Abstract
Genome sequencing has identified an extensive repertoire of single nucleotide polymorphisms among clinical isolates of Mycobacterium tuberculosis, but the extent to which these differences influence phenotypic properties of the bacteria remains to be elucidated. To determine whether these polymorphisms give rise to phenotypic diversity, we have integrated genome data sets with RNA sequencing to assess their impact on the comparative transcriptome profiles of strains belonging to M. tuberculosis Lineages 1 and 2. We observed clear correlations between genotype and transcriptional phenotype. These arose by three mechanisms. First, lineage-specific changes in amino acid sequence of transcriptional regulators were associated with alterations in their ability to control gene expression. Second, changes in nucleotide sequence were associated with alteration of promoter activity and generation of novel transcriptional start sites in intergenic regions and within coding sequences. We show that in some cases this mechanism is expected to generate functionally active truncated proteins involved in innate immune recognition. Finally, genes showing lineage-specific patterns of differential expression not linked directly to primary mutations were characterized by a striking overrepresentation of toxin–antitoxin pairs. Taken together, these findings advance our understanding of mycobacterial evolution, contribute to a systems level understanding of this important human pathogen, and more broadly demonstrate the application of state-of-the-art techniques to provide novel insight into mechanisms by which intergenic and silent mutations contribute to diversity.
Keywords: tuberculosis, genome evolution, RNA sequencing, lineage-specific mutations, transcriptional start sites
Introduction
Whole-genome sequencing demonstrates that patient isolates of Mycobacterium tuberculosis complex (MTBC) can be distinguished from each other by single nucleotide polymorphisms (SNPs) (Comas et al. 2010), allowing construction of a robust phylogeny comprising six genetically distinct lineages primarily associated with human tuberculosis (TB) (Hershberg et al. 2008; Comas and Gagneux 2009; Comas et al. 2010). However, the extent to which the genotypic diversity amongst clinical isolates influences phenotypic properties of the bacteria remains to be determined. The high frequency of nonsynonymous compared with synonymous changes suggests that SNPs are under relatively little constraint from purifying selection (Hershberg et al. 2008), raising the possibility that they may have phenotypic consequences for the bacteria.
Here, we explored this hypothesis by undertaking a combined genome and transcriptome comparison of strains belonging to two MTBC lineages, Lineages 1 and 2. Lineage 1 accompanied the early out-of-Africa migration of anatomically modern humans and has been classed as one of the evolutionarily “ancient” lineages (Hershberg et al. 2008). Lineage 2 is a “modern” lineage. It initially emerged in Asia and includes the “Beijing family” of strains (Parwati et al. 2010), which is of interest in the context of its high virulence in animal models (Coscolla and Gagneux 2010), its recent spread in human populations (Cowley et al. 2008), and its association with multidrug resistance (Borrell and Gagneux 2009).
Materials and Methods
Genome Data Set
Previously sequenced strains were accessed from publicly available databases (EBI ENA) and previously published work (Comas et al. 2010).
Genome Sequencing
Genomic DNA for N0031 was extracted using the cetyltrimethylammonium bromide method described previously (van Soolingen et al. 1991), and 2 μg DNA was used for sequencing on the Illumina platform. Sequencing libraries were constructed using the Epicentre Nextera DNA kit according to manufacturer’s instructions. Paired-end 75 base read sequencing was performed on a single lane as part of a multiplexed run. In total, 10.6 million reads were generated, corresponding to an average sequence depth of 180 reads.
Mapping Genome Data and SNP and Indel Calling
MAQ (Li et al. 2008) was used to map reads to the most recent common ancestor of the MTBC (Comas et al. 2010). This sequence is based on the H37Rv genome (AL123456) but substituting H37Rv alleles by those in the common ancestor of the lineages. Default MAQ parameters were used, removing SNPs with a Phred score <30, read depth <5, and nonunique matches. A nonredundant list of variable positions called with high confidence in at least one strain was constructed and used to recover the base call in all other strains. SNPs and indels called within repetitive regions (genes annotated as PE/PPE/insertions/phages) were removed.
Phylogenetic Analysis
Phylogenetic analysis was based on the high confidence SNPs, consisting of 13,086 variable genomic positions. A neighbor-joining tree based on a concatenate of these positions was generated with MEGA (Tamura et al. 2011), using 1,000 bootstrap replications and observed number of substitutions as a measure of genetic distance. In cases where SNP calls were missing from individual strains, pairwise deletion was performed and missing data in the comparison ignored.
Predicting the Functional Effect of Mutations
SNPs were categorized as nonsynonymous or synonymous using snpEff (Cingolani et al. 2012). We used the Sorting Intolerant From Tolerant (SIFT) algorithm to predict nonsynonymous SNPs likely to affect protein function based on sequence homology (Ng and Henikoff 2003). Briefly, SIFT looks for homologs in other bacteria of the gene of interest and 1) scores the conservation of the positions where mutations are found and 2) weights this score by the nature of the amino acid change. These measures are incorporated into a normalized probability score, and a SIFT score ≤0.05 indicates predicted functional impact. The recommended >3.5 conservation threshold was used to filter biased predictions. All available non-MTBC complete mycobacterial genomes were used for the Blast database (N = 13). Prediction of SNPs that cause a destabilization of the protein structure was made using CUPSAT (Parthiban et al. 2006). CUPSAT predicts the change in free energy of protein unfolding between wild-type and mutant proteins (ΔΔG). Protein stability is categorized as destabilizing (−ΔΔG), neutral (0 ΔΔG), or stabilizing (+ΔΔG); changes in stability of <0.5 ΔΔG are not significant. Where existing protein structures were not available, homology modeling was performed and the highest confidence model (>99%) with the greatest coverage chosen (Kelley and Sternberg 2009). Construction of homology models that covered the SNP position was not possible for four regulators.
RNA-seq Strains and Culture Conditions
Representative genome sequenced strains from Lineages 1 and 2 were selected based on the greatest within-lineage SNP distances. Strains were as follows: Lineage 1—N0072, N0153, N0157; Lineage 2—N0145, N0052, N0031. Single colony bacterial stocks were grown in Middlebrook 7H9 supplemented with 0.5% glycerol, 10% Middlebrook ADC, and 0.05% Tween-80 in roller bottle culture. Exponential phase cultures were harvested at an optical density of 0.4–0.8 (OD600); growth curves were performed to ensure calibration of exponential growth phase of the clinical strains. Stationary phase cultures were harvested 1 week after the cultures reached 1.0 OD600 as defined previously (Arnvig et al. 2011).
