Abstract
Autotransporters are virulence-related proteins of Gram-negative bacteria that are secreted via an outer-membrane-based C-terminal extension, the translocator domain. This domain supposedly is sufficient for the transport of the N-terminal passenger domain across the outer membrane. We present here the crystal structure of the in vitro-folded translocator domain of the autotransporter NalP from Neisseria meningitidis, which reveals a 12-stranded β-barrel with a hydrophilic pore of 10 × 12.5 Å that is filled by an N-terminal α-helix. The domain has pore activity in vivo and in vitro. Our data are consistent with the model of passenger-domain transport through the hydrophilic channel within the β-barrel, and inconsistent with a model for transport through a central channel formed by an oligomer of translocator domains. However, the dimensions of the pore imply translocation of the secreted domain in an unfolded form. An alternative model, possibly covering the transport of folded domains, is that passenger-domain transport involves the Omp85 complex, the machinery required for membrane insertion of outer-membrane proteins, on which autotransporters are dependent.
Keywords: autotransporters, crystal structure, Neisseria meningitidis, outer-membrane protein, protein secretion
Introduction
The cell envelope of Gram-negative bacteria is composed of two membranes, the inner and the outer membrane, which are separated by the peptidoglycan-containing periplasm. Many different pathways for the secretion of proteins into the extracellular medium have evolved in these bacteria (Lee and Schneewind, 2001). Some of these pathways are one-step mechanisms, whereby the proteins are transported in a single step from the cytoplasm into the extracellular medium without a periplasmic intermediate. The molecular machinery involved in these secretion processes varies widely in complexity. For example, the type I secretion system consists of only three proteins, whereas the type III system consists of over 20 proteins (Lee and Schneewind, 2001). In other pathways, the transport of proteins across the cell envelope is conducted in two separate steps. Such proteins are first transported across the inner membrane, usually via the Sec system (Driessen et al, 1998), whereas the subsequent transport of the periplasmic intermediate across the outer membrane is mediated by a different molecular machinery. This machinery can be very complex, as is the case in the type II secretion system, which consists of at least 12 different proteins (Filloux et al, 1998). Perhaps the simplest protein secretion mechanism is the autotransporter pathway (Henderson et al, 1998). Proteins secreted via this pathway often contribute to the virulence of pathogenic bacteria, for example by mediating adhesion to host cells or by mediating actin-promoted bacterial mobility (Henderson and Nataro, 2001). Autotransporters are synthesised with an N-terminal signal sequence for transport across the inner membrane, presumably via the Sec system (Sijbrandi et al, 2003). Furthermore, they consist of a secreted functional domain and a C-terminal extension, the translocator domain, which mediates the translocation of the functional domain (the passenger) across the outer membrane (Pohlner et al, 1987). The translocator domain is a 25–30 kDa protein fragment, the β-core, with the general characteristics of an integral β-barrel outer-membrane protein and may additionally contain a ‘linker domain', which is sensitive to extracellular proteases and therefore cell-surface exposed (Klauser et al, 1993; Konieczny et al, 2001). The autotransporter mechanism is dependent on the Omp85-containing outer-membrane-protein assembly machinery (Voulhoux et al, 2003). The passage of the passenger through the outer membrane is presumably independent of ATP hydrolysis or a proton gradient (Klauser et al, 1993). Two distinct models have been proposed for the protein-translocation process. Initially, it was proposed that the passenger domain is transported in an unfolded state through a pore formed by a single translocator domain (Pohlner et al, 1987; Jose et al, 1995). This mechanism was supported by the observation that disulphide bond formation in artificial passengers prevented translocation, suggesting that the translocation channel is narrow (Klauser et al, 1992; Jose et al, 1996). More recently, however, another model for translocation has been proposed. The observation of limited translocation of disulphide-bond-containing Fv domains fused to the IgA protease translocator domain from Neisseria gonorrhoeae (Veiga et al, 1999) suggested that the channel should be wider than the pore within the β-barrel of a translocator domain. Consistently, the solubilised translocator domain of IgA protease showed multimeric ring-like structures with a central cavity of ∼2 nm in electron micrographs (Veiga et al, 2002). Hence, it was suggested that the passenger domains are translocated through the central channel of this multimeric structure.
Translocation of the passenger across the outer membrane places the N-terminal passenger domain at the cell surface. This domain may stay attached to the cell surface, anchored by the translocator domain. However, many passengers contain a protease subdomain, which may mediate release by autocatalytic processing (Henderson et al, 1998). Alternatively, other outer-membrane proteins may be involved in processing.
The membrane-embedded β-cores of the translocators are predicted to form β-barrels, similar to known structures of outer-membrane proteins (Loveless and Saier, 1997; Yen et al, 2002). However, these known structures contain an even number of β-strands with both termini located at the periplasmic side of the membrane (Schulz, 2002), a configuration that would be inconsistent with the final destination of the passenger domain at the cell surface. Therefore, several alternative structures have been hypothesised, such as uneven-stranded β-barrels (Klauser et al, 1993; Suhr et al, 1996; Veiga et al, 2002), amphipathic transmembrane helices (Klauser et al, 1993; Hendrixson et al, 1997) or a protein domain located within the β-barrel similar to the plug domains observed in TonB-dependent receptors (Koebnik et al, 2000; Konieczny et al, 2001). To gain insight into the molecular mechanism of the autotransporter secretion, we are studying the autotransporter NalP, a protease from Neisseria meningitidis involved in the intermolecular processing of other autotransporters at the bacterial cell surface (van Ulsen et al, 2003). Here, we present the crystal structure of its translocator domain.
Results
Expression, refolding and purification of the NalP translocator domain
On the basis of sequence alignment of 120 autotransporters (Yen et al, 2002), the translocator domain of NalP was estimated to consist of residues D777 to F1084. A DNA segment encoding this protein fragment, designated NalPβ, was cloned after PCR amplification in pET11a and expressed in Escherichia coli, where the polypeptide accumulated as non-native protein in inclusion bodies. A protein of similar size (32 kDa) was detected on immunoblots in membrane preparations of N. meningitidis expressing nalP, suggesting that a similar domain is generated in vivo after cleavage of the secreted passenger domain (Figure 1A). Like many other outer-membrane proteins (Dekker et al, 1995; Konieczny et al, 2001), this neisserial 32 kDa protein displayed heat modifiability, that is, the heat-denatured form had a lower electrophoretic mobility than the nondenatured form (Figure 1A). This property was useful to monitor the in vitro folding of recombinant NalPβ. Thus, NalPβ was refolded in vitro in the presence of detergent micelles (Figure 1B) and subsequently purified by anion-exchange chromatography.
Figure 1.
