbers; (G) anaphase II stage with laggards extending along the axis of a spindle axis; (H) telophase II with three heterogeneous groups of decondensed chromosomes, and three elements being left aside. Bar, 10 μm.
Instead of the typical 10 or 20 chromosomal entities that can usually be defined in mutant dmc1 meiosis, we observed figures that were indistinguishable from the pDMC1∷RNAi/BRCA2-transformed plants. The 123 meiotic figures examined in these five independent transformants had the following characteristics: 31 prophase I to metaphase I stages had no synapsed chromosomes or bivalent structures (Figures 6A–C); 61 meiotic figures were similar to the previously defined ‘failed anaphase I' (Figures 6D–F); 17 meiocytes presented 10–20 independent chromosomal structures; 14 meiotic figures were ‘failed anaphase II' (Figure 6G).
The brca2 phenotype is alleviated in spo11-1 but not in dmc1 mutant plants
To examine whether the Brca2 function was dependent upon meiotic DSBs, the pDMC1∷RNAi/BRCA2 construct was introduced into spo11-1+/− and dmc1−/− heterozygous plants. Meiosis in spo11-1 mutant plants presents the same characteristics as in dmc1 plants at the cytological level (Grelon et al, 2001; see Figures 7A and G). While the brca2 phenotype was prevalent in all spo11-1+/−, dmc1+/− and dmc1−/− transformed plants examined (Figures 7B and H, D and J, and E and K), spo11-1−/− plants were not affected by the presence of a pDMC1∷RNAi/BRCA2 transgene (Figures 7C and I). This was observed whether the spo11-1−/− mutant plants were T1 transformants (two independent plants), or whether they were T2 progeny derived from three independent T1 spo11-1+/− transformed plants. Divergence between the spo11-1−/− and dmc1−/− mutant phenotypes upon introduction of a pDMC1∷RNAi/BRCA2 transgene suggests that a structural difference exists in the chromosomes of these mutants, thus bringing indirect evidence for Spo11-dependent DSB formation at meiosis in Arabidopsis. Finally, we crossed dmc1+/− (pDMC1∷RNAi/BRCA2) plants with dmc1+/− (pDMC1∷RNAi/RAD51) plants to obtain triple dmc1−/− (pDMC1∷RNAi/RAD51; pDMC1∷RNAi/BRCA2) plants. In these plants, meiosis was similar to meiosis in their related parental dmc1−/− (pDMC1∷RNAi/BRCA2) or dmc1−/− (pDMC1∷RNAi/RAD51), that is, they presented a typical ‘Brca2-depletion' phenotype (Figures 7F and L).
