Abstract
Pollination is an early and critical step in plant reproduction, leading to successful fertilization. It consists of many sequential processes, including adhesion of pollen grains onto the surface of stigmatic papilla cells, foot formation to strengthen pollen–stigma interaction, pollen hydration and germination, and pollen tube elongation and penetration. We have focused on an examination of the expressed genes in papilla cells, to increase understanding of the molecular systems of pollination. From three representative species of Brassicaceae (Arabidopsis thaliana, A. halleri and Brassica rapa), stigmatic papilla cells were isolated precisely by laser microdissection, and cell type-specific gene expression in papilla cells was determined by RNA sequencing. As a result, 17,240, 19,260 and 21,026 unigenes were defined in papilla cells of A. thaliana, A. halleri and B. rapa, respectively, and, among these, 12,311 genes were common to all three species. Among the17,240 genes predicted in A. thaliana, one-third were papilla specific while approximately half of the genes were detected in all tissues examined. Bioinformatics analysis revealed that genes related to a wide range of reproduction and development functions are expressed in papilla cells, particularly metabolism, transcription and membrane-mediated information exchange. These results reflect the conserved features of general cellular function and also the specific reproductive role of papilla cells, highlighting a complex cellular system regulated by a diverse range of molecules in these cells. This study provides fundamental biological knowledge to dissect the molecular mechanisms of pollination in papilla cells and will shed light on our understanding of plant reproduction mechanisms.
Keywords: Arabidopsis halleri, Arabidopsis thaliana, Brassica rapa, Laser microdissection, Papilla cell, RNA sequencing
Introduction
In flowering plants, several steps occur from anthesis to fertilization during sexual reproduction. The fertilization process begins when the pollen grains land on the stigma surface, that is the papilla cells. For pollen to adhere to the stigma, proteins and lipids are provided from the stigma and pollen; these mix to form a structure called the foot (Gaude and Dumas 1984, Dickinson 1995). In particular, proteinaceous components and lipids of the pollen coat are essential for both foot formation and subsequent pollen hydration, termed ‘coat conversion’ (Elleman and Dickinson 1986). After foot formation between pollen and papilla cells, water and resources for germination and pollen tube elongation are provided from the stigma to pollen, through a capillary system (Zuberi and Dickinson 1985, Zinkl et al. 1999). Once the pollen grain has germinated, the pollen tube directionally penetrates into a papilla cell, elongates through the stigma and style, and enters into the ovary (Cheung et al. 1995, Franklin-Tong 1999, Lord 2000, Shimizu and Okada 2000). The pollen tube eventually arrives at the embryo sac in the ovule, and finally, in the ovary, two sperm cells are released from the pollen tube tip and fertilize an egg cell and central cell, respectively, resulting in seed development (Dickinson and Elleman 1994, Stephenson et al. 1997).
In these plant reproduction processes, pollination is an early and important step for successful fertilization. Papilla cells have various roles in this step, such as discrimination of pollen grains for inter- and intraspecies barriers, pollen hydration, supply of stigmatic resources for pollen germination and pollen tube elongation, and pollen tube guidance (de Nettancourt 2001, Goto et al. 2011). However, the molecular basis of these mechanisms of pollination is still largely unclear (Hiroi et al. 2013).
Flowering plants have evolved exquisite systems for the control of mating interactions before fertilization, and the most extensively studied of these is self-incompatibility (SI) (Suwabe et al. 2010, Watanabe et al. 2012). SI is a genetic system for intraspecific pollen selectivity and is recognized as the most sophisticated system to prevent selfing (Bateman 1995). It is controlled by a single, highly polymorphic S locus and is manifested during the interaction of pollen grain with the papilla cells of the stigma epidermis (Watanabe et al. 1994, Suzuki et al. 1999). In Brassicaceae, S-locus receptor kinase (SRK) encodes a Ser/Thr receptor kinase, which localizes to the stigmatic plasma membrane and interacts with the pollen coat cysteine-rich protein (SP11/SCR) to induce the SI reaction (Schopfer et al. 1999, Takasaki et al. 2000, Takayama et al. 2000, Takayama et al. 2001). Downstream factors of the SI system, M-locus receptor kinase (MLPK), arm repeat containing 1 (ARC1) and Exo70A1, a subunit of the exocyst complex, have been identified, which mediate pollen–stigma interactions (Gu et al. 1998, Stone et al. 1999, Stone et al. 2003, Murase et al. 2004, Kakita et al. 2007a, Kakita et al. 2007b, Samuel et al. 2009). However, while some of these factors have been identified in the SI signaling cascade, the overall systems of SI and pollen–stigma interaction at the molecular level are still largely unclear, even in the much studied Brassicaceae.
For establishment of a knowledge base to aid understanding of the molecular mechanisms of pollination in the stigmatic papilla cell, including SI, transcriptome analysis is undoubtedly informative (Matsuo et al. 2011, Bai et al. 2013). So far, genome-wide transcriptome analysis of the stigma has been conducted in Arabidopsis, rice and maize, to study pollen–stigma communication and pollen tube guidance, by microarray technology (Swanson et al. 2004, Li et al. 2007). However, the microarray system has several limitations: high background levels due to cross-hybridization; lack of sensitivity at low expression levels; and reliance upon existing knowledge of genome sequences (Huang et al. 2013). In contrast, RNA sequencing (RNA-seq) has the potential to overcome these limitations (Wang et al. 2009) and offers a variety of new possibilities in biology, such as whole-genome sequencing, global identification of expressed genes, gene expression profiling and identification of novel genes, alternative splicing and sequence variation (Marioni et al. 2008, Garg et al. 2011).
Another problem in studying gene expression is that because it is difficult to isolate single cell types specifically from plant organs, many microarray studies have been conducted using RNAs extracted from a mixture of tissues and/or cell types (Autran et al. 2011). This limitation makes it difficult to analyze expressed genes at high precision in the target cell types. To overcome this, laser microdissection (LM) is a valuable tool to isolate specific cell types from sectioned specimens of heterogeneous tissues (Girke et al. 2000, Asano et al. 2002, Nakazono et al. 2003). LM allows the isolation of individual cells directly from the surrounding tissue, based on histological identification, by laser capture and laser cutting (Kerk et al. 2003, Suwabe et al. 2008, Kubo et al. 2013). Given the advantages and opportunities offered by RNA-seq and LM, this combination has the potential for precise and efficient analysis of transcriptional gene expression profiles of individual cell types.
Here, we report the establishment and characterization of papilla cell expressed gene data sets, among existing gene data in public databases, in Brassicaceae species Arabidopsis thaliana, A. halleri and Brassica rapa, by LM–RNA-seq analysis. Arabidopsis thaliana is a model plant species and A. halleri is a close relative. Brassica rapa, a vegetable member of the Brassicaceae, is a model species of the genus Brassica, and the annotated draft genome sequence has recently been completed (Wang et al. 2011). By using LM technology, the papilla cells of these species were isolated precisely, and papilla cell-specific gene expression profiles were obtained by RNA-seq. Using papilla cell-specific gene expression data sets from A. thaliana, A. halleri and B. rapa, we investigated the molecular biology of papilla cells by bioinformatics analysis, in terms of pollination mechanisms and papilla cell development.
Results and Discussion
Optimal conditions for paraffin-embedded sectioning of papilla cells
It is known that high integrity RNA is critical for gene expression analysis, and thus we first evaluated the optimal histological fixation conditions for paraffin embedding papilla cells for RNA extraction. The use of four fixatives (100% ethanol, 75% ethanol/25% acetate, 60% ethanol/40% acetate and 100% acetone) was compared for fixation of stigmas, including papilla cells, of A. thaliana, A. halleri and B. rapa. The quality of total RNA of LM papilla cells in each fixative was assessed using an Agilent 2100 Bioanalyzer. In A. thaliana, RNA quality expressed as the RNA integrity number (RIN) = 7.0, 6.9, 6.2 and 6.5 in 100% ethanol, 75% ethanol/25% acetate, 60% ethanol/40% acetate and 100% acetone, respectively. In A. halleri, the RIN = 7.1, 6.8, 7.1 and 6.3 in 100% ethanol, 75% ethanol/25% acetate, 60% ethanol/40% acetate and 100% acetone, respectively; and in B. rapa, the RIN = 5.5, 3.7, 3.5 and 6.2 in 100% ethanol, 75% ethanol/25% acetate, 60% ethanol/40% acetate and 100% acetone, respectively (Table 1). Thus, optimal fixation conditions varied depending on the plant species. In addition, morphological disintegration of tissue sections, especially in the ovary, was occasionally observed in 100% ethanol and 100% acetone (data not shown). Taken together, the best fixative conditions in terms of RNA quality and morphology was 75% ethanol/25% acetate in A. thaliana, 60% ethanol/40% acetate in A. halleri and 100% acetone in B. rapa.
