Abstract
Horizontal gene transfer (HGT) of DNA from the plastid to the nuclear and mitochondrial genomes of higher plants is a common phenomenon; however, plastid genomes (plastomes) are highly conserved and have generally been regarded as impervious to HGT. We sequenced the 158 kb plastome and the 690 kb mitochondrial genome of common milkweed (Asclepias syriaca [Apocynaceae]) and found evidence of intracellular HGT for a 2.4-kb segment of mitochondrial DNA to the rps2–rpoC2 intergenic spacer of the plastome. The transferred region contains an rpl2 pseudogene and is flanked by plastid sequence in the mitochondrial genome, including an rpoC2 pseudogene, which likely provided the mechanism for HGT back to the plastome through double-strand break repair involving homologous recombination. The plastome insertion is restricted to tribe Asclepiadeae of subfamily Asclepiadoideae, whereas the mitochondrial rpoC2 pseudogene is present throughout the subfamily, which confirms that the plastid to mitochondrial HGT event preceded the HGT to the plastome. Although the plastome insertion has been maintained in all lineages of Asclepiadoideae, it shows minimal evidence of transcription in A. syriaca and is likely nonfunctional. Furthermore, we found recent gene conversion of the mitochondrial rpoC2 pseudogene in Asclepias by the plastid gene, which reflects continued interaction of these genomes.
Keywords: Asclepias syriaca, gene conversion, horizontal gene transfer, mitochondrial genome, phylogeny, plastome
Introduction
Horizontal gene transfer (HGT) is the phenomenon in which genetic material is transmitted laterally between organisms or between genomes within organisms, rather than vertically though sexual reproduction (Keeling and Palmer 2008; Bock 2010; Renner and Bellot 2012). This process can occur irrespective of the relatedness of the organisms and occurs frequently between prokaryotes and eukaryotes (Keeling and Palmer 2008; Bock 2010). HGT between genomes in plants takes place through intracellular transfer of DNA among the nuclear, plastid, and mitochondrial genomes. In land plants, HGT of plastid DNA (ptDNA) to the mitochondrial genome and transfer of ptDNA and mitochondrial DNA (mtDNA) to the nuclear genome are well documented (Richardson and Palmer 2007; Keeling and Palmer 2008; Bock 2010; Smith 2011; Renner and Bellot 2012). However, the highly conserved land plant plastome has long been considered “essentially immune” to HGT (Richardson and Palmer 2007, p. 7; Smith 2011), until the recent reports of a single case of movement of mtDNA into the plastome (Goremykin et al. 2009; Iorizzo et al. 2012a, 2012b).
In the single confirmed case of mitochondrial to plastid HGT, mtDNA including a putative mitochondrial cox1 pseudogene was discovered in the cultivated carrot (Daucus carota) plastome by Blast analyses during the characterization of the grape mitochondrial genome (Goremykin et al. 2009) and confirmed by sequencing of the carrot mitochondrial genome (Iorizzo et al. 2012b). Iorizzo et al. (2012a) proposed transposition via a non-long terminal repeat (LTR) retrotransposon as the mechanism of HGT and by surveying the plastomes of other members of Apiaceae found that six additional species of Daucus and its close relative, cumin (Cuminum cyminum), share the DNA transfer from the mitochondrial to plastid genomes.
A putative mitochondrial to plastid HGT of an rpl2 pseudogene was detected by Ku et al. (2013) in the common milkweed (Asclepias syriaca L.: Apocynaceae) based on Blast similarity searches. Here, we confirm transfer of mtDNA, including the rpl2 pseudogene (ψrpl2), into the plastome of A. syriaca based on the full genome sequences of the plastome and mitochondrial genome for this species. Furthermore, we suggest that homologous recombination mechanisms that function to repair double-strand breaks in plastomes enabled the movement of mtDNA into a highly conserved plastome, a process that was facilitated by the presence of plastid sequence in the mitochondrial genome from earlier HGT events. We place this gene transfer event in an evolutionary context by showing the phylogenetic distribution of the insertion across subfamily Asclepiadoideae and determine that it occurred in the common ancestor of tribe Asclepiadeae and demonstrate that the exogenous ptDNA has been retained relatively intact in all daughter lineages. Through additional phylogenetic analyses of homologous sequences in the plastid and mitochondrial genomes, we determined that there has been ongoing exchange between the two genomes leading to gene conversion of a mitochondrial pseudogene (ψrpoC2) in Asclepias. Finally, by means of transcriptome sequencing for A. syriaca, we show that only a small portion of the horizontally transferred segment, not including ψrpl2, is transcribed leading us to suggest that most of the segment lacks function, although the expressed ends may have some regulatory role.
Materials and Methods
Illumina Library Preparation and Sequencing
We extracted DNA using one of the following kits or methods: FASTDNA kit (MP Bio), DNeasy kit (Qiagen), or the cetyl trimethylammonium bromide method (Doyle and Doyle 1987). We prepared Illumina libraries for A. syriaca and 11 other Asclepiadoideae (table 1) following (Straub et al. 2011) or using the gel-free size selection protocol described in Straub et al. (2012) for Asclepias nivea and Secamone afzelii. We obtained mate pair libraries for A. syriaca with insert sizes of 2, 3.6, 4.5, and 5 kb from Global Biologics (Columbia, MO). We sequenced either single-end or paired-end Illumina short reads (36, 80, or 101 bp) for each library on Illumina HiSeq 2000, Illumina GAIIx, or Illumina MiSeq sequencers at either the Oregon State University Center for Genome Research and Biocomputing (OSU-CGRB) or the Oregon Health and Science University.
Table 1.
Survey of Species of Apocynaceae for the Presence of the mtDNA Insert in the Plastome and the Presence of ψrpoC2 in the Mitochondrial Genome Downstream of rpl2
| Subfamily | Tribe | Subtribe | Species | Voucher | rps2–rpoC2 Intergenic Spacer Size Similar to A. syriaca | rps2–rpoC2 Intergenic Spacer Method of Confirmation | PCR Product for rpl2–ψrpoC2 | Informatically Determined ψrpoC2 |
|---|---|---|---|---|---|---|---|---|
| Apocynoideae | Baisseeae | Oncinotis tenuiloba Stapf | S. Liede 3621 (UBT) | No | PCR | No | n/a | |
| Asclepiadoideae | Asclepiadeae | Asclepiadinae | Asclepias nivea L. | M. Fishbein 6061 (OKLA) | Yes | PCR, NGS | Yes | Yesa |
| Asclepias syriaca L. | W. Phippen 5 (OSC) | Yes | PCR, NGS | Yes | Yes | |||
| Astephaninae | Astephanus triflorus R. Br. | P. Bruyns 8901E (BOL) | Yes | NGS | Yes | Yes | ||
| Cynanchinae | Cynanchum leave (Michx.) Pers. | M. Fishbein 5228 (OKLA) | Yes | PCR | Yes | No | ||
| Raphistemma pulchellum Wall. | D. Middleton, P. Karaket, S. Lindsay, T. Phutthai, and S. Suddee 5103 (E) | Yes | PCR | Yes | No | |||
| Seutera angustifolia (Pers.) Fishbein & W.D. Stevens | M. Fishbein 5604 (OKLA) | Yes | PCR | Yes | No | |||
| Eustegiinae | Eustegia minuta (L.f.) N.E.Br. | P. Bruyns 4357 (BOL) | Yes | PCR, NGS | Yesb | Yes | ||
| Gonolobinae | Dictyanthus pavonii Decne. | M. Fishbein 5842 (OKLA) | Yes | PCR | Yes | No | ||
| Matelea biflora (Raf.) Woodson | A. Rein 39 (OKLA) | Yes | NGS | Yes | Yes | |||
| Metastelmatinae | Metastelma californicum Benth. | M. Fishbein 6436 (OKLA) | Yes | PCR | Yes | No | ||
| Orthosiineae | Orthosia scoparia (Nutt.) Liede & Meve | M. Fishbein 5280 (OKLA) | Yes | NGS | Yes | Yes | ||
| Oxypetalinae | Araujia sericifera Brot. | M. Fishbein 6070 (OKLA) | Yes | NGS | No | Yesa | ||
| Funastrum arenarium (Decne. ex Benth.) Liede | M. Fishbein 6442 (OKLA) | Yes | PCR | Yes | No | |||
| Tylophorinae | Vincetoxicum rossicum (Kleopow) Barbar. | T. Livshultz 03-31 (BH) | Yes | PCR, NGS | Yes | Yes | ||
| Ceropegieae | Aniostominae | Sisyranthus trichostomus K. Schum. | C. Peter 3760c | No | PCR, NGS | No | Yes | |
| Heterostemminae | Heterostemma piperifolium King & Gamble | D.J. Middleton 194 (A) | No | PCR, NGS | Yes | No | ||
| Fockeeae | Fockea edulis K. Schum. | T. Livshultz s.n. (BH) | No | PCR | Yes | No | ||
| Marsdenieae | Marsdenia astephanoides (A.Gray) Woodson | M. Fishbein 5104 (OKLA) | No | PCR, NGS | Yes | Yes | ||
| Telosma cordata (Burm. f.) Merr. | T. Livshultz 01-33 (BH) | No | PCR, NGS | Yesb | Yes | |||
| Periplocoideae | Periploca sepium Bunge | T. Livshultz s.n. (PH) | No | PCR | No | n/a | ||
| Secamonoideae | S. afzelii (Roem. & Schult.) K.Schum. | F. Billiet S2211 (BR) | No | PCR, NGS | No | Yes |
Note.—n/a, not applicable.