Figure 7.
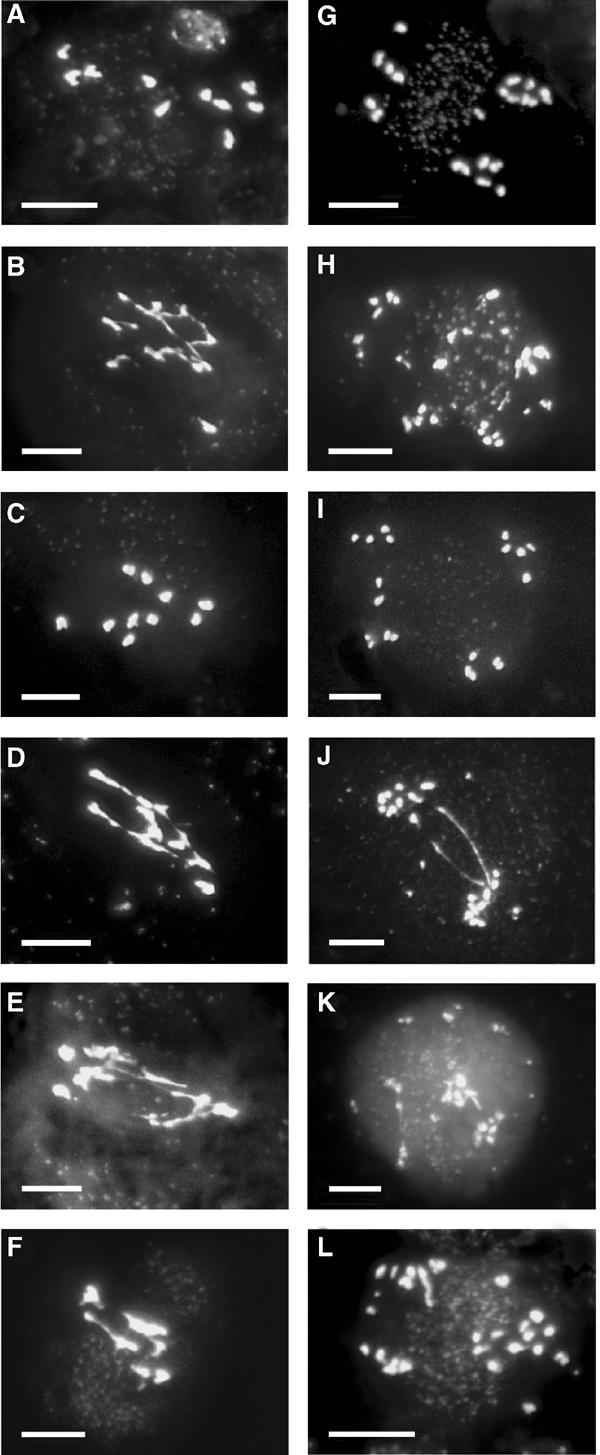
Meiosis in different mutant contexts following the introduction of pDMC1∷RNAi/BRCA2. (A, G) Meiosis in spo11-1−/− mutant plants: (A) anaphase I showing 10 chromatids; (G) anaphase II showing 20 chromosomes. (B, H) Meiosis in spo11-1+/− pDMC1∷RNAi/BRCA2-transformed plants showing typical ‘Brca2-deficient' features: (B) ‘failed anaphase I' with chromosomes stretching; (H) anaphase II with more than 20 chromosomal pieces. (C, I) Meiosis in spo11-1−/− pDMC1∷RNAi/BRCA2-transformed plants showing typical spo11-1−/− features: (C) anaphase I with 10 chromatids; (I) anaphase II with 20 chromosomes. (D, J) Meiosis in dmc1+/− pDMC1∷RNAi/BRCA2-transformed plants showing typical ‘Brca2-deficient' features: (D) ‘failed anaphase I' with chromosomes stretching; (J) anaphase II with more than 20 chromosomal pieces. (E, K) Meiosis in dmc1−/− pDMC1∷RNAi/BRCA2-transformed plants showing typical ‘Brca2-deficient' features: (E) ‘failed anaphase I'; (K) anaphase II with more than 20 chromosomal pieces. (F, L) Meiosis in dmc1−/− pDMC1∷RNAi/RAD51 pDMC1∷RNAi/BRCA2-transformed plants showing typical ‘Brca2-deficient' features: (F) ‘failed anaphase I'; (L) anaphase II with more than 20 chromosomal pieces.
Discussion
With these results, we show that meiosis is profoundly disturbed following the specific and timely silencing of the BRCA2 genes in Arabidopsis. Recently, partial complementation of a brca2 knockout mouse (alive but sterile) by the human BRCA2 gene allowed Sharan et al (2003) to examine the involvement of Brca2 at meiosis. Incomplete synapsis of the chromosomes and altered distribution of Rad51 and Dmc1 foci in these mouse spermatocytes indicate a role for Brca2 in recruiting Rad51 and eventually Dmc1 (via Rad51?) onto the meiotic chromosomes. In the course of our experiments, we found that not only Rad51, but also Dmc1, the meiotic RecA homolog in eucaryotes, are partners of the Arabidopsis Brca2 protein. These interactions may indicate a functional link otherwise suggested by the synthetic brca2 phenotype when both Rad51 and Dmc1 are depleted in Arabidopsis meiocytes. These latter data also allow us to hypothesize a role for Rad51 at meiosis in the dmc1 context.
