Abstract
Cytokinin activity in plants is closely related to nitrogen availability, and an Arabidopsis gene for adenosine phosphate-isopentenyltransferase (IPT), IPT3, is regulated by inorganic nitrogen sources in a nitrate-specific manner. In this study, we have identified another regulatory system of cytokinin de novo biosynthesis in response to nitrogen status. In rice, OsIPT4, OsIPT5, OsIPT7 and OsIPT8 were up-regulated in response to exogenously applied nitrate and ammonium, with accompanying accumulation of cytokinins. Pre-treatment of roots with l-methionine sulfoximine, a potent inhibitor of glutamine synthetase, abolished the nitrate- and ammonium-dependent induction of OsIPT4 and OsIPT5, while glutamine application induced their expression. Thus, neither nitrate nor ammonium, but glutamine or a related metabolite, is essential for the induction of these IPT genes in rice. On the other hand, glutamine-dependent induction of IPT3 occurs in Arabidopsis, at least to some extent. In transgenic lines repressing the expression of OsIPT4, which is the dominant IPT in rice roots, the nitrogen-dependent increase of cytokinin in the xylem sap was significantly reduced, and seedling shoot growth was retarded despite sufficient nitrogen. We conclude that plants possess multiple regulation systems for nitrogen-dependent cytokinin biosynthesis to modulate growth in response to nitrogen availability.
Keywords: Ammonium, Arabidopsis thaliana, Cytokinin, Glutamine, Nitrate, Oryza sativa
The nucleotide sequence reported in this paper has been submitted to DDBJ under accession number AB853903 (OsIPT8).
Introduction
Nitrogen is an essential but often limiting nutrient for plant growth and development. To regulate growth under limited nitrogen supply, plants sense the internal and external nitrogen status and coordinate various metabolic processes and developmental programs. To enable such an elaborate response, a wide range of genes are regulated in response to the nitrogen status and to intracellular, intercellular and interorgan signals.
Inorganic nitrogen sources differ depending on soil conditions: nitrate is the major form in most natural soils, whereas ammonium may dominate under reducing conditions, such as in paddy fields. In terms of nitrogen source-dependent gene regulation, two response modes were characterized in previous studies: the nitrate-specific response and the nitrogen-non-specific response (Sakakibara 2003b, Sakakibara et al. 2006, Castaings et al. 2011). As for the former, transcriptome analysis and other studies have identified a wide range of nitrate-responsive genes including nitrate-assimilatory and related genes (Sakakibara et al. 1996, Matsumura et al. 1997, Redinbaugh and Campbell 1998, Wang et al. 2000, Wang et al. 2003, Scheible et al. 2004, Sakakibara et al. 2006, Okamoto et al. 2009). This response is rapid and does not require de novo protein synthesis since it is cycloheximide insensitive (Gowri et al. 1992, Sakakibara et al. 1997, Sakakibara 2003a). Key transcription factors and cis-element sequences for the nitrate-specific response recently have been described (Castaings et al. 2009, Konishi and Yanagisawa 2010, Konishi and Yanagisawa 2011, Konishi and Yanagisawa 2013, Sawaki et al. 2013). On the other hand, the nitrogen-non-specific response is observed following exogenous application of nitrate, ammonium and various amino acids, and a wide spectrum of genes have been reported to be up- or down-regulated during the response (Hirose et al. 1997, Sivasankar et al. 1997, Stitt 1999, Stitt et al. 2002, Sonoda et al. 2003, Tabuchi et al. 2007, Konishi and Yanagisawa 2010, Kiba et al. 2012).
Recent studies indicated a linkage between the nitrogen signaling network and phytohormones including cytokinin, ABA and auxin (Kiba et al. 2011, Krouk et al. 2011, Ruffel et al. 2011). Cytokinin metabolism and signaling are closely related to nitrogen availability (Sakakibara et al. 1998, Taniguchi et al. 1998, Takei et al. 2001, Miyawaki et al. 2004, Takei et al. 2004, Ruffel et al. 2011). Cytokinin regulates a variety of processes in plant growth and development, such as shoot and root growth, apical dominance, leaf longevity, sink capacity and stress responses (Werner et al. 2003, Ashikari et al. 2005, Kim et al. 2006, Tanaka et al. 2006, Kitazawa et al. 2008, Bartrina et al. 2011, Kondo et al. 2011, F. Qin et al. 2011a; H. Qin et al. 2011b). trans-Zeatin (tZ), cis-zeatin (cZ) and N6-(Δ2-isopentenyl)adenine (iP) are common natural cytokinins in plants (Mok and Mok 2001, Sakakibara 2006). The concentrations of tZ, iP and their conjugates (tZ-type cytokinins and iP-type cytokinins, respectively) correlate with the supplied amounts of nitrogen (Samuelson and Larsson 1993, Kiba et al. 2011); nitrogen supplements elevate the cytokinin levels in Zea mays (maize) (Takei et al. 2001), Hordeum vulgare (barley) (Samuelson and Larsson 1993), Arabidopsis thaliana (Arabidopsis) (Takei et al. 2004), Triticum aestivum (wheat) (Garnica et al. 2010) and Urtica dioica (stinging nettle) (Wagner and Beck 1993). According to current understanding, cytokinins are synthesized in various parts of the plant body and act locally and at a distance. They may be systemically transported via the vasculature to coordinate shoot and root development (Matsumoto-Kitano et al. 2008, Kudo et al. 2010, Bishopp et al. 2011). This suggests that cytokinins could function as long-range signals communicating the rhizospheric nitrogen status.
The initial step of the de novo biosynthesis of iP- and tZ-type cytokinins is catalyzed by adenosine phosphate-isopentenyltransferase (IPT), producing the nucleotide precursors (Sakakibara 2006). In Arabidopsis, IPT3, which is expressed in phloem tissue, is up-regulated by exogenous nitrate (Miyawaki et al. 2004, Takei et al. 2004). The up-regulation is nitrate specific, and a loss-of-function mutation severely diminished the nitrate-dependent accumulation of cytokinin. This indicated that IPT is a key regulator of the nitrate-responsive modulation of cytokinin activity (Takei et al. 2004).
The nitrate-specific up-regulation of Arabidopsis IPT3 is not the only mode of nitrogen-dependent regulation of cytokinin levels. For instance, the expression of the cytokinin-responsive maize response regulator genes, ZmRR1 and ZmRR2, apparently is induced by exogenous nitrate as well as ammonium (Sakakibara et al. 1998, Sakakibara et al. 1999). In barley, pre-treatment with inhibitors of nitrate reductase (NR) and glutamine synthetase (GS) diminished the nitrate-dependent accumulation of cytokinin (Samuelson and Larsson 1993).
Paddy rice (Oryza sativa) utilizes ammonium as its major inorganic nitrogen source. Eight IPT genes (OsIPT1–OsIPT8) have been identified; their different organ specificities (Sakamoto et al. 2006) suggest functional differentiation. The dominant cytokinin species in rice are different from those in Arabidopsis: cZ and its conjugates (cZ-type cytokinins) are more abundant than tZ-type and iP-type cytokinins, although cZ activity is generally weak and equivalent to that of tZ only in specific contexts, such as the inhibition of seminal root elongation (Kojima et al. 2009, Choi et al. 2012, Kudo et al. 2012, Kudo et al. 2013). Therefore, insights into the regulation of cytokinin metabolism in response to nitrogen nutrition in rice may foster a deeper general understanding of the nitrogen-dependent regulation of cytokinin activity. In this study, we identified nitrogen-responsive IPT genes in rice and characterized their regulation. We found that a key signal for gene induction is glutamine or a related metabolite. Our results reveal mechanistic differences and commonalities in the nitrogen-dependent regulation of de novo cytokinin biosynthesis between rice and Arabidopsis.
Results
Changes in cytokinin concentrations in response to inorganic nitrogen sources
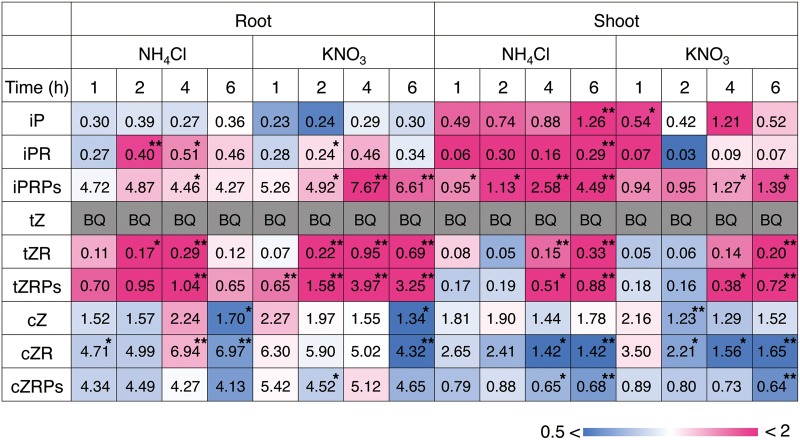
To examine whether nitrogen nutrition affects cytokinin metabolism in rice, cytokinins and their conjugates were quantified in rice seedlings after supplement of nitrate or ammonium. In roots and shoots, iP riboside 5′-phosphates (iPRPs) and tZ riboside 5′-phosphates (tZRPs), early products of de novo cytokinin synthesis, accumulated following exposure to both nitrogen sources (Fig. 1; Supplementary Tables S1, S2). Similar patterns were observed in the nucleosides and active-form nucleobases, iP riboside (iPR), iP and tZ riboside (tZR), whereas tZ was below the quantification limit at all times. The accumulation of transcripts of cytokinin-responsive type-A response regulator genes was also increased in roots (Supplementary Fig. S1). On the other hand, there was no apparent increase in cZ, cZ riboside (cZR) and cZR 5′-phosphates (cZRPs). These results suggested that exposure to inorganic nitrogen sources activated the de novo synthesis of tZ- and iP-type cytokinins in rice.
