Abstract
The chromosomal distribution of genes with sex-biased expression is often nonrandom, and in species with XY sex chromosome systems, it is common to observe a deficit of X-linked male-biased genes and an excess of X-linked female-biased genes. One explanation for this pattern is that sex-specific selection has shaped the gene content of the X. Alternatively, the deficit of male-biased and excess of female-biased genes could be an artifact of differences between the sexes in the global expression level of their X chromosome(s), perhaps brought about by a lack of dosage compensation in males and hyperexpression in females. In the montium fruit fly, Drosophila serrata, both these explanations can account for a deficit of male-biased and excess of female-biased X-linked genes. Using genome-wide expression data from multiple male and female tissues (n = 176 hybridizations), we found that testis- and accessory gland-specific genes are underrepresented whereas female ovary-specific genes are overrepresented on the X chromosome, suggesting that X-linkage is disfavored for male function genes but favored for female function genes. However, genes with such sex-specific functions did not fully account for the deficit of male-biased and excess of female-biased X-linked genes. We did, however, observe sex differences in the global expression level of the X chromosome and autosomes. Surprisingly, and in contrast to other species where a lack of dosage compensation in males is responsible, we found that hyperexpression of X-linked genes in both sexes leads to this imbalance in D. serrata. Our results highlight how common genomic distributions of sex-biased genes, even among closely related species, may arise via quite different evolutionary processes.
Keywords: sex-biased gene expression, X chromosome, dosage compensation, Drosophila
Introduction
An intriguing feature of genes with sex-biased expression is their apparent nonrandom chromosomal distribution. For example, the X chromosomes of several Drosophila species exhibit a striking paucity of male-biased and an excess of female-biased genes (Parisi et al. 2003; Sturgill et al. 2007; Mikhaylova and Nurminsky 2011; Meisel et al. 2012), whereas in mice and humans, the X chromosome harbors an excess of certain male-biased genes (Wang et al. 2001; Lercher et al. 2003; Yang et al. 2006). Two broad classes of explanation have been proposed to explain these genomic patterns. On one hand, chromosome-wide differences in the expression of X-linked and autosomal genes may create apparent differences in the distribution of sex-biased genes on these chromosomes. Alternatively, the gene content on the X and the autosomes may have been shaped by a history of sex-specific selection.
Hypotheses featuring sex-specific selection suggest that the X chromosome is often a maladaptive location for genes with male-specific functions and in some cases a favored location for genes with female-specific functions (Parisi et al. 2003; Gupta et al. 2006; Sturgill et al. 2007). As a consequence, the X chromosome becomes “demasculinized” or “feminized” through mechanisms such as compensatory gene duplication or translocation between the X and autosomes or changes in sex-biased expression (Connallon and Clark 2011; Gallach and Betran 2011). At least three major sources of sex-specific selection have been identified. First, for mutations with sexually antagonistic fitness effects, depending on the degree of dominance and the direction and magnitude of opposing selection coefficients in males and females, selection can either favor X-linkage of female-benefit alleles or disfavor X-linkage of male-benefit alleles (Rice 1984). Second, and specific to the case of X-chromosome demasculinization, selection may act against X-linkage of testis-specific genes due to male meiotic sex chromosome inactivation (MSCI) (Betran et al. 2002; Khil et al. 2004; Hense et al. 2007). Although selection to escape the effects of X-chromosome inactivation during spermatogenesis can possibly explain the lack of X-linked testis-specific male-biased genes, the hypothesis cannot explain the observed lack of other types of male-biased genes on the X in Drosophila melanogaster (Parisi et al. 2003; Sturgill et al. 2007; Meiklejohn et al. 2011). A third source of sex-specific selection can arise due to insufficient dosage compensation in males. Here, the mechanism that balances expression between X-linked and autosomal genes in males (Ohno 1967), which are heterogametic in XY systems, fails to express male-biased genes at an optimally high level, making the X chromosome a maladaptive location for such genes (Vicoso and Charlesworth 2009; Bachtrog et al. 2010; Meisel et al. 2012).
Rather than reflecting differences in evolved gene content, a statistical deficit of male-biased X-linked genes can occur when the X and autosomes simply differ in chromosome-wide expression levels (Prince et al. 2010; Meiklejohn and Presgraves 2012). For example, in D. melanogaster, although dosage compensation certainly operates in somatic tissue (Gupta et al. 2006; Sturgill et al. 2007; Meiklejohn and Presgraves 2012), it may not occur in the testes (Meiklejohn et al. 2011). Indeed, when the average 1.5-fold lower expression of X-linked genes in testes is accounted for, a previously observed underrepresentation of male-biased genes on the X chromosome was no longer found for this tissue (Meiklejohn and Presgraves 2012). Moreover, studies of ZW sex chromosome systems in birds suggest that a lack of complete dosage compensation in somatic tissue (Ellegren et al. 2007; Mank and Ellegren 2009) may explain the excess of male-biased Z-linked genes in these species (Ellegren et al. 2007). Interestingly, the differences between X and autosomal global expression levels need not be due to inadequate dosage compensation in the heterogametic sex; in the flour beetle, Tribolium castaneum, both sexes hyperexpress the X chromosome (Prince et al. 2010). In this case, hyperexpression in females may provide an explanation for not only the deficit of male-biased X-linked genes but also the excess of female-biased X-linked genes.