RNA-seq
Isolation of RNA was performed using the FastRNA Pro blue kit from QBiogene/MP Bio according to manufacturer’s instructions. Cultures were first rapidly cooled by addition of ice directly to the culture prior to centrifugation. All RNA samples were treated with Turbo DNase free (Ambion) until any DNA contamination removed. Concentration and quality control of RNA samples was measured by Nanodrop (ND-1000, Labtech) and Agilent RNA chip (2100 Bioanalyser). Construction of strand-specific cDNA libraries was carried out with 2–3 μg total RNA using the Illumina directional mRNA-Seq protocol (Part # 15018460 Rev. A), but with exclusion of polyA-tail and size selection to capture all RNA species. Terminator-5′-phosphate-dependent exonuclease (Epicentre Biotechnologies) was used to deplete processed RNAs in cDNA samples used in transcriptional start site (TSS) mapping analysis (Vertis Biotechnologie AG). Single-end read sequencing was performed on Illumina Genome Analyser (GA) and HiSeq (HS) sequencers, using a single flow cell lane per library. Supplementary table S3, Supplementary Material online, details run metrics.
Raw RNA-seq Read Filtering and Mapping
Raw reads were first filtered to discard low-quality reads and FastQC (Babraham Bioinformatics) used to inspect read base qualities. Poor-quality reads were trimmed using the SolexaQA package (Cox et al. 2010) using default parameters, trimming bases with confidences P > 0.05, and removing reads <25 bases. A reference-based assembly using the reference genome H37Rv was performed using the Burrows–Wheeler Alignment (BWA) tool (Li and Durbin 2009).
TSS Mapping
Custom Perl scripts were written for TSS calling. Briefly, the increment in reads from one genome position to the next consecutive base was calculated for all genomic positions, with an increment significantly above the average background coverage defined as candidate TSS. TSS peak height was considered as representative of the level of expression of the TSS. To build a genome-wide TSS map for M. tuberculosis, automated annotation of the putative TSS detected according to genomic distribution was performed similar to Sharma et al. (2010).
Transcriptome Clustering
Hierarchical cluster analysis of the transcriptomes was performed using the hclust function in R (R Development Core Team 2008) by the complete linkage method. Spearman distances were calculated from the dissimilarity matrix of pairwise correlations of total gene expression (N = 4,015 genes), expressed as Reads Per Kilobase per Million mapped reads (RPKM). Clade support using 1,000 bootstrap replications was performed using the R function pvclust.
Lineage Differential Expression
Genome coverage of reads mapping to genes, antisense, and sRNAs were calculated using BEDtools (Quinlan and Hall 2010). Statistical testing for differential expression was performed using DESeq (Anders and Huber 2010), a method based on the negative binomial distribution and implemented in the R statistical environment (R Development Core Team 2008). Raw reads were normalized using DESeq to adjust for differences in library sizes; figures displaying quantified RNA expression levels are based on this normalization. Reads from technical replicates were combined and treated as one sample. Gene deletions at either strain or lineage level were removed from the analysis by a Perl script (N = 223 genes); deletions were identified based on genome coverage using the respective strains genome, with a threshold of <90% gene coverage to define a deletion. Read counts were normalized using DESeq. Normalized expression of features (annotated genes, antisense, or sRNAs) that overlapped with strains from different lineages due to strain-specific expression were filtered and removed, with 1,606 features entered into the analysis. For the purpose of testing for lineage-specific differential expression in DESeq, strains from the same lineage were treated as biological replicates and the mean expression from the two lineages compared. Significant differential expression was defined as P < 0.05 (P value adjusted for multiple testing using Benjamini–Hochberg method). All heatmaps were produced in R, using the fold change of normalized reads versus mean expression of the six strains.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction
cDNA for quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) was made using random primers and Superscript III according to manufacturer’s instructions (Invitrogen). qRT-PCR was carried out on a 7500 Fast Real-Time PCR System (Applied Biosystems) using Fast SYBR Green Master Mix (Applied Biosystems). Each 96-well plate consisted of a closed experimental plate design, consisting of the six RNA-seq study strains. RNA without RT (RT−) was analyzed alongside cDNA (RT+). Standard curves were performed for each gene analyzed and the cycle threshold values used to quantify cDNA level. Data were averaged, adjusted for chromosomal DNA contamination (RT+ minus RT−), and normalized to corresponding 16S rRNA values. Three biological replicates were tested in three independent qRT-PCR experiments per gene.
Statistical Analysis
All χ2 tests were two-tailed and performed using GraphPad Prism v5.03c. Where multiple χ2 tests were performed, multiple testing correction was applied using the false discovery rate (FDR) method and implemented in R. Analysis of qRT-PCR experiments were performed using two-tailed unpaired t-test. For the rest of the analyses, only selected categories were tested and no test for multiple correction was required.
Results
Genome Diversity
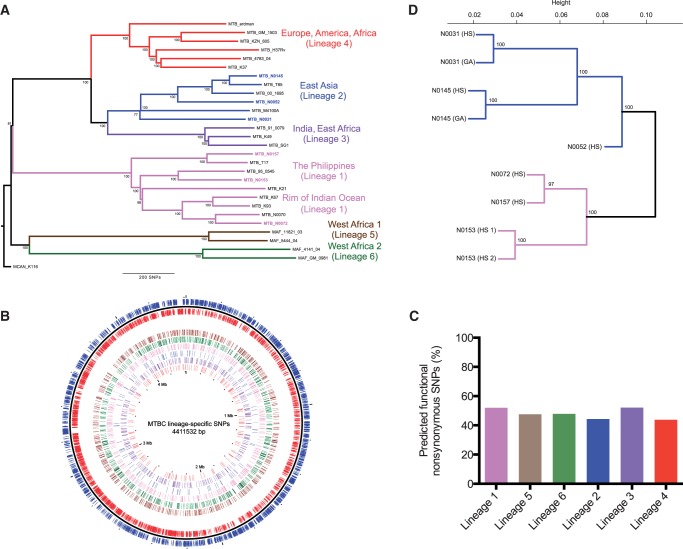
SNPs were identified in the genome sequences of 28 representative human-associated MTBC isolates belonging to Lineages 1–6 (fig. 1A). We focused the analysis on SNPs conserved among all strains from the same lineage (referred to as “lineage-specific SNPs”), with inclusion of SNPs in the internal phylogenetic branch that defines the modern lineages (Lineages 2, 3, and 4) (Hershberg et al. 2008). In total, there were 2,772 SNPs (supplementary table S1, Supplementary Material online), which were evenly distributed genome-wide (fig. 1B). As previously described at the total genome level (Fleischmann et al. 2002; Hershberg et al. 2008), more nonsynonymous than synonymous SNPs were found in all lineages (1.8 ± 0.3 times more nonsynonymous SNPs), corresponding to a mean of 64.2% coding SNPs resulting in amino acid change. Grouping the genes by function, SNPs were present across all functional categories, but a significantly higher ratio of nonsynonymous SNPs (>3-fold) was detected in the regulatory protein category (χ2, P = 0.03) (table 1).