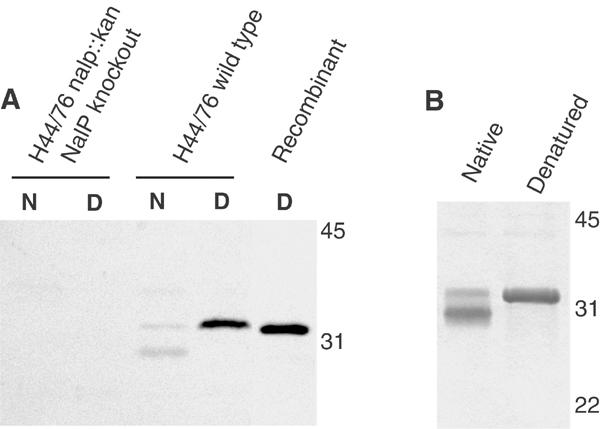
Biochemical characterisation of the NalP translocator domain. (A) Western blot analysis with NalPβ antiserum of membrane preparations of a N. meningitidis nalP knockout mutant and of the wild-type strain H44/76, incubated at room temperature (N) or 100°C (D) before electrophoresis, and of recombinant NalPβ produced in inclusion bodies in E. coli. (B) Heat modifiability of refolded NalPβ was analysed on Coomassie-stained SDS–PAGE gels. Samples were incubated at room temperature (native) or 100°C (denatured) prior to electrophoresis. The positions of molecular size markers (in kDa) are indicated at the right in both panels. These figures demonstrate that recombinant NalPβ is similar to the processed translocator domain of NalP that remains in the meningococcal outer membrane after secretion of the passenger domain.
Pore activity of in vitro-refolded NalPβ
Since NalPβ is predicted to form protein-conducting channels, the refolded protein was reconstituted in planar lipid bilayers to measure pore activity. The experiments revealed openings and closings of pores of two sizes with single-channel conductances of 0.15 nS (frequent) and 1.3 nS (infrequent), respectively (Figure 2). Although smaller than the 3.0 nS single-channel conductance measured in the case of the recombinant translocator domain of BrkA, an autotransporter of Bordetella pertussis (Shannon and Fernandez, 1999), the openings of 1.3 nS indicate that the recombinant NalPβ protein forms pores in the membrane that could correspond to the protein-translocation channels. Stepwise increase of the applied potential up to 250 mV did not result in closure of the observed channels, indicating that these channels are not voltage gated. Assuming that the protein forms a perfect cylinder, the pore dimensions calculated from the observed conductance steps would be 8.4 Å for the 1.3 nS channels and 2.4 Å for the 0.15 nS channels.
Figure 2.
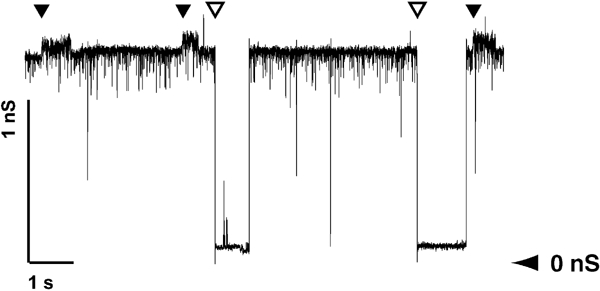
Recording of NalPβ pores formed in planar lipid bilayers at an applied potential of 100 mV. The recordings show conductance steps of 1.3 nS (open arrowhead) and 0.15 nS (filled arrowhead). The horizontal arrowhead shows the zero-conductance level.
Crystallisation and structure determination
The purified refolded NalPβ protein was crystallised in space groups P6122 and C2221. The structure was solved using multiple wavelength anomalous dispersion (MAD) of a selenium–methionine derivative NalPβ crystal of space group P6122. The structure was refined to 2.6 Å resolution (Figures 3 and 4) and used for molecular replacement in space group C2221 at 3.2 Å resolution (Table I). The two crystal forms yielded very similar structures (with r.m.s.d. of 1.2 Å for all backbone and Cβ atoms; Figure 5) with the distinct difference that loop L1 is pushed inwards in space group P6122 (over 6.6 Å for V837 Cα) due to a crystal contact. This difference in conformation may indicate that the extracellular loops are flexible, which is a general feature of outer-membrane proteins (Koebnik et al, 2000).
Figure 3.
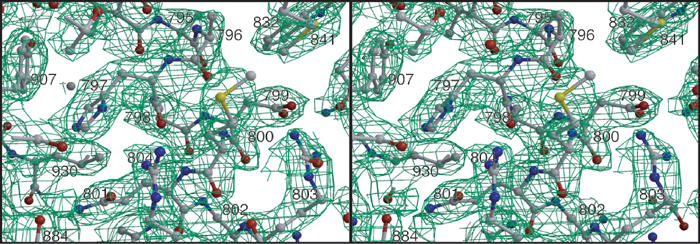
Stereo picture of the final 2m∣Fo∣-D∣Fc∣ electron density map of space group P6122 at a σ-level of 1.0 around the α-helix. The protein model is depicted in ball-and-stick representation.
Figure 4.
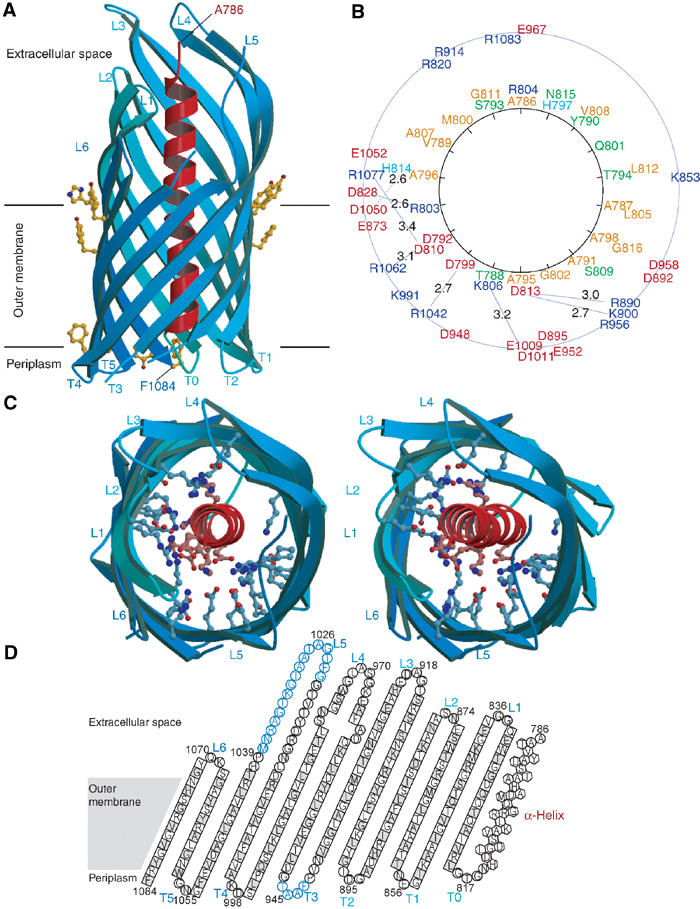
Crystal structure of NalPβ. (A) Side view of NalPβ in space group P6122 shows a 12-stranded β-barrel (blue ribbon representation) with a shear number of 14. The hydrophobic membrane-embedded region is flanked by aromatic residues (yellow ball-and-stick). The periplasmic side is characterised by short turns (T0–T5) and the extracellular side by longer loops (L1–L6) connecting the alternating β-strands. An α-helical ‘plug' (red ribbon representation) is connected to the barrel via T0 and positions the N-terminus of the translocator domain (Ala 786) at the extracellular side. (B) Schematic representation of the mixed character of the α-helix and its interactions with the β-barrel wall with the α-helical residues depicted on a helical wheel. Colour coding: positively charged residues in blue, negatively charged residues in red, hydrophilic residues in green and nonpolar residues in orange. The distances between charged groups of the helix and the barrel wall are indicated in Å. (C) Stereo top view of the NalPβ β-barrel in the same orientation as in (B). The barrel interior is highly hydrophilic due to the presence of many charged amino acids (ball-and-stick representation). (D) Topology model of NalPβ; residues pointing outwards from the β-barrel are indicated in grey. Amino acids in β-strands are indicated as squares, in α-helix as hexagons and in loops as circles. Amino acids not visible in the electron density of space group P6122 are indicated in blue.