Although the nonrandom genomic distributions of sex-biased genes seen across multiple species and sex chromosome systems suggest a role for varied forms of sex-specific selection, some of the patterns may also be accounted for by differences in global expression level between chromosomes. It remains evident that as new genomes are studied, novel phenomena are uncovered, which expose variation in the specific assumptions underlying some of these hypotheses (e.g., Prince et al. 2010). Thus, further studies of sex-biased expression in additional taxa are required (Kaiser and Bachtrog 2010; Meiklejohn and Presgraves 2012). For this reason, we analyzed the genomic distribution of sex-biased genes in D. serrata, a member of the highly diverse but less-studied montium subgroup, which diverged from the group containing D. melanogaster approximately 40 Ma (Tamura et al. 2004). Using a custom microarray platform and assaying samples derived from multiple tissues in both sexes, we show a significant deficit of male-biased and excess of female-biased genes on the X chromosome. The pattern appears not only consistent with sex-specific selection but surprisingly, global expression differences due to hyperexpression of the X chromosome in both sexes.
Results and Discussion
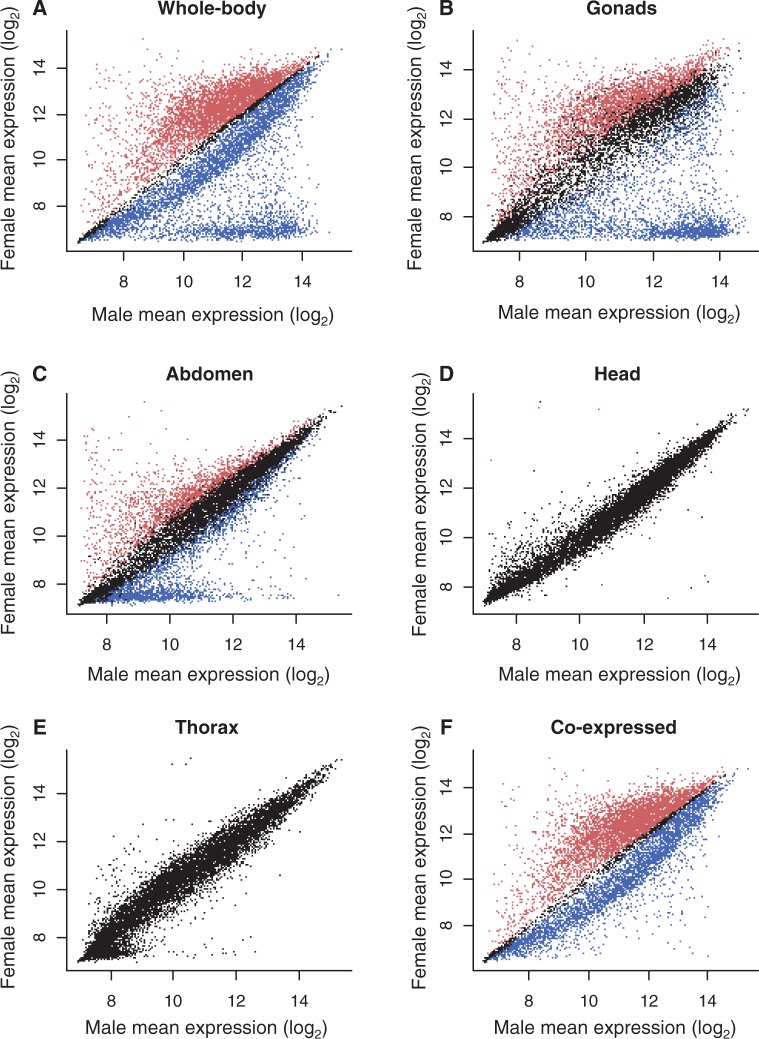
A total of 10,867 genes (93.4% of genes on the array) were sex-biased in the whole-body samples (Welch’s two-sample t-test; t40–81 FDR < 0.05; fig. 1A), 5,031 were female-biased, 5,836 were male-biased, and the remaining 749 were classified as unbiased. To account for the possibility that the genome of an inbred line and/or an interaction between line and sex could affect the detection of sex-biased genes, we also ran mixed effects analyses of variance (ANOVAs) where sex was fitted as a fixed effect and line and the sex × line interaction were random effects. Because the results were very similar (10,862 were sex-biased in both analyses, 5 were unique to Welch’s t-test, and 116 were unique to ANOVA), we report only Welch’s t-test results. In the tissue-specific samples, most sex-biased genes were expressed in the male and female reproductive tissues. Many sex-biased genes were restricted to the gonads (3,890 female-biased and 3,298 male-biased) (Welch’s two-sample t-test; t2–5 FDR < 0.05; fig. 1B) and gonadectomized abdomen (2,447 female-biased and 3,181 male-biased) (Welch’s two-sample t-test; t3–6 FDR < 0.05; fig. 1C), which still contained numerous reproductive organs except for the accessory glands, testes, and ovaries. There were very few sex-biased genes in the nonreproductive tissues. Although sample sizes for these tissues were much smaller than for whole body, where we detected a large number of sex-biased genes, they were of similar size to the gonad and abdomen samples, where many sex-biased genes were also detected. For the head samples, only nine genes were sex-biased, five were female-biased, and four were male-biased (Welch’s two-sample t-test; t3–6 FDR < 0.05; fig. 1D), and no sex-biased genes were identified in the thorax (Welch’s two-sample t-test; t2–5 FDR > 0.5; fig. 1E).
