Abstract
Eukaryotic genomes contain numerous DNA transposons that move by a cut-and-paste mechanism. The majority of these elements are self-insufficient and dependent on their autonomous relatives to transpose. Miniature inverted repeat transposable elements (MITEs) are often the most numerous nonautonomous DNA elements in a higher eukaryotic genome. Little is known about the origin of these MITE families as few of them are accompanied by their direct ancestral elements in a genome. Analyses of MITEs in the yellow fever mosquito identified its youngest MITE family, designated as Gnome, that contains at least 116 identical copies. Genome-wide search for direct ancestral autonomous elements of Gnome revealed an elusive single copy Tc1/Mariner-like element, named as Ozma, that encodes a transposase with a DD37E triad motif. Strikingly, Ozma also gave rise to two additional MITE families, designated as Elf and Goblin. These three MITE families were derived at different times during evolution and bear internal sequences originated from different regions of Ozma. Upon close inspection of the sequence junctions, the internal deletions during the formation of these three MITE families always occurred between two microhomologous sites (6–8 bp). These results suggest that multiple MITE families may originate from a single ancestral autonomous element, and formation of MITEs can be mediated by sequence microhomology. Ozma and its related MITEs are exceptional candidates for the long sought-after endogenous active transposon tool in genetic control of mosquitoes.
Keywords: transposable elements, MITEs, microhomology, origin
Introduction
Transposable elements (TEs) are integral components of eukaryotic genomes. They made important contributions to host genomes during evolution (Slotkin and Martienssen 2007; Pritham 2009; Rebollo et al. 2012; Dooner and Weil 2013). Their intimate interactions with genic contents in genomes kindled an array of major evolutionary steps leading to current life forms (Zhou et al. 2004; Lin et al. 2007; Baucom et al. 2009; Gonzalez et al. 2009; Hollister et al. 2011; Jiang et al. 2011). DNA TEs are a major type of mobile genetic material in eukaryotic genomes. They use a cut-and-paste mechanism to move from one genomic location to another. Even though these elements are abundant in eukaryotic genomes, few of them are active in transposition probably because many of them are subject to purifying selection (Petrov et al. 2011). An element that can produce transposases, proteins required for transposition, to mobilize itself is an autonomous element. However, nonautonomous elements that do not encode functional transposases are often much more abundant than autonomous elements. Some nonautonomous elements are very similar to autonomous elements in sequence except that their transposase coding sequences are disrupted by mutations such as deletions and frameshifts. An extreme type of nonautonomous elements is collectively called miniature inverted repeat transposable element (MITE). Compared with autonomous elements and canonical nonautonomous elements, MITEs are much shorter and rarely bear apparent transposase coding sequences (Feschotte et al. 2002; Jiang et al. 2004; Piriyapongsa and Jordan 2007; Fattash et al. 2013).
MITEs are abundant in eukaryotic genomes. A higher eukaryotic genome typically contains dozens to hundreds of different MITE families (Jiang et al. 2004; Nene et al. 2007; Piskurek et al. 2009; Schnable et al. 2009; International Aphid Genomics Consortium 2010; Han et al. 2010; Yaakov et al. 2012). A single MITE family can often reach hundreds of copies in a genome. Some are capable of achieving much higher copy numbers (Charrier et al. 1999; Lepetit et al. 2000; Hikosaka and Kawahara 2004; Macas et al. 2005; Ray et al. 2005; Remigereau et al. 2006). A MITE family may be related to a TE superfamily if its terminal inverted repeat (TIR) sequence is similar to that of an autonomous element in the same superfamily, and it generates the same sized target site duplication (TSD) as that of an autonomous element. For example, the Tourist type MITE families share TIRs with those of the PIF/Harbinger elements and they generate TSDs of three base pairs (Yang et al. 2001; Zhang et al. 2001; Jiang et al. 2003). Also, Stowaway MITEs share TIRs with Tc1/mariner elements and they always generate a duplication of the dinucleotide target site “TA” (Bureau and Wessler 1994; Feschotte et al. 2003). Similarly, MITE families can belong to other superfamilies including hAT, Mutator, P, and PiggyBac (Yang and Hall 2003b; Osborne et al. 2006; Quesneville et al. 2006; Wang et al. 2010). However, different MITE families within the same superfamily may or may not share similar TIRs, depending on whether the autonomous elements they were derived from have similar or different TIRs. For example, all of the rice Stowaway MITEs share similar TIRs because all of their related autonomous elements Osmars share similar TIRs, whereas most of the Stowaway-like MITEs in the yellow fever mosquito genome bear different TIRs because most of the Tc1/mariner families in the genome bear different TIRs (Tu 2000; Feschotte et al. 2003; Nene et al. 2007).