Table 1.
Data collectiona and refinement statistics
| Data collection | |||
| Space group | P6122 | P6122 | C2221 |
| Beam line | ID14EH1 | BW7A | X11 |
| Derivative | Native | Se-Met | Se-Met |
| Wavelength (Å) | 0.934 | 0.9758 | 0.811 |
| 0.9762 | |||
| 0.93113 | |||
| Cell dimensions (Å) | |||
| a | 58.0 | 57.9 | 57.4 |
| b | 58.0 | 57.9 | 84.9 |
| c | 346.4 | 345.4 | 123.0 |
| Resolution (Å) | 2.6 | 3.1 | 3.2 |
| Observations | 97 916 | 99 681 | 45 309 |
| 74 903 | |||
| 98 377 | |||
| Unique reflections | 10 715 | 7173 | 5100 |
| 7108 | |||
| 6898 | |||
| Completeness (%) | 98.5 (88.7) | 98.2 (94.7) | 98.6 (94.8) |
| 97.8 (91.1) | |||
| 98.6 (94.0) | |||
| Rsym (%) | 6.4 (19.1) | 12.1 (36.6) | 15.0 (35.9) |
| 10.6 (33.5) | |||
| 9.8 (28.8) | |||
| Mean I/σ(I) | 24.0 (9.2) | 17.3 (4.5) | 9.4 (3.0) |
| 16.3 (4.3) | |||
| 18.8 (5.1) | |||
| Refinement | |||
| Protein atoms | 2060 | 1959 | |
| Detergent atoms | 26 | 10 | |
| Water atoms | 29 | 0 | |
| Resolution range (Å) | 30–2.6 | 30–3.2 | |
| R (%) | 22.8 | 21.5 | |
| Rfree (%) | 28.5 | 29.8 | |
| Mean B-factor (Å2) | 24.2 | 22.7 | |
| r.m.s.d. bonds (Å) | 0.020 | 0.017 | |
| r.m.s.d. angles (deg) | 1.80 | 1.92 | |
| Residues in Ramachandran plot (%) | |||
| Most favoured | 88.1 | 81.5 | |
| Additional allowed | 11.9 | 16.1 | |
| Generously allowed | 0 | 2.4 | |
| Disallowed | 0 | 0 | |
| For the seleno-methionine protein, three data sets were measured at peak (first line), inflection (second line) and remote (third line) wavelengths of the selenium. | |||
| aNumbers in parentheses correspond to values in the highest resolution shell. | |||
Figure 5.
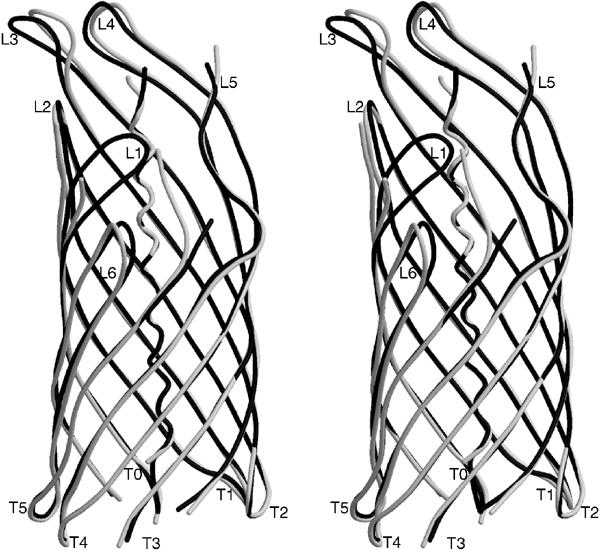
Stereo view of the differences in backbone conformation for space groups P6122 (black coil representation) and C2221 (white coil representation). In space group C2221, no interpretable electron density was observed for residues 816–817, 834–836, 915–917, 943–945, 972–973 and 1024–1036. The channel size in space group P6122 is reduced to 6.5 × 12.5 Å (10.5 × 12.5 Å in space group C2221), because loop L1 is pushed inwards due to a crystal packing contact.
Overall structure of NalPβ
The overall architecture of the NalPβ structure is characterised by a β-barrel, which forms a pore of 10 × 12.5 Å (in space group C2221) that is occupied by an N-terminal α-helix (Figure 4). The β-barrel (residues 819–1084) is a regular structure with 12 antiparallel β-strands, showing all features common to outer-membrane proteins (Schulz, 2002). Importantly, both termini of the barrel are located on the same, presumably periplasmic, side of the molecule that is characterised by short turns. The N-terminal β-strand is connected via a short periplasmic turn, T0, to the preceding α-helix (residues 786–814), which is positioned in the pore with the N-terminus on the extracellular side of the membrane. This results in a configuration that is compatible with the protein state after translocation of the passenger domain (Figure 6).
Figure 6.
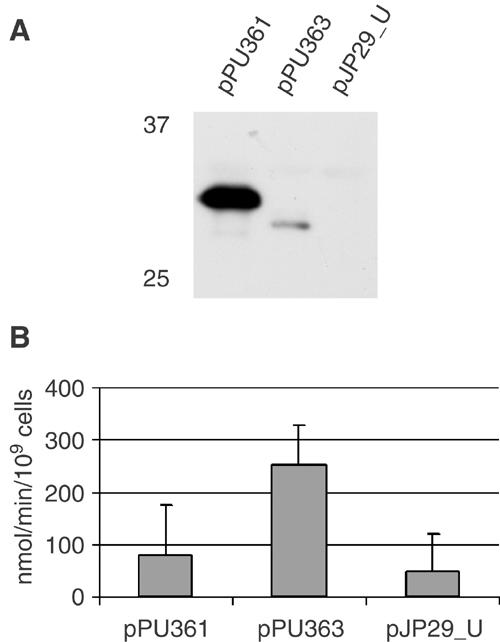
Uptake of the β-lactam antibiotic nitrocefin by E. coli CE1265 cells expressing NalPβ variants. CE1265 (pPU361) and CE1265 (pPU363) express NalPβ and NalPβΔhelix, respectively, both fused to the signal sequence of PhoE, and CE1265 (pJP29_U) expresses just the signal sequence of PhoE. (A) Western blot analysis with NalPβ antiserum of CE1265 cells expressing the NalPβ variants. Plasmids used are indicated above the lanes, and molecular weight markers on the left of the panel. Only the relevant part of the gel, showing that expression of NalPβΔhelix is much lower than that of NalPβ, is shown. (B) The rate of nitrocefin uptake mediated by the NalPβ variants in CE1265 is given in nmol/min/109 cells. Experiments were performed three times in triplicate, and the values given are the means and the standard deviation. Differences between CE1265 (pPU363) on the one hand and CE1265 (pPU361) and CE1265 (pJP29_U) on the other hand were statistically significant (P<0.005).