Table 1.
Optimal fixative conditions for different Brassicaceae species
| Plant species | RNA quality (RIN) |
|||
|---|---|---|---|---|
| 100% ethanol | Ethanol/acetic acid |
100% acetone | ||
| 75%/25% | 60%/40% | |||
| A. thaliana | 7.0 | 6.9 | 6.2 | 6.5 |
| A. halleri | 7.1 | 6.8 | 7.1 | 6.3 |
| B. rapa | 5.5 | 3.7 | 3.5 | 6.2 |
RNA-seq and identification of papilla-expressed genes
Using optimal fixation conditions for pistils, embedded specimens were prepared using 2–6 floral buds from each plant species. Serial paraffin sections of pistil specimens were prepared at 6–8 µm in thickness and papilla cells were isolated by LM, using approximately 60 pistil sections from each plant sample (Fig. 1). Total RNAs were extracted from the LM-collected papilla cells, confirmed to be acceptable quality for RNA-seq analysis (Supplementary Fig. S1), and 1.5 rounds of mRNA linear amplification were performed for each sample. Subsequently, 50–200 ng µl−1 cDNAs were obtained from each sample, and cDNA fragment libraries were subjected to SOLiD5500xl sequencing. As a result of the RNA-seq analysis, 65,690,858, 101,509,276 and 74,175,637 raw reads of 75 bp in length were obtained in A. thaliana, A. halleri and B. rapa, respectively. On the alignment of raw reads to the A. thaliana and B. rapa reference genomes, a total of 21,310, 23,414 and 26,852 unigenes were defined, respectively, after removing low-quality raw reads (MapQ ≤10) and adaptor sequences. Of these, 17,240, 19,260 and 21,026 unigenes were found to be >10 read tags and could be assigned an annotation based on database sequence assembly in A. thaliana, A. halleri and B. rapa, respectively (Table 2; Supplementary Tables S1–S3). In A. thaliana, approximately 50–60% of the genes have previously been predicted to be expressed in stigmas (Suwanson et al. 2005), and the remaining ∼50% of the genes have never been predicted to be expressed in stigmas, and thus they are likely to be novel stigma-expressed genes, including papilla-expressed genes. The fact that approximately half of the annotated genes from the RNA-seq have not been detected previously in microarray and cDNA subtraction analyses highlights the advantages, including greater sensitivity, of the combination of LM and RNA-seq analysis for the tissue of interest.
Fig. 1.
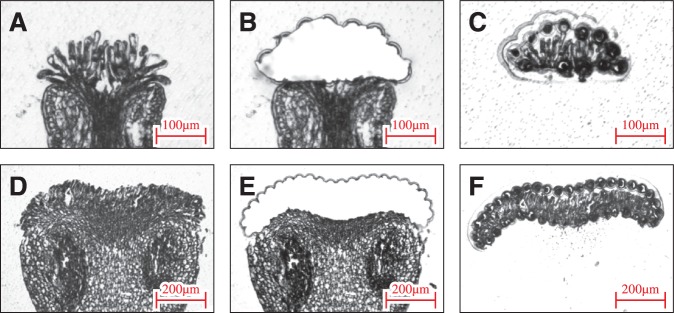
Isolation of a papilla cell by LM in Arabidopsis and Brassica. The paraffin-embedded stigmas were cut into 6–8 µm thick sections using a microtome and mounted on PEN membrane frame slides. After removing the paraffin, LM was performed using an Arcturus XT Laser Capture Microdissection System. (A–C) Arabidopsis thaliana, (D–F) B. rapa, (A, D) before LM, (B, E) after LM, and (C, F) isolated papilla cells.
Table 2.
Summary of experimental tags sequenced in reference A. thaliana and B. rapa genomes
| Plant species | Total reads | Mapped reads | MapQ ≥10 | Total genes | Unigenes | |
|---|---|---|---|---|---|---|
| Reads >0 | Reads ≥10 | |||||
| A. thaliana | 65,690,858 | 38,955,652 | 35,266,802 | 33,602a | 21,310 | 17,240 |
| 59.30% | 53.69% | 63.42%d | 51.31%d | |||
| A. hallerib | 101,509,276 | 33,613,230 | 25,431,815 | – | 23,414 | 19,260 |
| 33.11% | 25.05% | 69.68%d | 57.32%d | |||
| B. rapa | 74,175,637 | 62,343,265 | 54,037,867 | 41,173c | 26,852 | 21,026 |
| 84.05% | 72.85% | 65.22%d | 51.07%d |
a Total gene number from A. thaliana deposited in the database (http://www.arabidopsis.org/).
b The total gene number from A. halleri was not determined.
c Total gene number from B.rapa deposited in the database (http://brassicadb.org/brad/).
d Unigenes/total genes×100 (%). For both A. thaliana and A. halleri, the total gene number used for calculation was 33,602.
Among the top 100 most highly expressed genes of the A. thaliana papilla cell transcriptome (Supplementary Table S1), gene ontology (GO) annotation analysis showed that genes in the categories membrane-related (54/100), cell wall (25/100) and metabolism (24/100) were significantly enriched and some of these have already been proved to be involved in plant reproduction systems. For example, among these genes, the SCA-like Arabidopsis LTP, AT2G38540, has been shown to have various roles both in plant reproduction and in growth (Chae et al. 2010). PDIL2-1, AT2G47470, which encodes a cell wall-related protein disulfide isomerase, has been verified to be important in embryo sac maturation (Wang et al. 2008). This demonstrates that these categories include genes that are expected to be particularly important in papilla cell processes, such as regulation of papilla cell formation and pollen–papilla communication.
Validation of the LM–RNA-seq transcriptome in papilla cells
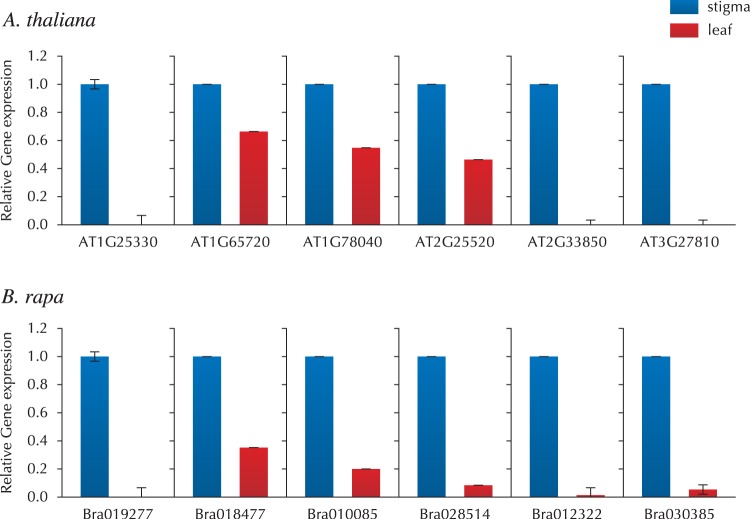
To assess the validity and reliability of our LM–RNA-seq data from papilla cells, quantitative real-time PCR (qRT-PCR) analysis was performed on 12 representative genes belonging to different classification groups (Supplementary Table S4). Expression levels of the 12 genes were compared between papilla cells and the leaf to determine whether these genes were expressed in papilla specifically or preferentially (Fig. 2). The results from qRT-PCR showed that these 12 genes were expressed exclusively in papilla cells, and this result correlated with those obtained by LM–RNA-seq. To confirm further the expression and localization of the genes in papilla cells, in situ hybridizations were performed with six representative genes, of stress-related, metal tolerance and unknown functions (Supplementary Table S5). Probes from all six genes exhibited hybridization signals in papilla cells and the transmitting tract of the stigma, confirming that the corresponding gene transcripts were detected in papilla cells by in situ hybridization (Fig. 3).