aPartial sequence determined (see main text).
bConfirmed by Sanger sequencing.
cPersonal spirit-preserved collection of Dr Craig Peter, Department of Botany, Rhodes University, Grahamstown, South Africa.
Sequence Assembly and Analysis
We used Alignreads v. 2.25 (Straub et al. 2011) and the A. syriaca reference plastome we previously sequenced (GenBank: JF433943.1) with one copy of the inverted repeat removed for reference-guided plastome assembly of a random subset of 5 million reads from the A. syriaca data set. We conducted Blast (Altschul et al. 1997) searches (BlastN) against the GenBank nucleotide database to identify regions of similarity to plant mitochondrial genomes in the insert region. We used Velvet 1.0.12 (Zerbino and Birney 2008) and Velvet Optimiser (http://bioinformatics.net.au/software.velvetoptimiser.shtml, last accessed July 4, 2013) with a hash length of 71, expected coverage of 31, and coverage cutoff of 0.326 to perform de novo assembly of the mitochondrial genome of A. syriaca. We trimmed the A. syriaca mate pair reads to 30 bp (2, 3.6, 4.5 kb libraries—101 bp original length) or removed bases below Q30 on either end of the reads and retained reads ≥30 bp (5 kb library—36 bp original length) using Trimmomatic v. 0.20 (Lohse et al. 2012). The trimmed mate pair reads were utilized in SSPACE 2.0 (Boetzer et al. 2011) to scaffold the mitochondrial de novo contigs. We used Geneious 6.1.4 (Biomatters Ltd.) to design polymerase chain reaction (PCR) primers (supplementary table S1, Supplementary Material online) to fill gaps between scaffolds using standard Sanger dye-termination sequencing (Applied Biosystems). We further used the PCR results to order scaffolds into a master circle. Undetermined sequence in the Velvet assembly was ascertained either by read mapping in Geneious or reference-guided assembly in Alignreads.
We identified homologous regions between the plastome and mitochondrial genome using BLAT (Kent 2002) and annotated these regions using Mitofy (Alverson et al. 2010). We aligned assembly contigs and consensus sequences with a selection of asterid plastome sequences available in GenBank (table 2) using MAFFT v. 6.857 (Katoh and Toh 2008) and compared them in BioEdit v. 7.0.5.3 (Hall 1999) to determine insert sizes (length of nonhomologous regions). We confirmed the 5′ and 3′ ends of the insert sequence in the A. syriaca plastome by PCR (1× Phusion® Flash High-Fidelity PCR Master Mix [Finnzymes], 0.2 µM forward primer 5′-ACACTCTCGTAGCGCCGTATAGTCTT-3′, 0.2 µM reverse primer 5′-GGTTCAAAGGATTAGTGCACCCTTCA-3′, cycling conditions: 30 s 98 °C, 25 cycles of 10 s 98 °C, 30 s 61 °C, 90 s 72 °C, extension 10 min 72 °C) followed by standard Sanger dye-termination sequencing.
Table 2.
List of Asterid Plastome Sequences Downloaded from NCBI and Aligned to Determine the Size of the Typical Asterid rps2–rpoC2 Intergenic Spacer and Stop Codon
| Order | Family | Species | NCBI Accession Number | Size ofrps2–rpoC2 (bp) | Canonical Stop Codon Utilized? |
|---|---|---|---|---|---|
| Apiales | Apiaceae | Anthriscus cerefolium | NC_015113.1 | 220 | Yes |
| Daucus carota | NC_008325.1 | 209 | Yes | ||
| Araliaceae | Panax ginseng | NC_006290.1 | 212 | Yes | |
| Asterales | Asteraceae | Ageratina adenophora | NC_015621.1 | 260 | No |
| Guizotia abyssinica | NC_010601.1 | 246 | No | ||
| Helianthus annuus | NC_007977.1 | 327 | No | ||
| Jacobaea vulgaris | NC_015543.1 | 221 | No | ||
| Lactuca sativa | NC_007578.1 | 241 | No | ||
| Parthenium argentatum | NC_013553.1 | 286 | No | ||
| Ericales | Ericaceae | Vaccinium macrocarpon | NC_019616.1 | 262 | No |
| Primulaceae | Ardisia polysticta | NC_021121.1 | 221 | Yes | |
| Theaceae | Camellia sinensis | NC_020019.1 | 247 | No | |
| Gentianales | Rubiaceae | Coffea arabica | NC_008535.1 | 213 | Yes |
| Lamiales | Lamiaceae | Salvia miltiorrhiza | NC_020431.1 | 208 | Yes |
| Tectona grandis | HF567869.1 | 208 | Yes | ||
| Oleaceae | Jasminum nudiflorum | NC_008407.1 | 258 | Yes | |
| Olea europaea | NC_013707.2 | 208 | Yes | ||
| Olea woodiana | NC_015608.1 | 208 | Yes | ||
| Pedaliaceae | Sesamum indicum | KC569603.1 | 209 | Yes | |
| Solanales | Convolvulaceae | Ipomoea purpurea | NC_009808.1 | 213 | Yes |
| Solanaceae | Atropa belladonna | NC_004561.1 | 221 | Yes | |
| Nicotiana sylvestris | NC_007500.1 | 227 | Yes | ||
| Nicotiana tabacum | NC_001879.2 | 227 | Yes | ||
| Nicotiana tomentosiformis | NC_007602.1 | 222 | Yes | ||
| Nicotiana undulata | NC_016068.1 | 227 | Yes | ||
| Solanum bulbocastanum | NC_007943.1 | 228 | Yes | ||
| Solanum lycopersicum | NC_007898.2 | 225 | Yes | ||
| Solanum tuberosum | NC_008096.2 | 225 | Yes | ||
We assembled the A. nivea plastome sequence using a combination of de novo and reference-guided assembly following (Straub et al. 2011) and using the A. syriaca reference (GenBank: JF433943.1). For other Asclepiadoideae, we made reduced random read pools providing approximately 125× sequencing depth of the plastome, or used all reads if <125× depth was obtained, and then conducted reference-guided assembly in Alignreads using the A. nivea sequence as a reference, the medium similarity setting, and the masking parameters and evaluation criteria of Straub et al. (2012). We performed a second iteration of this analysis using the result of the first assembly as the reference in cases where the whole insert region was not assembled in a single contig during the first round of assembly.