Fig. 1.—
The influence of genomic diversity at the transcriptomic level. (A) Neighbor-joining phylogeny based on 28 representative MTBC strains, using 13,086 variable positions. The six main lineages are named and branches colored as defined previously (Gagneux et al. 2006; Hershberg et al. 2008). Node support after 1,000 bootstrap replications is shown on branches and the tree is rooted by the outgroup Mycobacterium canetti. (B) Genome distribution of 2,772 lineage-specific SNPs. Moving from the outer to innermost ring: forward (blue) and reverse (red) genes, lineage-specific SNPs (Lineage 6, 5, 1, 2, 3, 4). (C) Percentage SIFT predicted functional nonsynonymous SNPs per lineage. (D) Unsupervised hierarchical clustering of total gene expression of all annotated genes (4,015). Strain replicates are also shown (strain N0153, N0145, and N0031). Node support after 1,000 bootstrap replications is shown for each branch.
Table 1.
Summary of SNP Distribution and SNP Predictions among Functional Categories for the Lineage-Specific SNPs
| Lineage SNPs |
|||||
|---|---|---|---|---|---|
| Functional Categorya | Nonsynon | Synon | Nonsynon/Synon | χ2 P Value | Predicted Functional SNPsb |
| 1,543 | 840 | 1.8 | 368 | ||
| Information pathways | 96 | 73 | 1.3 | 0.11 | 12 |
| Lipid metabolism | 168 | 99 | 1.7 | 0.63 | 66 |
| Intermediary metabolism and respiration | 394 | 237 | 1.7 | 0.40 | 88 |
| Cell wall and cell processes | 377 | 197 | 1.9 | 0.63 | 91 |
| Conserved hypotheticals | 343 | 174 | 2.0 | 0.63 | 63 |
| Virulence, detoxification, adaptation | 63 | 29 | 2.2 | 0.63 | 17 |
| Regulatory proteins | 102 | 31 | 3.3 | 0.03 | 31 |
| Intergenic | 382 | N/A | N/A | N/A | |
Note.—A 3-fold overrepresentation of nonsynonymous (nonsynon) to synonymous (synon) SNPs was found within the category of transcriptional regulators (χ2, P = 0.03).
aCategories are based on Tuberculist annotations.
bFunctional SNPs predicted using SIFT (SIFT score ≤ 0.05).
High Percentage of Predicted Functional SNPs
As an initial assessment of potential phenotypic consequences associated with individual SNPs, we used a computational method—SIFT (Ng and Henikoff 2003). This uses sequence alignments of homologous proteins to predict amino acid substitutions likely to be tolerated without loss of function. We identified 368 lineage-specific SNPs that were predicted to impact protein function, out of 844 nonsynonymous SNPs for which predictions could be made (table 1; complete list in supplementary table S2, Supplementary Material online). Functional SNPs were again distributed across all gene function categories. Broken down by lineage, an average of 44.4% of the nonsynonymous SNPs were predicted functional (fig. 1C). Although significantly fewer predicted functional (54, 14.7%) than tolerated (105, 22.7%) SNPs occurred within genes classed as essential for growth on the basis of transposon screens (χ2, P = 0.003) (Sassetti and Rubin 2003; Sassetti et al. 2003), this percentage (14.7%) may suggest a false-positive error rate for the predictions. This is close to the previously described error rate for SIFT (Ng and Henikoff 2003).
Functional Impairment of Transcriptional Regulators
To test for phenotypic consequences of predicted functional mutations, we focused on nonsynonymous SNPs in genes encoding transcriptional regulators in Lineages 1 and 2. We identified 11 genes with lineage-specific SNPs predicted by SIFT analysis as likely to impair protein function and a further three genes with nonsense or frameshift insertion/deletion (indel) mutations (table 2). We refined our prediction of functional impairment by examining the location of each mutation with respect to functionally important domains in the protein (Punta et al. 2012) and the predicted effect on protein stability (ΔΔG) using existing protein structures or homology models (Parthiban et al. 2006; Kelley and Sternberg 2009). Five of the SNPs were predicted by all three criteria to have a deleterious impact and we classed these, together with frameshift mutations and a protein truncation, as having a high likelihood of causing functional impairment. Of the remaining six SNPs highlighted by SIFT, two (in mprA and zur) occur in the modern lineage branch, and so are present in M. tuberculosis H37Rv, a common laboratory strain belonging to Lineage 4 (Hershberg et al. 2008; Comas et al. 2010). The observation that deletion of the corresponding genes from H37Rv generates phenotypic changes (Maciag et al. 2007; Pang et al. 2007) suggests that the altered proteins retain biological function and that these may represent false-positive SIFT predictions.
Table 2.
Regulatory Proteins with Predicted Functional SNPs and Indels in Lineages 1 and 2
| Gene | Regulator Type | SNPa | Mutation | Lineage | SIFT Score | Domain | Protein Stabilityb |
|---|---|---|---|---|---|---|---|
| High predictive score | |||||||
| Rv0275 | TetR | T 331588 C | L24S | Modern | 0.00 | HTH | −3.18 |
| NarL | 2-component regulator | G 940602 C | G169R | 2 | 0.00 | HTH | −4.66 |
| KdpD | 2-component sensor | Indel | H67 frameshift | 1 | n/a | 2-component sensor | n/a |
| BlaI | penicillinase repressor | T 2096430 G | L57R | 1 | 0.05 | HTH | −8.72 |
| SirR | Fe-dependent repressor | C 3097349 | Q131X | 1 | n/a | Fe-dependent repressor | n/a |
| VirS | AraC | T 3447480 G | L316R | 1 | 0.01 | HTH | −2.03 |
| Rv3167c | TetR | C 3536008 A | P17Q | 1 | 0.02 | HTH | -1.21 |
| Rv3830c | TetR | Indel | S208 frameshift fusion | 2 | n/a | Low complexity | n/a |
| Low predictive score | |||||||
| RamB | HTH-XRE | A 555945 G | Q121R | 1 | 0.02 | Low complexity | n/a |
| Rv0377 | LysR | G 455325 C | R302P | 2 | 0.00 | Low complexity | n/a |
| MprA | 2-component regulator | A 1097023 G | S70G | Modern | 0.04 | cheY | +2.83 |
| TcrS | 2-component sensor | C 1157771 G | S62C | 1 | 0.01 | Low complexity | n/a |
| Zur | Fur | G 2641840 A | R64H | Modern | 0.02 | HTH | +0.47 |
| Rv3736 | AraC | G 4187063 A | G144R | 1 | 0.01 | Arabinose-binding | n/a |
Note.—High predictive score: SNPs map to HTH DNA-binding domains of regulator and predicted to cause a loss of protein stability. Low predictive score: SNPs outside of HTH domain or in low-complexity regions and no loss of protein stability.
aAlleles are the ancestral and mutant with genomic position based on H37Rv.
bn/a: change in stability not possible due to stop codon or indel mutation or no protein structure over position.