Chromosome behavior was profoundly disorganized at meiosis in the pDMC1∷RNAi/BRCA2-transformed plants. No bivalents were formed, chromosomes appeared either as a unique and dense body in the center of the cell as if being completely entangled, or stretched across the cell as if separating with difficulty; chromosome fragments were often visible. An identical phenotype was obtained when both Rad51 and Dmc1 were inactivated. Indeed, while it was shown that the Rad51/Dmc1 recombinases require Hop2 to perform fully homologous recombination in yeast (Tsubouchi and Roeder, 2003), disruption of a Hop2 ortholog (AHP2) in Arabidopsis led to meiotic figures that are reminiscent of the pDMC1∷RNAi/BRCA2 or dmc1(pDMC1∷RNAi/RAD51) phenotypes (Schommer et al, 2003). We thus propose that, following the introduction of DSBs by Spo11, an essential step in repair by homologous recombination requires either Brca2 or Rad51 and Dmc1. These functions may also act in a common pathway as inactivation of all three did not aggravate the brca2 or dmc1(pDMC1∷RNAi/RAD51) phenotype. In their absence, the Spo11 DSBs may become prey for alternate DNA repair pathways. Our hypothesis predicts that the pDMC1∷RNAi/BRCA2 phenotype should be dependent upon the Spo11 function, which we were able to demonstrate. These latter data indicate that Brca2 acts downstream of Spo11, which was not the case for the recently described Arabidopsis Mei1 and Cdc45 functions. In mei1 mutant or RNAi/CDC45 plants, chromosome fragmentation at meiosis was not eliminated in a spo11-1 background (Grelon et al, 2003; Stevens et al, 2003).
The aberrant chromosome interactions observed at meiosis under Brca2 or Dmc1 plus Rad51 depletion could result from unresolved interhomolog interactions or from assembly of the chromosomes into unrecognized chromosome structures. However, unresolved interhomolog interactions do not seem to prevent synapsis and bivalent formation in an Arabidopsis xrcc3 mutant (Bleuyard and White, 2004). Moreover, since bivalent formation is eliminated in dmc1 plants where only distinct univalents can be observed, further depletion of Rad51 should not restore interhomolog interactions in this context. The same may apply to the brca2 phenotype if it is to result from a similar defect in homologous recombination. While homologous recombination is defective, nonhomologous end joining (NHEJ) is still functional in brca2−/− mammalian cell lines (Xia et al, 2001; Merel et al, 2002; Moynahan et al, 2002). The altered chromosome structures found in vegetative brca2 or rad51 mouse cells, or in the Ustilago brh2 mutant, are, thus, due to repair of chromosome breaks by NHEJ (Lim and Hasty, 1996; Tirkkonen et al, 1997; Patel et al, 1998; Yu et al, 2000; Kojic et al, 2002). We propose that, similarly, upon combined Rad51 and Dmc1 or Brca2 depletion at meiosis in Arabidopsis, DSB repair occurs via NHEJ instead of homologous recombination, which can result in translocation. Consequently, the resulting dicentric chromosomes are stretched and eventually break when centromeres start migrating toward opposite poles at anaphase I.