Sequencing Depth Estimation in A. syriaca
We removed duplicate sequences from the read pool to eliminate PCR duplicates that could potentially inflate the observed sequencing depth using custom python scripts for paired-end reads and the FASTX-toolkit (http://codex.cshl.org/labmembers/gordon/fastx_toolkit/, last accessed July 24, 2013) for single-end reads. We then used the assembled A. syriaca plastome and mitochondrial genome sequences to map reads in BWA v. 0.5.7 (Li and Durbin 2009). To exclude reads that may have originated in the other genome, we used only perfect matches to construct read pileups using SAMtools v. 0.1.7 (Li et al. 2009). We used the pileups to determine the number of reads with starts mapping in the insert regions. To ensure that only reads from the correct genome were counted, in regions of 100% sequence similarity we excluded the read starts for reads wholly fitting into the window of similarity from the counts. This estimate is conservative because reads that did originate from the genome in question will have been eliminated. We then sampled six sequence regions of the same size as the inserted segment from the surrounding areas of the plastome. Each was separated from the insert segment and each other sample by the read length (80 bp), so that no read was present in two windows. We counted read starts in these windows and used them to construct a 95% confidence interval for the number of reads expected to be found for each genome per sequence window.
PCR Screening for the Insert in Asclepiadoideae Plastomes and the rpoC2 Pseudogene in Asclepiadoideae Mitochondrial Genomes
To screen for the presence of the insert in the rps2–rpoC2 intergenic spacer in other Asclepiadoideae, we used the same PCR primers used in A. syriaca and the same PCR conditions with the exception of lowering the annealing temperature to either 55 °C or 50 °C to achieve amplification in some species.
To screen for the presence of an rpoC2 pseudogene in the mitochondrial genomes of Asclepiadoideae, we designed one PCR primer in the most divergent region between the plastid rpl2 pseudogene and mitochondrial rpl2 (5′-GCGATTGGTCTAAACCTTCGA-3′). To further increase specificity, we incorporated a penultimate mismatch (Cha et al. 1992). We designed three reverse primers using the A. syriaca mitochondrial rpoC2 as a guide (R1: 5′-ACCATTGATCATTTTGTTATCATCCA-3′; R2: 5′-ACATATGAAATAGCGGACGGTCTA-3′; R3: 5′-TGTCAAAGAGGCGAAGAAATCT-3′). We used the PCR conditions given above modified by decreasing the annealing temperature to 55 °C and increasing the number of cycles to 30.
Sequencing and Assembly of rpoC2/ψrpoC2
To informatically assemble ψrpoC2, we used BLAT to identify reads that hit the plastid rpoC2 sequence of each species for which we collected Illumina data. We determined the sequence of ψrpoC2 by using the plastid sequence as a reference and mapping reads with one or more mismatches or those supporting indels versus the plastid sequence in Geneious using the Geneious assembler with custom sensitivity settings to allow gaps with a maximum of 50% per read and size of 60 bp, a word length of 24 with words repeated more than eight times ignored, a maximum of 25% mismatches per read, an index word length of 14, and a maximum ambiguity of four. We mapped multiple best matches randomly and did not use the fine tuning option. We detected gaps larger than those recovered by read mapping with BLAT. We checked portions of the informatically determined sequence for accuracy using primers R1 and R2 as sequencing primers in Eustegia and Telosma, and R3 and an additional primer (R4: 5′-TCTAATGGAAAAAGCAAATTGAATGA-3′) in A. syriaca, and employed the ψrpoC2 PCR protocol and standard Sanger dye-termination sequencing described above. We used the same read mapping approach to detect variants of the rps2–rpoC2 intergenic spacer across Asclepiadoideae that could correspond to the homologous region of the mitochondrial genome if present in species other than A. syriaca.
Phylogenetic Analyses
We prepared a matrix for phylogenetic analysis by aligning plastome sequences in MAFFT v. 6.864b. Due to the difficulty in assembling putative pseudogenes accD and ycf1 in Asclepias (Straub et al. 2011), we excised these regions from the alignment. We used GBlocks v. 0.91b (Talavera and Castresana 2007) with default settings (minimum of seven and ten sequences for a conserved position and a flank position, respectively; maximum of eight nonconserved positions; minimum length of 10 bp in a block), modified by allowing positions where half of the sequences contained gaps, to remove unalignable and poorly aligned regions of the matrix. We analyzed this final matrix (supplementary data set S1, Supplementary Material online) by utilizing CIPRES resources (Miller et al. 2010) to run RAxML v. 7.6.3 (Stamatakis 2006; Stamatakis et al. 2008) with the following settings: -p 6540444 -x 18884 -N 1000 -k -f a -m GTRCAT. The maximum likelihood (ML) search and 1,000 rapid bootstrapping replicates were performed in a single run. The tree was rooted using S. afzelii, a member of subfamily Secamonoideae, the sister subfamily of Asclepiadoideae, as an outgroup.
We prepared a second matrix for phylogenetic analysis by aligning the sequences of rpoC2 from the plastomes of each species, the informatically assembled ψrpoC2 sequence from each species, and an outgroup plastid rpoC2 sequence from Nerium oleander (GenBank: GQ997692.1), a member of Apocynoideae (another subfamily of Apocynaceae), using MAFFT. We conducted a phylogenetic analysis of the final matrix (supplementary data set S2, Supplementary Material online) using RAxML through CIPRES with the following settings: -p 1648144 -x 2465477 -N 5000 -k -f a -m GTRGAMMA. The ML search and 5,000 rapid bootstrapping replicates were performed in a single run.
Chloroplast Transcriptome Sequencing and Analysis
We mechanically homogenized fresh frozen leaves (∼200 mg) from A. syriaca (W. Phippen 5 (OSC)) on dry ice in a FastPrep-24 bead mill (MP Bio). We added 1.5 ml of cold extraction buffer (3 M LiCl/8 M urea; 1% PVP K-60; 0.1 M dithiothreitol [Tai et al. 2004]) to the ground tissue, homogenized the tissue, and pelleted cellular debris at 200 × g for 10 min at 4 °C. The supernatant was incubated overnight at 4 °C, RNA was pelleted by centrifugation (20,000 × g for 30 min at 4 °C), and cleaned using the ZR Plant RNA MiniPrep kit (Zymo Research). We prepared an RNA-Seq library using 2 µg total RNA, followed by ribosomal RNA subtraction (RiboMinus™; Life Technologies), and standard TruSeq reagents (Illumina Inc.) modified to enable strand-specific sequencing by dUTP incorporation (Parkhomchuk et al. 2009). In this approach, second strand synthesis is supplemented with a dUTP/dNTP mixture (Fermentas ThermoScientific) in place of standard dNTPs. Prior to enrichment PCR, uracil-containing sites are degraded with a uracil-specific excision reagent mixture (New England Biolabs). We enriched the library using standard TruSeq amplification primers, and a 12-pM aliquot was sequenced at OSU-CGRB using an Illumina HiSeq 2000 to obtain 101 bp single-end reads.
We removed reads not passing the Illumina chastity and purity filters and trimmed all reads to 100 bp. We used Trimmomatic to trim leading and trailing bases less than Q20, to trim the rest of the sequence when the average quality in a sliding window of 5 bp fell below Q30, and to exclude reads <36 bp following trimming, which resulted in a final data set of 24.6 M reads. We determined expression of the insert region and flanking operons by mapping the quality-filtered reads back to the plastome reference sequence using BWA. To estimate sequencing depth, we calculated the average depth in a sliding window of 20 bp across the atpI–rpoC2 region. We also calculated reads per kilobase of exon model per million mapped reads (RPKM) values (Mortazavi et al. 2008) separately for sense and antisense reads mapped to the insert region and flanking genes using Artemis v. 15.1.1 (Carver et al. 2012) and BamView v. 1.2.9 (Carver et al. 2013).