Position of the α-helix in the β-barrel channel
The interior of the β-barrel is highly hydrophilic with 20 charged residues pointing inwards. The charged residues form patches axially along the barrel wall (Figure 4B), reminiscent of the axially aligned charged residues that can be observed in the crystal structure of the outer-membrane channel TolC of the type I secretion system (Koronakis et al, 2000). The α-helix, with charges predominantly clustered on one side, interacts with the barrel wall through seven salt bridges as well as through 16 hydrogen bonds and numerous van der Waals contacts. Although the helix seems to obstruct the pore in the crystal structure, there is a small water-filled channel that runs from the periplasm to the medium. The planar lipid bilayer experiments suggest that ions could leak alongside the α-helix resulting in the observed 0.15 nS conductance steps. Displacement of the α-helix from the pore, perhaps facilitated by the detergent and the high salt concentration used in the planar lipid bilayer experiments, would then result in an open channel that may correspond to the observed 1.3 nS conductance steps (Figure 2).
NalPβ pore activity in vivo
To test the pore activity of NalPβ in vivo as well as the influence of the α-helix on this activity, two variants of the NalPβ were constructed. Plasmids pPU361 and pPU363 encode hybrid proteins consisting of the signal sequence of the E. coli outer-membrane porin PhoE fused to the NalP translocator domain starting from either D777 (NalPβ) or G818 (NalPβΔhelix), respectively. As a negative control, a plasmid was constructed that only encodes the signal sequence of PhoE (pJP29_U). The hybrid genes were under the control of the phoE promoter, which is constitutively expressed in the phoR mutant E. coli strain CE1265. This strain does not produce the major porins OmpF, OmpC and PhoE and could therefore be used to assess the pore-forming activity of the two NalPβ variants. Both proteins were correctly targeted to the outer membrane (results not shown), but the expression level of NalPβΔhelix was considerably lower than that of NalPβ with α-helix (Figure 6A).
The formation of pores in the outer membrane was determined in an antibiotic sensitivity assay, in which the zone of growth inhibition around a disc containing the antibiotic tested is a measure for the diffusion of the antibiotic through the pores. Expression of NalPβ resulted in an increased sensitivity to several antibiotics (Table II), demonstrating that NalPβ forms pores in vivo. Moreover, despite its drastically lower expression level (Figure 6A), NalPβΔhelix caused an even higher antibiotic sensitivity, indicating that the removal of the α-helix increases the pore activity. Additionally, the rate of uptake of the β-lactam antibiotic nitrocefin was measured (Figure 6B). This antibiotic was taken up significantly faster in cells expressing NalPβΔhelix, despite the low expression level of this protein. The difference in nitrocefin uptake between cells expressing NalPβ and the negative control was not significant. Taken together, these results clearly indicate that NalPβ can function as a pore in vivo and that deletion of the central α-helix enhances the pore activity.
Table 2.
Antibiotic sensitivity of E. coli CE1265 expressing the signal sequence of PhoE (pJP29_U), NalPβ (pPU361) or NalPβΔhelix (pPU363)
| Antibiotic | Concentration | MW | pJP29_Ua | pPU361 | pPU363 |
|---|---|---|---|---|---|
| Cycloserin | 1 mg/ml | 102 | 0 | 0 | 0 |
| Nalidixin | 1 mg/ml | 254 | 3 | 6 | 8 |
| Ampicillin | 1.5 mg/ml | 371 | 0 | 3 | 7 |
| Tetracycline | 2 mg/ml | 444 | 7 | 8 | 9 |
| Rifampicin | 0.3 mg/ml | 823 | 1 | 6 | 12 |
| Bacitracine | 690 IE/ml | 1421 | 0 | 5 | 8 |
| Vancomycin | 5 mg/ml | 1485 | 1 | 8 | 11 |
| SDS | 1 mg/ml | 288 | 0 | 0 | 0 |
| aThe growth inhibition zone, caused by the sensitivity to the antibiotic, was measured in mm from the rim of the disc. Values are the mean of three measurements of independent transformants. | |||||
Discussion
We present the crystal structure of the translocator, or β-domain of the neisserial autotransporter NalP, which inserts into the outer membrane to mediate translocation of the N-terminal passenger domain. The structure reveals a 12-stranded β-barrel in a conformation that is classical for outer-membrane proteins. A striking and novel feature is an α-helix that runs through the hydrophilic pore formed by the β-barrel. This helix, which in the primary sequence is located N-terminally to the barrel domain, positions the N-terminus of the translocator domain at the extracellular side of the outer membrane, a configuration consistent with the final stage of translocation, when the passenger domain has been secreted into the extracellular medium. Moreover, NalPβ showed pore activity in vivo and in vitro, and removing the α-helix enhanced this activity, suggesting that the helix functions in blocking the barrel pore.
Sequence alignment of autotransporter-translocator domains (Yen et al, 2002) suggests that their general structural features are conserved. Furthermore, as in NalP, secondary structure predictions suggest the presence of a long α-helix directly preceding the β-core in many other autotransporters, including AidaI of E. coli, BrkA of B. pertussis, Hap of Hemophilus influenzae and IgA protease and App of N. meningitidis, even though the sequence conservation in this segment is low (data not shown). Consequently, the NalPβ crystal structure may be used to model autotransporter secretion. The models for secretion that have been discussed in the literature are the classical model, in which the passenger domain is translocated through the pore within the β-barrel (Pohlner et al, 1987; Klauser et al, 1992; Jose et al, 1996), and the more recently proposed model, in which translocation occurs via a central pore formed by a multimer of translocator domains (Veiga et al, 2002).
Multimer model
Translocation through a central multimeric pore was proposed, based upon two observations (Veiga et al, 2002): (i) translocation, albeit at a low efficiency, of a folded, disulphide-bond-containing artificial passenger, that is, a single-chain antibody (scFv), by the translocator domain of IgA protease of N. gonorrhoeae (Veiga et al, 1999); (ii) the ring-like structures in electron micrographs of the IgA protease translocator domain after its overproduction and purification from E. coli outer membranes (Veiga et al, 2002). However, the position of the α-helix in the in vitro-folded NalPβ domain is inconsistent with the translocation of the passenger through a central pore of a multimer. Attempts to trap NalPβ oligomers by chemical crosslinking after overexpression in N. meningitidis failed, suggesting that NalPβ does not form oligomers in vivo either (unpublished results). In addition, we observed that the monomers of NalPβ in the crystal are arranged in nonphysiological layers of alternating upward and downward oriented molecules. Finally, the membrane interface of NalPβ is hydrophobic, and thus a central pore in a multimer would also be hydrophobic and probably filled with lipids from the outer membrane.