The observation that meiosis was not affected, at least at the cytological level, in pDMC1∷RNAi/RAD51 plants may be peculiar to Arabidopsis. Disruption of RAD51 expression in other organisms affects meiosis (Shinohara et al, 1992; Rinaldo et al, 2002) or is embryo–lethal (Lim and Hasty, 1996; Tsuzuki et al, 1996). However, the same pDMC1∷RNAi/RAD51 transgenes clearly altered meiosis in all dmc1−/− mutant plants examined but not in heterozygous dmc1+/− sister plants, thus indicating that (i) our constructs were active, (ii) the Rad51 function may not be evidenced when Dmc1 is present and (iii) Dmc1 can promote bivalent formation without Rad51. The existence of overlapping as well as separate Rad51- and Dmc1-dependent pathways of meiotic recombination in yeast was previously suggested by Dresser et al (1997). In yeast, Rad51 or Rad54 overexpression largely suppresses the meiotic defects of dmc1 mutations, possibly explaining the moderate dmc1 phenotype observed in some yeast backgrounds (Bishop et al, 1999; Tsubouchi and Roeder, 2003). We suggest that Rad51 can effect some homology-dependent DSB repair in dmc1 mutant plants perhaps by driving the repair of the meiotic DNA DSBs toward intersister recombination. This would explain the startling observation that only intact univalents can be seen in the Arabidopsis dmc1 mutant meiocytes, the same phenotype as described for the spo11-1 mutant in which we may presume that no DSB occurs.
Materials and methods
BRCA2 cDNA cloning
In all, 1 μg of total RNA extracted from a 2-day freshly subcultured cell suspension of A. thaliana was reverse transcribed by the MMLV reverse transcriptase with oligodT primers and in the presence of dNTPs in a final volume of 20 μl. PCR was performed with 5 μl of the reverse transcription reaction, using BRCA2 specific primers (IV Nter ATTCCCGGGATGTCGACGTGGCAATTATTT; V Nter ATTCCCGGGATGTCGACGTGGCATTTATTT; IV/V Cter ATTCCCGGGTCAGCATGAAGGTGATTTACA), in a final volume of 100 μl, and in the presence of dNTP and Pfx polymerase (Invitrogen). The PCR products were cloned into the SmaI site of pUC18. Bacterial strains and media were as described previously (Doutriaux et al, 1998).
Real-time quantitative PCR (RT-qPCR): Total RNA was extracted from plant tissues and cultures with Trizol (Invitrogen). A measure of 5 μg of RNAse-free DNAse I-treated (Amersham Biosciences) total RNA was reverse transcribed using the SuperScript system for RT–PCR (Invitrogen) and oligodT as a primer. Specific primers were designed for each gene expression to be quantified: Dmc1-up GTTCATATCAGACCCAAAAAAGCC; Dmc1-do AGATTCGGAGCATCGTAGACTTTG; Rad51-up CTTAGGGATGCTGGTCTCTGTAC; Rad51-do GTCAACCTTGGCATCACTAATTC; Brca2IV-up TTCATTCCAAGAGGTAGGCAACC; Brca2IV-do CAGAGCATGTTTCTTTGAAGGAG; Brca2V-up CTATAAACAATGGAAGTGTCAACAC; Brca2V-do ATCTAAATTGCTTTGAAGATGAATG, Act-up GGTAACATTGTGCTCAGTGGTGG; Act-do AACGACCTTAATCTTCATGCTGC. Amplifications were performed in a GeneAmp 9600 thermocycler (Applied Biosystems) with ‘qPCR Mastermix Plus for SYBR Green I' (Eurogentec), with 1/50 of the RT reaction in a total volume of 25 μl. The reactions were incubated at 95°C for 10 min to activate the hot-start recombinant Taq DNA polymerase, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min, except for the BRCA2 genes where a 55°C for 15 s step was added. RT–PCR experiments were conducted in triplicate, and mean copy numbers of individual mRNA species were calculated using a genomic DNA standard of known molar concentration as in Charrier et al (2002). To control the oligonucleotides specificity, a dissociation curve was established following each PCR amplification. Genomic DNA contamination was assessed using intron-specific primers. Genomic copy numbers, ranging from 0 to 8 per ng of total RNA used for the RT, were included to establish the final values. Data were standardized by normalizing absolute levels of each gene to the absolute level of ACTIN 2/8 as in Charrier et al (2002).