To test our predictions, we compared transcriptional profiles of Lineage 1 and 2 strains by RNA sequencing (RNA-seq). We anticipated that impaired protein function would result in altered expression of genes encoding the transcription factors themselves (in the case of autoregulated proteins) and/or their regulated targets.
Transcriptome Diversity
Three strains were selected from different phylogenetic branches of the lineages and total RNA extracted from exponentially growing cultures was analyzed by RNA-seq (run details and complete data set of normalized read counts shown in supplementary tables S3, S4, and S12, Supplementary Material online). For two of the strains (Lineage 1 N0153 and Lineage 2 N0145), additional RNA-seq was performed after a 5′ phosphate-dependent exonuclease digestion step to facilitate mapping of TSSs (full list in supplementary table S5, Supplementary Material online) (Sharma et al. 2010). Transcriptome diversity paralleled genome diversity. Figure 1D illustrates the relationship inferred by unsupervised clustering of RNA-seq data, showing a close match to that established by standard whole-genome-based phylogenetic analysis (fig. 1A). Interestingly, RNA-seq data for the laboratory-adapted reference strain H37Rv grown in the same conditions and growth phase (Arnvig et al. 2011) identified this as an outlier from the clinical strains with respect to its transcriptional profile (supplementary fig. S1, Supplementary Material online). Three transcriptomes were sequenced on further technical runs, and expression between the technical replicates was highly correlated (supplementary fig. S2, Supplementary Material online).
A total of 112 genes were identified as having a lineage-specific pattern of differential expression with fold change ranging from 1.9 to 39.3 (based on a statistical cut-off of P < 0.05); 88 (78.6%) were higher in Lineage 1, and 24 (21.4%) more highly expressed in Lineage 2 strains (supplementary table S6A; genes of particular interest are summarized in supplementary table S7, Supplementary Material online). Twenty-six of the genes were identified as differentially expressed in previous microarray comparisons of ancient versus modern lineages or M. tuberculosis H37Rv versus Mycobacterium bovis (within Lineage 6) (Golby et al. 2007; Homolka et al. 2010). A parallel analysis of antisense transcription identified similar conservation by lineage (supplementary fig. S3, Supplementary Material online), with a differential expression pattern for 56 genes; 23 were higher in Lineage 1 and 33 in Lineage 2 (supplementary table S6B, Supplementary Material online).
Transcriptional Regulators
Three of the regulatory proteins with predicted functional SNPs—Rv0275c, Rv3082c (VirS), and Rv3167c—were included in the set of lineage-specific differentially expressed genes (supplementary table S7A, Supplementary Material online). VirS has previously been shown to act as an inhibitor of its own transcription and as a positive regulator of the adjacent divergently expressed MymA locus (Rv3083–3089) (Singh et al. 2003). Consistent with its predicted functional impairment by mutation of the helix-turn-helix (HTH) DNA-binding domain, virS expression was 17-fold higher in Lineage 1, with no effect on expression of MymA. Targets of transcriptional regulators Rv0275c and Rv3167c are unknown, but the proximity of TSSs suggests that binding of the regulators to upstream sequences will repress transcription of the adjacent divergent genes Rv0276 and Rv3168. Expression of Rv0276 follows Rv0275c in being 10-fold higher in Lineage 2, although it falls outside of the statistical cut-off (P = 0.08), and Rv3178 expression is 5-fold higher in Lineage 1 (P = 0.12). A frameshift mutation in Lineage 2 was predicted to inactivate Rv3830c by generating a fusion with Rv3829c. Although we observed no significant change in expression of Rv3830c itself, 14-fold and 21-fold increased expression of flanking genes (Rv3829c, a phytoene dehydrogenase, and Rv3131, a hypothetical protein) in Lineage 2 suggests that the functional protein may act as a repressor and that this regulation is lost in the case of mutant allele.
We were unable to identify a lineage-specific transcriptional signature for the remaining four regulators for which we had predicted functional impairment (narL, blaI, sirR, and kdpD). This may be due to an incorrect prediction or alternative culture conditions other than exponential growth, such as the presence of beta-lactams (blaI) (Sala et al. 2009) or low potassium (kdpD) (Walderhaug et al. 1992), may be required to uncover defects in associated regulatory responses.
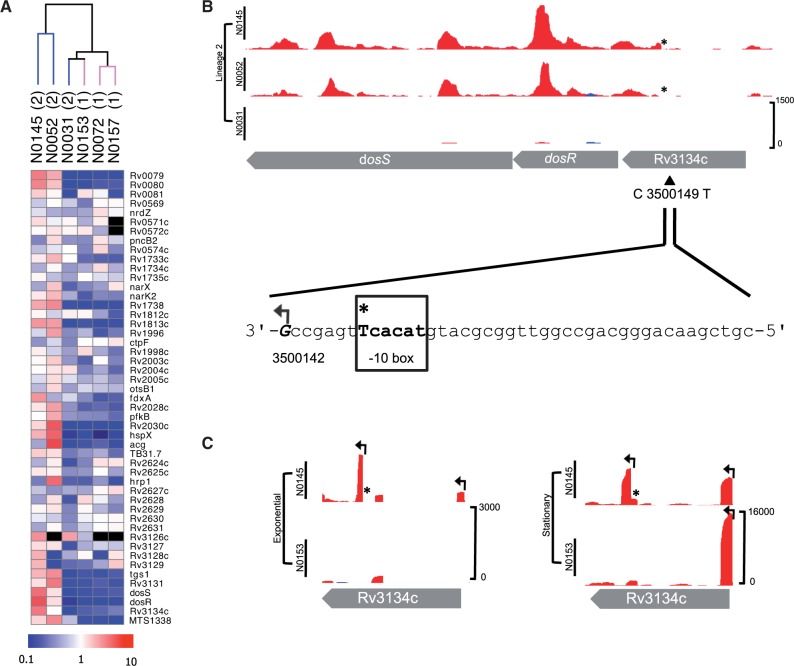
DosR Regulon
It has previously been reported that genes belonging to the DosR regulon are overexpressed during exponential growth in strains belonging to the Beijing family (Reed et al. 2007; Homolka et al. 2010). Among these genes, only Rv1733c met statistical criteria for upregulation in Lineage 2 in the lineage comparison (supplementary table S6A, Supplementary Material online), but an enhanced DosR response was clearly seen in the individual strains N0145 and N0052 (fig. 2A). The outlying strain, N0031, belongs to a basal branch of Lineage 2 that diversified prior to expansion of the major Beijing branches represented by N0145 and N0052 (fig. 1A). A 350 kb genomic duplication that includes the DosR operon has been suggested to contribute to increased constitutive expression of the regulon in Beijing strains (Domenech et al. 2010). This duplication is present in N0145 and N0031, but absent from N0052, and therefore cannot account for the observed differential pattern of DosR expression in our study (supplementary fig. S4, Supplementary Material online). A previously identified indel in dosT linked to the Beijing strains was also not found to be responsible for the observed DosR regulon phenotype (Fallow et al. 2010).