Fig. 1.
Accumulation pattern of cytokinins in rice roots and shoots in response to nitrate and ammonium. Rice seedlings were hydroponically grown in tap water for 11 d after sowing and transferred to nitrogen-free culture medium for 3 d. Then, the roots were dipped into culture media containing 1 mM NH4Cl, 1 mM KNO3 or 1 mM KCl. After the time indicated, roots and shoots were separately harvested in triplicate, and the cytokinin contents were quantified. The value in each block indicates the concentration (mean values) as pmol g−1 FW. The relative accumulation level of each compound compared with the accumulation in the KCl treatment is color-coded. BQ, below the quantification limit. *P < 0.05; **P < 0.01 (Student’s t-test, comparison with the KCl treatment). The complete data set is presented in Supplementary Tables S1 and S2.
Identification of nitrogen-responsive OsIPT genes
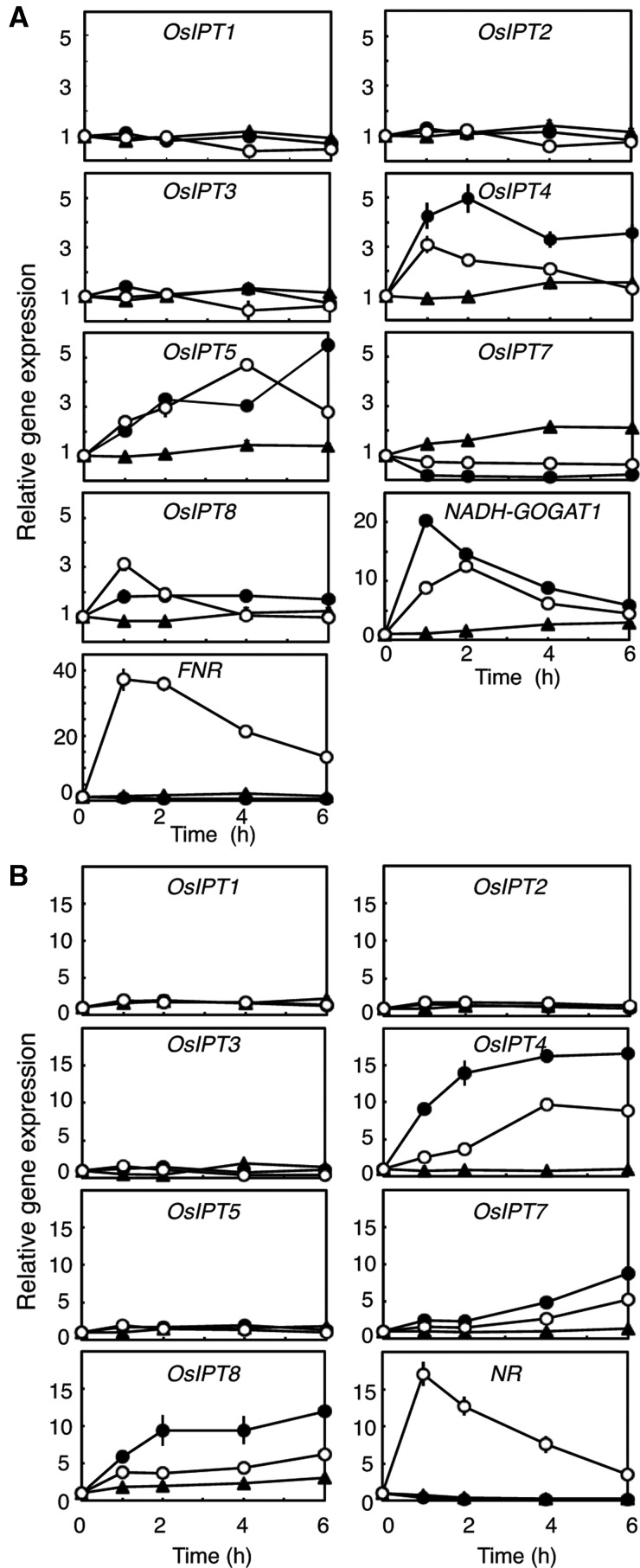
Since nucleotide precursors of cytokinins were increased by the nitrogen supplements, we examined the accumulation levels of OsIPT transcripts in roots and shoots (Fig. 2). Since OsIPT6 seems to be non-functional in the Nipponbare cultivar (Sakamoto et al. 2006), we excluded it from analysis. To discriminate between nitrate-specific and nitrogen-non-specific responses, genes for non-photosynthetic-type ferredoxin-NADP+ oxidoreductase (FNR) (Aoki and Ida 1994) and NADH-dependent glutamate synthase 1 (NADH-GOGAT1) (Hirose et al. 1997) were used as indicators for the two response types, respectively, in roots (Fig. 2A). The NR gene (Hamat et al. 1989) served as an indicator for nitrate-specific responses in shoots (Fig. 2B). In roots, the accumulation of OsIPT4 and OsIPT5 transcripts was clearly increased by both ammonium and nitrate (Fig. 2A). Ammonium had a more potent effect on the induction of OsIPT4 expression than nitrate. On the other hand, the level of the OsIPT7 transcript was decreased by both treatments. In shoots, the accumulation levels of OsIPT4, OsIPT7 and OsIPT8 transcripts were increased by both nitrogen sources, with ammonium having a stronger effect than nitrate (Fig. 2B). The expression patterns of the indicator genes did not suggest any nitrate effects caused by ammonium-containing media.
Fig. 2.
Changes in the accumulation of OsIPT transcripts in response to nitrogen sources in roots (A) and shoots (B). Rice seedlings were hydroponically grown and treated with 1 mM NH4Cl (filled circles), 1 mM KNO3 (open circles) or 1 mM KCl (filled triangles) in the same manner as in Fig. 1, and roots and shoots were harvested at the indicated times. Total RNA prepared from the samples was subjected to qPCR. The amounts of transcripts were normalized to the value at 0 min. qPCR was performed in triplicate, and mean values with the SD are shown.
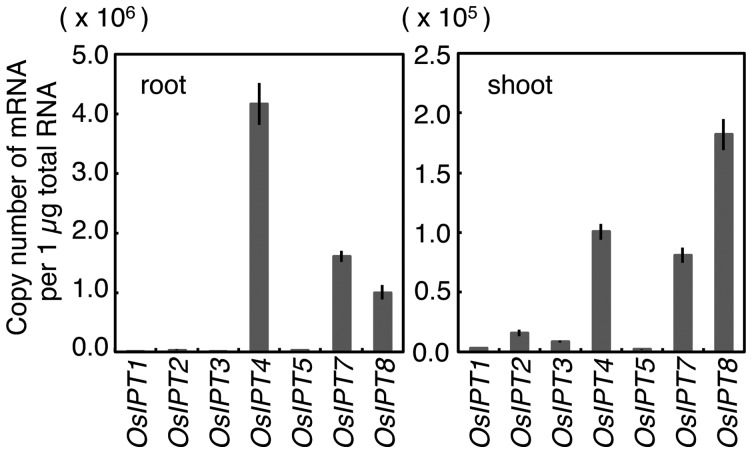
When we analyzed the steady-state accumulation levels of OsIPT genes in rice seedlings before nitrogen application, we found OsIPT4, OsIPT7 and OsIPT8 transcripts abundantly accumulated in roots and shoots (Fig. 3). The OsIPT4 transcript accumulated mostly in roots, while the OsIPT8 transcript was most abundant in shoots. Accumulation levels of the OsIPT5 transcript were much lower than those of other nitrogen-responsive IPT genes. These results suggested that the expression of OsIPT4 on the one hand, and of OsIPT4, OsIPT7 and OsIPT8 on the other affected nitrogen-dependent cytokinin synthesis in roots and shoots, respectively.
Fig. 3.
Accumulation of OsIPT transcripts in roots (left) and shoots (right) of rice seedlings hydroponically grown for 2 weeks in tap water. Total RNAs prepared from each organ were subjected to qPCR. The accumulations of transcripts are indicated as amounts per total RNA. qPCR was performed in triplicate, and mean values with the SD are shown.