Yeast two-hybrid assay
The BRCA2, RAD51 and DMC1 cDNA sequences were fused in frame to pGBT9 Gal4 binding domain (BD) and pGAD424 Gal4 activating domain (AD) (Clontech). Yeast two-hybrid assays were conducted in strains PJ69-4a (MATa and MATα) by assessing transcription of two different reporter genes: ade4 and his3 (Finley and Brent, 1994; James et al, 1996). pGBT9 or pGAD424 vectors containing the fused cDNAs were transformed into yeast strains PJ69-4a of opposite mating types. Subsequent mating of adequate combinations of these strains allowed selection of Leu+, Trp+ diploids in which pairs of vectors with candidate gene fusions were present. Interactions allowing the reconstitution of a functional Gal4 transcription factor were assessed by plating the diploids on adequate media (−Leu, −Trp, −His or −Leu, −Trp, −Ade) to reveal reporter gene expression.
RNAi constructs and plants
RNAi constructs aimed at silencing the DMC1, BRCA2 and RAD51 genes were established by cloning pairs of PCR-amplified short cDNA fragments specific for each gene in diverging orientation in pKannibal (Figure 2B; Helliwell and Waterhouse, 2003). Primer pairs were designed to PCR-amplify two identical 425 bp fragments for the RNAi/EMB506 construct (506-XhoI-XbaI-up GACTCGAGTCTAGACGAGGAAGAGCTTGCAAAAG and 5O6-EcoRI-ClaI-do GAGAATTCATCGATATCAGCACCATTGGTCAACA), two identical 410 bp fragments for the RNAi/DMC1 construct (D-XhoI-up AGTCTCGAGTGATGGCTTCTCTTAAAGC and D-KpnI-do AGTGGTACCAGACCTAAATTCCCCAAAAG; D-XbaI-up AGTTCTAGATGATGGCTTCTCTTAAAGC and D-ClaI-do AGTATCGATAGACCTAAATTCCCCAAAAG), two identical 501 bp fragments for the RNAi/BRCA2 construct (B-KpnI-do AGTGGTACCTGAAAAAGACTGTTGGGAAC and B-XhoI-up AGTCTCGAGGGCATTTATTTTCCGATTC; B-BamHI-up AGTGGATCCGGCATTTATTTTCCGATTC and B-ClaI-do AGTATCGATTGAAAAAGACTGTTGGGAAC) and two identical 398 bp fragments for the RNAi/RAD51 construct (R-XhoI-XbaI-up GTTCTCGAGTCTAGAAGGAGGTATTGAAAC and R-EcoRI-ClaI-do GTGAATTCATCGATGCATTTGTCGAGCCGAA). Adequate restriction enzyme sites allowed their subsequent two-step cloning in opposite orientations into pKannibal. The RNAi constructs were then cloned into pPF408. pPF408 was derived from pPZP100, which contains the T-DNA left and right borders (LB and RB) for transfer of the T-DNA into plants (Hajdukiewicz et al, 1994). A Basta resistance (BastaR) cassette was introduced into pPZP100 to produce vector pPF111; the DMC1 promoter, as defined by Klimyuk and Jones (1997), was PCR-amplified and cloned into pPF111 to produce vector pPF408. RNAi constructs were isolated from pKannibal following XhoI–SpeI digestion and introduced into the XhoI–SpeI-restricted pPF408 to be set under the pDMC1 promoter control. RNAi constructs containing T-DNA vectors were electroporated into Agrobacterium strain GV3101 (Koncz and Schell, 1986). Arabidopsis, ecotype WS, transformation was carried out as described previously (Clough and Bent, 1998). Transformed T1 plants were selected by Basta under greenhouse conditions. Presence of the RNAi construct in the BastaR plants was checked by PCR amplification of their genomic DNA using one oligonucleotide specific for the pKannibal intron region (Oligo41 cttcttcgtcttacacatcac) and another specific for the relevant construct. The Arabidopsis dmc1 and spo11-1-2 mutations have been described previously (Couteau et al, 1999; Grelon et al, 2000).