Fig. 1.—
Sex-biased expression of 11,631 genes of D. serrata: (A) whole-body (n = 71 hybridizations per sex), (B) gonads (nfemale = 3, nmale = 4), (C) gonadectomized abdomen (nfemale = 4, nmale = 4), (D) head (nfemale = 4, nmale = 4), (E) thorax (nfemale = 3, nmale = 4), and (F) whole-body excluding sex-specific genes (n = 71 per sex). Red represents female-biased genes, blue are male-biased genes, and black are unbiased genes (Welch’s t-test, FDR < 0.05).
Demasculinization and Feminization of the X Chromosome
The X chromosome contained fewer male-biased but more female-biased genes than expected by chance (1,000 permutations of chromosome location: P < 0.001; fig. 2A and B). Permutation tests also indicated that three of the four autosomal chromosome arms had slightly more male-biased genes than expected (2L, P = 0.004; 2R, P < 0.001; 3R, P = 0.027), and that chromosomes 2L (P = 0.001) and 2R (P = 0.004) had slightly fewer female-biased genes than expected. The pattern of masculinization and defeminization of chromosome 2L has also been observed in D. melanogaster (Parisi et al. 2003; Meisel et al. 2012). The number of sex-biased genes on chromosome 3L did not differ from the random expectation for either sex (males: P = 0.290; females: P = 0.278). Notably, we had unusually high power to detect sex differences in expression (as low as a fold change of 1.03) in whole bodies. As a complementary approach, we also assessed the genomic distribution of genes classified as sex-biased using the classic fold-change threshold of 2, which is less likely to be influenced by large sample sizes. This subset of highly sex-biased genes had a very similar genomic distribution to the full sample of genes (supplementary fig. S1, Supplementary Material online).
Fig. 2.—
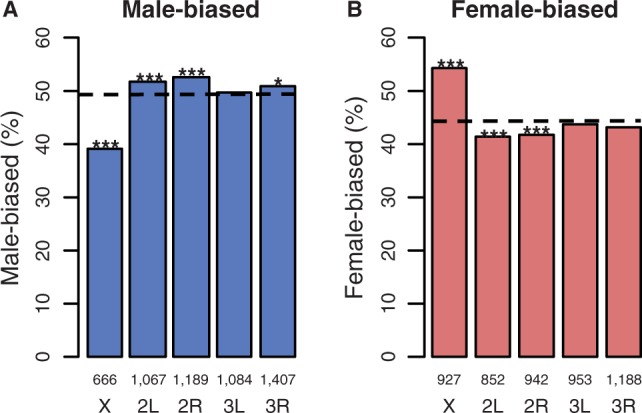
The genomic distribution of sex-biased genes in D. serrata: (A) male-biased and (B) female-biased. The dotted line is the expected percentage of sex-biased genes per chromosome from 1,000 random permutations of the data; numbers above chromosome labels indicate the number of genes in each bar, and asterisk indicates probablilty that observed value does not differ from expected: ***P < 0.001, **P < 0.01, *P < 0.05.
Using a metric of tissue specificity (τ > threshold of 0.9, Yanai et al. 2005; Meiklejohn and Presgraves 2012; Meisel et al. 2012), we categorized 1,315 genes as sex-specific (88 female-specific and 1,227 male-specific; table 1). Exclusion of genes categorized as sex-specific based on whole-body data removed only the most extremely sex-biased genes, supporting the idea that these genes are indeed sex-specific (fig. 1F). As expected, sex-specific genes were predominantly expressed in the gonads, including 77 (87.5%) of female-specific genes and 1,147 (93.5%) of male-specific genes (table 1).
Table 1.