Spatial expression patterns of nitrogen-inducible OsIPT genes
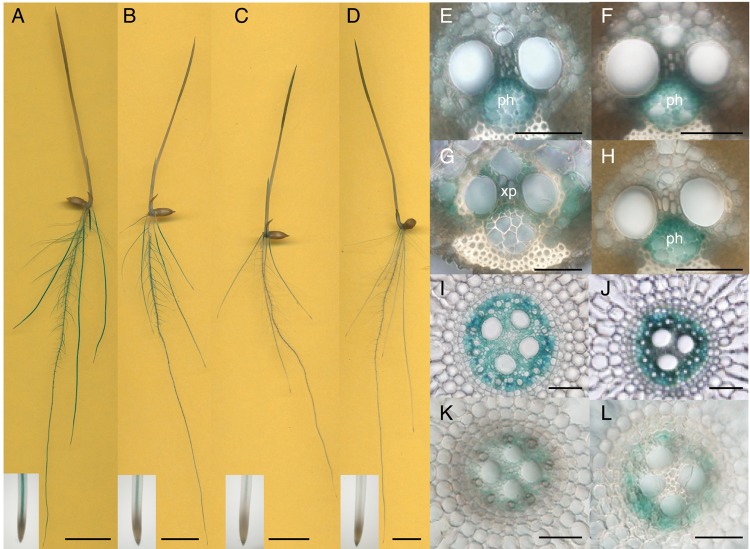
To reveal the tissue specificities of the nitrogen-dependent cytokinin biosynthesis genes, DNA fragments containing about 3 kb of the upstream sequences including the N-terminal regions of OsIPT4, OsIPT5, OsIPT7 and OsIPT8 were fused with the β-glucuronidase (GUS) coding sequence, and the constructs (designated OsIPT4pro:GUS, OsIPT5pro:GUS, OsIPT7pro:GUS and OsIPT8pro:GUS) were transformed into rice. We detected similar GUS staining patterns with 10–13 independent T1 transgenic lines of each construct, and T3 representative lines were further analyzed in detail (Fig. 4). In these transgenic lines, GUS activity was detected in roots, and less so in shoots (Fig. 4A–D). This finding was consistent with the results in Fig. 3. In roots, GUS staining patterns were essentially the same for all IPT promoters: staining was observed in vascular bundles of seminal and crown roots (Fig. 4I–L) but not in the apical meristems (Fig. 4A–D, insets). The staining patterns in the transformants did not change with different nitrogen sources (data not shown).
Fig. 4.
Distribution of GUS activity under the control of the 5′ upstream region of the OsIPT4 (A, E and I), OsIPT5 (B, F and J), OsIPT7 (C, G and K) and OsIPT8 (D, H and L) genes. (A–D) Whole transformant seedlings. Close-ups of the root apex are shown in the insets at the bottom. (E–L) Cross-sections of vascular bundles in mature leaf blades (E–H) and seedling roots (I–L). ph, phloem; xp, xylem parenchyma. Scale bars, 1 cm (A–D) and 50 µm (E–L).
In mature leaf blades of the transformants, GUS activity was also detected in the vascular bundles, but showed tissue specificity. In OsIPT4pro:GUS, OsIPT5pro:GUS and OsIPT8pro:GUS transformants, GUS activity was detected in the phloem, while in OsIPT7pro:GUS transformants it was found in xylem parenchyma cells (Fig. 4E–H).
Intracellular localization of rice IPTs
In Arabidopsis, nitrogen-responsive IPT3 is localized in plastids (Kasahara et al. 2004). We examined the intracellular localization of OsIPTs in rice. Prior to this analysis, we conducted a 5′-rapid amplification of cDNA ends (RACE) analysis to predict the translation start site of OsIPT8 because of the discrepancy in the predicted lengths of the reading frame between Sakamoto et al. (2006) and a public database (Rice Genome Annotation Project; http://rice.plantbiology.msu.edu/). In our analysis, the first ATG codon of amplified cDNA appeared 45 bp upstream of that of the public database and 126 bp upstream of that reported by Sakamoto et al. (2006) (Supplementary Fig. S2).
To examine the intracellular localization of OsIPT proteins, translational fusions were made with green fluorescent protein (GFP) at the C-termini (designated as OsIPT1–GFP to OsIPT8–GFP) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. The constructs were introduced into Arabidopsis cells by particle bombardment (Fig. 5). Since rice tissues are mechanically rigid and it was difficult to observe subcellular localizations, we used Arabidopsis as a heterologous system. GFP fluorescence of OsIPT4–GFP, OsIPT5–GFP and OsIPT8–GFP appeared as small dots on plastids (Fig. 5J–L). When we co-introduced DsRed2-fused pAtFSD3, a plastid nucleoid-associated protein (Myouga et al. 2008), the fluorescence signals fully overlapped (Fig. 5M–U). The GFP fluorescence of OsIPT7–GFP co-localized with DsRed2-tagged Arabidopsis geranylgeranyl diphosphate synthase 6 (GGPS6) (Okada et al. 2000), a control marker for mitochondria (Fig. 5G–I). These results strongly suggested that nitrogen-inducible OsIPT4, OsIPT5 and OsIPT8 are localized in plastids, whereas OsIPT7 is localized in mitochondria. On the other hand, the GFP fluorescence from OsIPT1–GFP, OsIPT2–GFP and OsIPT3–GFP and their translational fusions at the N-termini (GFP–OsIPT1, GFP–OsIPT2 and GFP–OsIPT3) were all observed in the cytoplasm (Fig. 5A–F).
Fig. 5.
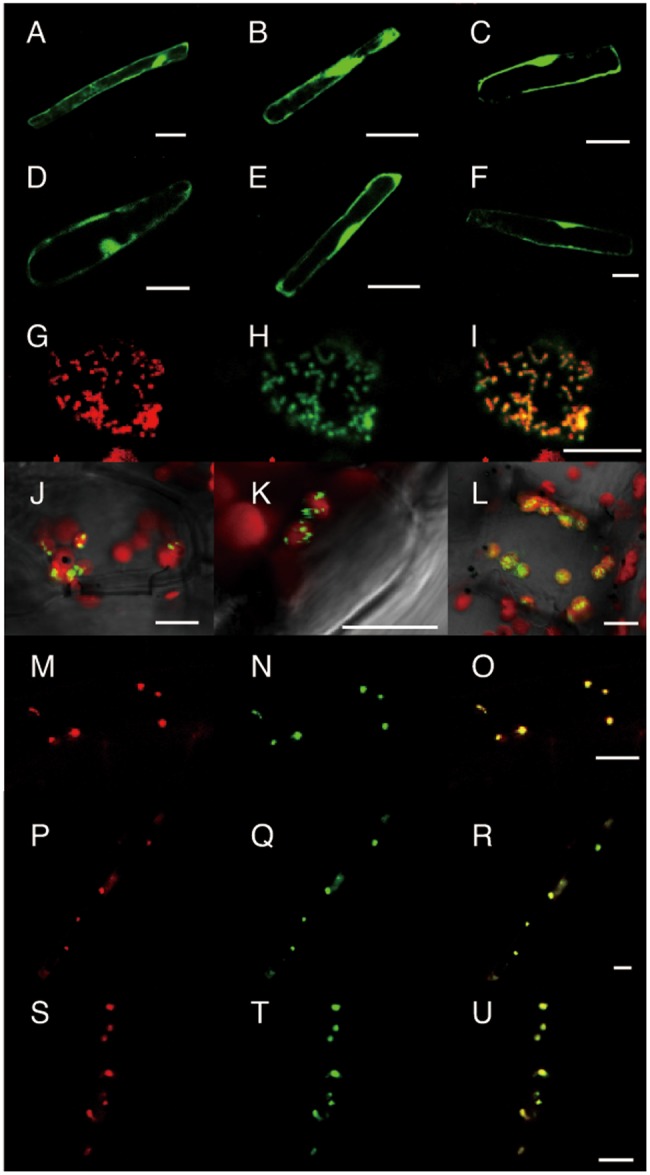
Subcellular localization of GFP-tagged OsIPT proteins in Arabidopsis observed by confocal laser-scanning microscopy. The translational fusion genes OsIPT1-GFP (A), OsIPT2-GFP (B), OsIPT3-GFP (C), GFP-OsIPT1 (D), GFP-OsIPT2 (E) and GFP-OsIPT3 (F) were transiently expressed in root epidermal cells by particle bombardment. The fusion genes pGGPS6-DsRed2 (G), a control for mitochondrial localization, and OsIPT7-GFP (H) were co-introduced into a leaf mesophyll cell. (I) Merged image of (G) and (H). The fusion genes OsIPT4-GFP (J), OsIPT5-GFP (K) and OsIPT8-GFP (L) were introduced into leaf mesophyll cells and superimposed on Chl autofluorescence (red). The fusion gene AtFSD3-DsRed2 (M, P and S), a control for nucleoid localization, was co-introduced into root epidermal cells with either OsIPT4-GFP (N), OsIPT5-GFP (Q) or OsIPT8-GFP (T). (O, R and U) Merged images of (M) and (N), (P) and (Q), and (S) and (T), respectively. Scale bars, 20 µm (A–F) and 10 µm (G–U).