Cytological observations
Cytological observations were conducted as previously described by Couteau et al (1999). Images were captured with a 3CCD Sony color video camera. Since their phenotypes were identical, observations from two or three plants independently transformed with the same RNAi constructs were pooled.
Acknowledgments
We specially acknowledge Prof. FW Stahl and Dr R Devoret for the careful attention they gave to our manuscript. We are grateful to Drs A Nicolas, Y Henry, C Mézard and I Small for their comments and critical reading of the manuscript, Dr P Waterhouse (CSIRO, Australia) for providing the pKannibal vector, Dr M Grelon for providing the spo11-1 Arabidopsis mutant and Dr A Betzner for cloning the DMC1 promoter according to Klimyuk and Jones (1997). We also thank JP Bares and G Santé for plant maintenance, S Domenichi for cytology technical support and R Boyer for photographic work. This work was supported by CNRS, Génoplante contracts AF1999085 (E Gérard) and AF2001051. N Siaud and E Dray are the recipients of a Ministère de l'Education Nationale, de la Recherche et de la Technologie fellowship.
References
- Albert S, Despres B, Guilleminot J, Bechtold N, Pelletier G, Delseny M, Devic M (1999) The EMB 506 gene encodes a novel ankyrin repeat containing protein that is essential for the normal development of Arabidopsis embryos. Plant J 17: 169–179 [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P (1997) An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417 [DOI] [PubMed] [Google Scholar]
- Bignell G, Micklem G, Stratton MR, Ashworth A, Wooster R (1997) The BRC repeats are conserved in mammalian BRCA2 proteins. Hum Mol Genet 6: 53–58 [DOI] [PubMed] [Google Scholar]
- Bishop DK, Nikolski Y, Oshiro J, Chon J, Shinohara M, Chen X (1999) High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells 4: 425–444 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, White CI (2004) The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J 23: 439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Blomberg N, Nilges M (1996) Internal repeats in the BRCA2 protein sequence. Nat Genet 13: 22–23 [DOI] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase–polymerase chain reaction. Plant Physiol 130: 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, Scully R (1998) Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell 2: 317–328 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Connor F, Bertwistle D, Mee PJ, Ross GM, Swift S, Grigioreva E, Tybulewicz VLJ, Ashworth A (1997) Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat Genet 17: 423–430 [DOI] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux M (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doutriaux M-P, Couteau F, Bergounioux C, White C (1998) Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol Gen Genet 257: 283–291 [DOI] [PubMed] [Google Scholar]
- Dresser ME, Ewing DJ, Conrad MN, Dominguez AM, Barstead R, Jiang H, Kodadek T (1997) DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147: 533–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley RL, Brent R (1994) Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc Natl Acad Sci USA 91: 12980–12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M, Gendrot G, Vezon D, Pelletier G (2003) The Arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J 35: 465–475 [DOI] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G (2001) AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J 20: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Helliwell C, Waterhouse P (2003) Constructs and methods for high-throughput gene silencing in plants. Methods 30: 289–295 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M 2002. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21: 8981–8993 [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Klimyuk VI, Jones JD (1997) AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J 11: 1–14 [DOI] [PubMed] [Google Scholar]
- Kojic M, Kostrub CF, Buchman AR, Holloman WK (2002) BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell 10: 683–691 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Lim DS, Hasty P (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol 16: 7133–7143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T, Pellegrini L, Venkitaraman AR, Blundell TL (2003) Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair 2: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Masson JY, West SC (2001) The Rad51 and Dmc1 recombinases: a non-identical twin relationship. Trends Biochem Sci 26: 131–136 [DOI] [PubMed] [Google Scholar]