Fig. 2.
Expression analysis of genes identified in the papilla by LM–RNA-seq. qRT-PCR was performed using the SYBR Green Real-Time PCR Master Mix with a Bio-Rad CFX96 Real-Time Detection System. cDNAs were prepared from total RNAs of stigma and leaf. Representation of the relative expression levels of the genes was normalized to the expression level of UBC21. Quantitative validation of three replicates was calculated using the delta–delta threshold cycle relative quantification method.
Fig. 3.
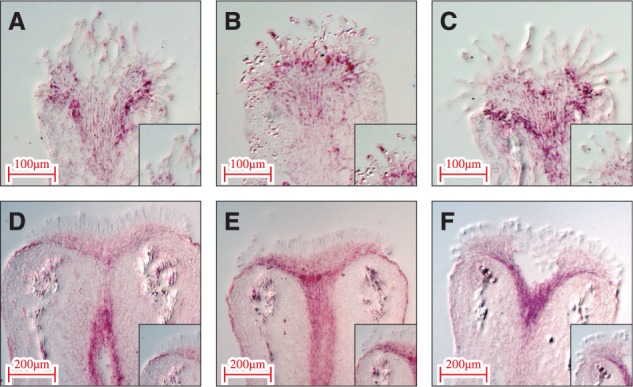
Confirmation of gene expression patterns by in situ hybridization. The pistils for in situ hybridization were collected 1 d before anthesis. Both antisense and sense probes were synthesized using a T7/SP6 digoxigenin RNA labeling kit. Insets show the signal area in papilla cells at high magnification. (A) AT1G09690, (B) AT1G53130, (C) AT2G41430, (D) Bra004382, (E) Bra019017 and (F) Bra028171.
Taken together, these results demonstrate the validity and reliability of our LM–RNA-seq data in papilla cells. This also highlights the potential capability of LM–RNA-seq as a technique for fine-scale gene expression analysis.
Comparison of expressed genes in papilla cells and other tissues in A. thaliana
To characterize the Arabidopsis papilla-specific genes, LM–RNA-seq data from papilla cells were compared with those from microarray analysis of stamen and leaf in A. thaliana (Schmid et al. 2005) (Fig. 4A). Among the 17,240 genes expressed in papilla cells, approximately half (8,538/17,240) were uniformly detected in all tissues examined here, suggesting that these are conserved genes for general cellular function common to all tissues. In contrast, one-third of the genes (5,668/17,240) were papilla specific. Because papilla cells have specialized functions, focusing on pollination, these genes are expected to be involved in the regulation and maintenance of the specific roles of papilla cells.
Fig. 4.
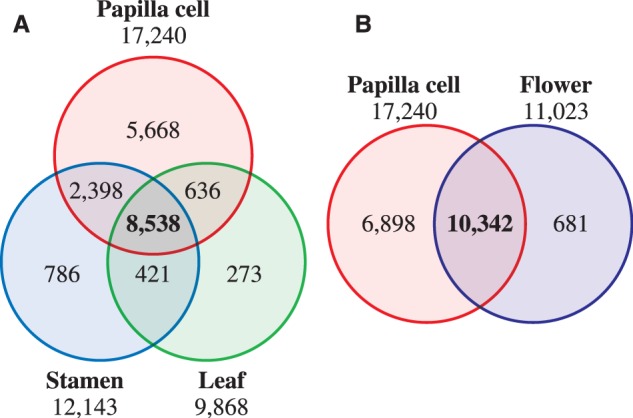
Comparison of transcriptome data sets from papilla cells, stamen, leaf and flower in A. thaliana. (A) Venn diagram for the number of genes expressed in papilla cells, stamen and leaf. Expression data for stamen and leaf are from Schmid et al. (2005). (B). Venn diagram for the number of genes expressed in papilla cells and flowers. Expression data for flowers are from Schmid et al. (2005).
It is noteworthy that 6,898 papilla cell-expressed genes from the present study were not identified in the previous microarray data from whole flowers (Schmid et al. 2005) (Fig. 4B). Furthermore, 40% of the genes (2,749/6,898) were also not contained on the Arabidopsis ATH1 Genome Array GeneChip, which includes >22,500 probe sets representing approximately 24,000 genes. This reflects the limitation of microarray techniques, and at the same time highlights the potential of LM–RNA-seq technology to increase greatly the accuracy of cell type-specific gene isolation and the scale of transcriptome analysis.
Comparison of papilla-expressed gene data sets from the three Brassicaceae species
To reveal the similarities and differences in papilla-expressed genes in Brassicaceae, the data sets from A. thaliana, A. halleri and B. rapa were compared (Fig. 5). Because the genome of Brassica has been triplicated during the course of evolution, every gene has essentially three copies, i.e. one orthologous and two paralogous genes, in the genome. Thus, in the case of B. rapa, 14,099 unigenes were further selected from the 21,026 unigenes by blast analysis with the e-value cut-off of 1 × E−5, to remove paralogous information and to build one-to-one relationships with the A. thaliana genes. It is noteworthy that there were 12,311 expressed genes common to the data sets from all three species, corresponding to 71% of the total expressed genes in A. thaliana, 64% in A. halleri and 87% in B. rapa, indicating that a large proportion of the expressed genes were common to papilla cells across the species boundary in Brassicaceae. Indeed, among the top 100 most highly expressed genes, 82, 82 and 78 genes in A. thaliana, A. halleri and B. rapa, respectively, were common to all three species (Supplementary Tables S1–S3).
Fig. 5.
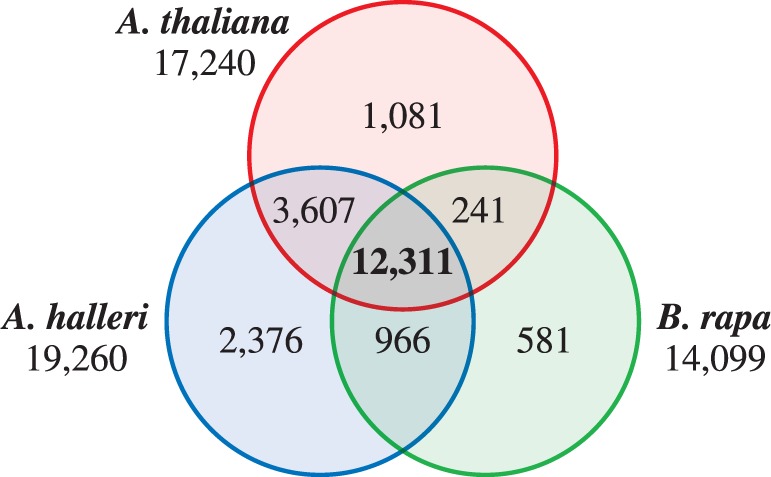
Conservation of papilla-expressed genes in Brassicaceae. Venn diagram for the number of genes in papilla cells from A. thaliana, A. halleri and B. rapa.
These gene sets were next sorted into six functional categories, taking into consideration several aspects of the biological role, function and physiology of stigmas, i.e. signal transduction, cell wall-related, stress/defense, metabolism, ion transport and transcription: this categorization is based on a functional classification of the pistil-expressed genes according to the GO annotations (Tung et al. 2005, Allen et al. 2010). From the 17,240, 19,260 and 14,099 genes expressed in papilla cells of A. thaliana, A. halleri and B. rapa, 6,468, 7,481 and 5,700 unigenes were classified into six groups according to their annotations and GO terms, respectively. General classification based on GO terms of the papilla-expressed gene data sets revealed that the proportion of categories showed broad similarities among species, in terms of functional classes (Fig. 6; Supplementary Table S6). These results reflect a degree of functional conservation of papilla cells between Arabidopsis and Brassica, and this is consistent with previous results in stigma (Tung et al. 2005, Allen et al. 2010). They also support the theory that mechanisms for stigmatic function are similar in different species, depending on the degree of species differentiation (Allen et al. 2010). A previous study indicated that the expression profiles of stigma-expressed genes in maize were more similar to those of rice than those of Arabidopsis (Swanson et al. 2004, Li et al. 2007, Xiao et al, 2012).