Metabolic signals for nitrogen-dependent OsIPT expression
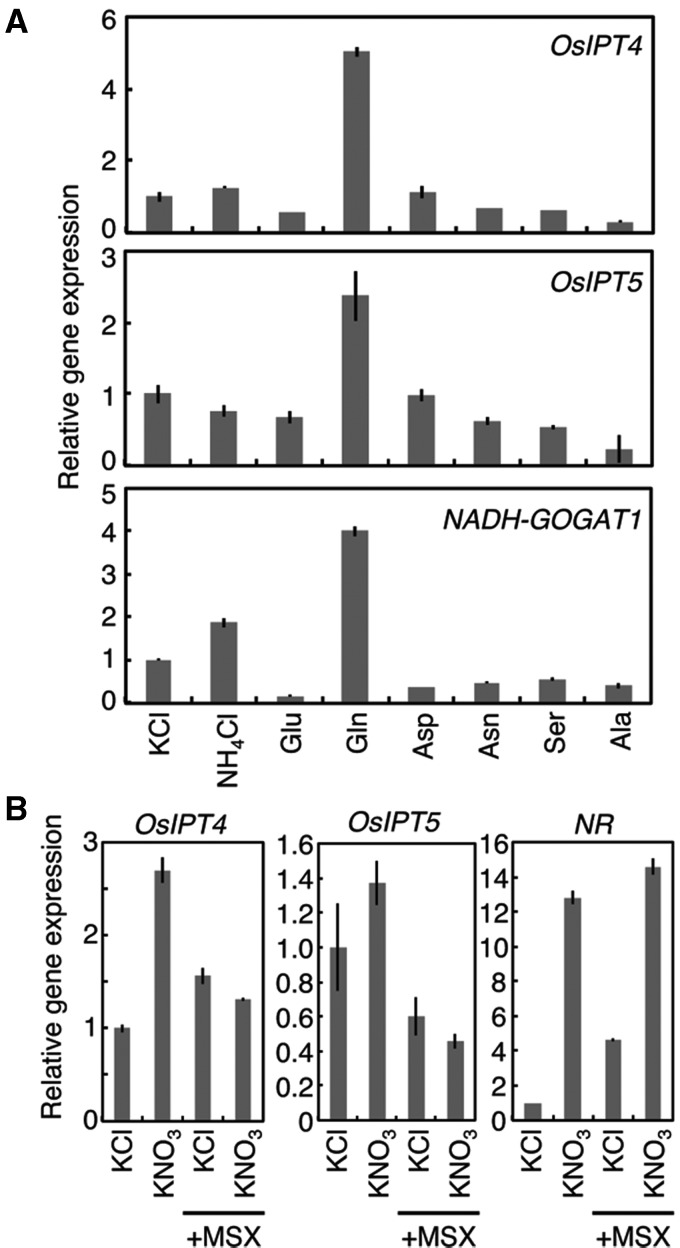
To obtain information about the regulation of nitrogen-dependent OsIPT expression, we examined the effects of l-methionine sulfoximine (MSX), an inhibitor of GS (Pace and Mcdermott 1952). Roots of rice seedlings grown for 2 weeks without nitrogen were pre-treated with MSX before ammonium or amino acids were applied. Pre-treatment with MSX completely inhibited the ammonium-induced accumulation of OsIPT4 and OsIPT5 transcripts (Fig. 6A), indicating that ammonium itself is not a direct inducing signal. On the other hand, application of glutamine induced expression of OsIPT genes as well as that of NADH-GOGAT1, a glutamine-responsive gene (Hirose et al. 1997). Other amino acids had no comparable effects. Application of glutamine increased the cytokinin concentration in roots (Supplementary Fig. S3). These results suggested that glutamine or a related metabolite regulates the cytokinin concentration via nitrogen-dependent OsIPT4 and OsIPT5 expression.
Fig. 6.
Effects of methionine sulfoximine (MSX) pre-treatment on the induction of OsIPT transcript accumulation by nitrogen compounds. Rice seedlings were hydroponically grown in tap water for 11 d after sowing and then on nitrogen-free culture medium for 3 d. The seedlings were pre-treated with 1 mM MSX for 2 h and then incubated with (A), 1 mM KCl, 1 mM NH4Cl or 50 mM of the indicated amino acids for 3 h; or (B), 10 mM KCl or 10 mM KNO3 for 3 h. In (B), samples without MSX pre-treatment were also prepared. Total RNA prepared from the roots was subjected to qPCR. The amounts of transcripts were normalized with respect to the value in the KCl treatment. qPCR was performed in triplicate, and mean values with the SD are shown.
In Arabidopsis, the nitrogen-responsive AtIPT3 is induced in a nitrate-specific manner (Takei et al. 2004). To see whether the nitrate-specific response is conserved in OsIPT gene induction in rice, we examined the effects of nitrate on the expression of OsIPT4, OsIPT5 and NR after MSX pre-treatment. The induction of OsIPT4 and OsIPT5 by nitrate was completely inhibited by the MSX pre-treatment, although NR induction was not affected (Fig. 6B). This implied that nitrate-specific regulation of OsIPT gene induction is negligible or very minor under the given conditions.
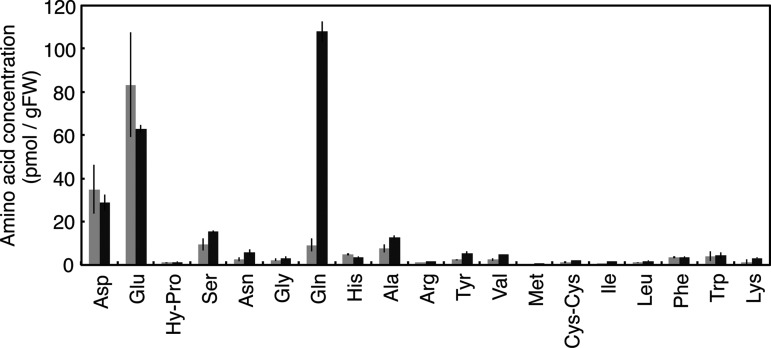
Next, we analyzed the responses of amino acid concentrations in rice roots to inorganic nitrogen sources. Rice seedlings were grown for 2 weeks after germination without a nitrogen source, before ammonium or nitrate were supplemented. Upon exposure to ammonium, glutamine accumulated drastically in the roots within 30 min, and increased further over 2 h (Fig. 7; Supplementary Table S3). On the other hand, concentrations of aspartic and glutamic acids were decreased, and other amino acids showed no significant effects. In the nitrate treatment, a small but increased concentration of glutamine was also observed (Supplementary Table S3). These results were consistent with the positive effects of exogenously applied glutamine on the induction of OsIPT4 and OsIPT5 expression.
Fig. 7.
Changes in the concentrations of free amino acids in rice roots after application of ammonium. Rice seedlings were hydroponically grown in tap water for 11 d after sowing and transferred to nitrogen-free culture medium for 3 d. Then, the roots were dipped into the culture medium containing 1 mM KCl (light gray) or 1 mM NH4Cl (dark gray) for 30 min. Roots were harvested in triplicate, and the amino acid contents were analyzed. Bars represent mean values with the SD. The detailed data are provided in Supplementary Table S3.
Characterization of OsIPT4 knock-down transgenic rice
To evaluate the contribution of OsIPT4 to nitrogen-dependent cytokinin accumulation, OsIPT4-repressed transgenic rice lines were generated through RNA interference (RNAi) techniques. We made two types of construct in which the targets of RNAi were designed to be in the 5′- and 3′-untranslated regions of OsIPT4 (Supplementary Fig. S4A). Two independent T1 transgenic lines in which the OsIPT4 expression levels were reduced were selected for each construct. In these RNAi lines, basal expression levels of OsIPT4 were reduced to about 50% and there was no induction in response to ammonium (Supplementary Fig. S4B). As for other OsIPT genes, the expression of OsIPT1, OsIPT2 and OsIPT3 was reduced in the roots of these RNAi lines (Supplementary Fig. S5). However, the basal expression levels of these three IPT genes in roots were very low (Fig. 3) and not affected by ammonium (Supplementary Fig. S5). Thus, the decrease in OsIPT4 expression was the dominant effect in these RNAi lines, and we evaluated OsIPT4 function in these lines.
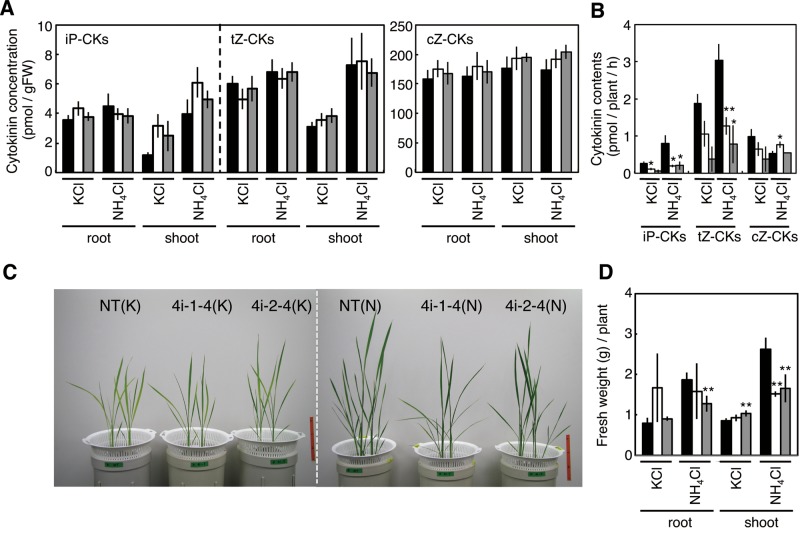
To examine whether OsIPT4 induction contributes to the nitrogen-responsive cytokinin accumulation, we established the T3 RNAi lines and monitored cytokinin concentrations following ammonium application. Contrary to our expectation, no significant differences were found between non-transformants and the RNAi lines at the whole-organ level (Fig. 8A). Since OsIPT4 is expressed in the vasculature (Fig. 4), and because major ammonium-induced changes occurred in the xylem-mobile nucleosides including iPR and tZR (Fig. 1; Supplementary Tables S1, S2), we analyzed cytokinin contents in stem base exudates which are derived mainly from xylem sap. In non-transformant control plants, the contents of tZ- and iP-type cytokinins in the exudate were increased following ammonium exposure (Fig. 8B). However, in both RNAi lines, the cytokinin contents in KCl-treated controls were decreased compared with non-transformants, and ammonium was ineffective. These results suggested that a reduction of the nitrogen-dependent induction of OsIPT4 expression affects cytokinin export from roots via the xylem.