Fig. 2.—
DosR regulon and SNP-associated TSS. (A) Heatmap of regulon (comprising of 48 genes and 1 sRNA) in the six strains. Unsupervised hierarchical clustering of strains by normalized gene expression of the regulon separates Lineage 2 Beijing subgroup strains (N0145 and N0052). Scale bar indicates fold change, from 10-fold downregulation (blue) to 10-fold upregulation (red). Genes not expressed are colored black. (B) Exponential phase mapped RNA-seq reads in Lineage 2 strains for dosR. Plot shows reads mapping to the forward (blue) and reverse (red) DNA strand. Plots are shown at an identical scale with scale bar indicating maximum read depth included in the bottom panel (this convention is kept for all plots). The C to T SNP in Beijing strains is indicated with an asterisk (*) and the new TSS 7 nucleotides from the created -10 box highlighted. (C) RNA-seq TSS mapping for strain N0145 (Lineage 2) and N0153 (Lineage 1) grown at exponential and stationary phases; TSS shown with arrows. The Beijing-specific TSS within Rv3134c is expressed in both phases.
Excluding indels, we were unable to identify any amino acid mutations that might alter the function of DosR or related regulatory components. However, a Beijing-specific synonymous SNP (C 3500149 T) was identified within Rv3134c, the gene immediately upstream of DosR that encodes a protein that is itself a member of the DosR regulon; this SNP was documented by Fallow et al. (2010). It was found here that the SNP generates a TAnnnT -10 consensus motif that is characteristic of actinomycetes and is found in association with ∼75% of all TSS mapped in M. tuberculosis (Zheng et al. 2011; Newton-Foot and Gey van Pittius 2012; Cortes et al. 2013) and is located seven nucleotides upstream of a novel TSS expressed in exponential and stationary phase samples of Beijing strain N0145 (fig. 2B). The new TSS is distinct from the standard Rv3134c intergenic TSS associated with growth-phase induction of the DosR regulon and from secondary promoters identified within the Rv3134c gene of M. tuberculosis H37Rv (fig. 2C) (Bagchi et al. 2005). The resulting transcript is clearly seen in the total RNA analysis and runs through dosR in the two Beijing strains (fig. 2B).
Another Beijing-specific SNP (C 3509626 A) similarly generates a TAnnnT consensus motif and associated TSS for the two-component sensor protein encoded by Rv3143. Increased expression is evident in total transcriptome profiles from the two Beijing strains, but in this case, the downstream targets of the regulator are unknown.
SNP-Associated TSS
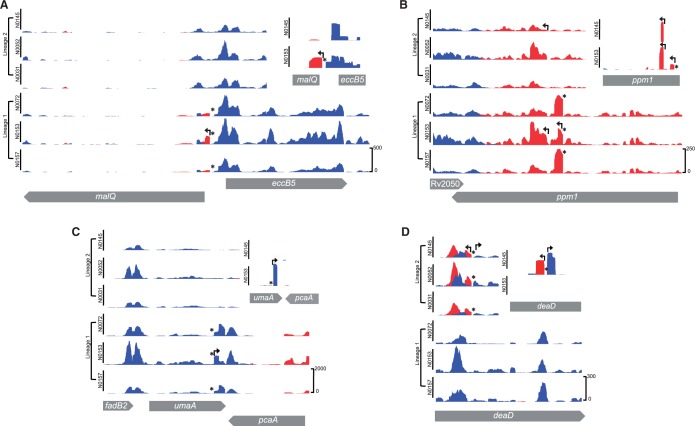
Alignment of lineage-specific SNPs with a total transcriptome map of M. tuberculosis H37Rv (Cortes et al. 2013) identified 94 instances (1.2% of 7,601 TSSs) in which a SNP fell within the 30-nucleotide region upstream of a TSS. This frequency was markedly higher amongst the 168 genes with differentially expressed sense and antisense transcripts, with 23 of the respective TSS harboring one or more SNPs in this upstream region (χ2, P < 0.0001) (supplementary table S7B, Supplementary Material online). In ten cases, lineage-specific SNPs created a TAnnnT consensus motif linked to a new TSS.
For three of the differentially expressed genes (malQ, Rv3680, and PE_PGRS62), the new TSS was located upstream of the predicted translational start, either within an intergenic region or the adjacent gene (fig. 3A). The remaining six new TSSs (umaA, mgtA, Rv0724A, ppm1, Rv2765, and spoU) were located within the differentially expressed gene itself and, if translated, would give rise to truncated protein products. In some cases, truncated proteins may retain biological function. Ppm1 (Rv2051c) is a bifunctional protein created by fusion of polyprenyl phosphomannose synthase and apolipoprotein N-acyltransferase activities (Gurcha et al. 2002). The C 2309356 T SNP in Lineage 1 is associated with a novel internal transcript that includes the intact C-terminal polyprenyl phosphomannose synthase domain and at the same time introduces a T467I mutation that is predicted by SIFT analysis to impair the function of the N-terminal N-acyl transferase domain (fig. 3B). A second internal ppm1 TSS is present in all strains at position 2309159; the resulting transcript again provides the option of dissociating the two enzymatic activities.
Fig. 3.—
Examples of SNP-associated TSS. For each example, the main panels show total transcriptome profiles for the six strains; insets in the top right show TSS mapping for strain N0145 (Lineage 2) and N0153 (Lineage 1). Lineage-specific SNPs are indicated with an asterisk. (A) Intergenic TSS: Lineage 1 SNP (T 2017560 A) is associated with a new TSS and increased expression of malQ in the respective strains. (B) Internal coding TSS: Lineage 1 SNP (C 2309356 T) within ppm1 is associated with a new TSS and upregulation of ppm1 transcription. A second internal TSS present in all strains also indicated in the TSS mapping inset. (C) Internal coding TSS and 3′ antisense: SNP-associated TSS in 3′ region of umaA in Lineage 1 strains is associated with higher umaA expression and pcaA antisense expression. (D) Internal coding TSS and antisense: SNP within deaD in all Lineage 2 strains is associated with a new TSS and increased antisense transcription. The SNP also creates a -10 consensus on the forward strand; this is associated with a new TSS but has no significant impact on the level of sense transcription.
SNPs that alter residues outside the -10 motif may also influence promoter activity. G 4092921 T is associated with a 100-fold increase in reads mapping to a TSS upstream of PE_PGRS60, for example. The mutation changes an existing -10 TAnnnT motif to an “extended -10” TGnTAnnnT consensus (Newton-Foot and Gey van Pittius 2012). This change is similar to that generated by a SNP that drives increased promoter activity and inhA expression in isoniazid-resistant strains of M. tuberculosis (Ramaswamy and Musser 1998; Ramaswamy et al. 2003).