Fig. 6.
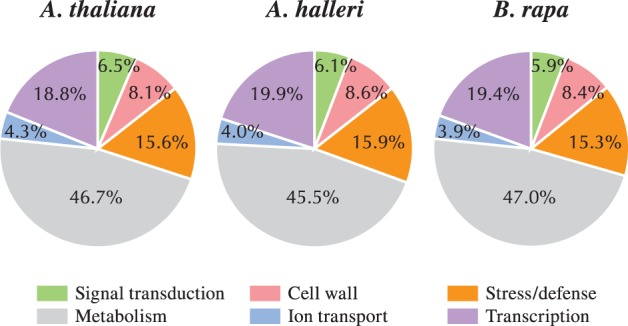
Classification of papilla-expressed genes by gene ontology, in A. thaliana, A. halleri and B. rapa. The pistil-expressed gene sets were sorted into six functional categories based on a functional classification of the genes, according to the gene ontology (GO) annotations (Tung et al. 2005, Allen et al. 2010): signal transduction, cell wall-related, stress/defense, metabolism, ion transport and transcription.
In contrast to the above consideration of the similarity between the three species, 1,081, 2,376 and 581 papilla-expressed genes from A. thaliana, A. halleri and B. rapa, respectively, were detected in one of the species but not the other two (Fig. 5). This is presumably a result of species differentiation in Brassicaceae.
Biological and functional characteristics of papilla cells in Brassicaceae
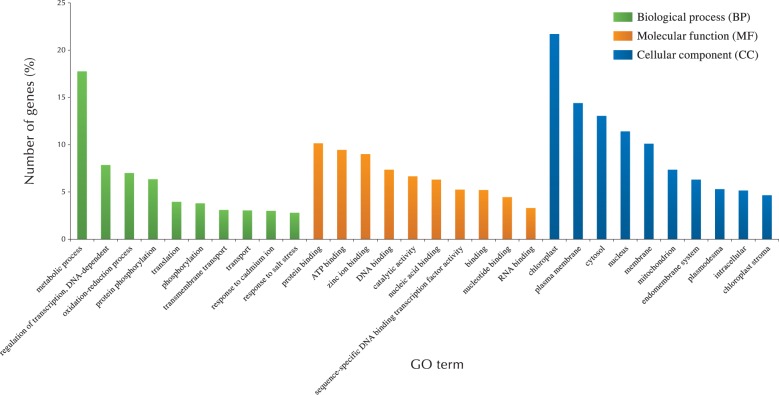
To characterize the biological and functional roles of papilla cells in Brassicaceae, GO terms were assigned and sorted for the 12,311 expressed genes common to all three species. Among these, 4,309 distinct GO terms were annotated, consisting of 2,214 biological processes, 416 cellular components and 1,679 molecular functions (Supplementary Table S7). The 10 most highly represented GO terms in each category of biological process, cellular components and molecular functions are shown in Fig. 7.
Fig. 7.
Functional categories of the 12,311 genes common to three Brassicaceae species. GO terms were assigned and sorted for 12,311 expressed genes common to three Brassicaceae species.
Metabolic process is the highest within biological processes and not the highest in all categories (Fig. 7). In ‘metabolic process’, highly represented GO terms included members of lipid metabolism. For example, among our data sets, Extracellular Lipase 4 (EXL4) was expressed almost identically in papilla cells in all the species studied (Supplementary Table S7). EXL4 is a member of the GDSL lipases, which are active in hydrolysis and synthesis of lipids and/or esters (Updegraff et al. 2009). GDSL lipases have multiple physiological roles such as development, flowering and seed germination in plants, and EXL4 is involved in the rapid initiation of pollen hydration. The exl4 disrupted mutation results in slower pollen hydration on the stigma and decreased competitiveness in pollination, suggesting that changes in lipid composition at the pollen–papilla interface are essential for pollen hydration on papilla cells. Lipids have a broad range of biological functions, such as a component of the cell membrane and cuticle layer in epidermal cells, a mediator of signal transduction across extracellular and intracellular membranes, and an energy source for cellular functions (Wolters-Arts et al. 1998, Wolters-Arts et al. 2002), and the high proportion of lipid metabolism genes highlights their important role both in general cellular function (papilla cell development) and specific functions (pollen adhesion, pollen tube growth and pollen tube directional guidance and signaling).
Another represented GO category within ‘metabolic process’ was associated with ‘cell wall-related’, including a member of the pectin methylesterase (PME), pectin lyase-like superfamily (PPME) and glycosyl hydrolase family (Bosch et al. 2005) (Supplementary Table S7). The plant cell wall is composed of polysaccharides and proteins, and the polysaccharides, such as cellulose, hemicellulose and pectin, constitute the major part of the cell wall (Lehner et al. 2010, Komaki and Sugimoto. 2012). The varying proportions and interactions of the polysaccharides ensure the strong and flexible properties of cell walls (Daher et al. 2011). In Brassicaceae, the cell wall of papilla cells is composed of two layers, layer I (cuticle) and layer II (pectocellulosic wall) (Elleman et al. 1988). PPME and PME are proposed to mediate cell wall loosening and expansion, thus enabling pollen tube growth through the papilla cell wall (Bosch and Hepler 2005, Jiang et al. 2005, Renault et al. 2011). Application of an exogenous PME induces thickening of the apical cell wall and dissipation of the intracellular tip-focused Ca2+ gradient in papilla cells (Hülskamp et al. 1995, Takamatsu and Takagi 2011). Thus, papilla cells in Brassicaceae have a broad range of metabolic turnover and cell wall remodeling systems, and genes in lipid metabolism and cell wall-related categories are expected to be important players in successful fertilization, through cell wall remodeling and cell–cell signaling interactions in papilla cells.
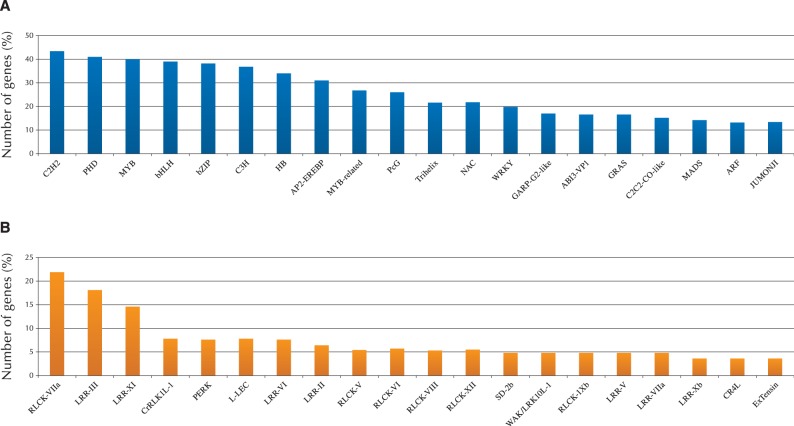
The second category of biological process represented in the data set was ‘regulation of transcription’. Based on the Database Arabidopsis Transcription Factors (DATF: http://datf.cbi.pku.edu.cn/), several families of transcription factors (TFs) were represented in the 12,311 common genes, and 754 TF genes were identified (Supplementary Table S8). Among these, six families of zinc finger (C2H2, C3H), PHD, MYB, bHLH and bZIP groups were represented (Fig. 8A). HECATE encodes a basic helix–loop–helix (bHLH) TF which controls the reproductive development of the transmitting tract and floral organ (Gremski et al. 2007). No Transmitting Tract (NTT) encodes a C2H2/C2HC zinc finger TF expressed in the transmitting tract, which is required for development of both the transmitting tract and the funiculus of ovules (Crawford et al. 2007). Other key genes were also identified: HALF FIELD (HAF) (Crawford et al. 2007, Crawford and Yanofsky 2011), Brassinosteroid enhanced expression 1 (BEE1) and BEE3 (Friedrichsen et al. 2002). HAF, BEE1 and BEE3 encode closely related bHLH TFs that act redundantly to specify reproductive tract tissues, where they are expressed in distinct but overlapping patterns (Crawford and Yanofsky 2011). Thus, although little is known about the TF-controlled genetic network in reproductive development, the data sets of TF are expected to be a valuable resource for the study of genetic networks in papilla cell development in Brassicaceae.