Number of Genes Categorized as Sex-Specific in D. serrata within Each Tissue (τ < 0.9)
| Female-Limited | Male-Limited | |
|---|---|---|
| Head | 0 | 0 |
| Thorax | 1 | 3 |
| Abdomen | 10 | 7 |
| Gonads | 77 | 1,147 |
| Accessory glands | — | 70 |
A deficit of male function genes on the X chromosome may reflect a history of selection for demasculinization (Parisi et al. 2003; Gupta et al. 2006; Sturgill et al. 2007). In support, we found deficits of both testis-specific genes (P < 0.001 from 1,000 permutations; fig. 3A) and accessory gland-specific genes (P < 0.001 from 1,000 permutations; fig. 3B) on the X chromosome in D. serrata. However, no bias in the genomic distribution of male-specific genes expressed in male abdomen was found (P > 0.05 from 1,000 permutations; fig. 3C). Although these results share broad similarities with other Drosophila species and even mosquitoes (Parisi et al. 2003; Sturgill et al. 2007; Zhang et al. 2007; Mikhaylova and Nurminsky 2011; Meiklejohn and Presgraves 2012; Meisel et al. 2012), there were some key differences. In the mosquito Anopheles gambiae, there is an excess instead of a deficit of accessory gland-specific X-linked genes (Meiklejohn and Presgraves 2012). On the D. melanogaster X chromosome, there is a deficit of sperm proteome-specific genes (Meisel et al. 2012) but not testis-specific genes in general (Meiklejohn and Presgraves 2012; Meisel et al. 2012). Because we used conserved synteny between D. serrata and D. melanogaster (Stocker et al. 2012) to place genes on chromosomes, it is possible that genes which have transposed from the X chromosome to an autosome, a move which has occurred more than expected by chance for testis-specific genes in D. melanogaster (Betran et al. 2002; Han and Hahn 2012), were incorrectly assigned to the X chromosome in D. serrata. If this were the case, our finding of a deficit of testis-specific genes in D. serrata is conservative because we may have assigned autosomal genes to the X chromosome. The observed excess of female-biased genes on the X was also consistent with enrichment for female-specific functions. There was an excess of ovary-specific X-linked genes (P = 0.013 from 1,000 permutations; fig. 3D) but no deviation from random for X-linked female abdomen genes (P > 0.05 from 1,000 permutations; fig. 3E). Very few sex-specific genes were found in the head and thorax (table 1).
Fig. 3.—
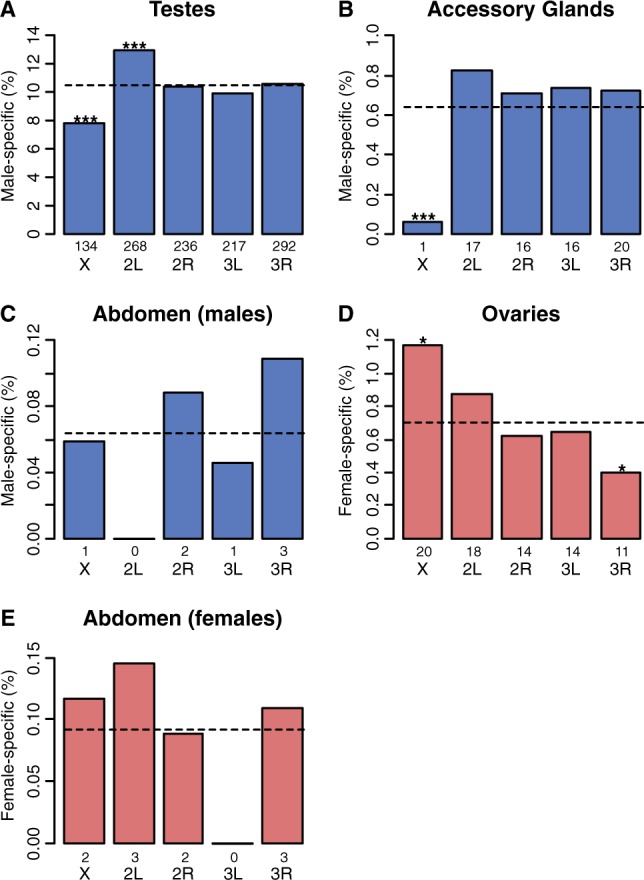
The X chromosome of D. serrata is lacking in male-biased and enriched for female-biased genes. Male-specific tissues are shown in blue and female-specific in red. The number of sex-specific genes per tissue is shown in table 1. The dotted line is the random expectation for the percentage of biased genes per chromosome estimated from 1,000 permutations; numbers above chromosome labels indicate the number of genes in each bar, and asterisk indicates probablilty that observed value does not differ from expected: ***P < 0.001, **P < 0.01, *P < 0.05.
The deficit of testis-specific genes on the X chromosome of D. serrata could reflect avoidance of MSCI (Betran et al. 2002). However, this hypothesis cannot explain the lack of X-linked accessory gland-biased genes, which do not experience MSCI. Alternatively, genes that are highly biased toward expression in male-limited tissues may have as yet unknown pleiotropic effects in females. If such effects are sexually antagonistic, then X-linkage may not be favored by selection (Rice 1984). Reasons for the excess of ovary-specific genes on the X chromosome are less obvious, although sexual antagonism could also play a role.