The origins of the majority of MITE families are mysterious. Unlike canonical nonautonomous elements, most MITE families are not direct deletion derivatives of existing autonomous elements or canonical nonautonomous elements. It was proposed that abortive gap repair (AGR) of the donor site of an excised autonomous element may be involved in the generation of MITEs (Feschotte et al. 2002). AGR is thought to be involved in the formation of canonical nonautonomous elements of Ac, P, Tc1, and Mutator (Engels et al. 1990; Doseff et al. 1991; Plasterk 1991; Nassif et al. 1994; Lisch et al. 1995; Rubin and Levy 1997). However, evidence for the involvement of AGR in MITE formation has been scarce. For example, short direct repeats commonly observed in AGR are not present at the break points for mPing and mPIF (Kurkulos et al. 1994; Hsia and Schnable 1996; Rubin and Levy 1997; Zhang et al. 2001; Jiang et al. 2003). Additionally, a number of MITE families have unusually long TIRs despite much shorter TIRs found on the autonomous elements of the superfamily. For example, Tc1/mariner elements typically have TIRs shorter than 50 bp, but a number of Stowaway MITEs such as Milord, Cele1, Cele2, CeleTc1/Tc7, Tc6, CeleTc2, and CeleTc5 bear much longer TIRs (Dreyfus and Emmons 1991; Oosumi et al. 1995; Feschotte et al. 2002; Jurka et al. 2005). Similarly, PIF/Harbinger, hAT, P, and PiggyBac elements typically bear TIRs shorter than 30 bp, but MITEs such as CbmPIF1a, Joey, PALTTAA2_CE, Snabo-2, Xfb, Galileo, and MathE3 bear much longer TIRs (Unsal and Morgan 1995; Besansky et al. 1996; Chen et al. 1997; Surzycki and Belknap 1999; Tu 2001; Zhang et al. 2001; Feschotte et al. 2002; Marzo et al. 2013). Even the whole sequences of some non-Mutator MITE families such as PALTA1_CE, PALTA2_CE, PALTA4_CE, Mirza, CeleTc2, CeleTc5, Cele7, PALTTAA1_CE, PALTTAA3_CE, and Hairpin are essentially foldback structures (Oosumi et al. 1995; Ade and Belzile 1999; Feschotte et al. 2002; Jurka et al. 2005). The formation of these MITEs with unexpectedly long TIRs cannot be explained with simple internal sequence deletion of the autonomous elements. In addition, multiple MITE families sharing similar TIRs can be present in a genome, and the number of these MITE families may exceed the number of canonical transposase coding elements bearing similar TIRs. For example, the rice genome has 36 Stowaway families sharing similar TIRs but has only 25 transposase coding Osmar families (Feschotte et al. 2003). Although the loss of the transposase coding elements may explain this difference, it is also possible that one autonomous element can give rise to multiple MITE families. To understand the formation of MITE families from autonomous elements, the co-presence of a MITE and its parental autonomous element is important. Because TE sequences particularly those with low copy numbers like autonomous elements tend to be lost relatively rapidly from a genome during evolution, the autonomous element of a newly formed MITE is more likely to be present in the genome. Therefore, newly formed MITE families are valuable materials to gain insights into the birth of MITEs. In addition, newly formed MITEs are potentially still active in the genome. Endogenous active transposons in mosquitoes are long sought-after tools for use in genetic control of mosquitoes to prevent diseases mediated by these vector insects.
This report describes detailed analyses of the MITE family, herein designated as Gnome, in the yellow fever mosquito. Gnome is the youngest MITE family newly derived from a single copy autonomous element. This autonomous element also gave birth to two additional MITE families (Elf and Goblin) independently during evolution, resulting from internal deletion of different regions of the autonomous element. Goblin carries much longer TIRs (54 bp) that involve inversely duplicated subterminal sequences of the autonomous element. Interestingly, the break points of internal deletion of the autonomous element clearly show microhomology of 6–8 bases, suggesting that the miniaturization of the autonomous elements involves AGR followed by microhomology-mediated end joining (MMEJ). These results demonstrate that one autonomous element can give rise to multiple MITE families and microhomologous sites on the autonomous elements play important roles in the formation of MITE families.
Materials and Methods
MITE Sequence Retrieval, Clustering, and Alignment
MITE sequences were used as input for the Member function of MITE Analysis Kit (MAK) (http://labs.csb.utoronto.ca/yang/MAK/, last accessed October 8, 2013) (Yang and Hall 2003a; Janicki et al. 2011) to retrieve all complete members with or without their flanking sequences from the Aedes aegypti genomic sequence database assembly AaegL1 downloaded from www.vectorbase.org (last accessed October 8, 2013). The Identicals function of MAK was used to identify clusters of elements with identical sequences. Sequences of the representative members from the largest 10 clusters were aligned using Muscle at EMBL-EBI (http://www.ebi.ac.uk/, last accessed October 8, 2013). The alignment was shaded with Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html, last accessed October 8, 2013).