Multimerisation, possibly mediated through interactions in the passenger domains, may play, however, an important role at a different stage in the autotransporter secretion process. For example, it has been shown that folding of passenger domains lacking an intramolecular folding domain is restored by coexpression of an autotransporter with this folding domain as a passenger (Ohnishi et al, 1994; Oliver et al, 2003). Furthermore, the autoproteolytic cleavage of the passenger of Hap from H. influenzae was shown to occur intermolecularly (Fink et al, 2001). Similarly, NalP of N. meningitidis has been demonstrated to process other neisserial autotransporters, including IgA protease and App (van Ulsen et al, 2003). Thus, the observed multimers of the translocator domain of IgA protease may be functional in processing rather than outer-membrane translocation.
Translocation through the β-barrel pore
The monomeric nature of the NalP translocator domain suggests that a single translocator domain is sufficient for translocation. After reconstitution in planar lipid bilayers, in vitro-folded NalPβ showed two sorts of pore activities. Although calculations of the pore dimensions based on single-channel conductance should be interpreted with caution (Van Gelder et al, 2000), they were consistent with the size of the pores observed in the crystal structure. Furthermore, the in vivo data showed that NalPβ facilitated the diffusion of large antibiotics, such as vancomycin and bacitracine. Therefore, at least a proportion of the recombinant NalPβ channels was in an open conformation. However, deletion of the α-helix enhanced the diffusion rate, indicating that the helix can plug the channel formed by NalPβ. Thus, the structural and biochemical data obtained for NalPβ show the presence of a pore within the β-barrel that can be obstructed by the α-helix and argue in favour of a translocation mechanism involving only one translocator domain. Secretion through the pore of a single β-barrel may take place in two distinct ways, that is, with the N-terminus first (the ‘threading' model) or with the C-terminus of the passenger first, which then should be facilitated by the presence of a hairpin loop (the ‘hairpin' model). In both models, the passenger domain folds extracellularly and must remain unfolded at the periplasmic side of the membrane, possibly with the help of chaperones (Henderson et al, 1998; Purdy et al, 2002).
Hairpin model
In the classical hairpin model (Pohlner et al, 1987; Jose et al, 1995; Henderson et al, 1998), the hairpin is a temporary fold, located at the translocator domain/passenger interface and formed in the pore of the translocator domain, possibly during the assembly of the β-barrel. This hairpin could possibly comprise the conserved sequence stretch called the folding domain that is found in many autotransporters (Oliver et al, 2003). The extracellular loop of the hairpin may initiate folding. The only solved structure of a secreted autotransporter passenger domain is that of pertactin P.69 from B. pertussis (Emsley et al, 1996), which shows a parallel β-helical structure. The ongoing folding of this β-roll structure might pull the passenger domain through the pore and provide the energy for translocation (Klauser et al, 1993). Interestingly, the program BetaWrap (Bradley et al, 2001) predicted a high propensity for a parallel β-helical structure in the majority of passenger domains tested (i.e., 72% P<0.01, 91% P<0.05, of 119 autotransporters aligned Yen et al (2002)).
The pore within the β-barrel is relatively narrow (10 × 12.5 Å) and would only just allow for the presence of two extended polypeptide chains simultaneously, while the α-helix can only be formed after the translocation of the passenger is completed. It is to be expected that the side chains of barrel-wall residues that form hydrogen bonds with the α-helix in the structure do not point inwards when the helix is not yet formed: they are either flexible or fold backwards and form hydrogen bonds with other residues from the barrel wall. This would result in a slightly larger pore that could accommodate the two extended strands passing through the narrow channel, although still with a narrow fit. Nevertheless, the translocation channel seems to be too narrow to allow for the passage of domains that contain periplasmically formed disulphide bridges, which appear to be present not only in some artificial passengers (Veiga et al, 1999), but even in natural passengers (Brandon and Goldberg, 2001). Furthermore, this model implies that folding and translocation of the passenger domains are coupled processes. However, it has been shown that the deletion of a folding domain in the passenger of BrkA of B. pertussis did not abrogate translocation and resulted in the exposure of a protease-sensitive unfolded passenger at the cell surface (Oliver et al, 2003).
Threading model
In the threading model, translocation starts with the N-terminus, and the β-barrel pore needs to accommodate only one extended strand at the time, for which the pore observed in the crystal structure is wide enough. However, in this model, the N-terminus of the passenger has to be targeted to the pore. Since artificial passengers fused to translocator domains can be secreted (Jose et al, 1996; Maurer et al, 1997), a specific targeting signal seems lacking. Furthermore, the folding domain identified in many autotransporters (Oliver et al, 2003) would be the last segment of the passenger to appear at the cell surface, and hence would not act as a folding initiator in the translocation process. Thus, although the threading model does solve the steric exclusion problem associated with the classical hairpin model, it raises its own questions with respect to targeting and energy source.
An alternative translocation model
Because of the intrinsic problems associated with each of the translocation models described above, there is a strong need for alternative views. An alternative model, we would like to propose here for consideration, implicates an important role for the Omp85 complex, the machinery required for the assembly of integral outer-membrane proteins. Recently, it was demonstrated that also autotransporter secretion depends on the Omp85 complex (Voulhoux et al, 2003). Integral outer-membrane proteins contain relatively large hydrophilic extracellular loops, which together may encompass over 50% of the total protein. The Omp85 machinery might assist in the membrane insertion of their barrel domain by forming a pore that allows for the passage of the hydrophilic loops across the membrane. Since the translocator domain of autotransporters is an integral outer-membrane protein, this domain could function primarily as a recognition signal for the Omp85 complex, and the passenger domain could be transported across the outer membrane through the Omp85 pore, rather than through a pore formed by the translocator domain. It is worthwhile to point out the similarity of this model to the two-partner secretion systems (Jacob-Dubuisson et al, 2001). In the two-partner secretion systems, the secreted proteins, such as the filamentous haemagglutinin of B. pertussis, are transported across the outer membrane via an outer-membrane protein with the general designation TpsB. Importantly, at least some of the proteins secreted via this system possess a similar β-helix structure as the passenger domains of the autotransporters (Kajava et al, 2001), whereas the TpsB proteins show similarity to Omp85 (Yen et al, 2002). Thus, when the TpsB proteins can transport proteins that can form β-helical structure to the cell surface, one can expect Omp85 to have the same capacity. Obviously, if this model is correct, the designation ‘autotransporter' is not appropriate anymore.