Differentially Expressed Antisense
Antisense transcriptomes for the two lineages display a broadly similar level of conservation to the sense transcriptomes, with a total of 56 genes showing lineage-specific differential expression (supplementary table S6, Supplementary Material online). Antisense transcripts arise either from internal TSS or from overlapping 3′-untranslated regions (UTRs) in convergent gene pairs (Arnvig et al. 2011). Three of the differentially expressed 3′-UTR antisense transcripts (pcaA, Rv1898, and ribD) were associated with SNPs that create a new TAnnnT-linked forward TSS in the adjacent divergent gene; in the case of pcaA, the transcript was also detected as a significant increase in umaA sense expression (fig. 3C). For a further six antisense transcripts (Rv0552, Rv0842, Rv0874c, deaD, Rv2672, and fadE20), introduction of a TAnnnT motif on the reverse strand was associated with new TSS arising within the gene itself (supplementary table S7C, Supplementary Material online). In the case of deaD, in Lineage 2 (and other modern lineages), a C to T SNP creates a new -10 motif and TSS on both forward and reverse strands of DNA (fig. 3D). A highly expressed antisense transcript that is present within the ino1 gene (Rv0046c) in one of the Lineage 1 strains (N0157) is associated with a new TAnnnT motif created by a C 50557 T SNP. Interestingly, this is a homoplasic SNP, generally rare in M. tuberculosis (Comas et al. 2009; Schürch et al. 2011), and occurs in a subbranch of Lineage 1 and a subbranch of Lineage 4, including strain H37Rv, which also expresses the antisense transcript (Arnvig and Young 2012).
Toxin–Antitoxin Modules
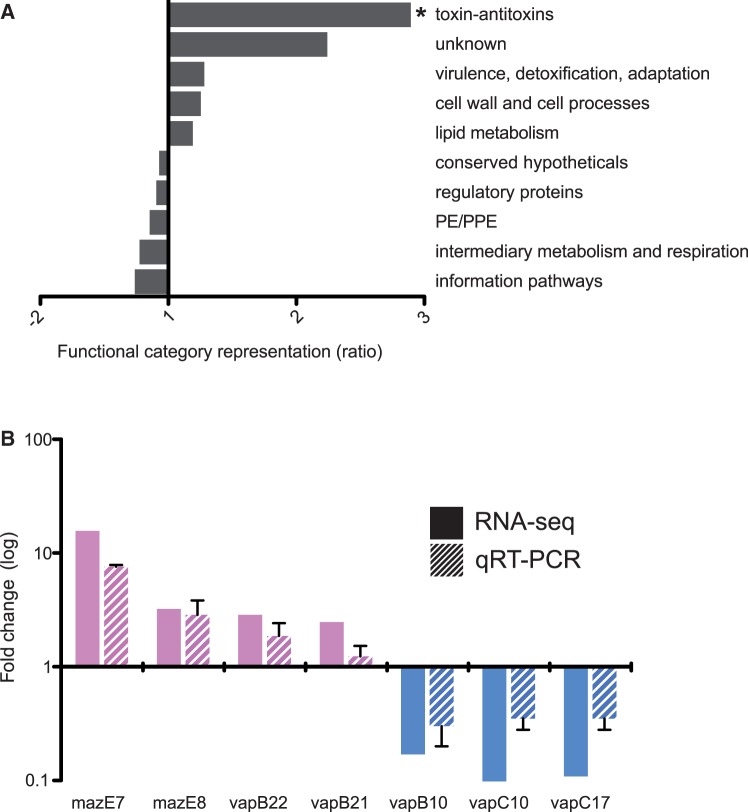
We were unable to identify direct SNP associations for the remainder of the genes showing lineage-specific patterns of differential expression (135 out of 168 differentially expressed genes and antisense). We anticipate that their differential expression reflects downstream consequences of primary mutations. Analysis of the panel of differentially expressed genes according to functional category identified a 2-fold overrepresentation of proteins involved in virulence, detoxification, and adaptation. This was driven by ten TA genes ( supplementary table S7D, Supplementary Material online), and separate classification of all TA as an independent category revealed 2.9-fold overrepresentation in the differentially expressed set compared with the genome representation (χ2, P = 0.03) (fig. 4A and supplementary table S8, Supplementary Material online). For selected TA, the pattern of differential gene expression seen by RNA-seq was confirmed by quantitative RT-PCR (fig. 4B and supplementary tables S9 and S10, Supplementary Material online).
Fig. 4.—
Overrepresentation of differentially expressed toxin–antitoxins (TAs). (A) Ratio of significant differential gene expression grouped by functional category, compared with the genome-wide representation of the category. Values on the x axis represents the difference as fold change, positive fold change indicates overrepresentation of a particular function category and negative values underrepresentation. There were 2.9-fold more toxin–antitoxins than expected (χ2, P = 0.03). (B) Validation of selected RNA-seq differentially expressed toxin–antitoxins (solid bars) by qRT-PCR (striped bars). Fold change relative to Lineage 1 expression on y axis (log10 scale) and bars colored by lineage with higher expression. Error bars for qRT-PCR indicate the standard deviation of three biological replicates.
Transcription of TA modules is generally repressed by binding of the cognate TA complex to the promoter region and activated when the antitoxin is degraded in response to signals associated with environmental stress (Buts et al. 2005). Differential expression could result from mutations that affect stability or repressor activity of the TA complex, mutations that alter promoter sequences, or mutations that alter the proteolytic activity in the cell. Two differentially expressed toxins have nonsynonymous lineage-specific SNPs, vapC10 (Lineage 2, G103D) and mazF7 (Lineage 1, R101P), but the SIFT algorithm was unable to predict functional consequences for these mutations. All TA pairs with detectable transcripts were expressed from a single major TSS. In two cases, the TSS was located within the annotated coding sequence, and we have suggested alternative translation start sites for these (supplementary table S11, Supplementary Material online). In most cases (31 out of 51 expressed TA pairs; 60.8%), the TA pairs were encoded by leaderless mRNAs. A single TSS-associated SNP was identified, with position -1 of the vapB22 (Rv2830c) TSS switched from G to A in Lineage 2. This may contribute to the decreased expression of vapB22 observed in these strains (supplementary table S7D, Supplementary Material online).
The lack of direct SNP associations leads us to infer that differential expression of TA genes reflects general differences in regulatory networks between the lineages. A series of genes that are preferentially expressed in Lineage 1 strains have previously been implicated in the H37Rv response to acid stress and cell wall damage, including ahpC and ahpD, fabD, and lpqS (Fisher et al. 2002) (supplementary table S6A, Supplementary Material online). Upregulation of these genes may be associated with the stress-related sigma factor sigB (Rv2710), which has 2-fold higher expression in Lineage 1 but falls outside the statistical cut-off (P = 0.06).