Results
Sequencing and Assembly of the A. syriaca Plastome and Mitochondrial Genome Confirm Intracellular HGT
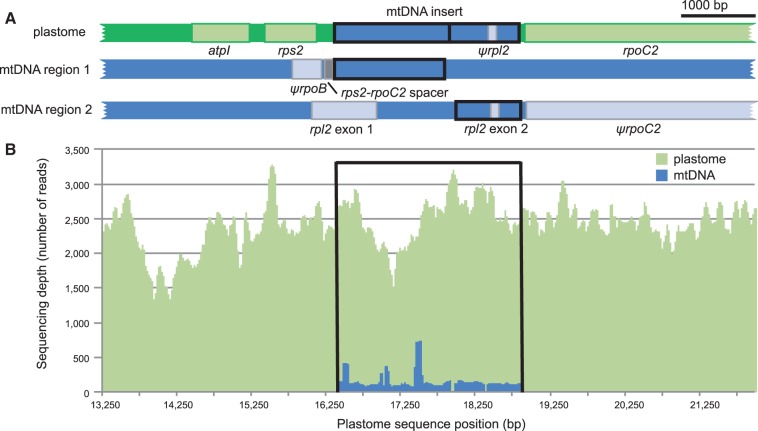
Sequencing and assembly of a 158,719-bp A. syriaca plastome (GenBank: KF386166) confirmed a 2,427-bp insertion in the rps2–rpoC2 intergenic spacer (2,677 bp total length; fig. 1A) relative to other angiosperms in the asterid clade (ca. 80,000 species). This insertion is also present in the plastome of a second A. syriaca individual (GenBank: JF433943.1) that we previously sequenced (Straub et al. 2011; Ku et al. 2013), and this region in the newly sequenced individual differs by the insertion of 21 bp, 16 of which are repeated upstream of the insertion, and deletion of 21 bp that comprises a direct repeat in the first individual. The rps2–rpoC2 intergenic spacer ranges from 208 to 327 bp in 28 other photosynthetic asterid species with plastomes in GenBank (table 2). Blast (Altschul et al. 1997) searches of the GenBank nucleotide database using the A. syriaca rps2–rpoC2 spacer region as a query returned high confidence hits to plant mitochondrial genomes, including grape (Vitis vinifera), papaya (Carica papaya), and watermelon (Citrullus lanatus). The putatively homologous mitochondrial region includes rpl2, confirming that the A. syriaca plastome contains a pseudogene (ψrpl2) consisting of the second exon of mitochondrial rpl2. Assembly of the approximately 690 kb milkweed mitochondrial genome from the same individual that we used for plastome sequencing revealed that it contains sequences homologous to the inserted segment in the plastome (fig. 1A), which are split between two regions of 1,091 and 1,401 bp, and separated by 55,845 bp in the master circle of the contemporary A. syriaca mitochondrial genome.
Fig. 1.—
Region of horizontal transfer from the mitochondrial genome to the plastome of Asclepias syriaca. (A) Diagram of the A. syriaca plastome and homologous regions of the mitochondrial genome. The mtDNA insert in the plastome and homologous regions in the mitochondrial genome are indicated by black boxes. (B) Sequencing depth of the A. syriaca plastome surrounding the mtDNA insert and the homologous region of the mitochondrial genome.
To confirm that the appearance of mitochondrial sequence in the plastome was not the product of misassembly, we utilized the quantitative nature of short read sequencing and the fact that ptDNA is typically an order of magnitude more abundant than mtDNA in Asclepias genomic DNA extractions (Straub et al. 2011; Straub et al. 2012). A total of 72,004 reads mapped to the mtDNA insert in the plastome, following a correction for reads that could have originated from the mitochondrial genome due to 100% sequence similarity in blocks that exceed the read length. This value falls within the 95% confidence interval (65,947–76,521) of reads mapped to equal-sized partitions of the surrounding single-copy portion of the plastome and differs markedly from the corrected value of 3,509 reads mapped to the homologous regions of the mitochondrial genome. The inserted region therefore has similar sequencing depth to the surrounding plastome, rather than the mitochondrial genome, which demonstrates that it is not an assembly artifact (fig. 1B).
The plastome insert and the mitochondrial sequence have a pairwise sequence divergence of 0.072 substitutions per site and differ by 29 indels with a total length of 251 bp. The pairwise sequence divergence between ψrpl2 and the mitochondrial rpl2 is 0.089 substitutions per site. Both the plastid and mitochondrial sequences have the conserved stop codon characteristic of core eudicots that separates the portion of the gene encoded in the mitochondrial genome (5′ rpl2 sensu Adams et al. 2001) and the portion presumably transferred to the nuclear genome (Adams et al. 2001). In the translated pseudogene sequence, five of the substitutions relative to mitochondrial rpl2 are nonsynonymous, and one is a nonsense mutation that would lead to a truncation 16 amino acids prior to the conserved end of 5′ rpl2. In addition, the homologous regions of the mitochondrial genome are flanked by sequence of plastid origin, a partial rps2–rpoC2 intergenic spacer on the 5′ end and an rpoC2 pseudogene (ψrpoC2) on the 3′ end (fig. 1A), which has numerous missense, nonsense, and frameshift mutations. We confirmed the presence of the insert in the plastome by PCR amplification and sequencing of the insert region using primers anchored in the flanking plastid genes, one of which (rps2) does not have a pseudogene in the milkweed mitochondrial genome.
Phylogenetic Distribution of the Mitochondrial to Plastid Genome HGT
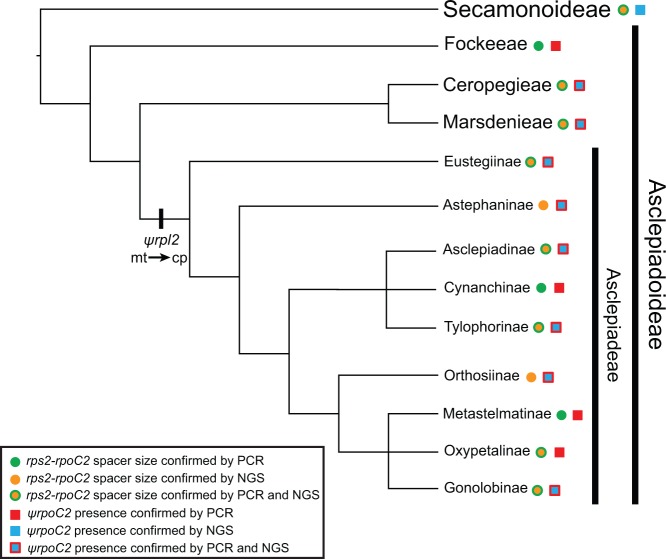
To place the transfer event into an evolutionary context, we surveyed 22 other species of Apocynaceae to determine whether they shared the mitochondrial segment of DNA in the rps2–rpoC2 intergenic spacer (table 1). First, we used PCR to screen 17 species spanning three of four subfamilies of Apocynaceae, with sampling concentrated in Asclepiadoideae, the subfamily to which A. syriaca belongs, to determine whether the spacer region was similar in size to that observed in A. syriaca, or whether it was more typical of other asterids (fig. 2). All members of tribe Asclepiadeae (subfamily Asclepiadoideae) with successful PCR amplification had spacer regions as large as or larger than A. syriaca. The large spacer region was also present in Eustegia (table 1), an enigmatic monotypic African genus that, while possessing synapomorphic features of tribe Asclepiadeae (Bruyns 1999), has been resolved in phylogenetic analyses of plastid sequence data alternatively as sister to the rest of the Asclepiadeae (Liede 2001) or as the sister group of the tribes Marsdenieae and Ceropegieae (Rapini et al. 2003; Meve and Liede 2004; Rapini et al. 2007). Members of tribes Fockeeae, Marsdenieae, and Ceropegieae of Asclepiadoideae and subfamilies Secamonoideae, Periplocoideae, and Apocynoideae did not show evidence of an insertion in the spacer region (table 1).
Fig. 2.—
Evolutionary relationships of the Asclepiadoideae (Apocynaceae) and results of PCR and NGS surveys of the size of the rps2–rpoC2 intergenic spacer in the plastome and for the presence of ψrpoC2 in the mitochondrial genome downstream of rpl2. The topology of the phylogenetic tree of the relationships among tribes of Asclepiadoideae and subtribes of Asclepiadeae is a summary of the plastome phylogeny obtained in this study (fig. 4) augmented with the relationships of major clades not sampled here, as indicated by previous phylogenetic studies of ptDNA sequences (Liede 2001; Liede and Täuber 2002; Rapini et al. 2003; Liede-Schumann et al. 2005; Rapini et al. 2007; Livshultz 2010). The hash mark represents horizontal transfer of DNA from the mitochondrial genome to the plastome in the common ancestor of Asclepiadeae. The circles and squares represent PCR and NGS screening of the size of the rps2–rpoC2 intergenic spacer and presence of ψrpoC2, respectively. The colors of the circles and squares indicate which method was used. Details of species sampling are given in table 1.