In conclusion, we present the first structure of a translocator domain of an autotransporter involved in the secretion of its attached passenger domain. Although this structure did not provide the definitive answer to the translocation mechanism involved, it clearly revealed problems with existing models and urged the necessity of new hypotheses. Thus, the structure serves as a starting point for further research on the outer-membrane translocation mechanism of this secretion system, which forms the largest group of secreted proteins of Gram-negative bacteria.
Materials and methods
Protein expression and purification
The DNA fragment encoding the translocator domain of NalP (residues D777 to F1084) was obtained from the nalP gene of N. meningitidis strain H44/76 (GenBank AY150284) by PCR using primers 5′-gcaattccatatggacggtgtacgcatcttcaacaatct-3′ and 5′-caagatctcagaaccggtagcctacgccga-3′. The NdeI and BglII sites included in the primers (underlined) facilitated cloning of the fragment into expression vector pET11a (Invitrogen), yielding plasmid pPU320, placing the coding sequence under the control of the T7 promoter. The domain, which does not contain an affinity tag, was overproduced in inclusion bodies in E. coli BL21(DE3) (Invitrogen). The cells were lysed by sonication, and the inclusion bodies were purified as described (Dekker et al, 1995), dissolved to a concentration of 10 mg/ml in 8 M urea, 0.1 M glycine and 20 mM Tris–HCl (pH 7.6) followed by 1 h ultracentrifugation at 200 000 g to remove membrane fragments. Purified solubilised inclusion bodies of the cloned translocator domain were used to obtain polyclonal antiserum (Eurogentec).
SDS–PAGE and Western blotting
Membranes were isolated as described (van Ulsen et al, 2001) and proteins were separated on SDS–PAGE with reduced amounts of SDS to detect heat modifiability (Dekker et al, 1995). Samples were dissolved in SDS–PAGE loading buffer (0.062 M Tris–HCl, pH 6.8, 10% (v/v) glycerol, 0.01% (w/v) bromophenol blue, 2.5% (v/v) β-mercaptoethanol and 2% (w/v) SDS) and incubated for 10 min at room temperature or at 100°C. Western blotting and immunodetection were performed as described (van Ulsen et al, 2001).
Refolding of NalPβ
Refolding of the recombinant translocator domain of NalP was initiated by rapid 10-fold dilution of the protein from the stock solution in urea into a buffer containing 0.5% (w/v) N-dodecyl-N,N-dimethyl-1-ammonio-3-propanesulphonate (SB-12), 20 mM Tris–HCl (pH 8.0) and 1 M NaCl. Refolding was carried out for 72 h at 37°C and monitored by SDS–PAGE. Se-Met NalPβ was produced in E. coli strain B834(DE3) (methionine-auxotroph, Novagen), which was grown in minimal medium (17 nM (NH4)2Fe(SO4)2, 0.2 mM MgSO4, 30 mM KH2PO4, 30 mM K2HPO4, 0.5% glucose) supplemented with 0.3 mM seleno-L-methionine (Acros). Refolded native and Se-Met NalPβ were dialysed against buffer A (20 mM Tris–HCl (pH 9.0), 0.2% (w/v) SB-12) and purified by anion-exchange chromatography on a MonoQ column (Amersham Pharmacia). The column was washed successively with buffer A and buffer B (20 mM Tris–HCl (pH 9.0), 0.06% (w/v) n–decylpentaoxyethylene (C10E5)) and eluted with buffer B supplemented with 0.5 M NaCl. The protein was subsequently dialysed against buffer B and concentrated on a centricon concentrator with 10 kDa cutoff (Amicon) and finally dialysed against 10 mM Tris–HCl (pH 7.5) and 0.06% (w/v) C10E5.
Planar lipid bilayer experiments
Planar black lipid bilayers were formed from 1% (v/v) L-α-phosphatidylcholine (Sigma: P-5638) in hexane/chloroform (9:1 v/v) across a pierced (∅ 150 μm) Teflon membrane pretreated with a solution of hexadecane/hexane (1:50 v/v). Bilayer formation and conductance measurements were performed as described (Van Gelder et al, 2000), with the exception that the analogous output voltage signal was converted by a Powerlab/4SP (ADInstruments) and recorded on a computer using the Chart4 program (ADInstruments). Purified refolded NalPβ, dialysed to 1% (v/v) octyl-polyoxyethylene (Alexis), was added to a buffer of 1 M KCl, 1 mM CaCl2 and 10 mM Tris–HCl (pH 7.4), and insertion into the membrane was monitored by measuring the current at an applied voltage of 100 mV. Both negative and positive potentials were tested and, to test for possible voltage gating, the potential was increased in steps of 5 mV up to 200–250 mV or to the breaking of the membrane. Conductance was calculated from measurements at a transmembrane potential of 100 mV. The pore size was calculated by applying.
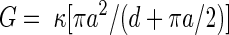 |
where G is the conductance, κ the bulk conductance (11.3 S/m), a the pore radius and d the membrane thickness (Van Gelder et al, 2000).
Crystallisation and X-ray analysis
Crystals of space group P6122 (cell dimensions a=b=58.0 Å, c=346.4 Å) were grown by the hanging-drop vapour diffusion method using equal parts of protein solution (17 mg/ml native NalPβ or 8.5 mg/ml Se-Met NalPβ, 10 mM Tris–HCl (pH 7.5), 0.06% (w/v) C10E5 and 0.5% (w/v) heptyl-β-glucopyranoside and, only in the case of Se-Met NalPβ, 10 mM dithiothreitol (DTT)) and 9% (w/v) polyethylene glycol 1000 (PEG1k), 200 mM lithium sulphate and 100 mM sodium citrate buffer (pH 4.0) at 28°C. P6122 crystals were stabilised in 200 mM lithium sulphate, 100 mM sodium citrate buffer (pH 4.0), 20% (w/v) PEG1k, 20% (v/v) 2-methyl-2,4-pentanediol (MPD) and 0.06% (w/v) C10E5 (and 10 mM DTT for the Se-Met NalPβ crystals) and rapidly frozen at 100 K for data collection. Crystals of space group C2221 (cell dimensions a=57.4, b=84.9, c=123.0 Å) were grown using 8.5 mg/ml Se-Met NalPβ (in 10 mM Tris–HCl (pH 7.5), 0.06% (w/v) C10E5 and 0.5% (w/v) heptyl-β-glucopyranoside) and 88 mM Tris–HCl (pH 7.0), 9% (w/v) polyethylene glycol 2000 monomethyl ether (PEG2k mme) and 18% (v/v) MPD at 28°C. C2221 crystals were stabilised in 88 mM Tris–HCl (pH 7.0), 20% (w/v) PEG2k mme, 25% (v/v) MPD, 0.06% (w/v) C10E5 and 10 mM DTT and flash-frozen at 100 K. Diffraction data were collected at 100 K at the ESRF and DESY synchrotrons and processed using DENZO and SCALEPACK (Otwinowski and Minor, 1997). MAD data of P6122 crystals were collected at the selenium edge (0.9758 Å), inflection (0.9762 Å) and a remote wavelength (0.9311 Å). Data were collected to 2.6 Å (P6122 native), 3.1 Å (P6122 Se-Met) and 3.2 Å (C2221 Se-Met) resolution with completeness in all cases higher than 97.8% and Rmerge from 6.1 to 15.0%. Data collection statistics and refinement statistics are found in Table I.