To examine the extent to which genes with sex-specific expression (which are likely targets of sex-specific selection) accounted for the genomic distribution of sex-biased genes in D. serrata, we excluded them and reanalyzed the data. Following exclusion, the distribution of sex-biased genes remained nonrandom, and the deficit/excess of male-biased/female-biased genes on the X chromosome remained significant (whole-body analysis excluding 1,315 genes: P < 0.001 from 1,000 permutations; supplementary fig. S2, Supplementary Material online, see also supplementary file S1, Supplementary Material online, for a list of genes excluded). Although we cannot reject hypotheses incorporating sexual antagonism to explain the nonrandom distribution of sex-biased genes, a conflict which can be present even when genes are only moderately sex-biased (Bonduriansky and Chenoweth 2009), our results suggest that factors additional to sex-specific selection may have shaped the genomic distribution of sex-biased genes in D. serrata.
Hyperexpression of Genes on the X Chromosome
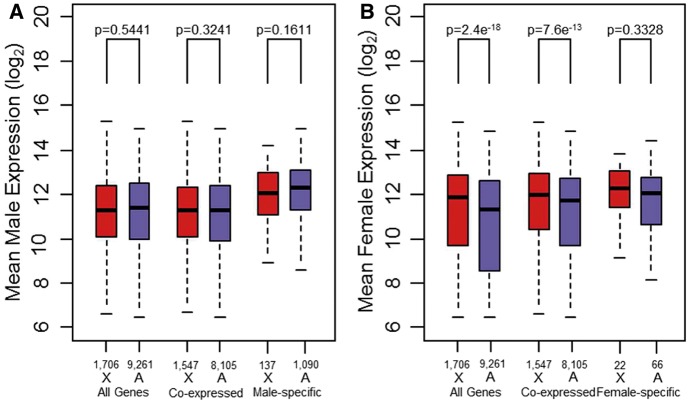
We also assessed whether the chromosomal distribution of sex-biased genes in D. serrata could be accounted for by differences in global expression between the X chromosome and autosomes (Prince et al. 2010; Meiklejohn et al. 2011; Meiklejohn and Presgraves 2012). We first examined all genes regardless of sex-specificity followed by both co-expressed and sex-specific genes. In males, there was no difference between the X chromosome and the autosomes (Mann–Whitney test: W1706, 9261 = 7,826,733, P = 0.5441), suggesting that dosage compensation was functional and should not create the appearance of a deficit/excess of male-/female-biased X-linked genes. However, to our surprise, females expressed X-linked genes at a considerably higher level than autosomal genes (Mann–Whitney test: W1706, 9261 = 8,949,732, P = 2.4e−18; fig. 4). Because females are homogametic and males heterogametic, this finding could explain the excess of female-biased and deficit of male-biased X-linked genes still observed after accounting for other factors such as sex-specific gene function. The same pattern was observed in the subset of genes that are co-expressed in both sexes (Mann–Whitney test: males, W1547, 8105 = 6,368,256, P = 0.3241; females, W1547, 8105 = 6,989,116, P = 7.6e−13). However, for sex-specific genes, female-specific X-linked genes no longer appeared to be significantly hyperexpressed (Mann–Whitney test: W22, 66 = 827, P = 0.3328), whereas for male-specific genes, there was still no deviation from a 1:1 X:Autosome expression ratio (Mann–Whitney test: W137, 1090 = 69,186, P = 0.1611).
Fig. 4.—
Dosage compensation via hyperexpression in both sexes. (A) Boxplots showing mean expression of X-linked genes (red) and autosomal genes (blue) for males. Plots are shown for all genes on the microarray (All Genes) and subsets containing either genes expressed in both sexes (Co-expressed) or genes expressed in one sex only (sex-specific). P values are from Mann–Whitney U tests comparing expression on the X chromosome versus the autosomes (see Materials and Methods). (B) As in (A) but for females. Numbers above chromosome labels indicate the number of genes.
Overexpression of X-linked genes in females presents the intriguing possibility that the apparent excess of female-biased genes on the D. serrata X may reflect differences in X–autosome global expression levels. Our data are consistent with D. serrata male dosage compensation via hyperexpression of the X chromosome to a level that balances the expression of autosomal genes (fig. 4A). Because females have two copies of X-linked genes, there is no need to hyperexpress these genes to achieve a balance with autosomal expression (Ohno 1967). However, it appears that females “overexpress” X-linked genes approximately 1.46-fold relative to autosomal genes (95% bootstrap confidence interval = 1.46–1.48, resampling n = 71 female samples with replacement 500 times) (fig. 4B). This is below the theoretical 2-fold expression difference expected if hyperexpression was of equal strength in males and females. Although hyperexpression appears to occur in females of seven other Drosophila species (Gupta et al. 2006; Sturgill et al. 2007; Zhang and Oliver 2010), its magnitude appears to be considerably greater in D. serrata. To our knowledge, a similar level of X chromosome hyperexpression (in both sexes) has only been reported in the red flour beetle T. castaneum (Prince et al. 2010).