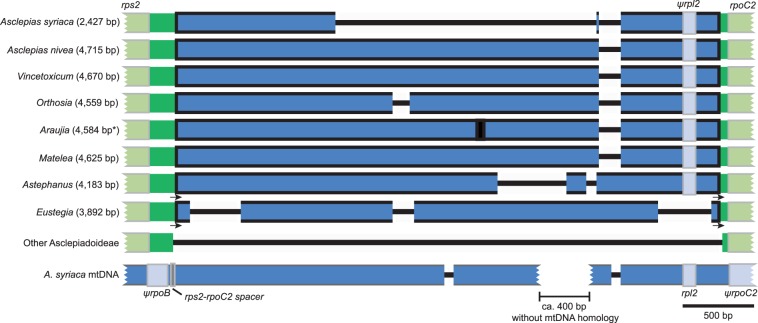
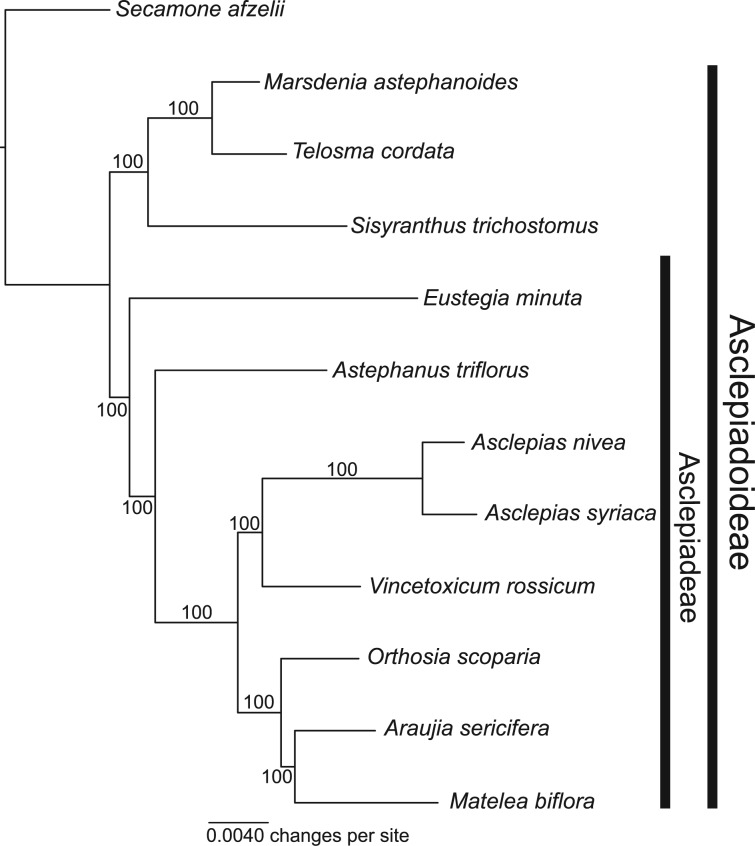
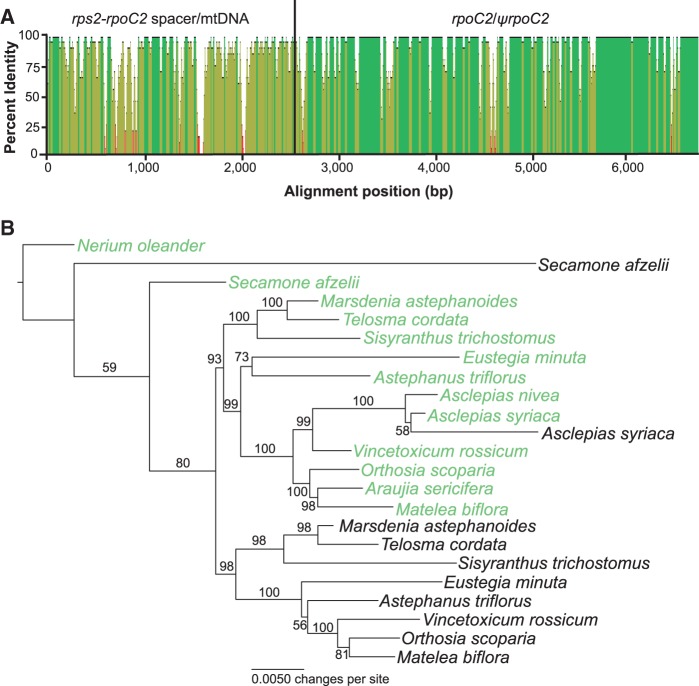
We selected 11 species for whole plastome sequencing and phylogenetic analysis to determine whether the large spacers of tribe Asclepiadeae contained mitochondrial inserts and the order of evolutionary events leading to the mtDNA transfer to the plastome. As expected, we detected inserts ranging in size from 2,427 to 4,715 bp in the rps2–rpoC2 intergenic spacer in the plastome assemblies of all species of Asclepiadeae, including Eustegia, but not in other Apocynaceae (figs. 2 and 3). Comparison of the inserts of other Asclepiadeae with the A. syriaca mitochondrial genome revealed a segment of approximately 400 bp that is absent from the plastome and mitochondrial genome of A. syriaca and does not have significant Blast hits in GenBank. The phylogenetic analysis of the plastome sequences (11,933 variable of 117,206 total characters; −ln likelihood −257,351.74) considering only substitutions, but not indel events, including the insertion from the mitochondrial genome, revealed that Eustegia is sister to the rest of Asclepiadeae (fig. 4) and may be retained in the tribe as its earliest diverging lineage. Therefore, the plastome insertion provides a rare genomic change that is a molecular synapomorphy for Asclepiadeae.
Fig. 3.—
Variability in the mtDNA inserts in the plastomes of Asclepiadeae. Plastome mtDNA inserts are shown as blue boxes outlined in black and their lengths are given in parentheses. Sequence deletions of 50 bp or more relative to the other sequences are shown as black lines. The black box represents undetermined sequence in the Araujia assembly and the asterisk after the length denotes that it is an estimate. The arrows (not drawn to scale) indicate 24 bp imperfect direct repeats observed in Astephanus and Eustegia. The 5′ repeat is identical in Astephanus and Eustegia and the 3′ repeat differs from the 5′ repeat by 1 bp in Astephanus and 2 bp in Eustegia. Representations of the conserved sequence in other sampled Asclepiadoideae (Marsdenia, Sisyranthus, and Telosma) and sequences of the two homologous regions of the mitochondrial genome of A. syriaca are shown for comparison.
Fig. 4.—
ML phylogeny of the Asclepiadoideae (Apocynaceae) based on plastome sequences of major lineages. The numbers shown above or below the branches are bootstrap support values.
Comparison of the 3′-end of aligned plastid rpoC2 sequences showed that multiple distinct mutations have occurred in Apocynaceae that truncate the protein by 2–18 amino acid residues (supplementary fig. S1, Supplementary Material online). The length of the protein is otherwise conserved across most asterids, although Asteraceae and Ericales also exhibit some length variation (table 2).
Phylogenetic Distribution of the Plastid to Mitochondrial HGT
To explore a possible mechanism for the DNA transfer from the mitochondrial genome to the plastome, we used PCR to screen the same set of species evaluated by PCR for the presence of the mtDNA insertion in the plastome for a ψrpoC2 gene flanking the mitochondrial rpl2 gene in the mitochondrial genome. Nearly all surveyed members of Asclepiadoideae showed the presence of the pseudogene based on amplification using a mitochondrial genome-specific primer anchored in rpl2 and three different primers designed from the A. syriaca ψrpoC2 sequence (fig. 2 and table 1). However, samples from subfamilies Secamonoideae and Periplocoideae did not amplify using these primers, indicating that either the pseudogene is not present or the PCR priming sites are divergent or absent. A ψrpoC2 sequence of 499 bp was informatically assembled for S. afzelii, indicating that all reverse primer sites are in fact absent in this species.