Fig. 8.
Characterization of OsIPT4-repressed transgenic lines. (A) Comparison of cytokinin concentrations. Non-transformant (black), 4i-1-4 (white) and 4i-2-4 (light gray) seedlings were hydroponically grown in tap water for 2 weeks and then transferred to culture medium containing 1 mM NH4Cl or 1 mM KCl. After 6 h, roots and shoots were harvested and cytokinin contents were quantified. (B) Comparison of cytokinin amounts exuded from the xylem. Asterisks indicate significant differences between non-transformants and the OsIPT4-repressed lines as established by Student’s t-test (**P < 0.01; *P < 0.05). (C and D) Comparison of growth. Rice plants were first grown in pH-controlled water for 2 weeks and then in liquid culture medium containing 1 mM NH4Cl (N) or 1 mM KCl (K) for 3 weeks. (C) Plants at 5 weeks after sowing. (D) Quantification of root and shoot fresh weight. Mean values ± SD from four plants of each treatment are shown. NT, non-transformant. Asterisks indicate significant differences between non-transformants and OsIPT4-repressed lines, according to Student’s t-test (**P < 0.01).
To study the effects of a deficiency of OsIPT4 function on growth, the T3 RNAi lines were hydroponically grown with 1 mM NH4Cl or KCl. The two independent OsIPT4-repressed lines showed retarded shoot growth compared with non-transformants in the presence of abundant nitrogen, whereas the differences in root growth were not significant (Fig. 8C, D). When the transgenic lines were grown for prolonged periods in a greenhouse, the retardation of shoot growth gradually declined and finally disappeared (data not shown). Thus, the effect of OsIPT4 repression on shoot growth is most pronounced during early growth stages.
Conservation of glutamine-dependent regulation of IPT in Arabidopsis
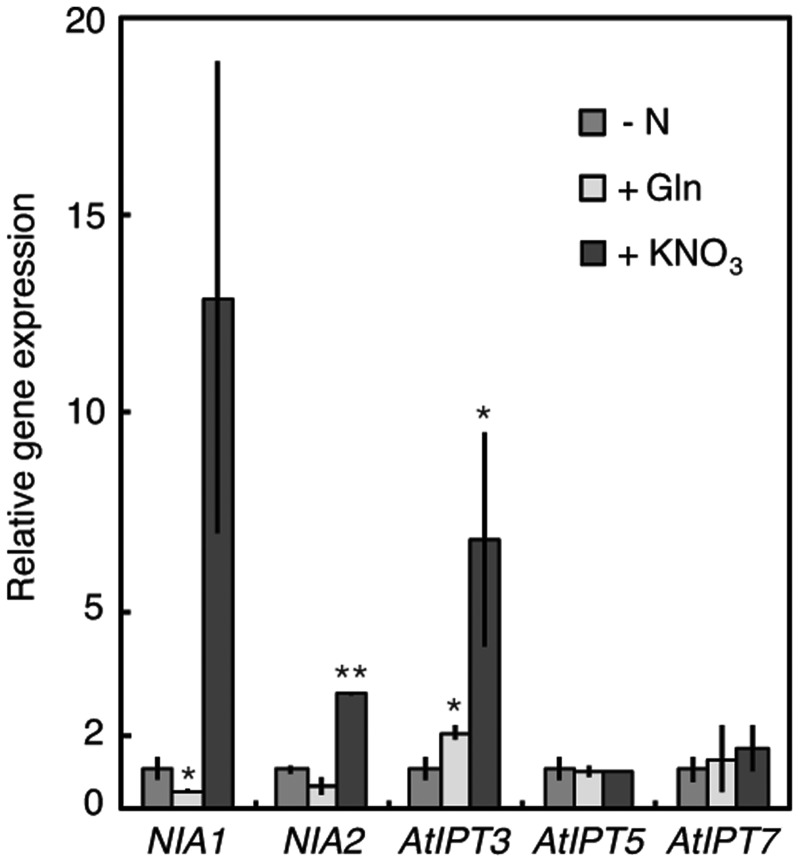
To see whether the glutamine-related regulation is conserved in Arabidopsis IPT regulation, we examined the effects of MSX pre-treatment followed by nitrate and glutamine exposure in Arabidopsis (Fig. 9). AtIPT3 expression in roots was induced by exogenous nitrate even after MSX pre-treatment. Glutamine application also increased the accumulation of AtIPT3 transcripts about 2-fold (Fig. 9). Although this increase seemed small compared with the nitrate response, it consistently occurred in two independent experiments (Fig. 9). Thus, the glutamine-related regulatory mechanism of IPT expression exists in rice as well as in Arabidopsis, although the nitrate-specific regulation is the major one in Arabidopsis.
Fig. 9.
Effects of MSX pre-treatment on the induction of AtIPT transcript accumulation by exogenous nitrogen compounds. Arabidopsis seedlings were grown under standard conditions for 2 weeks after germination and transferred to nitrogen-free medium for 5 d. The seedlings were pre-treated with 1 mM MSX for 2 h and then incubated for 2 h with nitrogen-free medium (–N), 50 mM glutamine (+Gln) or 10 mM KNO3 (+KNO3) solution containing 1 mM MSX. Total RNA prepared from roots was subjected to qPCR which was performed in triplicate. The amounts of transcripts were normalized to that of the Actin 2 transcript. The experiment was independently repeated twice. The values were normalized with respect to those obtained in the nitrogen-free medium, and mean values with the SD are shown. Asterisks indicate significant differences between the nitrogen-free treatment and the Gln or nitrate treatment, according to Student’s t-test (**P < 0.01; *P < 0.05).
Discussion
In previous studies on Arabidopsis, nitrogen-dependent de novo cytokinin biosynthesis was shown to be regulated in a nitrate-specific manner (Miyawaki et al. 2004, Takei et al. 2004). Here we demonstrated the involvement of glutamine metabolism in an additional regulatory system. We found that in rice, multiple IPT genes respond to exogenous inorganic nitrogen sources in a nitrogen-non-specific manner (Fig. 2), and that at least OsIPT4 and OsIPT5 respond to glutamine or a related compound as a metabolic signal (Fig. 6). In addition, the glutamine-related regulation of IPT expression appears to be functional also in Arabidopsis (Fig. 9). Since cytokinin activity was enhanced not only by nitrate but also by ammonium in barley and maize (Samuelson and Larsson 1993, Sakakibara et al. 1998, Sakakibara et al. 1999), the regulation of cytokinin biosynthesis by glutamine metabolism might be common in plants.
It is unlikely that the nitrate-specific regulation of cytokinin biosynthesis is restricted in Arabidopsis, because most plants preferentially utilize nitrate as the inorganic nitrogen source, and because a supply of nitrate to wheat (T. aestivum L.) that had been grown with ammonium further increased cytokinin levels (Garnica et al. 2010). At present, it is unclear whether the nitrate-specific regulation of IPT expression has been lost in rice. So far as we could determine, no data supporting the involvement of nitrate-specific regulatory steps in rice cytokinin metabolism are available. As ammonium is the major inorganic nitrogen source in paddy fields, the nitrate-specific regulation of cytokinin biosynthesis might have degenerated during domestication. Whatever the case, plants have evolved multiple modes of regulation of cytokinin de novo biosynthesis through nitrate, the basic nitrogen form taken up from the soil, and through glutamine metabolism, a key component of nitrogen assimilation. Thus the nitrogen-dependent control of cytokinin activity appears to be important for the modulation of plant growth and development.
At present, we do not know if glutamine itself functions as a genuine signal. The amido moiety of glutamine is utilized by glutamine-amidotransferase superfamily enzymes to form a wide variety of nitrogen-containing compounds (Massiere and Badet-Denisot 1998). Such metabolites might play a critical role in the control of IPT expression. We tried to evaluate the effects of 6-diazo-5-oxo-l-norleucine, a potent inhibitor of all glutamine-amidotransferases, in tests analogous to the MSX experiments reported above. However, the 6-diazo-5-oxo-l-norleucine pre-treatment itself severely affected the accumulation of transcripts, probably due to its cytotoxicity (data not shown), making it impossible to obtain unambiguous results. Glutamine is a major form of nitrogen in rice stem base sap (i.e. xylem sap) (Fukumorita and Chino 1982) and phloem sap (Hayashi and Chino 1990), and nitrogen-regulated IPTs are expressed in the vasculature (Fig. 4). Therefore, glutamine surrounding the translocation conduits could be an indicator of the internal nitrogen status for the regulation of cytokinin production. Exogenously supplied glutamine was demonstrated to affect the expression of a number of genes (Shiraishi et al. 1992, Vincentz et al. 1993, Watanabe et al. 1994, Hirose et al. 1997, Sonoda et al. 2003, Konishi and Yanagisawa 2010). While it remains unclear whether all of these genes are regulated by the same mechanism, it is intriguing that NADH-GOGAT1 and OsIPT4 apparently are. In sink organs, NADH-GOGAT1 plays an important role in the reutilization of nitrogen imported from source organs (Tabuchi et al. 2007, Tamura et al. 2010). In addition, GS1;2 and NADH-GOGAT1 are key elements in the primary assimilation of ammonium in roots (Tabuchi et al. 2007, Tamura et al. 2010, Funayama et al. 2013). GOGAT utilizes glutamine and supplies glutamate to GS to drive the GS/GOGAT cycle, an essential process in nitrogen assimilation (Miflin and Lea 1980). Nitrogen metabolic and developmental processes could be coordinated through a linkage between IPT regulation and GOGAT function.