Hyperexpression of the X chromosome in both sexes of D. serrata could have arisen through sex-specific selection. As the sex chromosomes evolve, the newly formed Y chromosome is expected to degenerate over time (Ohno 1967; Zhou and Bachtrog 2012). Thus, selection may favor hyperexpression of X-linked genes in males to restore the balance with autosomal genes (Ohno 1967). Because the sexes share a genome, selection for increased expression of X-linked genes in males could cause a correlated response in females (Lande 1980). Support for this idea was recently found in D. melanogaster where hyperexpression of the X chromosome in males requires expression of the dosage compensation complex and specific changes to chromatin structure (histone modifications) (Conrad and Akhtar 2011). Although the dosage compensation complex is primarily male-limited (Gladstein et al. 2010), changes to X chromosome structure that bring about hyperexpression in males also occur in females (Zhang and Oliver 2010). However there is currently no evidence for such a scenario in D. serrata, our finding of hyperexpression in both sexes suggests that a similar mechanism involving X chromosome structure and/or the dosage compensation complex may be involved. As our microarray does not contain the dosage compensation complex, the latter could not be assessed.
Unlike mammals, where hyperexpression occurs in both sexes but females avoid overexpression through X chromosome inactivation (Pessia et al. 2012), counter selection on D. serrata females may not have been strong enough for such a mechanism to evolve, at least for genes that are co-expressed in both sexes. For instance, the most dosage-sensitive genes are those involved in macromolecular complexes, transcription regulation, and signal transduction pathways, whereas over- or under-expression of “other classes” of genes may have limited fitness effects (Birchler 2012). It is possible that genes co-expressed in both sexes of D. serrata are of this other class and so a mechanism to inhibit hyperexpression in females may not be needed. It is interesting, however, that female-limited X-linked genes do not appear significantly overexpressed. The lack of significance may also be a statistical power issue given the relatively small number of genes in this subset of the data. However, if dosage compensation in D. serrata is gene-specific rather than chromosome-wide, as in birds (Mank et al. 2008; Itoh et al. 2010) and some insects (Kaiser and Bachtrog 2010), then overexpression of X-linked female-specific genes may never have occurred. In that case, there would have been no selection for increased expression of these genes in males and thus no correlated response in females.
As a further test for chromosome-level expression differences between X-linked and autosomal genes, we investigated expression of ribosomal protein-encoding genes (Parisi et al. 2003; Prince et al. 2010). Although ribosomal proteins are assumed to be in 1:1 stoichiometry (Voynow and Kurland 1971; Hardy 1975), global chromosomal differences in expression would cause X-linked ribosomal genes to be expressed at a higher level than autosomal genes. Consistent with this expectation, the overall pattern suggested higher X than autosomal expression, although the results did not quite reach statistical significance (females: Welch’s t28.842 = −1.6342, P = 0.0565; males: Welch’s t25.549 = −1.4316, P = 0.08219; see supplementary file S2, Supplementary Material online, for genes tested), perhaps as a result of low power given the small number of genes (X: 14, autosomes: 63 genes) as was the case for T. castaneum (Prince et al. 2010). Further, given that many ribosomal proteins have extraribosomal functions (Lindstrom 2009; Bhavsar et al. 2010), the 1:1 stoichiometry assumption may not always hold, and this may further weaken the power of such a test.
Conclusion
Although previous studies have examined the extent to which sex-specific selection (Parisi et al. 2003; Gupta et al. 2006; Sturgill et al. 2007) and/or global differences in expression between the X and the autosomes (Prince et al. 2010; Meiklejohn and Presgraves 2012) can account for patterns of X–autosome sex-biased gene expression, we could reject neither for D. serrata. We found evidence consistent with both demasculinization and feminization, which suggests a role for selective mechanisms, but also X chromosome hyperexpression in both sexes, which could create the statistical appearance of demasculinization/feminization. Although similar genomic patterns have been observed in many species for sex-biased genes, these patterns may not always share a common underlying cause.
Materials and Methods
Microarray and Experimental Design
A custom NimbleGen 12x135K microarray designed from D. serrata expressed sequence tags (ESTs) (Frentiu et al. 2009) was used to measure expression of 11,631 ESTs (supplementary file S3, Supplementary Material online). A panel of 43 wild-derived inbred lines of D. serrata from a single population (St Lucia, Brisbane, Australia) were used for the whole-body samples (n = 2 hybridizations per sex for each line for a total of 168), and a laboratory stock from the same location was used to obtain samples of several body parts: head (n = 4 per sex), thorax (female n = 3; male n = 4), gonadectomized abdomen (n = 4 per sex), ovaries (n = 3), testes (n = 4), and accessory glands (n = 4); all replicate hybridizations are biological replicates from independent RNA extractions of different groups of flies. RNA samples were randomly allocated to microarray slides and sectors. Flies were reared in 50 ml holding vials containing standard yeast medium and maintained at 25 °C with a 12-h day/night cycle. Offspring were collected as virgins with the use of light CO2 anesthesia and held for 3 days in same-sex groups of five flies. Two replicate pools of 30 flies per line per sex and four replicate pools of 100 flies for the whole-body and tissue samples, respectively, were snap-frozen in liquid nitrogen, without the use of CO2 anesthesia. RNA extractions were performed by using the Trizol procedure followed by RNA isolation using RNeasy minikits. cDNA synthesis, labeling, hybridization, and microarray scanning were performed by the Centre for Genomics and Bioinformatics, Bloomington, IN. Quality control of the array data was performed via the BioConductor “oligo package” using probe level models (Gentleman et al. 2004; Carvalho and Irizarry 2010; Draghici 2012) and the experimental metrics report provided by NimbleGen. For the whole-body samples, this reduced the data set from n = 168 to n = 142 hybridizations, but no hybridizations were excluded for the tissue-specific samples (n = 34 hybridizations). Expression measurements were normalized via a log2 transformation and summarized by taking the median of a probe set (Draghici 2012) where probe-level observations were the mean of the two replicates of that probe on the array. The expression data have been submitted to the NCBI Gene Expression Omnibus (GSE45801).