Pairwise divergence between rpoC2 and ψrpoC2 in A. syriaca was 0.011 substitutions per site, which is much less than the pairwise divergence observed between the plastome insert and the homologous regions of the mitochondrial genome (fig. 5A), indicating the possibility of more recent exchange between the two genomes. To test for evidence of gene conversion, we informatically assembled the ψrpoC2 sequences for species with sequenced plastomes and obtained Sanger sequences for portions of the pseudogene in A. syriaca, Eustegia, and Telosma to confirm the informatically assembled sequences. Low sequencing depth of the mitochondrial genome and/or the apparent presence of more than one rpoC2 pseudogene prevented the mitochondrial sequences of Araujia and A. nivea from being successfully assembled. Phylogenetic analysis of the successfully determined mitochondrial sequences and the plastid homologs (690 variable of 4,222 total characters; −ln likelihood −11896.01) indicated that there has indeed been recent recombination between the plastid gene and the mitochondrial pseudogene in A. syriaca, as the mitochondrial sequence is strongly supported to share more recent ancestry with plastid sequences than with mitochondrial sequences from other species (fig. 5B). Some of the other mitochondrial sequences share single-nucleotide polymorphisms with the plastid sequence from the same species and may be mosaics containing smaller tracts of gene conversion (Hao and Palmer 2009; Hao et al. 2010) than what we observed in A. syriaca and which are too short to affect the placement of the sequences in the phylogenetic analysis (Hao and Palmer 2011).
Fig. 5.—
Gene conversion between the plastid rpoC2 and mitochondrial ψrpoC2 genes of A. syriaca. (A) Plot of percent identity between homologous regions of the plastome and mitochondrial genomes of A. syriaca over a 20-bp sliding window. Green indicates 100% identity, brown <100% but at least 30% identity, and red <30% identity. Gap positions in the alignment were considered to have no identity. (B) ML phylogeny of plastid rpoC2 and mitochondrial ψrpoC2 sequences. The numbers above or below the branches are bootstrap support values. Plastid gene sequences are indicated in green and mitochondrial pseudogene sequences in black. In the absence of gene conversion or other confounding factors, both the plastid and mitochondrial sequences should produce monophyletic groups with identical branching orders; however, strongly supported placement of the A. syriaca mitochondrial pseudogene sequence in the plastid clade indicates sufficient gene conversion to affect the outcome of the phylogenetic analysis.
Due to the presence of the partial rps2–rpoC2 intergenic spacer observed in the A. syriaca mitochondrial genome, we used this sequence to look for evidence of more than one (i.e., a plastid and a putatively mitochondrial) copy of this sequence. Asclepias nivea, Araujia, Orthosia, Matelea, and Marsdenia all showed evidence of two sequences. The other species did not, but due to the short length of this sequence, it is possible that it is present in all species but has not accumulated mutations in some of them. The observation of two sequences, especially in Marsdenia (tribe Marsdenieae), leaves open the possibility that this sequence was present in the ancestral mitochondrial genome of Asclepiadeae along with ψrpoC2.
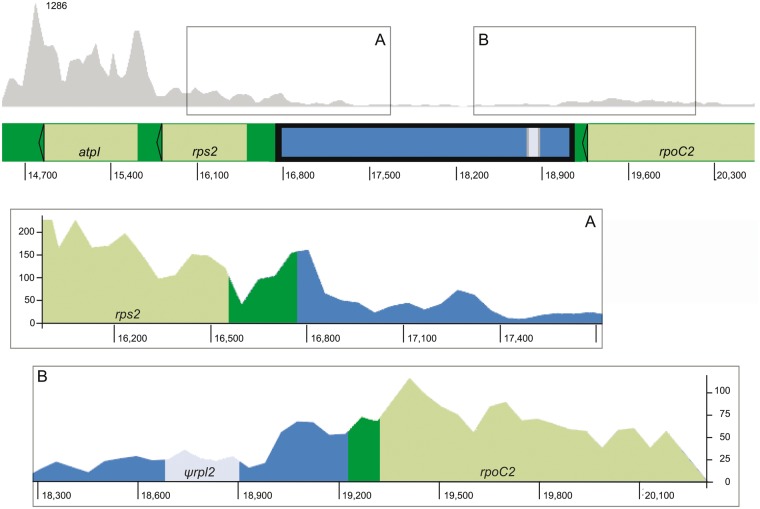
Expression Analysis of the Plastome Insert Region
We evaluated transcriptome sequences derived from leaf tissue of the same A. syriaca individual used in genomic sequencing to determine whether the ψrpl2 exon was expressed at the level of mRNA. Our results show that the flanking operons from rpoC2 (including rpoB, rpoC1, and rpoC2) and rps2 (including rps2, atpA, atpI, and atpH) are actively transcribed in leaf tissue, but that transcripts representing the insert and the ψrpl2 exon are not abundant (fig. 6). The RPKM values for sense (and antisense) mapped reads were 3,298 (6), 854 (1), 149 (12), and 254 (12) for atpI, rps2, the insert, and rpoC2, respectively.
Fig. 6.—
Gene expression levels of the plastome insert and flanking operons in A. syriaca. The gray plot shows relative sequencing depth for plastome regions flanking the rps2–rpoC2 intergenic spacer; color scheme for plastid genes, plastid spacers, and the mitochondrial insert follows figures 1 and 3. (A) Expression of the 5′-end of rps2 and the flanking insert sequence. (B) Expression of the 3′-end of rpoC2 and flanking insert sequence, including ψrpl2. Plastome coordinates are given on the x axis and read depth on the y axis for both (A) and (B). Note that the scale is not the same in (A) and (B).
Discussion
Plant plastomes are highly conserved, and intracellular gene transfer involving the plastome has been shown to be highly asymmetric. Although genetic material from plastid genomes is regularly transferred to plant mitochondrial and nuclear genomes, it has long been believed that plant plastomes do not tolerate incorporation of foreign DNA (Richardson and Palmer 2007; Keeling and Palmer 2008; Smith 2011). The power of the quantitative nature of next-generation sequencing (NGS) allowed confirmation of just such a rare event, transfer of mtDNA into the plastome, through sequencing and assembly of these two genomes in common milkweed. Further sampling of other species of Apocynaceae revealed that this transfer event uniquely occurred in the common ancestor of tribe Asclepiadeae. Because of the conserved nature of the plastome, evidence of this intracellular HGT has been preserved through the divergence of thousands of species, originating perhaps in the late Eocene (Rapini et al. 2007). Evidence for the mechanism of the HGT event, as well as of continued interaction of the plastid and mitochondrial genomes, is present in the DNA sequences obtained from representative species.
The Mechanism of the Mitochondrial to Plastome HGT in Milkweeds
A suite of DNA repair mechanisms functions to maintain the integrity of the plastome and contribute to the conservative evolution of this genome, which exists in a genotoxic environment due to photo-oxidative stress (Maréchal and Brisson 2010). Double-strand breaks in ptDNA can be repaired by homologous recombination through both the classical double-strand break repair model (DSBR) and the synthesis-dependent strand annealing model (SDSA), either of which can result in gene conversion if the template used differs from the DNA under repair (Odom et al. 2008; Maréchal and Brisson 2010). Homologous sequences as short as 50–150 bp are long enough to allow these mechanisms to occur (Singer et al. 1982; Watt et al. 1985; Maréchal and Brisson 2010). One of these ptDNA repair mechanisms is likely responsible for the incorporation of mtDNA into the plastid genome in the ancestor of Asclepiadeae following a double-strand break in the rps2–rpoC2 intergenic spacer.
All Asclepiadoideae (the subfamily containing Asclepiadeae) share the presence of ψrpoC2 flanking rpl2 in the mitochondrial genome, indicating that the ancestral mitochondrial genome of Asclepiadeae contained the necessary homologous sequence on one flank of the mtDNA that was inserted into the plastome. Presence of plastid rps2–rpoC2 flanking the other end of the inserted segment (as observed in the mitochondrial genome of A. syriaca and likely present in other Asclepiadoideae) in combination with ψrpoC2 could have facilitated repair of a double-strand break between rps2 and rpoC2 through either DSBR or SDSA that used the mtDNA as a template, rather than another copy of the plastome, of which there are multiple copies per cell in most higher plants (Smith 2011). There is also a plastid ψrpoB adjacent to the spacer sequence in the mitochondrial genome of A. syriaca (fig. 1A), but there is no evidence that it played a role in the HGT mechanism. It could represent an independent transfer of ptDNA to the mitochondrial genome or if it was part of a single transfer that included the rps2–rpoC2 spacer sequence, the intervening rpoC2 and rpoC1 genes and 3′ rpoB sequence (ca. 9,100 bp) have since been deleted.