In rice, cZ-type cytokinins are more abundant than iP- and tZ-type cytokinins (Kojima et al. 2009), but only the latter are produced in response to exogenous nitrogen (Fig. 1). In addition, levels of cZ-type cytokinins were not affected in OsIPT4-repressed lines (Fig. 8A, B). The biosynthesis pathway of cZ has not yet been elucidated in rice, but our results indicate that nitrogen-responsive IPTs are not directly involved in its de novo biosynthesis.
Our observations of GFP-tagged IPTs indicate that nitrogen-responsive IPTs are differentially localized in plastids and mitochondria in rice. Promoter::reporter analyses showed that OsIPT4, OsIPT5 and OsIPT8 were expressed in the phloem while OsIPT7 was expressed in the xylem parenchyma in leaves (Fig. 4). Mitochondria are abundant in xylem parenchyma cells in rice leaves (Botha et al. 2008), which is in line with the expression of mitochondria-directed OsIPT7 in this tissue. On the other hand, GFP-tagged OsIPT4, OsIPT5 and OsIPT8 were localized in plastids (Fig. 5) and so was the Arabidopsis nitrogen-responsive IPT3. Given that OsIPT4 and OsIPT8 are the major IPT genes expressed in leaves and roots, it seems that the major route providing prenyl for nitrogen-dependent cytokinin de novo biosynthesis is the methylerythritol phosphate pathway as it is in Arabidopsis (Kasahara et al. 2004).
When we fused full-length OsIPT4, OsIPT5 and OsIPT8 to GFP, GFP fluorescence was observed on nucleoids in plastids (Fig. 5). Since we used a heterologous transient expression system and a strong promoter, we cannot exclude the possibility of an artificial localization. Further detailed studies will be needed to clarify this point.
Our analysis at the seedling stage suggested that the repression of OsIPT4 affects cytokinin translocation in the xylem. The observed growth retardation could be attributed at least in part to the decreased cytokinin transfer. At the seedling stage, OsIPT4 transcripts are much more abundant in roots than in shoots (Fig. 3). Consequently, root-borne cytokinin synthesized mainly by OsIPT4 might be important for normal shoot growth during early growth stages. In older plants, other IPTs might compensate for any malfunction of OsIPT4. On the other hand, we could not detect significant changes in cytokinin concentrations at the whole-organ level (Fig. 8). At present, we do not have an explanation for this discrepancy, but homeostatic regulation of cytokinin metabolism might mask local concentration differences at the whole-organ level.
In this study, we characterized the dual regulation system of cytokinin biosynthesis by nitrogen in rice, but we still do not understand how plants organize this system to optimize their growth and development under conditions of limiting and variable nitrogen availability. Further studies should focus on functional differentiations of the regulatory system. As for the growth optimization, other phytohormones probably interact with cytokinin-mediated growth regulation. For instance, recent studies indicated an interaction between plant architecture, macronutrients and phytohormones such as strigolactone (Yoneyama et al. 2007, Minakuchi et al. 2010, Umehara et al. 2010, Yamaguchi and Kyozuka 2010, Seto et al. 2012, Yoneyama et al. 2012). For a deeper understanding at the whole-plant level, we will have to study the nitrogen-dependent dual regulation system in the context of hormone–hormone interactions.
Materials and Methods
Plant materials and growth conditions
Rice (O. sativa) cultivar Nipponbare and A. thaliana ecotype Columbia were used in this study. For experiments on nitrogen responses, rice seeds were incubated in distilled water at 30°C for 2 d in the dark. The germinated seeds were sown on mesh trays floating on tap water adjusted to pH 5.5 using HCl and grown for 11 d in an environment-controlled greenhouse with a 12 h light (30°C)/12 h dark (25°C) photoperiod. Seedlings then were transferred to one-quarter-strength nutrient solution (Makino et al. 1983) without nitrogen source and further grown for 3 d. The media for treatment with 1 mM NH4Cl, 1 mM KNO3, 1 mM KCl or 1 mM MSX were prepared using the one-quarter-strength nutrient solution without nitrogen source. For characterization of OsIPT4-repressed lines, non-transformants and OsIPT4-repressed lines were grown in the growth chamber for a total of 5 weeks: in the first 2 weeks, plants were grown in pH-controlled water, then transferred to the liquid culture medium with 1 mM NH4Cl or 1 mM KCl for 3 weeks. The liquid culture medium was renewed every day to avoid nutrient depletion. For xylem sap collection, rice plants were first grown with normal liquid medium for 5 weeks and then transferred to nitrogen-free medium for 2 weeks to impose nitrogen starvation. Then, the plants were transferred to liquid medium containing 1 mM NH4Cl or 1 mM KCl. After 3 h, the shoot was cut off about 10 mm above the root/shoot transition with a razor blade, and root-pressure exudate was collected for 2 h. Samples were dried using a vacuum dryer, and subjected to cytokinin analysis.
Arabidopsis was grown on Molecular Genetics Research Laboratory (MGRL) growth medium (Fujiwara et al. 1992) in vertical agar plates containing 1% sucrose under 100 µmol m−2 s−1 fluorescent light and long-day conditions (16 h light/8 h dark) at 22°C for 2 weeks. The seedlings were transferred to nitrogen-free MGRL-based vertical agar plates for 5 d, with CaCl2·2H2O and KCl instead of Ca(NO3)2·4H2O and KNO3 at equivalent quantities.
Quantification of cytokinins
Extraction and determination of cytokinins from about 100 mg of fresh rice tissues were performed as described previously using ultra-performance liquid chromatography (UPLC)-tandem mass spectrometry (AQUITY UPLC™ System/XEVO-TQS; Waters) with an ODS column (AQUITY UPLC BEH C18, 1.7 µm, 2.1×100 mm, Waters) (Kojima et al. 2009).
Quantitative PCR analysis
Total RNA was prepared from plant samples using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. Single-stranded cDNA was synthesized using the SuperScript® III First-Strand Synthesis System (Invitrogen) with oligo(dT)12–18 primers. Quantitative real-time PCR (qPCR) analysis was carried out with the StepOne Plus Real-Time PCR System (Applied Biosystem) using gene-specific primers (Supplementary Table S4) and the KAPA SYBR® FAST ABI Prism® 2X qPCR kit (Kapa Biosystems) according to the manufacturer’s protocol. To calculate the absolute quantity of each transcript, purified plasmids containing the target sequences of the primers were used to obtain linear standard curves.
Histochemical analysis
Genomic DNA containing the putative promoter regions of OsIPT4 (–3,051 to +296 bp from the first ATG codon), OsIPT5 (–2,722 to +257 bp), OsIPT7 (–3,001 to +198 bp) and OsIPT8 (–2,810 to +272 bp) were amplified by PCR with genomic DNA using gene-specific primers containing a specific restriction site (Supplementary Table S4). The PCR products were cloned into the pCAMBIA-GUS vector (Hirose et al. 2005) at the restriction site. The resulting constructs were named OsIPT4pro:GUS, OsIPT5pro:GUS, OsIPT7pro:GUS and OsIPT8pro:GUS. Transgenic rice plants were generated by the Agrobacterium tumefaciens-mediated method (Hiei et al. 1994) using strain EHA105. Histochemical analysis of GUS activity was performed by the method of Jefferson (1987), modified by Kosugi et al. (1990). Details of the staining procedure were described previously (Hirose et al. 2005). The sections were incubated at 37°C for between 10 min and several hours in GUS reaction buffer.
5′-RACE
Total RNA was prepared from leaf blades of mature rice using the RNeasy Plant Mini Kit, and 5′-RACE was performed using the GeneRacer™ Kit (Invitrogen) with OsIPT8-specific primers (Supplementary Table S4) according to the instruction manual.
Analyses of GFP fusion proteins
The coding regions of OsIPT1–OsIPT5, OsIPT7 and OsIPT8 were fused to the N-terminus of the sGFP(S65T) sequence of the pGWB5 vector (Nakagawa et al. 2007), whose expression is controlled by the CaMV 35S promoter. The expressed proteins were named OsIPT1–GFP to OsIPT5–GFP, OsIPT7–GFP and OsIPT8–GFP, respectively. The full length (with stop codon) of the coding sequence of OsIPT1–OsIPT3 was also fused to the C-terminus of the sGFP(S65T) sequence of the pGWB6 vector (Nakagawa et al. 2007). The expressed proteins were designated as GFP–OsIPT1 to GFP–OsIPT3, respectively. These constructs were introduced into Arabidopsis root cells or rosette leaf cells by bombardment (PDS-1000/He, Bio-Rad) with 1 µm gold particles as described in the supplier’s protocol. For co-introduction of multiple constructs, the N-terminal region of AtGGPS6 was fused to the N-terminus of DsRed2 (pGGPS6-DsRed2), and so was the full coding sequence of AtFSD3 (pAtFSD3-DsRed2); both constructs were under the control of the CaMV 35S promoter. After overnight incubation in the dark, transient expression was observed by confocal laser-scanning fluorescence microscopy (Zeiss LSM510 META, Carl Zeiss).