Structure determination and refinement
The NalPβ structure was solved using the MAD phases of the P6122 selenium-derivative crystal. Five of the expected six selenium sites were found, and initial phases were calculated with the program SOLVE (Terwilliger, 2002). Solvent flattening and phase extension to 2.6 Å was performed in CNS (Brunger et al, 1998). These data were used for automatic model building by RESOLVE (Terwilliger, 2002), which built 55% of the main chain. The model was manually corrected and supplemented to yield a final model that contained 279 out of 309 amino acids, 29 water molecules, two sulphate ions and one C10E5 detergent molecule and had good geometry (Table I). Refinement in refmac (CCP4, 1994) using one TLS group yielded a crystallographic R-factor of 22.8% (Rfree 28.5%). Extra electron density was seen for additional detergent molecules, but the density was not good enough to insert whole detergent molecules. The structure of the NalPβ in space group C2221 was solved by molecular replacement in CNS, using the model from the P6122 space group and further refined in refmac using one TLS group, with a final R-factor of 21.5% (Rfree 29.8%).
In vivo pore measurements
Primers 5′-cgtacgtacaccagcgtctggctgtcactca-3′ and 5′- actgcagttacgggatcccggcctgtacagatgcagatg ccaca-3′ were used in a PCR with plasmid pJP29 (Bosch et al, 1986) as the template to obtain a DNA fragment that encodes the PhoE signal sequence (ssPhoE) under control of the phoE promoter. The second primer introduced a BamHI site (underlined) followed by a stop codon (in italics) and a PstI site (underlined) in the DNA at the position of the signal sequence cleavage site in the protein. This PCR fragment was cut with PstI and SnaBI and used to replace the PstI–SnaBI fragment in pJP29, yielding pJP29_U. Due to the introduced stop codon, only the signal sequence of PhoE is expressed from this plasmid. DNA fragments encoding NalPβ starting from D777 and NalPβΔhelix starting form G818 were obtained by PCR, using chromosomal DNA of N. meningitidis strain H44/76 as the template and primers 5′-ggatccccgacggtgtacgcatcttcaacagtct-3′ and 5′-caagatctcagaaccggtagcctacgccga-3′ and 5′-ggatcccgggtctgcgcgtcatcgcgcaa-3′ and 5′-caagatctcagaaccggtagcctacgccga-3′, respectively. The BamHI and BglII sites (underlined) enabled cloning into pJP29_U, resulting in plasmids pPU361 encoding a fusion protein of ssPhoE and NalPβ, and pPU363 encoding a fusion protein of ssPhoE and NalPβΔhelix. Plasmids were used to transform E. coli strain CE1265 (Korteland et al, 1985) in which the major porins are not expressed due to phoE and ompR mutations. In addition, the strain carries the phoR18 mutation, resulting in constitutive expression of the phoE promoter. Sensitivity of bacteria to antibiotics was determined as described by Van Gelder et al (1997). Briefly, a lawn of bacteria was plated on LB-agar plates by pouring 3.5 ml of inoculated soft-agar. Filter discs (∅ 6 mm) were spotted with 5 μl of antibiotic solutions and placed on the top-agar layer, after which the plates were incubated for 18 h at 37°C. Antibiotic sensitivity was assessed by measuring the ring of growth inhibition around the discs, starting from their edge. The experiment was performed in triplicate using independent transformants of CE1265 with the respective plasmids.
The rate of permeation of nitrocefin through the outer membrane was assessed from the rate of its hydrolysis by periplasmic β-lactamase in intact cells. CE1265 cells containing pBR322 encoding β-lactamase and either pPU361, pPU363 or pJP29_U were grown in LB medium to mid-log phase, after which the cells were diluted 1:1 with fresh LB and the OD600 was measured. Then, 100 μl of 0.5 mg/ml nitrocefin in dimethyl sulphoxide was added to 1 ml of cells, and the change in OD486 was followed for 10 s. To correct for leakage of β-lactamase into the medium, cell cultures were filtered through 0.2 μm filters (Schleicher and Schuell), after which nitrocefin hydrolysis was measured in 1 ml of the filtrate. The uptake of nitrocefin is expressed as nmol nitrocefin hydrolysed/min/109 cells, whereby a difference in OD486 of 1.0 corresponds to the hydrolysis of 68.1 nmol nitrocefin. The values given are the means of nine cultures for each plasmid, measured in groups of three on different days, whereby the value for each culture is the mean of three measurements. The significance of the differences in the values obtained was tested using a one-tailed Student's t-test.
Acknowledgments
We thank Loek van Alphen, Peter van der Ley and Lucy Vandeputte-Rutten for stimulating discussions, Ria Tommassen-van Boxtel for technical assistance and the beamline scientists at the European Synchrotron Radiation Facility (ESRF), Grenoble, France and at the European Molecular Biology Laboratory (EMBL), Hamburg, Germany for assistance during data collection. This research was supported by the ‘European Community-Access to Research Infrastructure Action of the Improving Human Potential Program', and the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW). Coordinates and structure factors are deposited to the Protein Data Bank with accession Nos 1 uyn and 1 uyo.