Genomic Location of Sex-Biased Genes
Genes were tested for sex-biased expression using Welch’s t-tests and applying a false discovery rate (FDR) of 0.05 (Benjamini and Hochberg 1995). The chromosomal location of ESTs was established based on homology with D. melanogaster. As observed among other Drosophila species (Bhutkar et al. 2008), there is strong chromosome level conservation of orthologous genes between D. serrata and D. melanogaster (Stocker et al. 2012). Stand-alone Blast (version 2.2.27+) was used to perform tBlastx (default settings) between our EST sequences and D. melanogaster chromosome, coding, gene, transcript, and pseudogene sequences obtained from Flybase. Genes associated with chromosomes 2 (4,323 genes, 37%), 3 (4,938 genes, 43%), and X (1,706 genes, 15%) were used in the analysis; these chromosomes accounted for 10,967 (95%) of the genes in our microarray. A further 547 (4.7%) of the genes in our array had poor-quality Blast hits (e-value > 0.1) and were therefore omitted from further analyses.
Identification of Sex-Specific Genes via Tissue Specificity
Due to intrinsic variability in microarray data, it is difficult to define genes as not expressed. To overcome this issue, we identified genes that are likely expressed exclusively in a single sex by using a sex-specific metric of tissue specificity:
| (1) |
where Ei is mean expression of tissue i and Emax is the maximum of the tissue-specific mean expression across all tissues (Yanai et al. 2005). This metric ranges from 0, for genes expressed at the same level in all tissues, to 1, for genes that are highly expressed in one tissue only, and has previously been used to classify genes as sex-limited if expression was highly biased toward sex-limited tissues such as the ovaries and testes (Meiklejohn and Presgraves 2012; Meisel et al. 2012). We calculated τ using measures of gene expression for nine different tissue types (male- and female-only dissections of head, thorax, and abdomen plus ovaries, testes, and accessory gland). Using a stringent τ threshold of 0.9, genes were classified as sex-specific if expression was highly biased for any of the sex-limited tissues or, for example, head of females only. Likewise, genes were classified as co-expressed in both sexes if they fell below the τ threshold.
Statistical Analyses
To test whether sex-biased genes were distributed nonrandomly across chromosomes, we used permutation tests where the chromosomal location of each gene was permuted 1,000 times (Meisel et al. 2012). The number of sex-biased genes per chromosome was deemed to not differ from random if more than 5% of the permuted estimates were greater/less than the observed value. The same permutation approach was used to assess demasculinization and feminization of the X chromosome by testing whether the chromosomal distribution of testis-, accessory gland-, abdomen-, and ovary-specific genes was nonrandom. To test for differences in global expression between the X chromosome and autosomal genes, Mann–Whitney tests were performed on males and females separately (Meisel et al. 2012). Mann–Whitney tests were further performed on sex-specific and co-expressed subsets of the male and female data to determine whether observed X–autosome expression differences were specific to one or both of these gene classes.
Supplementary Material
Supplementary files S1–S3 and figures S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank H. Rundle for supplying a proportion of the fly lines used; P. Innocenti for help with the EST collation; and A. Reddiex, C. Latimer, B. Rusuwa, and T. Gosden for technical assistance in the laboratory. They also thank M. Blows, D. Ortiz-Barrientos, and two anonymous reviewers for comments and suggestions on an earlier version of the manuscript. This work was supported by funding from the Australian Research Council awarded to S.F.C. and R.B.