Determination of free amino acids
Rice plants hydroponically grown for 14 d after germination as described above were transferred to one-quarter-strength nutrient solution with 1 mM NH4Cl or 1 mM KCl. After 30 min and 2 h, roots were harvested and rapidly frozen in liquid nitrogen. Three biological replicates were prepared. The frozen roots were powdered in liquid nitrogen and then homogenized in 10 vols. of 10 mM HCl with 0.2 mM methionine sulfone as an internal control. The homogenate was centrifuged and the supernatant was filtered through Ultrafree-MC filters (Millipore). Amino acid contents in the resulting filtrate were determined using Pico·Tag® (Waters) with an HPLC System (Waters Alliance 2695 HPLC system/2475) according to the instruction manual.
Generation of OsIPT4-repressed lines
Target regions for RNAi were amplified by PCR from rice cDNAs with specific primers (Supplementary Table S4). The resulting PCR products were cloned into the pENTR/D-TOPO vector (Invitrogen) according to the instruction manual. The plasmids were used for LR reactions using the Gateway LR clonase Enzyme Mix (Invitrogen) and the pANDA vector (Miki and Shimamoto 2004). Transgenic rice plants were generated by the A. tumefaciens-mediated method using strain EHA105.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan [a Grant-in-Aid for Scientific Research on Innovative Areas (No. 21114005) and an NC-CARP project].
Supplementary Material
Acknowledgments
We are grateful to Professor Ko Shimamoto (Nara Institute of Science and Technology) for providing the pANDA vector. We also thank Drs. Takatoshi Kiba (RIKEN) and Toru Kudo (RIKEN) for their evaluation of final growth of OsIPT4-repressed lines, Ms. Hiromi Ojima for technical assistance, and Dr. Yasuo Niwa (University of Shizuoka, Shizuoka, Japan) for providing the 35Spro:GFP (S65T) vector. The hormone analysis was supported by the Japan Advanced Plant Science Network.
Glossary
Abbreviations
- CaMV
Cauliflower mosaic virus
- cZ
cis-zeatin
- cZR
cZ riboside
- cZRP
cZR 5′-phosphate
- FNR
ferredoxin-NADP+ oxidoreductase
- GFP
green fluorescent protein
- GS
glutamine synthetase
- GUS
β-glucuronidase
- iP
N6-(Δ2-isopentenyl)adenine
- iPR
iP riboside
- iPRP
iPR 5′-phosphate
- IPT
adenosine phosphate-isopentenyltransferase
- MSX
l-methionine sulfoximine
- NADH-GOGAT1
NADH-dependent glutamate synthase 1
- NR
nitrate reductase
- qPCR
quantitative real-time PCR
- 5′-RACE
5′-rapid amplification of cDNA ends
- RNAi
RNA interference
- tZ
trans-zeatin
- tZR
tZ riboside
- tZRP
tZR 5′-phosphate
Disclosures
The authors have no conflicts of interest to declare.
References
- Aoki H, Ida S. Nucleotide sequence of a rice root ferredoxin-NADP+ reductase cDNA and its induction by nitrate. Biochim. Biophys. Acta. 1994;1183:552–556. doi: 10.1016/0005-2728(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell. 2011;23:69–80. doi: 10.1105/tpc.110.079079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A, Lehesranta S, Vaten A, Help H, El-Showk S, Scheres B, et al. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 2011;21:927–932. doi: 10.1016/j.cub.2011.04.049. [DOI] [PubMed] [Google Scholar]
- Botha CEJ, Aoki N, Scofield GN, Liu L, Furbank RT, White RG. A xylem sap retrieval pathway in rice leaf blades: evidence of a role for endocytosis? J. Exp. Bot. 2008;59:2945–2954. doi: 10.1093/jxb/ern150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009;57:426–435. doi: 10.1111/j.1365-313X.2008.03695.x. [DOI] [PubMed] [Google Scholar]
- Castaings L, Marchive C, Meyer C, Krapp A. Nitrogen signalling in Arabidopsis: how to obtain insights into a complex signalling network. J. Exp. Bot. 2011;62:1391–1397. doi: 10.1093/jxb/erq375. [DOI] [PubMed] [Google Scholar]
- Choi J, Lee J, Kim K, Cho M, Ryu H, An G, et al. Functional identification of OsHk6 as a homotypic cytokinin receptor in rice with preferential affinity for iP. Plant Cell Physiol. 2012;53:1334–1343. doi: 10.1093/pcp/pcs079. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Yokota-Hirai M, Chino M, Komeda Y, Naito S. Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 1992;99:263–268. doi: 10.1104/pp.99.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumorita T, Chino M. Sugar, amino-acid and inorganic contents in rice phloem sap. Plant Cell Physiol. 1982;23:273–283. [Google Scholar]
- Funayama K, Kojima S, Tabuchi-Kobayashi M, Sawa Y, Nakayama Y, Hayakawa T, et al. Cytosolic glutamine synthetase1;2 is responsible for the primary assimilation of ammonium in rice roots. Plant Cell Physiol. 2013;54:934–943. doi: 10.1093/pcp/pct046. [DOI] [PubMed] [Google Scholar]
- Garnica M, Houdusse F, Zamarreno AM, Garcia-Mina JM. The signal effect of nitrate supply enhances active forms of cytokinins and indole acetic content and reduces abscisic acid in wheat plants grown with ammonium. J. Plant Physiol. 2010;167:1264–1272. doi: 10.1016/j.jplph.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Gowri G, Kenis JD, Ingemarsson B, Redinbaugh MG, Campbell WH. Nitrate reductase transcript is expressed in the primary response of maize to environmental nitrate. Plant Mol. Biol. 1992;18:55–64. doi: 10.1007/BF00018456. [DOI] [PubMed] [Google Scholar]
- Hamat HB, Kleinhofs A, Warner RL. Nitrate reductase induction and molecular characterization in rice (Oryza sativa L.) Mol. Gen. Genet. 1989;218:93–98. [Google Scholar]
- Hayashi H, Chino M. Chemical-composition of phloem sap from the uppermost internode of the rice plant. Plant Cell Physiol. 1990;31:247–251. [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Hirose N, Hayakawa T, Yamya T. Inducible accumulation of mRNA for NADH-dependent glutamate synthase in rice roots in response to ammonium ions. Plant Cell Physiol. 1997;38:1295–1297. [Google Scholar]
- Hirose N, Makita N, Yamaya T, Sakakibara H. Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiol. 2005;138:196–206. doi: 10.1104/pp.105.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 1987;5:387–405. [Google Scholar]
- Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T, Kamiya Y, et al. Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J. Biol. Chem. 2004;279:14049–14054. doi: 10.1074/jbc.M314195200. [DOI] [PubMed] [Google Scholar]
- Kiba T, Feria-Bourrellier AB, Lafouge F, Lezhneva L, Boutet-Mercey S, Orsel M, et al. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell. 2012;24:245–258. doi: 10.1105/tpc.111.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Kudo T, Kojima M, Sakakibara H. Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 2011;62:1399–1409. doi: 10.1093/jxb/erq410. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, et al. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl Acad. Sci. USA. 2006;103:814–819. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa D, Miyazawa Y, Fujii N, Hoshino A, Iida S, Nitasaka E, et al. The gravity-regulated growth of axillary buds is mediated by a mechanism different from decapitation-induced release. Plant Cell Physiol. 2008;49:891–900. doi: 10.1093/pcp/pcn063. [DOI] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography–tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 2009;50:1201–1214. doi: 10.1093/pcp/pcp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Hirakawa Y, Kieber JJ, Fukuda H. CLE peptides can negatively regulate protoxylem vessel formation via cytokinin signaling. Plant Cell Physiol. 2011;52:37–48. doi: 10.1093/pcp/pcq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. Identification of a nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. Plant J. 2010;63:269–282. doi: 10.1111/j.1365-313X.2010.04239.x. [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. The regulatory region controlling the nitrate-responsive expression of a nitrate reductase gene, NIA1, in Arabidopsis. Plant Cell Physiol. 2011;52:824–836. doi: 10.1093/pcp/pcr033. [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013;4:1617. doi: 10.1038/ncomms2621. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y, Nakajima K, Arai Y. An improved assay for beta-glucuronidase in transformed-cells—methanol almost completely suppresses a putative endogenous beta-glucuronidase activity. Plant Sci. 1990;70:133–140. [Google Scholar]
- Krouk G, Ruffel S, Gutierrez RA, Gojon A, Crawford NM, Coruzzi GM, et al. A framework integrating plant growth with hormones and nutrients. Trends Plant Sci. 2011;16:178–182. doi: 10.1016/j.tplants.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Kudo T, Akiyama K, Kojima M, Makita N, Sakurai T, Sakakibara H. UniVIO: a multiple omics database with hormonome and transcriptome data from rice. Plant Cell Physiol. 2013;54:e9. doi: 10.1093/pcp/pct003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Kiba T, Sakakibara H. Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 2010;52:53–60. doi: 10.1111/j.1744-7909.2010.00898.x. [DOI] [PubMed] [Google Scholar]