Fig. 8.
Distribution of TFs and receptor-like kinases (RLKs) in the 12,311 genes common to three Brassicaceae species. TFs were assigned by the Database Arabidopsis Transcription Factors (DATF: http://datf.cbi.pku.edu.cn/) for the 12,311 common genes. RLKs were identified and categorized among genes that have GO terms ‘plasma membrane’ within cellular components.
The predominant GO terms in the cellular components domain were ‘chloroplast’, ‘plasma membrane’ and ‘cytosol’. A high proportion of the category ‘chloroplast’ is not likely to be papilla specific, because the proportion of the GO term ‘chloroplast’ was uniformly high in other tissues such as leaf and stamen (data not shown). Several studies have shown that proteins integral to the plasma membrane, i.e. receptor-like kinases, play important roles in pollination, disease resistance, development and hormone response (Gaude and Dumas 1984, Hiscock and Allen 2008, Kato et al. 2010). The plant receptor-like kinases are transmembrane proteins that transduce external messages into the cell through their extracellular domains and intracellular kinase domain (Takasaki et al. 2000, Chen et al. 2009, Shinya et al. 2010). In our papilla-expressed gene data sets, protein kinase families and leucine-rich repeat receptor-like protein kinases (LRR-RLKs) were the most highly represented in the ‘plasma membrane’, many of which also corresponded to ‘binding (protein binding, ATP binding, zinc ion binding, DNA binding)’ in molecular function. Thus, it is likely that kinase families function in many aspects of papilla cells, such as coordination of pistil development, defense response and facilitation of pollen–pistil communication, with downstream mechanisms in the cytosol.
To understand further the role of protein kinase in papilla cell development and reproduction, we focus here on LRR-RLK in the papilla-expressed gene data set. Among 12,311 common genes, 220 LRR-RLKs were classified into 56 subfamilies (Fig. 8B; Supplementary Table S9). The first and fourth subfamilies included 22 cytoplasmic receptor-like kinases (RLCKs), which may modulate pollen tube growth and pollen–pistil interaction. Botrytis-induced kinase 1 (BIK1) is a member of the RLCK VIIa subfamily and BRI1-associated receptor kinase mediating brassinosteroid (BR) signaling (Clouse and Sasse 1998, Muschietti et al. 1998, Zuhua et al. 2000). The expression of BIK1 was highly represented in the common genes. Furthermore, the BR-responsive genes BEE1 and BEE3, downstream factors of the BRI1 receptor complex, were also represented. As mentioned above, these are TFs that regulate stigmatic cell development. Thus, BR signaling contributes to the pathway of papilla cell development, with a coordinated system of BIK1, BEE1, BEE3 and other as yet unidentified factors (Crawford and Yanofsky 2011). The second and third subfamilies included genes encoding LRR-RLKs that may be involved in the regulation of pollen tube growth. In tomato, Solanum lycopersicum, for example, pollen receptor kinases (PRKs) act in pollen–pistil interactions, mainly perceiving signals from pistil to regulate pollen tube growth (Muschietti et al. 1998, Tang et al. 2002, Johnson and Preuss 2003). Among the papilla cell-expressed genes, we identified several kinases, and these may play a role in coordinating pollination events and processes in pollen–papilla communication. Further analysis of this protein family will provide new insights into how pollen–papilla interactions are regulated and which are necessary for the signal cascade of pollination in Brassicaceae.
Conclusion
Pollination is an essential step for sexual reproduction, and its regulation is an important mechanism to maintain genetic barriers and diversity in plant species. Because correct functioning of papilla cells is critical for successful reproduction in flowering plants, we focused on the establishment and characterization, on the basis of gene expression, of the molecular components of cellular events in papilla cells. From this approach, >60% of the expressed genes were found to be common to papilla cells of three Brassicaceae species, highlighting the functional commonalities of papilla cells of Brassicaceae in terms of development and reproduction. These mechanisms included, in particular, production of certain metabolites, TFs, receptor kinases and signaling cascades, which requires a precise gene network and molecular control to ensure correct physiological, cellular, developmental and reproductive processes in papilla cells of Brassicaceae. Thus, papilla cells have complex cellular systems, which are regulated by a diversity of molecular players. In particular, LM–RNA-seq data reveal the wide variety of genes in each functional class that are involved in these processes. These data will be especially useful in studies of the spatial and temporal balance of gene expression that is critical for successful plant reproduction and development.
The LM–RNA-seq data reported here highlight the effectiveness of cell type-specific gene expression analysis, and these global gene expression data from papilla cells will be a valuable knowledge base for future research into plant reproduction, especially in pollination biology. This will shed light on the dissection of molecular mechanisms and components of pollination in papilla cells, aiding our understanding of the plant reproduction mechanism.
Materials and Methods
Plant materials
Arabidopsis thaliana ecotype Columbia (Col-0) and its close relative A. halleri were grown in a growth chamber (BIOTRON LH-240S, NK system) at 22°C under 16 h light/8 h dark photoperiod conditions. The individual of A. halleri subsp. gemmifera (Matsumura) O’Kane & Al-Shehbaz used in this study was collected from a population located in Tadaginzan, Inagawa, Osaka, Japan, by K.K.S. [population of halgem2 described by Shimizu-Inatsugi et al. (2009) and Tsuchimatsu et al. (2012)]. It was subjected to five rounds of self-fertilization by bud pollination in the laboratory to reduce heterozygosity, and is the clone of the individual W302 with S-haplogroup A used by Tsuchimatsu et al. (2010). Brassica rapa accession Chiifu-401-42, a DH Chinese cabbage line, was grown in a greenhouse under normal conditions, at Chungnam National University, Daejeon, Korea.
Tissue fixation and paraffin embedding
Pre-chilled chemical fixatives (100% ethanol, 75% ethanol/25% acetate, 60% ethanol/40% acetate and 100% acetone) were used for fixation. Pistils were collected from floral buds before anthesis, and 3–4 mm tips including the stigma were picked and placed immediately in each fixative solution, with a vacuum on ice for 5 min twice. Tissue fixation and paraffin embedding were performed using a microwave processor (LabPulse H2850, Energy Beam Sciences), as described by Takahashi et al. (2010), with minor modifications. Each fixative was replaced with a fresh aliquot, and samples on ice were microwaved at 150 W, 37°C for 15 min three times. Fixed samples were dehydrated by serial dilutions of 70, 80, 90, 100% and absolute ethanol, by microwaving at 250 W, 58°C for 90 s. In the case of samples in 100% ethanol and 100% acetone, the dehydration step was omitted and was replaced with 50% ethanol/50% 2-propanol or 50% acetone/50% n-butanol, respectively, in infiltration steps. For paraffin embedding, each fixative was replaced by 50% paraffin/50% 2-propanol by microwaving at 250 W 58°C for 10 min. In the case of samples fixed in 100% acetone, the fixative was replaced by 50% paraffin/50% n-butanol. All samples were then microwaved in 100% paraffin wax (Paraplast X-TRA, Fisher Scientific) at 150 W, 58°C for 10 min, followed by microwaving at 150 W, 58°C for 30 min four times. The specimens were then placed on a plastic dish filled with melted paraffin wax. The embedded specimens were cooled to room temperature and the paraffin blocks were stored at 4°C.