Literature Cited
- Bachtrog D, Toda NR, Lockton S. Dosage compensation and demasculinization of X chromosomes in Drosophila. Curr Biol. 2010;20:1476–1481. doi: 10.1016/j.cub.2010.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Betran E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar RB, Makley LN, Tsonis PA. The other lives of ribosomal proteins. Hum Genomics. 2010;4:327–344. doi: 10.1186/1479-7364-4-5-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutkar A, et al. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics. 2008;179:1657–1680. doi: 10.1534/genetics.107.086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA. Claims and counterclaims of X-chromosome compensation. Nat Struct Mol Biol. 2012;19:3–5. doi: 10.1038/nsmb.2218. [DOI] [PubMed] [Google Scholar]
- Bonduriansky R, Chenoweth SF. Intralocus sexual conflict. Trends Ecol Evol. 2009;24:280–288. doi: 10.1016/j.tree.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Clark AG. The resolution of sexual antagonism by gene duplication. Genetics. 2011;187:919–937. doi: 10.1534/genetics.110.123729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 2011;13:123–134. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- Draghici S. Statistics and data analysis for microarrays using R and bioconductor. Boca Raton, FL: CRC Press; 2012. [Google Scholar]
- Ellegren H, et al. Faced with inequality: chicken do not have a general dosage compensation of sex-linked genes. BMC Biol. 2007;5:40. doi: 10.1186/1741-7007-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu FD, Adamski M, McGraw EA, Blows MW, Chenoweth SF. An expressed sequence tag (EST) library for Drosophila serrata, a model system for sexual selection and climatic adaptation studies. BMC Genomics. 2009;10:40. doi: 10.1186/1471-2164-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Betran E. Intralocus sexual conflict resolved through gene duplication. Trends Ecol Evol. 2011;26:222–228. doi: 10.1016/j.tree.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladstein N, McKeon MN, Horabin JI. Requirement of male-specific dosage compensation in Drosophila females—implications of early X chromosome gene expression. PLoS Genet. 2010;6:e1001041. doi: 10.1371/journal.pgen.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, et al. Global analysis of X-chromosome dosage compensation. J Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Hahn MW. Inferring the history of interchromosomal gene transposition in Drosophila using n-dimensional parsimony. Genetics. 2012;190:813–825. doi: 10.1534/genetics.111.135947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy SJ. The stoichiometry of the ribosomal proteins of Escherichia coli. Mol Gen Genet. 1975;140:253–274. doi: 10.1007/BF00334270. [DOI] [PubMed] [Google Scholar]
- Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 2007;5:e273. doi: 10.1371/journal.pbio.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, et al. Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res. 2010;20:512–518. doi: 10.1101/gr.102343.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Bachtrog D. Evolution of sex chromosomes in insects. Annu Rev Genet. 2010;44:91–112. doi: 10.1146/annurev-genet-102209-163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- Lande R. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution. 1980;34:292–305. doi: 10.1111/j.1558-5646.1980.tb04817.x. [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. Evidence that the human X chromosome is enriched for male-specific but not female-specific genes. Mol Biol Evol. 2003;20:1113–1116. doi: 10.1093/molbev/msg131. [DOI] [PubMed] [Google Scholar]
- Lindstrom MS. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem Biophys Res Commun. 2009;379:167–170. doi: 10.1016/j.bbrc.2008.12.083. [DOI] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. All dosage compensation is local: gene-by-gene regulation of sex-biased expression on the chicken Z chromosome. Heredity (Edinb) 2009;102:312–320. doi: 10.1038/hdy.2008.116. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hultin-Rosenberg L, Webster MT, Ellegren H. The unique genomic properties of sex-biased genes: insights from avian microarray data. BMC Genomics. 2008;9:148. doi: 10.1186/1471-2164-9-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Landeen EL, Cook JM, Kingan SB, Presgraves DC. Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol. 2011;9:e1001126. doi: 10.1371/journal.pbio.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Presgraves DC. Little evidence for demasculinization of the Drosophila X chromosome among genes expressed in the male germline. Genome Biol Evol. 2012;4:1007–1016. doi: 10.1093/gbe/evs077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Malone JH, Clark AG. Disentangling the relationship between sex-biased gene expression and X-linkage. Genome Res. 2012;22:1255–1265. doi: 10.1101/gr.132100.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova LM, Nurminsky DI. Lack of global meiotic sex chromosome inactivation, and paucity of tissue-specific gene expression on the Drosophila X chromosome. BMC Biol. 2011;9:29. doi: 10.1186/1741-7007-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Sex chromosomes and sex-linked genes. Berlin, Heidelberg, New York: Springer-Verlag; 1967. [Google Scholar]
- Parisi M, et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GA. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 2012;109:5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince EG, Kirkland D, Demuth JP. Hyperexpression of the X chromosome in both sexes results in extensive female bias of X-linked genes in the flour beetle. Genome Biol Evol. 2010;2:336–346. doi: 10.1093/gbe/evq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Sex-chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Stocker AJ, et al. Physical and linkage maps for Drosophila serrata, a model species for studies of clinal adaptation and sexual selection. G3 (Bethesda) 2012;2:287–297. doi: 10.1534/g3.111.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: a consequence of dosage compensation? J Mol Evol. 2009;68:576–583. doi: 10.1007/s00239-009-9235-4. [DOI] [PubMed] [Google Scholar]
- Voynow P, Kurland CG. Stoichiometry of the 30S ribosomal proteins of Escherichia coli. Biochemistry. 1971;10:517–524. doi: 10.1021/bi00779a026. [DOI] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- Yang X, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Oliver B. An evolutionary consequence of dosage compensation on Drosophila melanogaster female X-chromatin structure? BMC Genomics. 2010;11:6. doi: 10.1186/1471-2164-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–237. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D. Chromosome-wide gene silencing initiates Y degeneration in Drosophila. Curr Biol. 2012;22:522–525. doi: 10.1016/j.cub.2012.01.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.