Discussion
The importance of the present study is in establishment of direct links between genetic differences observed among clinical isolates of the MTBC and phenotypic consequences at the level of transcription. This required the identification of all lineage-specific SNPs, which was performed for all six phylogenetic lineages, and is provided here for the TB community as an important future resource, necessary for the lineage typing of clinical isolates. Bioinformatic analyses of this data set showed a high percentage of nonsynonymous SNPs identified across the MTBC, which are likely to impair protein function. It has been suggested that this reflects a low frequency of purifying selection and has the potential to generate substantial functional diversity (Hershberg et al. 2008). Interestingly, a similar phenomenon has been observed in humans, where recent demographic expansions have led to the accumulation of low-frequency genetic variants associated with strong functional effects (Keinan and Clark 2012; Tennessen et al. 2012). Considering the tight link between the MTBC and its human host, with parallel population dynamics (Comas et al. 2013), it is interesting to speculate that these human expansions might have had a similar effect on the genetic diversity of the MTBC (Hershberg et al. 2008).
Focusing on Lineages 1 and 2, we predicted functional impairment of eight transcriptional regulators; transcriptional profiling provided confirmatory evidence in four cases. Elevated expression of virS in Lineage 1 recapitulates results of a previous microarray comparison of modern and ancient lineages (Homolka et al. 2010), with the absence of activation of the associated MymA regulon providing further evidence that the mutant virS lacks functional activity. Experimental deletion of virS in M. tuberculosis H37Rv resulted in pleiotropic cell wall defects and reduced growth in the spleen of guinea pigs (Singh et al. 2005), raising the possibility that this mutation may reduce the virulence of Lineage 1 strains.
Genes with lineage-specific patterns of differential expression were characterized by a high frequency of SNPs associated with TSSs. A striking observation was that SNPs generating a -10 consensus motif (TAnnnT) were frequently associated with the emergence of a new TSS. SNP-created -10 motifs accounted for 19 of the 168 (11%) lineage-specific differentially expressed transcripts. In addition to their effect on expression of downstream genes, as in the case of Rv3134c/DosR , TSS arising within coding regions may also play a role in generating functionally active truncated proteins. Ppm1 is a bifunctional enzyme, fusing an N-terminal apolipoprotein N-acyltransferase with a polyprenyl phosphomannose synthase that are encoded by separate genes in other mycobacteria (Gurcha et al. 2002; Rana et al. 2012). The C-terminal domain has recently been shown to be essential for optimal growth, whereas the N-terminal is unessential (Zhang et al. 2012). An internal TSS provides the option of separating the two activities, freeing the polyprenyl phosphomannose synthase to participate in other glycosylation pathways. A conserved internal TSS suggests that this option is retained by the MTBC, with additional flexibility in Lineage 1 provided by a new SNP-associated TSS and coincident predicted impairment of N-acyltransferase activity. It has been proposed that changes in the mannosylation of cell surface components have an important impact on recognition of mycobacteria by receptors on innate immune cells (Torrelles and Schlesinger 2010), and redistribution of mannose between lipoglycans and lipoproteins represents an attractive hypothesis to account for the differential inflammatory response to Lineage 1 and Lineage 2 strains (Portevin et al. 2011). Enhanced Lineage 1 transcription of mgtA (Rv0557) could also contribute to differences in macrophage phenotype (Torrelles et al. 2009).
New TSSs associated with SNP-generated TAnnnT motifs were also observed at a similar frequency in antisense orientation. With the introduction of RNA-seq approaches, pervasive expression of antisense transcripts has been recognized as a common feature of bacterial transcriptomes (Lasa et al. 2011; Raghavan et al. 2012). Interspecies comparison of upstream sequences in Escherichia coli and Salmonella typhimurium suggests that selective pressure for conservation of antisense promoters is lower than in the case of sense promoters (Raghavan et al. 2012). Whether this applies to intraspecific comparisons with much smaller evolutionary distances is not known, but we observed a broadly similar pattern of sense and antisense diversity in our MTBC lineage comparison, which may reflect the reduced purifying selection and increased genetic drift within MTBC (Hershberg et al. 2008). The biological significance of antisense transcripts is unknown; it is possible that double-stranded RNA molecules differ from single-stranded mRNAs in their efficiency of translation and susceptibility to degradation. The new TSS in umaA that generates antisense to the adjacent pcaA raises the intriguing possibility of a mechanism for co-ordinated regulation of the two genes. Both proteins are involved in modification of mycolic acids, and lineage-specific differential expression could again contribute to variation in innate immune reactivity (Rao et al. 2006; Barkan et al. 2012).
For the remaining differentially expressed genes, we were unable to identify any direct genotypic link, and we presume that they reflect downstream secondary effects of the primary mutations. The most striking feature is the overrepresentation of TA gene pairs, contributing to 10% of the total set of differentially expressed genes. TAs were noted in previous microarray studies comparing M. bovis with M. tuberculosis (Golby et al. 2007) and ancient with modern strains (Homolka et al. 2010). TA systems were originally identified by their role in plasmid maintenance but are now recognized as a common feature of bacterial genomes (Pandey and Gerdes 2005). With 62 annotated TA pairs (Lew et al. 2011), M. tuberculosis has more TAs than any other bacterium (Pandey and Gerdes 2005; Makarova et al. 2009). The toxin component is typically an endonuclease, with activity directed toward ribosome-associated mRNAs, rRNAs, and tmRNA, resulting in blockage of translation. An attractive hypothesis is that the role of TAs in M. tuberculosis is to drive the bacteria into reversible growth arrest in unfavorable environments, by responding to changes in antitoxin stability and proteolytic activities. Based on this model, we interpret their differential expression as a read-out of lineage differences in environmental sensing. Comparison of the overall TA transcription response suggests that the core lineage pattern is overlaid by strain-specific responses, and it can be envisaged that variability in the combined proteolytic and transcriptional regulatory network could readily generate heterogeneity within clonal populations.
Although it is clear that genotypic diversity generates transcriptional diversity between the two MTBC lineages, it remains to be shown whether this has biological and clinical consequences during infection. Both lineages are highly successful pathogens with proven long-term ability to maintain transmission cycles and it is likely that phenotypic diversity will reflect evolution under different circumstances rather than loss or gain of ability to cause disease. The differences that we have detected suggest that strains from the two lineages may present alternative ligand repertoires to host cells and respond differently to environmental changes generated by the host immune response. This in turn may confer varying degrees of fitness in different epidemiological settings.
Supplementary Material
Supplementary figures S1–S4 and tables S1–S12 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to the High-Throughput Sequencing team at the National Institute for Medical Research for library sequencing. This work was supported by the Medical Research Council (grant number U117581288), the European 7th Framework Program SysteMTb, the Swiss National Science Foundation (grant number PP0033–119205), and the National Institutes of Health (grant numbers AI090928 and HHSN266200700022C). I.C. is supported by European Union funding from the Marie Curie Framework Programme 7 actions (project 272086).