Preparation of paraffin-embedded sections and LM
The paraffin-embedded samples were cut into 6–8 µm thick sections using a microtome (RV240, YamatoKoki). Serial paraffin-embedded sections were mounted on PEN membrane frame slides (Applied Biosystems, Life Technologies) with RNAsecure Reagent (Ambion, Life Technologies), dried on a heating plate at 57°C for 30 s to 1 min, and then further dried at 4°C for 1 h. To remove paraffin, slides were immersed in Histo-clear II (National Diagnostics) for 5 min at room temperature twice, and dried completely at room temperature. LM was performed using an Arcturus XT Laser Capture Microdissection System (Applied Biosystems, Life Technologies). The procedure of LM is described in Suwabe et al. (2008). Briefly, selected areas were captured by an infrared laser onto CapSure Macro LCM Caps (Applied Biosystems, Life Technologies) and were subsequently cut by UV laser. The papilla cells that fused to the LCM cap were collected by removing the cap from the tissue section.
RNA extraction, amplification and cDNA preparation
Total RNAs of papilla cells collected by LM were extracted using the Pico-Pure RNA isolation kit (Applied Biosystems, Life Technologies). The quantity and quality of total RNA were assessed using an Agilent 2100 Bioanalyzer and RNA 6000 Pico kit (Agilent Technologies). The quality of the total RNA was judged by the RIN, which was calculated by 2100 Expert Software (Agilent Technologies). The RIN evaluates the quality of total RNA on a scale from 1 to 10, in which 10 corresponds to intact RNA and 1 corresponds to extremely degraded RNA. In plants, RNA with a quality value of more than RIN = 6 is acceptable for gene expression analysis (Takahashi et al. 2010). Thereafter, the mRNA was linear-amplified by a RiboAmp HS PLUS RNA amplification kit (Applied Biosystems, Life Technologies), according to the manufacturer’s instruction, through 1.5 cycles. An average of 5 ng of total RNA from each sample was used for linear amplification. cDNAs were synthesized from the linear-amplified mRNA using the SuperScript III First-Strand Synthesis system (Invitrogen, Life Technologies). The quantity and quality of cDNA were confirmed using Qubit fluorometer and Qubit dsDNA HS Assay kits (Invitrogen, Life Technologies).
RNA-seq by SOLiD 5500xl
Preparation of a fragment library for SOLiD sequencing was carried out following the manufacturer’s instructions (Applied Biosystems, Life Technologies). Briefly, cDNAs were amplified by 10 cycles of PCR and a fragment library prepared using the 5500 SOLiD Fragment Library Core kit. Adaptor ligation was conducted using 5500 SOLiD Fragment Library Barcode Adaptors. The quality of the cDNA fragment library was confirmed with a fragment peak of 200–300 bp and the absence of unreacting adaptors, by an Agilent 2100 Bioanalyzer and Agilent High Sensitivity DNA kit. Approximately 0.7–1.0 pmol of the cDNA fragment library of each papilla cell sample was applied to the SOLiD EZ Bead System, and then subjected to sequencing by the SOLiD 5500xl system (Applied Biosystems, Life Technologies), with a 1/6 plate scale per sample. Because of the linear amplification of mRNA, SOLiD RNA-seq was not directional.
Sequence data processing and gene ontology analysis for unigenes
Processing of sequence data for assembly was conducted using LifeScope Genomic Analysis Software (Applied Biosystems, Life Technologies), with a default parameter. Sequence reads with a quality of >Q10 were assembled to reference genome sequences (TAIR10 for Arabidopsis; http://www.arabidopsis.org/, BRAD for Brassica; http://brassicadb.org/brad/). The Arabidopsis unigenes were obtained from the TAIR and the B. rapa unigenes were searched using the BLASTX program against TAIR10, with a cut-off e-value of 1 × E−5, and assigned GO terms. Examples of visualization of mapping reads were displayed by Integrative Genomics Viewer (IGV) 2.3, for three representative genes, AT1G09690, AT1G53130 and AT2G41430, which were used for in situ hybridization (Supplementary Figs. S2–S4).
qRT-PCR
Total RNAs were extracted from stigma and leaf samples according to the method described above. The concentration and purity of total RNAs were determined by spectrophotometric analysis. Total RNA was reverse-transcribed using the SuperScript III First-Strand Synthesis system with oligo(dT) primer, according to the manufacturer’s instructions. qRT-PCR was performed using the SYBR Green Real-Time PCR Master Mix (Toyobo) with a Bio-Rad CFX96 Real-Time Detection System. Quantitative validation in three replicates was calculated using the delta–delta threshold cycle relative quantification method. A gene coding for ubiquitin-conjugating enzyme 21, UBC21, was used as a control. Sequence information of all primers is listed in Supplementary Table S10.
In situ hybridization
Pistils for in situ hybridization were collected 1 d before anthesis. Both antisense and sense probes were synthesized using a T7/SP6 digoxigenin (DIG) RNA labeling kit (Roche), according to the manufacturer’s instructions. In situ hybridization was performed according to Masuko et al. (2006), and tissue sections were observed under a light microscope by using SteREO Discovery V20 (Carl Zeiss, Jena, Germany).
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) [Grants-in-Aid for Scientific Research on Innovative Areas (Nos. 23113001 and 23113006 to G.S., K.S. and M.W., and No. 24113518 to K.Y.)]; the Japan Society for Promotion of Science (JSPS) [a Grant-in-Aid for Scientific Research (C) (No. 25450515 to G.S.), Grant-in-Aid for Scientific Research (B) (No. 25292005 to K.S. and M.W.)]; Meiji University [Research Funding for Computational Software Supporting Program (to K.Y.)].
Supplementary Material
Acknowledgments
The authors thank Dr. Yuhko Kobayashi and Professor Issei Kobayashi (Mie University) for supporting RNA sequencing. The authors are also grateful to Masumi Miyano, Kana Ito, Momoko Ishikawa, Yoshie Higuchi, Nanase Kon, Ayako Tsushima, Mizuho Kawagishi, Reina Shida and Ayumi Sadaike (Tohoku University) for technical assistance.
Glossary
Abbreviations
- bHLH
basic helix–loop–helix
- BR
brassinosteroid
- GO
gene ontology
- LM
laser microdissection
- LRR-RLK
leucine-rich repeat receptor-like protein kinase
- PME
pectin metylesterase
- qRT-PCR
quantitative real-time PCR
- RIN
RNA integrity number
- RNA-seq
RNA sequencing
- SI
self-incompatibility
- TF
transcription factor
Disclosures
The authors have no conflicts of interest to declare.