References
- Bosch D, Leunissen J, Verbakel J, de Jong M, van Erp H, Tommassen J (1986) Periplasmic accumulation of truncated forms of outer-membrane PhoE protein of Escherichia coli K-12. J Mol Biol 189: 449–455 [DOI] [PubMed] [Google Scholar]
- Bradley P, Cowen L, Menke M, King J, Berger B (2001) BETAWRAP: successful prediction of parallel β-helices from primary sequence reveals an association with many microbial pathogens. Proc Natl Acad Sci USA 98: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon LB, Goldberg MB (2001) Periplasmic transit and disulfidebond formation of the autotransported Shigella protein IcsA. J Bacteriol 183: 951–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Dekker N, Merck K, Tommassen J, Verheij HM (1995) In vitro folding of Escherichia coli outer-membrane phospholipase A. Eur J Biochem 232: 214–219 [DOI] [PubMed] [Google Scholar]
- Driessen AJ, Fekkes P, van der Wolk JP (1998) The Sec system. Curr Opin Microbiol 1: 216–222 [DOI] [PubMed] [Google Scholar]
- Emsley P, Charles IG, Fairweather NF, Isaacs NW (1996) Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature 381: 90–92, [DOI] [PubMed] [Google Scholar]
- Filloux A, Michel G, Bally M (1998) GSP-dependent protein secretion in Gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol Rev 22: 177–198 [DOI] [PubMed] [Google Scholar]
- Fink DL, Cope LD, Hansen EJ, StGeme JW III (2001) The Hemophilus influenzae Hap autotransporter is a chymotrypsin clan serine protease and undergoes autoproteolysis via an intermolecular mechanism. J Biol Chem 276: 39492–39500 [DOI] [PubMed] [Google Scholar]
- Henderson IR, Nataro JP (2001) Virulence functions of autotransporter proteins. Infect Immun 69: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Nataro JP (1998) The great escape: structure and function of the autotransporter proteins. Trends Microbiol 6: 370–378 [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, de la Morena ML, Stathopoulos C, StGeme JW III (1997) Structural determinants of processing and secretion of the Haemophilus influenzae hap protein. Mol Microbiol 26: 505–518 [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Locht C, Antoine R (2001) Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol Microbiol 40: 306–313 [DOI] [PubMed] [Google Scholar]
- Jose J, Jähnig F, Meyer TF (1995) Common structural features of IgA1 protease-like outer membrane protein autotransporters. Mol Microbiol 18: 378–380 [DOI] [PubMed] [Google Scholar]
- Jose J, Kramer J, Klauser T, Pohlner J, Meyer TF (1996) Absence of periplasmic DsbA oxidoreductase facilitates export of cysteine-containing passenger proteins to the Escherichia coli cell surface via the Iga beta autotransporter pathway. Gene 178: 107–110 [DOI] [PubMed] [Google Scholar]
- Kajava AV, Chen N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC (2001) Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol 42: 279–292 [DOI] [PubMed] [Google Scholar]
- Klauser T, Krämer J, Otzelberger K, Pohlner J, Meyer TF (1993) Characterization of the Neisseria Igaβ-core. The essential unit for outer membrane targeting and extracellular protein secretion. J Mol Biol 234: 579–593 [DOI] [PubMed] [Google Scholar]
- Klauser T, Pohlner J, Meyer TF (1992) Selective extracellular release of cholera toxin B subunit by Escherichia coli: dissection of Neisseria Igaβ-mediated outer membrane transport. EMBO J 11: 2327–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik R, Locher KP, Van Gelder P (2000) Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol 37: 239–253 [DOI] [PubMed] [Google Scholar]
- Konieczny MPJ, Benz I, Hollinderbäumer B, Beinke C, Niederweis M, Schmidt MA (2001) Modular organization of the AIDA autotransporter translocator: the N-terminal β1-domain is surface-exposed and stabilizes the transmembrane β2-domain. Antonie van Leeuwenhoek 80: 19–34 [DOI] [PubMed] [Google Scholar]
- Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C (2000) Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405: 914–919 [DOI] [PubMed] [Google Scholar]
- Korteland J, Overbeeke N, de Graaff P, Overduin P, Lugtenberg B (1985) Role of the Arg158 residue of the outer membrane PhoE pore protein of Escherichia coli K12 in bacteriophage TC45 recognition and in channel characteristics. Eur J Biochem 152: 691–697 [DOI] [PubMed] [Google Scholar]
- Lee VT, Schneewind O (2001) Protein secretion and the pathogenesis of bacterial infections. Genes Dev 15: 1725–1752 [DOI] [PubMed] [Google Scholar]
- Loveless BJ, Saier MH Jr (1997) A novel family of channelforming, autotransporting, bacterial virulence factors. Mol Membr Biol 14: 113–123 [DOI] [PubMed] [Google Scholar]
- Maurer J, Jose J, Meyer TF (1997) Autodisplay: one-component system for efficient surface display and release of soluble recombinant proteins from Escherichia coli. J Bacteriol 179: 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Nishiyama M, Horinouchi S, Beppu T (1994) Involvement of the COOH-terminal pro-sequence of Serratia marcescens serine protease in the folding of the mature enzyme. J Biol Chem 269: 32800–32806 [PubMed] [Google Scholar]
- Oliver DC, Huang G, Nodel E, Pleasance S, Fernandez RC (2003) A conserved region within the Bordetella pertussis autotransporter BrkA is necessary for folding of its passenger domain. Mol Microbiol 47: 1367–1383 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pohlner J, Halter R, Beyreuther K, Meyer TF (1987) Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325: 458–462 [DOI] [PubMed] [Google Scholar]
- Purdy GE, Hong M, Payne SM (2002) Shigella flexneri DegP facilitates IcsA surface expression and is required for efficient intercellular spread. Infect Immun 70: 6355–6364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz GE (2002) The structure of bacterial outer membrane proteins. Biochim Biophys Acta 1565: 308–317 [DOI] [PubMed] [Google Scholar]
- Shannon JL, Fernandez RC (1999) The C-terminal domain of the Bordetella pertussis autotransporter BrkA forms a pore in lipid bilayer membranes. J Bacteriol 181: 5838–5842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijbrandi R, Urbanus ML, ten Hagen-Jongman CM, Bernstein HD, Oudega B, Otto BR, Luirink J (2003) SRP-mediated targeting and Sec-dependent translocation of an extracellular Escherichia coli protein. J Biol Chem 278: 4654–4659 [DOI] [PubMed] [Google Scholar]
- Suhr M, Benz I, Schmidt MA (1996) Processing of the AIDA-I precursor: removal of AIDAc and evidence for the outer membrane anchoring as a beta-barrel structure. Mol Microbiol 22: 31–42 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2002) Automated structure solution, density modification and model building. Acta Crystallogr D 58: 1937–1940 [DOI] [PubMed] [Google Scholar]
- Van Gelder P, Dumas F, Winterhalter M (2000) Understanding the function of bacterial outer membrane channels by reconstitution into black lipid membranes. Biophys Chem 85: 153–167 [DOI] [PubMed] [Google Scholar]
- Van Gelder P, Saint N, van Boxtel R, Rosenbusch JP, Tommassen J (1997) Pore functioning of the outer membrane protein PhoE of Escherichia coli: mutagenesis of the constriction loop L3. Protein Eng 10: 699–706 [DOI] [PubMed] [Google Scholar]
- van Ulsen P, van Alphen L, Hopman CT, van der Ende A, Tommassen J (2001) In vivo expression of Neisseria meningitidis proteins homologous to the Haemophilus influenzae Hap and Hia autotransporters. FEMS Immunol Med Microbiol 32: 53–64 [DOI] [PubMed] [Google Scholar]
- van Ulsen P, van Alphen L, ten Hove J, Fransen F, van der Ley P, Tommassen J (2003) A Neisserial autotransporter NalP modulating the processing of other autotransporters. Mol Microbiol 50: 1017–1030 [DOI] [PubMed] [Google Scholar]
- Veiga E, de Lorenzo V, Fernandez LA (1999) Probing secretion and translocation of a β-autotransporter using a reporter single-chain Fv as a cognate passenger domain. Mol Microbiol 33: 1232–1243 [DOI] [PubMed] [Google Scholar]
- Veiga E, Sugawara E, Nikaido H, de Lorenzo V, Fernandez LA (2002) Export of autotransported proteins proceeds through an oligomeric ring shaped by C-terminal domains. EMBO J 21: 2122–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J (2003) Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299: 262–265 [DOI] [PubMed] [Google Scholar]
- Yen MR, Peabody CR, Partovi SM, Zhai Y, Tseng YH, Saier MH Jr (2002) Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim Biophys Acta 1562: 6–31 [DOI] [PubMed] [Google Scholar]