Literature Cited
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnvig K, Young D. Non-coding RNA and its potential role in Mycobacterium tuberculosis pathogenesis. RNA Biol. 2012;9:427–36. doi: 10.4161/rna.20105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnvig KB, et al. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002342. doi: 10.1371/journal.ppat.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi G, Chauhan S, Sharma D, Tyagi JS. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology. 2005;151:4045–4053. doi: 10.1099/mic.0.28333-0. [DOI] [PubMed] [Google Scholar]
- Barkan D, Hedhli D, Yan HG, Huygen K, Glickman MS. Mycobacterium tuberculosis lacking all mycolic acid cyclopropanation is viable but highly attenuated and hyperinflammatory in mice. Infect Immun. 2012;80:1958–1968. doi: 10.1128/IAI.00021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell S, Gagneux S. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2009;13:1456–1466. [PubMed] [Google Scholar]
- Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci. 2005;30:672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, Gagneux S. The past and future of tuberculosis research. PLoS Pathog. 2009;5: e1000600. doi: 10.1371/journal.ppat.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, Homolka S, Niemann S, Gagneux S. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One. 2009;4:e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, et al. Out-of-Africa migration and Neolithic co-expansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–82. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes T, et al. Genome-Wide Mapping of Transcriptional Start Sites Defines an Extensive Leaderless Transcriptome in Mycobacterium tuberculosis. Cell Reports. 2013 doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscolla M, Gagneux S. Does M tuberculosis genomic diversity explain disease diversity? Drug Discov Today Dis Mech. 2010;7:e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley D, et al. Recent and rapid emergence of W-Beijing strains of Mycobacterium tuberculosis in Cape Town, South Africa. Clin Infect Dis. 2008;47:1252–1259. doi: 10.1086/592575. [DOI] [PubMed] [Google Scholar]
- Cox MP, Peterson DA, Biggs PJ. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P, Kolly GS, Leon-Solis L, Fallow A, Reed MB. Massive gene duplication event among clinical isolates of the Mycobacterium tuberculosis W/Beijing family. J Bacteriol. 2010;192:4562–4570. doi: 10.1128/JB.00536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallow A, Domenech P, Reed MB. Strains of the East Asian (W/Beijing) lineage of Mycobacterium tuberculosis are natural mutants in the DosS/DosT-DosR two-component regulatory system. J Bacteriol. 2010;12:2228–2238. doi: 10.1128/JB.01597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MA, Plikaytis BB, Shinnick TM. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol. 2002;184:4025–4032. doi: 10.1128/JB.184.14.4025-4032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann RD, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golby P, et al. Comparative transcriptomics reveals key gene expression differences between the human and bovine pathogens of the Mycobacterium tuberculosis complex. Microbiology. 2007;153:3323–3336. doi: 10.1099/mic.0.2007/009894-0. [DOI] [PubMed] [Google Scholar]
- Gurcha SS, et al. Ppm1, a novel polyprenol monophosphomannose synthase from Mycobacterium tuberculosis. Biochem J. 2002;365:441–450. doi: 10.1042/BJ20020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolka S, Niemann S, Russell DG, Rohde KH. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 2010;6:e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Lasa I, et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci U S A. 2011;108:20172–20177. doi: 10.1073/pnas.1113521108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList—10 years after. Tuberculosis (Edinb) 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciag A, et al. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J Bacteriol. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Foot M, Gey van Pittius NC. The complex architecture of mycobacterial promoters. Tuberculosis (Edinb) 2013;93:60–74. doi: 10.1016/j.tube.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X, et al. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology. 2007;153:1229–1242. doi: 10.1099/mic.0.29281-0. [DOI] [PubMed] [Google Scholar]
- Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parwati I, van Crevel R, van Soolingen D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis. 2010;10:103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- Portevin D, Gagneux S, Comas I, Young D. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 2011;7:e1001307. doi: 10.1371/journal.ppat.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40: D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan R, Sloan DB, Ochman H. Antisense transcription is pervasive but rarely conserved in enteric bacteria. MBio. 2012;3:e00156-12. doi: 10.1128/mBio.00156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- Ramaswamy SV, et al. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003;47:1241–1250. doi: 10.1128/AAC.47.4.1241-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana AK, et al. Ppm1-encoded polyprenyl monophosphomannose synthase activity is essential for lipoglycan synthesis and survival in mycobacteria. PLoS One. 2012;7:e48211. doi: 10.1371/journal.pone.0048211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V, Gao F, Chen B, Jacobs WR, Jr, Glickman MS. Trans-cyclopropanation of mycolic acids on trehalose dimycolate suppresses Mycobacterium tuberculosis-induced inflammation and virulence. J Clin Invest. 2006;116:1660–1667. doi: 10.1172/JCI27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2008. [cited 2013 Oct 5]. Available from: http://www.R-project.org. [Google Scholar]
- Reed MB, Gagneux S, Deriemer K, Small PM, Barry CE., 3rd The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. J Bacteriol. 2007;189:2583–2589. doi: 10.1128/JB.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, et al. Genome-wide regulon and crystal structure of BlaI (Rv1846c) from Mycobacterium tuberculosis. Mol Microbiol. 2009;71: 1102–1116. doi: 10.1111/j.1365-2958.2008.06583.x. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch AC, et al. Mutations in the regulatory network underlie the recent clonal expansion of a dominant subclone of the Mycobacterium tuberculosis Beijing genotype. Infect Genet Evol. 2011;11:587–597. doi: 10.1016/j.meegid.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Singh A, Jain S, Gupta S, Das T, Tyagi AK. mymA operon of Mycobacterium tuberculosis: its regulation and importance in the cell envelope. FEMS Microbiol Lett. 2003;227:53–63. doi: 10.1016/S0378-1097(03)00648-7. [DOI] [PubMed] [Google Scholar]
- Singh A, et al. Requirement of the mymA operon for appropriate cell wall ultrastructure and persistence of Mycobacterium tuberculosis in the spleens of guinea pigs. J Bacteriol. 2005;187:4173–4186. doi: 10.1128/JB.187.12.4173-4186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrelles JB, Schlesinger LS. Diversity in Mycobacterium tuberculosis mannosylated cell wall determinants impacts adaptation to the host. Tuberculosis (Edinb) 2010;90:84–93. doi: 10.1016/j.tube.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrelles JB, et al. Inactivation of Mycobacterium tuberculosis mannosyltransferase pimB reduces the cell wall lipoarabinomannan and lipomannan content and increases the rate of bacterial-induced human macrophage cell death. Glycobiology. 2009;19:743–755. doi: 10.1093/glycob/cwp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walderhaug MO, et al. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J Bacteriol. 1992;174:2152–2159. doi: 10.1128/jb.174.7.2152-2159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, et al. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 2012;8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Hu GQ, She ZS, Zhu H. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics. 2011;12:361. doi: 10.1186/1471-2164-12-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.