References
- Allen AM, Lexer C, Hiscock SJ. Comparative analysis of pistil transcriptomes reveals conserved and novel genes expressed in dry, wet, and semidry stigmas. Plant Physiol. 2010;154:1347–1360. doi: 10.1104/pp.110.162172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H. Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J. 2002;32:401–408. doi: 10.1046/j.1365-313x.2002.01423.x. [DOI] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell. 2011;145:707–719. doi: 10.1016/j.cell.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Bai S, Saito T, Sakamoto D, Ito A, Fujii H, Moriguchi T. Transcriptome analysis of Japanese pear (Pyrus pyriflolia Nakai.) flower buds transitioning through endodormancy. Plant Cell Physiol. 2013;54:1132–1151. doi: 10.1093/pcp/pct067. [DOI] [PubMed] [Google Scholar]
- Bateman AJ. Self-incompatibility systems in angiosperms. III. Cruciferae. Heredity. 1995;6:285–310. [Google Scholar]
- Bosch M, Cheung AY, Hepler PK. Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 2005;138:1334–1346. doi: 10.1104/pp.105.059865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Hepler PK. Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell. 2005;17:3219–3226. doi: 10.1105/tpc.105.037473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Gonong BJ, Kim SC, Kieslich CA, Morikis D, Balasubramanian S, et al. A multifaceted study of stigma/style cysteine-rich adhesin (SCA)-like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction. J. Exp. Bot. 2010;61:4277–4290. doi: 10.1093/jxb/erq228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Liu J, Lei G, Liu YF, Li ZG, Tao JJ, et al. Effects of tobacco ethylene receptor mutations on receptor kinase activity, plant growth and stress responses. Plant Cell Physiol. 2009;50:1636–1650. doi: 10.1093/pcp/pcp107. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wang H, Wu HM. A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell. 1995;82:383–393. doi: 10.1016/0092-8674(95)90427-1. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu. Rev. Plant. Physiol. Plant Mol. Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Crawford BCW, Ditta G, Yanofsky MF. The NTT gene is required for transmitting-tract development in carpels of Arabidopsis thaliana. Curr. Biol. 2007;17:1101–1108. doi: 10.1016/j.cub.2007.05.079. [DOI] [PubMed] [Google Scholar]
- Crawford BCW, Yanofsky MF. HALF FIELD promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana. Development. 2011;138:2999–3009. doi: 10.1242/dev.067793. [DOI] [PubMed] [Google Scholar]
- de Nettancourt D. Incompatibility and Incongruity in Wild and Cultivated Plants. 2nd edn. Berlin: Springer-Verlag; 2001. [Google Scholar]
- Daher FB, Oostende C, Geitmann A. Spatial and temporal expression of actin depolymerizing factors ADF7 and ADF10 during male gametophyte development in Arabidopsis thaliana. Plant Cell Physiol. 2011;52:1177–1192. doi: 10.1093/pcp/pcr068. [DOI] [PubMed] [Google Scholar]
- Dickinson HG. Dry stigmas, water and self-incompatibility in Brassica. Sex. Plant Reprod. 1995;8:1–10. [Google Scholar]
- Dickinson HG, Elleman CJ. Pollen hydrodynamics and self-incompatibility in Brassica oleracea. In: Stephenson AG, Kao T-h, editors. Pollen–Pistil Interactions and Pollen Tube Growth. Rockville, MD: American Society of Plant Physiologists; 1994. pp. 45–61. [Google Scholar]
- Elleman CJ, Dickinson HG. Pollen–stigma interactions in Brassica. Structural reorganization in the pollen grains during hydration. J. Cell Sci. 1986;80:141–157. doi: 10.1242/jcs.80.1.141. [DOI] [PubMed] [Google Scholar]
- Elleman C, Willson CW, Sarker RH, Dickinson HG. Interaction between the pollen tube and stigmatic cell wall following pollination in Brassica oleracea. New Phytol. 1988;109:111–117. [Google Scholar]
- Franklin-Tong VE. Signaling in pollination. Curr. Opin. Plant Biol. 1999;2:490–495. doi: 10.1016/s1369-5266(99)00017-5. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, et al. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;102:1445–1450. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Patel RK, Tyagi AK, Jain M. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 2011;18:53–63. doi: 10.1093/dnares/dsq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude T, Dumas C. A membrane-like structure on the pollen wall surface in Brassica. Ann. Bot. 1984;54:821–825. [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J. Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 2000;124:1570–1581. doi: 10.1104/pp.124.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Okuda S, Mizukami A, Mori H, Sasaki N, Kurihara D, et al. Chemical visualization of an attractant peptide, LURE. Plant Cell Physiol. 2011;52:49–58. doi: 10.1093/pcp/pcq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development. 2007;134:3593–3601. doi: 10.1242/dev.011510. [DOI] [PubMed] [Google Scholar]
- Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl Acad. Sci. USA. 1998;95:382–387. doi: 10.1073/pnas.95.1.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi K, Sone M, Sakazono S, Osaka M, Masuko-Suzuki H, Matsuda T, et al. Time-lapse imaging of self- and cross-pollination in Brassica rapa. Ann. Bot. 2013;112:115–122. doi: 10.1093/aob/mct102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscock SJ, Allen AM. Diverse cell signalling pathways regulate pollen–stigma interactions: the search for consensus. New Phytol. 2008;179:286–317. doi: 10.1111/j.1469-8137.2008.02457.x. [DOI] [PubMed] [Google Scholar]
- Huang JC, Chang LC, Wang ML, Guo CL, Chung MC, Jauh GY. Identification and exploration of pollen tube small proteins encoded by pollination-induced transcripts. Plant Cell Physiol. 2013;52:1546–1559. doi: 10.1093/pcp/pcr095. [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Kopczak SD, Horejsi TF, Kihl BK, Pruitt RE. Identification of genes required for pollen–stigma recognition in Arabidopsis thaliana. Plant J. 1995;8:703–714. doi: 10.1046/j.1365-313x.1995.08050703.x. [DOI] [PubMed] [Google Scholar]
- Jiang LX, Yang SL, Xie LF. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell. 2005;17:584–596. doi: 10.1105/tpc.104.027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Preuss D. On your mark, get set, GROW! LePRK2–LAT52 interactions regulate pollen tube growth. Trends Plant Sci. 2003;8:97–99. doi: 10.1016/S1360-1385(03)00009-8. [DOI] [PubMed] [Google Scholar]
- Kakita M, Murase K, Iwano M, Matsumoto T, Watanabe M, Shiba H, et al. Two distinct forms of M locus protein kinase localize to the plasma membrane and interact directly with S locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell. 2007a;19:3961–3973. doi: 10.1105/tpc.106.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakita M, Shimosato H, Murase K, Isogai A, Takayama S. Direct interaction between S-locus receptor kinase and M-locus protein kinase involved in Brassica self-incompatibility signaling. Plant Biotechnol. 2007b;24:185–190. [Google Scholar]
- Kato M, Nagasaki-Takeuchi N, Ide Y, Maeshima M. An Arabidopsis hydrophilic Ca2+-binding protein with a PEVK-rich domain, PCaP2, is associated with the plasma membrane and interacts with calmodulin and phosphatidylinositol phosphates. Plant Cell Physiol. 2010;51:366–379. doi: 10.1093/pcp/pcq003. [DOI] [PubMed] [Google Scholar]
- Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM. Laser capture microdissection of cells from plant tissues. Plant Physiol. 2003;132:27–35. doi: 10.1104/pp.102.018127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki S, Sugimoto K. Control of the plant cell cycle by developmental and environmental cues. Plant Cell Physiol. 2012;53:953–964. doi: 10.1093/pcp/pcs070. [DOI] [PubMed] [Google Scholar]
- Kubo T, Fujita M, Takahashi H, Nakazono N, Tsutsumi N, Kurata N. Transcriptome analysis of developing ovules in rice isolated by laser microdissection. Plant Cell Physiol. 2013;54:750–765. doi: 10.1093/pcp/pct029. [DOI] [PubMed] [Google Scholar]
- Lehner A, Dardelle F, Soret-Morvan O, Lerouqe P, Driouch A, Mollet JC. Pectins in the cell wall of Arabidopsis thaliana pollen tube and pistil. Plant Signal Behav. 2010;5:1282–1285. doi: 10.4161/psb.5.10.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Xu W, Yang W, Kong Z, Xue Y. Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol. 2007;144:1797–1812. doi: 10.1104/pp.107.101600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord E. Adhesion and cell movement during pollination: cherchez la femme. Trends Plant Sci. 2000;5:368–373. doi: 10.1016/s1360-1385(00)01744-1. [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuko H, Endo M, Saito H, Hakozaki H, Park JI, Kawagishi-Kobayashi M, et al. Anther-specific genes, which expressed through microsporogenesis, are temporally and spatially regulated in model legume, Lotus japonicus. Genes Genet. Syst. 2006;81:57–62. doi: 10.1266/ggs.81.57. [DOI] [PubMed] [Google Scholar]