- McAllister KA, Bennett LM, Houle CD, Ward T, Malphurs J, Collins NK, Cachafeiro C, Haseman J, Goulding EH, Bunch D, Eddy EM, Davis BJ, Wiseman RW (2002) Cancer susceptibility of mice with a homozygous deletion in the COOH-terminal domain of the Brca2 gene. Cancer Res 62: 990–994 [PubMed] [Google Scholar]
- Merel P, Prieur A, Pfeiffer P, Delattre O (2002) Absence of major defects in non-homologous DNA end joining in human breast cancer cell lines. Oncogene 21: 5654–5659 [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M (2002) BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell 10: 1262–1263 [DOI] [PubMed] [Google Scholar]
- Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR (1998) Involvement of Brca2 in DNA repair. Mol Cell 1: 347–357 [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, Anand S, Lee, Blundell TL, Venkitaraman AR (2002) Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature 420: 287–293 [DOI] [PubMed] [Google Scholar]
- Rinaldo C, Bazzicalupo P, Ederle S, Hilliard M, La Volpe A (2002) Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics 160: 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder GS (1997) Meiotic chromosomes: it takes two to tango. Genes Dev 11: 2600–2621 [DOI] [PubMed] [Google Scholar]
- Schommer C, Beven A, Lawrenson T, Shaw P, Sablowski R (2003) AHP2 is required for bivalent formation and for segregation of homologous chromosomes in Arabidopsis meiosis. Plant J 36: 1–11 [DOI] [PubMed] [Google Scholar]
- Scully R, Livingston DM (2000) In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408: 429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A (1997) Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386: 804–810 [DOI] [PubMed] [Google Scholar]
- Sharan SK, Pyle A, Coppola V, Babus J, Swaminathan S, Benedict J, Swing D, Martin BK, Tessarollo L, Evans JP, Flaws JA, Handel MA (2003) BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131: 131–142 [DOI] [PubMed] [Google Scholar]
- Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, Volkman N, Tsuruta H, Blundell TL, Venkitaraman AR, Tainer JA (2003) Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J 22: 4566–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69: 457–470 [DOI] [PubMed] [Google Scholar]
- Stevens R, Grelon M, Vezon D, Oh J, Meyer P, Perennes C, Domenichini S, Bergounioux C (2003) A CDC45 homolog in Arabidopsis is essential for meiosis, as shown by RNA interference-induced gene silencing. Plant Cell 16: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirkkonen M, Johannsson O, Agnarsson BA, Olsson H, Ingvarsson S, Karhu R, Tanner M, Isola J, Barkardottir RB, Borg A, Kallioniemi OP (1997) Distinct somatic genetic changes associated with tumor progression in carriers of BRCA1 and BRCA2 germ-line mutations. Cancer Res 57: 1222–1227 [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS (2003) The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev Cell 5: 915–925 [DOI] [PubMed] [Google Scholar]
- Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA 93: 6236–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren M, Smith A, Partridge N, Masabanda J, Griffin D, Ashworth A (2002) Structural analysis of the chicken BRCA2 gene facilitates identification of functional domains and disease causing mutations. Hum Mol Genet 11: 841–851 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Helliwell CA (2003) Exploring plant genomes by RNA-induced gene silencing. Nat Rev Genet 4: 29–38 [DOI] [PubMed] [Google Scholar]
- West SC (2003) Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4: 435–445 [DOI] [PubMed] [Google Scholar]
- Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL (1997) RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem 272: 31941–31944 [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378: 789–792 [DOI] [PubMed] [Google Scholar]
- Xia F, Taghian DG, DeFrank JS, Zeng ZC, Willers H, Iliakis G, Powell SN (2001) Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal non homologous end joining. Proc Natl Acad Sci USA 98: 8644–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP (2002) BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297: 1837–1848 [DOI] [PubMed] [Google Scholar]
- Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, West SC, Venkitaraman AR (2000) Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev 14: 1400–1406 [PMC free article] [PubMed] [Google Scholar]