The two segments of the mitochondrial genome that were inserted into the plastome are not currently contiguous in A. syriaca. The arrangement observed in the plastome insert likely represents the ancestral mitochondrial genome sequence that has since been rearranged. This would be consistent with observations showing slow nucleotide substitutions in plant mitochondrial genomes but a rapid pace of rearrangement (Wolfe et al. 1987; Palmer and Herbon 1988; Maréchal and Brisson 2010; Knoop 2012). The presence of sequence in the plastome inserts without homology to the Asclepias mitochondrial genome or sequence in GenBank could be due to the deletion of this sequence in the Asclepias mitochondrial genome, though it was present in the ancestral mitochondrial genome, but also leaves open the possibility that the mechanism of transfer may be more complicated and has involved multiple steps or sources of DNA, such as the nuclear genome.
Due to the presence of homologous flanking sequence in the mitochondrial genomes of milkweeds, homologous recombination double strand break repair mechanisms (DSBR and SDSA) seem most likely as causal processes in the HGT. However, other mechanisms are known to operate in plastome repair. Single-strand annealing and microhomology-mediated end joining, also occur; however, these alternative pathways generally cause deletions, not insertions (Kwon et al. 2010; Maréchal and Brisson 2010). A mechanism thought to be similar to nonhomologous end joining, which frequently occurs during repair of nuclear DNA in plants, cannot be ruled out but has only rarely been observed in plastids, and due to its rarity has been cited as a reason that HGT had not been observed in plastomes (Odom et al. 2008; Kwon et al. 2010). A final possibility is that a transposable element could have facilitated the horizontal transfer event, which is thought to be the mechanism in the carrot mtDNA horizontal transfer to the plastome (Iorizzo et al. 2012a). Non-LTR retrotransposons characteristically produce short direct repeats due to target site duplication (Han 2010). Direct repeats are observed at either end of the plastome insertions in Eustegia and Astephanus, which belong to the earliest diverging lineages of the Asclepiadeae; however, this mechanism is unlikely given the absence of sequence in the insert region homologous to known transposable elements.
Regardless of how the mtDNA was incorporated into the plastome, it is still unknown how that DNA could have entered the plastid. There is no known native DNA uptake mechanism in plastids, and in the absence of such a mechanism, the double membrane of plastids and their lack of ability to fuse, as mitochondria do, may provide sufficient barriers to exogenous DNA (Richardson and Palmer 2007; Keeling and Palmer 2008; Bock 2010; Kwon et al. 2010; Smith 2011) in all but the most exceptional cases. The uptake of foreign DNA by plastids can be induced through stress treatments (Cerutti and Jagendorf 1995) and transformation accomplished using various methods (Maliga 2004), which further demonstrates that once the double membrane is breached, incorporation of foreign DNA is not a difficult step and can readily occur by homologous recombination (Maréchal and Brisson 2010).
Maintenance of the Insert over Evolutionary Time
In higher plants, horizontally transferred DNA is generally not functional in the recipient genome, even if the segment contains a gene or genes, and horizontally transferred DNA is usually only transiently maintained in the recipient genome on an evolutionary time scale (Richardson and Palmer 2007; Keeling and Palmer 2008; Bock 2010). Even considering the slower rate of evolution of the plastome than the nuclear genome, this large insertion has been maintained, relatively intact, since approximately the late Eocene in all subtribes of tribe Asclepiadeae. Sequence comparison across these subtribes indicates that the insert is beginning to degrade, and this is most notable in A. syriaca, in which approximately half of the insert sequence has been deleted. The general maintenance of this foreign insertion raises the question of whether or not it confers a selective advantage that caused that plastome variant to drive to fixation in the ancestor of Asclepiadeae, or whether it defied long odds as a neutral variant and was fixed by genetic drift.
In its current form, fitness advantages conferred by the horizontally transferred DNA cannot be realized at the protein level because there is minimal transcriptional activity in A. syriaca. The lack of abundance of primary or processed transcripts for the ψrpl2 exon indicates that the exon is likely not functional as a translated protein. This is not surprising given that rpl2 exon 2 is likely not essential for function even in the mitochondrial genome (Colas des Francs-Small et al. 2012). The insertion may still possess a less obvious role in post-transcriptional processing, as the mature RNAs from both flanking operons show large differences in final accumulated mRNA levels. Both the 5′ untranslated region of the rps2–atpA operon and the 3′ untranslated region of the rpoB–rpoC2 operon appear to extend approximately 100 bp into the insert (fig. 6), creating a more robust target for transcriptional modification making it possible that the insert behaves as a novel transcriptional enhancer by stimulating translational efficiency, either due to direct interactions with 70S ribosomes (Pfalz et al. 2009) or by stabilizing specific isoforms that are more readily translatable (Felder et al. 2001). These extensions may also serve as targets for PPR-class genes or sRNAs, both of which target the 5′ and 3′ termini of transcripts and stabilize RNAs by blocking degradation by chloroplast exonucleases (Felder et al. 2001; Pfalz et al. 2009; Barkan et al. 2012). It could be telling that Eustegia, which has had multiple large deletions in the insert, retains the short regions of the insert sequence where transcription is extended in A. syriaca (fig. 3). However, if the insert segment is indeed neutral, its success is likely a consequence of “lucky insertion” into an intergenic spacer between two transcriptional operons (rpoB–rpoC2 and rps2–atpA), rather than into a location that would have caused disruption of an essential gene in the gene-dense plastome. In this case, the observed minimal transcription is likely a function of the frequency of promoters throughout the plastome and inefficient, stochastic transcription termination in plastids which contribute to transcription of virtually the entire plastome (Hotto et al. 2012; Zhelyazkova et al. 2012).
Continued Exchange with the Mitochondrial Genome through Gene Conversion
The simultaneous phylogenetic analysis of mitochondrial ψrpoC2 and plastid rpoC2 sequences revealed relatively recent gene conversion, such that mitochondrial ψrpoC2 in Asclepias is more closely related to plastid rpoC2 than to ψrpoC2 in other Asclepiadeae. The position of the S. afzelii (subfamily Secamonoideae) mitochondrial sequence sister to all other Asclepiadoideae mitochondrial and plastid sequences could be because the ψrpoC2 in this species is of independent origin. Another possibility is that the pseudogenes are of common evolutionary origin, but there was plastid to mitochondrial gene conversion in the common ancestor of Asclepiadoideae after the divergence of Secamonoideae, which at that time caused the mitochondrial pseudogene to become more closely related to the plastid gene than the mitochondrial pseudogene in the sister lineage of Asclepiadoideae (i.e., Secamonoideae). Taken together, these results illustrate that the relationship between the two genomes is dynamic over evolutionary time.
This phenomenon has been observed before. A comparison of the maize and rice mitochondrial and plastid genomes that revealed that there has been ongoing intergenomic exchange through gene conversion (Clifton et al. 2004) since an ancient transfer of ptDNA to the mitochondrial genome that likely occurred before the divergence of gymnosperms and angiosperms (Wang et al. 2007). Gene conversion among native mitochondrial genes, those of plastid origin, and with exogenous DNA has been observed on a finer scale leading to mosaicism in mitochondrial genes, which can be a significant generator of diversity in plant mitochondrial genomes (Hao and Palmer 2009; Hao et al. 2010) and may be a cause of phylogenetic incongruence (Hao and Palmer 2011). It is possible that such interplay between genomes has affected the evolution of the plastome as well and could be a contributor to the variability we observed at the 3′ end of plastid rpoC2 in Apocynaceae.
Conclusion
The evidence provided in this study for HGT between the mitochondrial and plastid genomes of milkweeds, combined with the results of Iorizzo et al. (2012b) showing a transposable element-mediated HGT of mtDNA into the plastome of carrot, confirms that DNA transfer from land plant mitochondrial genomes to plastomes is possible and can occur by more than one mechanism. The sequencing of additional land plant plastomes may reveal that horizontal or intracellular gene transfer into these genomes is not as rare as was previously thought and gives further insight into these and other mechanisms of DNA transfer between plant mitochondrial and plastid genomes.