- Matsuo M, Hachisu R, Tabata S, Fukuzawa H, Obokata J. Transcriptome analysis of respiration-responsive genes in Chlamydomonas reinhardtii: mitochondrial retrograde signaling coordinates the genes for cell proliferation with energy-producing metabolism. Plant Cell Physiol. 2011;52:333–343. doi: 10.1093/pcp/pcq192. [DOI] [PubMed] [Google Scholar]
- Murase K, Shiba H, Iwano M, Che FS, Watanabe M, Isogai A, et al. A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science. 2004;303:1516–1519. doi: 10.1126/science.1093586. [DOI] [PubMed] [Google Scholar]
- Muschietti J, Eyal Y, McCormick S. Pollen tube localization implies a role in pollen–pistil interactions for the tomato receptor-like protein kinases LePRK1 and LePRK2. Plant Cell. 1998;10:319–330. doi: 10.1105/tpc.10.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazono M, Qui F, Brsuk LA, Schnable PS. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cell or vascular tissues of maize. Plant Cell. 2003;15:583–596. doi: 10.1105/tpc.008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault H, Amrani AE, Palanivelu P, Updegraff EP, Yu A, Renou JP, et al. GABA accumulation causes cell elongation defects and a decrease in expression of genes encoding secreted and cell wall-related proteins in Arabidopsis thaliana. Plant Cell Physiol. 2011;52:894–908. doi: 10.1093/pcp/pcr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Chong YT, Haasen KE, Aldea-Brydges MG, Stone SL, Goring DR. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell. 2009;21:2655–2671. doi: 10.1105/tpc.109.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, et al. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schopfer CR, Nasrallah ME, Nasrallah JB. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- Shimizu-Inatsugi R, Lihova J, Iwanaga H, Kudoh H, Marhold K, Savolainen O, et al. The allopolyploid Arabidopsis kamchatica originated from multiple individuals of Arabidopsis lyrata and Arabidopsis halleri. Mol. Ecol. 2009;18:4024–4028. doi: 10.1111/j.1365-294X.2009.04329.x. [DOI] [PubMed] [Google Scholar]
- Shimizu KK, Okada K. Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development. 2000;127:4511–4518. doi: 10.1242/dev.127.20.4511. [DOI] [PubMed] [Google Scholar]
- Shinya T, Motoyama N, Ikeda A, Wada M, Kamiya K, Hayafune M, et al. Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 2010;53:1696–1706. doi: 10.1093/pcp/pcs113. [DOI] [PubMed] [Google Scholar]
- Stephenson AG, Doughty J, Dixon S, Elleman C, Hiscock S, Dickinson HG. The male determinant of self-incompatibility in Brassica oleracea is located in the pollen coating. Plant J. 1997;12:1351–1359. [Google Scholar]
- Stone SL, Anderson EM, Mullen RT, Goring DR. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell. 2003;15:885–898. doi: 10.1105/tpc.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Arnoldo MA, Goring DR. A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science. 1999;286:1729–1731. doi: 10.1126/science.286.5445.1729. [DOI] [PubMed] [Google Scholar]
- Suwabe K, Suzuki G, Takahashi H, Shiono K, Endo M, Yano K, et al. Separated transcriptome of male gametophyte and tapetum in rice: validity of a laser microdissection (LM) microarray. Plant Cell Physiol. 2008;49:1407–1416. doi: 10.1093/pcp/pcn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwabe K, Suzuki G, Watanabe M. Achievement of genetics in plant reproduction research: the past decade for the coming decade. Genes Genet. Syst. 2010;85:297–310. doi: 10.1266/ggs.85.297. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Kai N, Hirose T, Fukui T, Nishio T, Takayama S, et al. Genomic organization of the S locus: identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa) Genetics. 1999;153:391–400. doi: 10.1093/genetics/153.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Edlund AF, Preuss D. Species specificity in pollen–pistil interactions. Annu. Rev. Genet. 2004;38:793–818. doi: 10.1146/annurev.genet.38.072902.092356. [DOI] [PubMed] [Google Scholar]
- Swanson R, Edlund AF, Preuss D. Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sex. Plant Reprod. 2005;18:163–171. [Google Scholar]
- Takahashi H, Kamakura H, Sato Y, Shiono K, Abiko T, Tsutsumi N, et al. A method for obtaining high quality RNA from paraffin sections of plant tissues by laser microdissection. J. Plant Res. 2010;123:807–813. doi: 10.1007/s10265-010-0319-4. [DOI] [PubMed] [Google Scholar]
- Takamatsu H, Takagi S. Actin-dependent chloroplast anchoring regulated by Ca2+-calmodulin in spinach mesophyll cell. Plant Cell Physiol. 2011;52:1973–1982. doi: 10.1093/pcp/pcr130. [DOI] [PubMed] [Google Scholar]
- Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature. 2000;403:913–916. doi: 10.1038/35002628. [DOI] [PubMed] [Google Scholar]
- Takayama S, Shiba H, Iwano M, Shimosato H, Che F-S, Kai N, et al. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl Acad. Sci. USA. 2000;97:1920–1925. doi: 10.1073/pnas.040556397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Shimosato H, Shiba H, Funato M, Che F-S, Watanabe M, et al. Direct ligand–receptor complex interaction controls Brassica self-incompatibility. Nature. 2001;413:534–538. doi: 10.1038/35097104. [DOI] [PubMed] [Google Scholar]
- Tang W, Ezcurra I, Muschietti J, McCormick S. A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell. 2002;14:2277–2287. doi: 10.1105/tpc.003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimatsu T, Kaiser P, Yew CL, Bachelier JB, Shimizu KK. Recent loss of self-incompatibility by degradation of the male component in allotetraploid Arabidopsis kamchatica. PLoS Genet. 2012;8:e1002838. doi: 10.1371/journal.pgen.1002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimatsu T, Suwabe K, Shimizu-Inatsugi R, Isokawa S, Pavidis P, Stadler T, et al. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature. 2010;464:1342–1346. doi: 10.1038/nature08927. [DOI] [PubMed] [Google Scholar]
- Tung CW, Dwyer KG, Nasrallah ME, Nasrallah JB. Genome-wide identification of genes expressed in Arabidopsis pistils specifically along the path of pollen tube growth. Plant Physiol. 2005;138:977–989. doi: 10.1104/pp.105.060558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff EP, Zhao F, Preuss D. The extracellular lipase EXL4 is required for efficient hydration of Arabidopsis pollen. Sex. Plant Reprod. 2009;22:197–204. doi: 10.1007/s00497-009-0104-5. [DOI] [PubMed] [Google Scholar]
- Wang H, Boavida LC, Ron M, McCormick S. Truncation of a protein disulfide isomerase, PDIL2-1, delays embryo sac maturation and disrupts pollen tube guidance in Arabidopsis thaliana. Plant Cell. 2008;20:3300–3311. doi: 10.1105/tpc.108.062919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Suwabe K, Suzuki G. Molecular genetics, physiology and biology of self-incompatibility in Brassicaceae. Proc. Jpn. Acad. Ser. B. 2012;88:519–535. doi: 10.2183/pjab.88.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Takasaki T, Toriyama K, Yamakawa S, Isogai A, Suzuki A, et al. A high degree of homology exists between the protein encoded by SLG and the S receptor domain encoded by SRK in self-incompatible Brassica campestris L. Plant Cell Physiol. 1994;35:1221–1229. doi: 10.1093/oxfordjournals.pcp.a078716. [DOI] [PubMed] [Google Scholar]
- Wolters-Arts M, Lush WM, Mariani C. Lipids are required for directional pollen-tube growth. Nature. 1998;392:818–821. doi: 10.1038/33929. [DOI] [PubMed] [Google Scholar]
- Wolters-Arts M, Van Der Weerd L, Van Aelst AC, Van Der Weerd J, Van As H, Mariani C. Water-conducting properties of lipids during pollen hydration. Plant Cell Environ. 2002;25:513–519. [Google Scholar]
- Xiao HX, Hao C, Ya LS, Fang W, Jun PM, Xin-Qi G, et al. Identification of genes specifically or preferentially expressed in maize silk reveals similarity and diversity in transcript abundance of different dry stigmas. BMC Genomics. 2012;13:294–311. doi: 10.1186/1471-2164-13-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D. Pollen–stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development. 1999;126:5431–5440. doi: 10.1242/dev.126.23.5431. [DOI] [PubMed] [Google Scholar]
- Zuberi MI, Dickinson HG. Pollen–stigma interaction in Brassica. III. Hydration of the pollen grains. J. Cell Sci. 1985;76:321–336. doi: 10.1242/jcs.76.1.321. [DOI] [PubMed] [Google Scholar]
- Zuhua H, Zhi-Yong W, Jianming L, Qun Z, Chris L, Pamela R, et al. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.