- Kudo T, Makita N, Kojima M, Tokunaga H, Sakakibara H. Cytokinin activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-zeatin-O-glucosyltransferase in rice. Plant Physiol. 2012;160:319–331. doi: 10.1104/pp.112.196733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Mae T, Ohira K. Photosynthesis and ribulose 1,5-bisphosphate carboxylase in rice leaves: changes in photosynthesis and enzymes involved in carbon assimilation from leaf development through senescence. Plant Physiol. 1983;73:1002–1007. doi: 10.1104/pp.73.4.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massiere F, Badet-Denisot MA. The mechanism of glutamine-dependent amidotransferases. Cell Mol. Life Sci. 1998;54:205–222. doi: 10.1007/s000180050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Kitano M, Kusumoto T, Tarkowski P, Kinoshita-Tsujimura K, Vaclavikova K, Miyawaki K, et al. Cytokinins are central regulators of cambial activity. Proc. Natl Acad. Sci. USA. 2008;105:20027–20031. doi: 10.1073/pnas.0805619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura T, Sakakibara H, Nakano R, Kimata Y, Sugiyama T, Hase T. A nitrate-inducible ferredoxin in maize roots: genomic organization and differential expression of two non-photosynthetic ferredoxin isoproteins. Plant Physiol. 1997;114:653–660. doi: 10.1104/pp.114.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miflin BJ, Lea PJ. Ammonia assimilation. In: Walker JM, editor. The Biochemistry of Plants. New York: Academic Press, Inc; 1980. pp. 169–202. [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, et al. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 2010;51:1127–1135. doi: 10.1093/pcp/pcq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004;37:128–138. doi: 10.1046/j.1365-313x.2003.01945.x. [DOI] [PubMed] [Google Scholar]
- Mok DW, Mok MC. Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, et al. A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell. 2008;20:3148–3162. doi: 10.1105/tpc.108.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y. Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 2000;122:1045–1056. doi: 10.1104/pp.122.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, et al. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009;50:67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]
- Pace J, Mcdermott EE. Methionine sulphoximine and some enzyme systems involving glutamine. Nature. 1952;169:415–416. doi: 10.1038/169415a0. [DOI] [PubMed] [Google Scholar]
- Qin F, Shinozaki K, Yamaguchi-Shinozaki K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011a;52:1569–1582. doi: 10.1093/pcp/pcr106. [DOI] [PubMed] [Google Scholar]
- Qin H, Gu Q, Zhang JL, Sun L, Kuppu S, Zhang YZ, et al. Regulated expression of an isopentenyltransferase gene (IPT) in peanut significantly improves drought tolerance and increases yield under field conditions. Plant Cell Physiol. 2011b;52:1904–1914. doi: 10.1093/pcp/pcr125. [DOI] [PubMed] [Google Scholar]
- Redinbaugh MG, Campbell WH. Nitrate regulation of the oxidative pentose phosphate pathway in maize root plastids: induction of 6-phosphogluconate dehydrogenase activity, protein and transcript levels. Plant Sci. 1998;134:129–140. [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. Nitrogen economics of root foraging: transitive closure of the nitrate–cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc. Natl Acad. Sci. USA. 2011;108:18524–18529. doi: 10.1073/pnas.1108684108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. Differential response of genes for ferredoxin and ferredoxin:NADP+ oxidoreductase to nitrate and light in maize leaves. J. Plant Physiol. 2003a;160:65–70. doi: 10.1078/0176-1617-00919. [DOI] [PubMed] [Google Scholar]
- Sakakibara H. Nitrate-specific and cytokinin-mediated nitrogen signaling pathways in plants. J. Plant Res. 2003b;116:253–257. doi: 10.1007/s10265-003-0097-3. [DOI] [PubMed] [Google Scholar]
- Sakakibara H. Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Hayakawa A, Deji A, Gawronski S, Sugiyama T. His–Asp phosphotransfer possibly involved in a nitrogen signal transduction mediated by cytokinin in maize: molecular cloning of cDNAs for two-component regulatory factors and demonstration of phosphotransfer activity in vitro. Plant Mol. Biol. 1999;41:563–573. doi: 10.1023/a:1006391304881. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kobayashi K, Deji A, Sugiyama T. Partial characterization of signalling pathway of nitrate-dependent expression of the genes for nitrogen-assimilatory enzymes using detached maize leaves. Plant Cell Physiol. 1997;38:837–843. [Google Scholar]
- Sakakibara H, Suzuki M, Takei K, Deji A, Taniguchi M, Sugiyama T. A response-regulator homolog possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J. 1998;14:337–344. doi: 10.1046/j.1365-313x.1998.00134.x. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N. Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 2006;11:440–448. doi: 10.1016/j.tplants.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Sugiyama T. Isolation and characterization of a cDNA that encodes maize uroporphyrinogen III methyltransferase, an enzyme involved in the synthesis of siroheme, which is a prosthetic group of nitrite reductase. Plant J. 1996;10:883–892. doi: 10.1046/j.1365-313x.1996.10050883.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Sakakibara H, Kojima M, Nagasaki H, Yamamoto Y, Inukai Y, et al. Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis gene in rice. Plant Physiol. 2006;142:54–62. doi: 10.1104/pp.106.085811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson ME, Larsson C-M. Nitrate regulation of zeatin riboside levels in barley roots: effects of inhibitors of N assimilation and comparison with ammonium. Plant Sci. 1993;93:77–84. [Google Scholar]
- Sawaki N, Tsujimoto R, Shigyo M, Konishi M, Toki S, Fujiwara T, et al. A nitrate-inducible GARP family gene encodes an auto-repressible transcriptional repressor in rice. Plant Cell Physiol. 2013;54:506–517. doi: 10.1093/pcp/pct007. [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, et al. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y, Kameoka H, Yamaguchi S, Kyozuka J. Recent advances in strigolactone research: chemical and biological aspects. Plant Cell Physiol. 2012;53:1843–1853. doi: 10.1093/pcp/pcs142. [DOI] [PubMed] [Google Scholar]
- Shiraishi N, Sato T, Ogura N, Nakagawa H. Control by glutamine of the synthesis of nitrate reductase in cultured spinach cells. Plant Cell Physiol. 1992;33:727–731. [Google Scholar]
- Sivasankar S, Rothstein S, Oaks A. Regulation of the accumulation and reduction of nitrate by nitrogen and carbon metabolites in maize seedlings. Plant Physiol. 1997;114:583–589. doi: 10.1104/pp.114.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda Y, Ikeda A, Saiki S, Wiren N, Yamaya T, Yamaguchi J. Distinct expression and function of three ammonium transporter genes (OsAMT1;1–1;3) in rice. Plant Cell Physiol. 2003;44:726–734. doi: 10.1093/pcp/pcg083. [DOI] [PubMed] [Google Scholar]
- Stitt M. Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 1999;2:178–186. doi: 10.1016/S1369-5266(99)80033-8. [DOI] [PubMed] [Google Scholar]
- Stitt M, Müller C, Matt P, Gibon Y, Carillo P, Morcuende R, et al. Steps towards an integrated view of nitrogen metabolism. J. Exp. Bot. 2002;53:959–970. doi: 10.1093/jexbot/53.370.959. [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Abiko T, Yamaya T. Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.) J. Exp. Bot. 2007;58:2319–2327. doi: 10.1093/jxb/erm016. [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T. Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol. 2001;42:85–93. doi: 10.1093/pcp/pce009. [DOI] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, et al. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 2004;45:1053–1062. doi: 10.1093/pcp/pch119. [DOI] [PubMed] [Google Scholar]
- Tamura W, Hidaka Y, Tabuchi M, Kojima S, Hayakawa T, Sato T, et al. Reverse genetics approach to characterize a function of NADH-glutamate synthase1 in rice plants. Amino Acids. 2010;39:1003–1012. doi: 10.1007/s00726-010-0531-5. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006;45:1028–1036. doi: 10.1111/j.1365-313X.2006.02656.x. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Kiba T, Sakakibara H, Ueguchi C, Mizuno T, Sugiyama T. Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett. 1998;429:259–262. doi: 10.1016/s0014-5793(98)00611-5. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S. Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 2010;51:1118–1126. doi: 10.1093/pcp/pcq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz M, Moureaux T, Leydecker M-T, Vaucheret H, Caboche M. Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J. 1993;3:315–324. doi: 10.1111/j.1365-313x.1993.tb00183.x. [DOI] [PubMed] [Google Scholar]
- Wagner BM, Beck E. Cytokinins in the perennial herb Urtica dioica L. as influenced by its nitrogen status. Planta. 1993;190:511–518. [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM. Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell. 2000;12:1491–1509. doi: 10.1105/tpc.12.8.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003;132:556–567. doi: 10.1104/pp.103.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Hamada K, Yokoi H, Watanabe A. Biphasic and differential expression of cytosolic glutamine synthetase genes of radish during seed germination and senescence of cotyledons. Plant Mol. Biol. 1994;26:1807–1817. doi: 10.1007/BF00019494. [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15:2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kyozuka J. Branching hormone is busy both underground and overground. Plant Cell Physiol. 2010;51:1091–1094. doi: 10.1093/pcp/pcq088. [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kim HI, Kisugi T, Nomura T, Sekimoto H, et al. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta. 2012;235:1197–1207. doi: 10.1007/s00425-011-1568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Kusumoto D, Sekimoto H, Sugimoto Y, Takeuchi Y, et al. Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta. 2007;227:125–132. doi: 10.1007/s00425-007-0600-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.