Supplementary Material
Supplementary figure S1, table S1, data sets S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank P. Bruyns, T. Livshultz, W. Phippen, and A. Rein for providing tissue or DNA samples; B. Haack, K. Hansen, M. Parks, and L. Ziemian for laboratory assistance; K. Weitemier for data analysis and programming assistance; M. Dasenko, S. Jogdeo, M. Peterson, and C. Sullivan for computational and sequencing support; and Y. Di for statistical consultation. This work was supported by the National Science Foundation (DEB 0919583 to R.C.C., M.F., and A.L.).
Literature Cited
- Adams KL, Ong HC, Palmer JD. Mitochondrial gene transfer in pieces: fission of the ribosomal protein gene rpl2 and partial or complete gene transfer to the nucleus. Mol Biol Evol. 2001;18:2289–2297. doi: 10.1093/oxfordjournals.molbev.a003775. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ, et al. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae) Mol Biol Evol. 2010;27:1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci. 2010;15:11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- Bruyns P. The systematic position of Eustegia R.Br.(Apocynaceae-Asclepiadoideae) Bot Jahrb Syst. 1999;121:19–44. [Google Scholar]
- Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, et al. BamView: visualizing and interpretation of next-generation sequencing read alignments. Brief Bioinform. 2013;14:203–212. doi: 10.1093/bib/bbr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Jagendorf A. Movement of DNA across the chloroplast envelope: implications for the transfer of promiscuous DNA. Photosynth Res. 1995;46:329–337. doi: 10.1007/BF00020448. [DOI] [PubMed] [Google Scholar]
- Cha RS, Zarbl H, Keohavong P, Thilly WG. Mismatch amplification mutation assay (MAMA): application to the cH-ras gene. Genome Res. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- Clifton SW, et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004;136:3486–3503. doi: 10.1104/pp.104.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas des Francs-Small C, et al. A PORR domain protein required for rpl2 and ccmFc intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. Plant J. 2012;69:996–1005. doi: 10.1111/j.1365-313X.2011.04849.x. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Felder S, et al. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell. 2001;13:2127–2141. doi: 10.1105/TPC.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goremykin VV, Salamini F, Velasco R, Viola R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol. 2009;26:99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Han JS. Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions. Mob DNA. 2010;1:15. doi: 10.1186/1759-8753-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Palmer JD. Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc Natl Acad Sci U S A. 2009;106:16728–16733. doi: 10.1073/pnas.0908766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Palmer JD. HGT turbulence: confounding phylogenetic influence of duplicative horizontal transfer and differential gene conversion. Mob Genet Elements. 2011;1:256–304. doi: 10.4161/mge.19030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Richardson AO, Zheng Y, Palmer JD. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc Natl Acad Sci U S A. 2010;107:21576–21581. doi: 10.1073/pnas.1016295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto AM, Germain A, Stern DB. Plastid non-coding RNAs: emerging candidates for gene regulation. Trends Plant Sci. 2012;17:737–744. doi: 10.1016/j.tplants.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Iorizzo M, et al. Against the traffic: the first evidence for mitochondrial DNA transfer into the plastid genome. Mob Genet Elements. 2012a;2:261–266. doi: 10.4161/mge.23088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorizzo M, et al. De novo assembly of the carrot mitochondrial genome using next generation sequencing of whole genomic DNA provides first evidence of DNA transfer into an angiosperm plastid genome. BMC Plant Biol. 2012b;12:61. doi: 10.1186/1471-2229-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V. Seed plant mitochondrial genomes: complexity evolving. In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Dordrecht (The Netherlands): Springer; 2012. pp. 175–200. [Google Scholar]
- Ku C, Chung W-C, Chen L-L, Kuo C-H. The complete plastid genome sequence of Madagascar periwinkle Catharanthus roseus (L.) G. Don: plastid genome evolution, molecular marker identification, and phylogenetic implications in asterids. PLoS One. 2013;8:e68518. doi: 10.1371/journal.pone.0068518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T, Huq E, Herrin DL. Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc Natl Acad Sci U S A. 2010;107:13954–13959. doi: 10.1073/pnas.1004326107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liede-Schumann S, Rapini A, Goyder DJ, Chase MW. Phylogenetics of the new world subtribes of Asclepiadeae (Apocynaceae-Asclepiadoideae): Metastelmatinae, Oxypetalinae, and Gonolobinae. Syst Bot. 2005;30:184–195. [Google Scholar]
- Liede S. Subtribe Astephaninae (Apocynaceae-Asclepiadoideae) reconsidered: new evidence based on cpDNA spacers. Ann Missouri Bot Gard. 2001;88:657–668. [Google Scholar]
- Liede S, Täuber A. Circumscription of the genus Cynanchum (Apocynaceae-Asclepiadoideae) Syst Bot. 2002;27:789–800. [Google Scholar]
- Livshultz T. The phylogenetic position of milkweeds (Apocynaceae subfamilies Secamonoideae and Asclepiadoideae): evidence from the nucleus and chloroplast. Taxon. 2010;59:1016–1030. [Google Scholar]
- Lohse M, et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P. Plastid transformation in higher plants. Annu Rev Plant Biol. 2004;55:289–313. doi: 10.1146/annurev.arplant.55.031903.141633. [DOI] [PubMed] [Google Scholar]
- Maréchal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186:299–317. doi: 10.1111/j.1469-8137.2010.03195.x. [DOI] [PubMed] [Google Scholar]
- Meve U, Liede S. Subtribal division of Ceropegieae (Apocynaceae-Asclepiadoideae) Taxon. 2004;53:61–72. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); 2010 Nov 14; New Orleans (LA). p. 1–8. [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Odom OW, Baek KH, Dani RN, Herrin DL. Chlamydomonas chloroplasts can use short dispersed repeats and multiple pathways to repair a double-strand break in the genome. Plant J. 2008;53:842–853. doi: 10.1111/j.1365-313X.2007.03376.x. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Herbon L. Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J Mol Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- Parkhomchuk D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapini A, Chase MW, Goyder DJ, Griffiths J. Asclepiadeae classification: evaluating the phylogenetic relationships of New World Asclepiadoideae (Apocynaceae) Taxon. 2003;52:33–50. [Google Scholar]
- Rapini A, van den Berg C, Liede-Schumann S. Diversification of Asclepiadoideae (Apocynaceae) in the New World. Ann Missouri Bot Gard. 2007;94:407–422. [Google Scholar]
- Renner S, Bellot S. Horizontal gene transfer in eukaryotes: fungi-to-plant and plant-to-plant transfers of organellar DNA. In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Dordrecht (The Netherlands): Springer; 2012. pp. 223–235. [Google Scholar]
- Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot. 2007;58:1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- Singer BS, Gold L, Gauss P, Doherty DH. Determination of the amount of homology required for recombination in bacteriophage T4. Cell. 1982;31:25–33. doi: 10.1016/0092-8674(82)90401-9. [DOI] [PubMed] [Google Scholar]
- Smith DR. Extending the limited transfer window hypothesis to inter-organelle DNA migration. Genome Biol Evol. 2011;3:743. doi: 10.1093/gbe/evr068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Straub SCK, et al. Building a model: developing genomic resources for common milkweed (Asclepias syriaca) with low coverage genome sequencing. BMC Genomics. 2011;12:211. doi: 10.1186/1471-2164-12-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub SCK, et al. Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am J Bot. 2012;99:349–364. doi: 10.3732/ajb.1100335. [DOI] [PubMed] [Google Scholar]
- Tai H, Pelletier C, Beardmore T. Total RNA isolation from Picea mariana dry seed. Plant Mol Biol Rep. 2004;22:93a–93e. [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Wang D, et al. Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 MYA. Mol Biol Evol. 2007;24:2040–2048. doi: 10.1093/molbev/msm133. [DOI] [PubMed] [Google Scholar]
- Watt VM, Ingles CJ, Urdea MS, Rutter WJ. Homology requirements for recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:4768–4772. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K, Li W, Sharp P. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P, et al. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell. 2012;24:123–136. doi: 10.1105/tpc.111.089441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.