Autonomous Element Retrieval and Analysis
Gnome sequence was used as the input for the Anchor function of MAK to retrieve longer elements bearing similar terminal sequences and Tc1/mariner-like transposase coding sequences. The transposase database was compiled from the transposase entries in GenBank. The output was manually inspected to remove false output entries. To identify similar autonomous elements of Ozma in the genome, the transposase amino acid sequence was used as the input sequence for the TpTE function of MAK to identify elements bearing closely related transposase coding sequences and also terminal structures (inverted repeats flanked by direct repeats). The output was manually inspected to select the entries with the best matches in the TIR regions. These sequences were grouped according to the TIR sequences, and three best representative elements (Ozana, Ozga, and Ozgana) were chosen for further analyses. Transposase sequences of these elements were aligned together with that of Mos1 on the EMBL-EBI server and shaded with Boxshade. A phylogenetic tree of the transposases rooted with Mos1 was constructed and visualized with Tree Top of GeneBee with 1,000 boot strap iterations (http://www.genebee.msu.su/services/phtree_reduced.html, last accessed October 8, 2013). Helix turn helix domains were predicted with NPS (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_hth.html, last accessed October 8, 2013) (Dodd and Egan 1990). The A. aegypti TEfam database was available at http://tefam.biochem.vt.edu (last accessed October 8, 2013) (Nene et al. 2007).
Sequence Divergence of MITE Families
To calculate the average sequence divergence of a MITE family, the consensus sequence of each family was constructed. The consensus sequence was used as the input for the Divergence function of MAK. Each divergence value is the complementary percentage of the similarity value in the pairwise alignment of a copy and the consensus sequence. The output contains the sequence divergence values for each member. The average divergence for each MITE family was calculated. To plot the number of elements against divergence, values of individual divergence were grouped into bins of 0.5% and the number of elements in each bin was counted. The overall sequence similarity for a MITE family is calculated as the complement of the average sequence divergence.
Analysis of Break Points and Junction Sequences
Each MITE sequence was aligned with Ozma with NCBI BLAST program to reveal break points and junction sequences. Ten nucleotides flanking each break point were retrieved and the break point sequences of a junction were compared to identify sequence features such as microhomology or insertion. Because of the reversed internal sequence in Goblin, the left break point sequence of Goblin was the reverse complementary sequence of the corresponding region on the 3′ subterminal sequence of Ozma.
Results
Gnome is a Newly Formed MITE Family
The genome of the yellow fever mosquito (A. aegypti) is particularly rich in MITEs, constituting 16% (∼225 Mb) of the mosquito genome (Tu 1999, 2000, 2001; Nene et al. 2007). The genome contains at least 142 MITE families, of which 108 can be roughly grouped with five superfamilies of DNA elements based on TSD size and limited similarity in the TIRs. Among the annotated MITE families, 56 are Stowaway-like families, 9 are Tourist-like, 20 are PiggyBac-like, 21 are hAT-like, and two are Mutator-like (Nene et al. 2007). Unlike the Stowaway elements in rice where all of families share similar TIRs (Feschotte et al. 2003), few of the 56 Stowaway-like families in yellow fever mosquito share similar TIRs, indicating that these MITEs were derived from autonomous elements belonging to different subgroups of the superfamily.
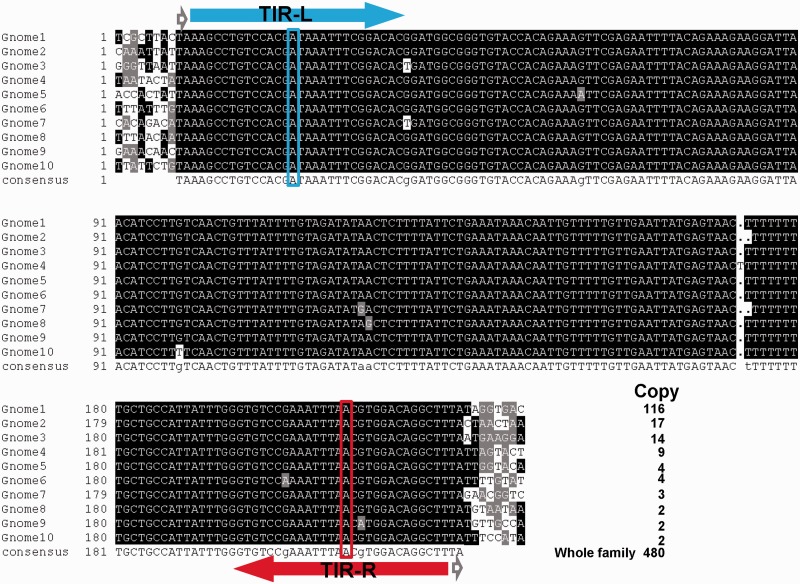
Newly formed MITE families are important both to understand the origin of MITEs and to understand the transposition activity of MITEs. The most apparent indicator of newly formed MITE families is the presence of highly similar or even identical copies resulting from recent transposition activity. By analyzing MITE families of the yellow fever mosquito genome for identical copies, a Stowaway-like MITE (TF000728 in TEfam) was found to have very high intrafamily sequence similarity and was designated as Gnome (fig. 1) (Yang et al. 2012). The genome contains 480 Gnome elements bearing TIRs on both ends. These elements share an overall sequence similarity of 99.4%. Importantly, there are five clusters of Gnome elements with greater than five identical copies. The largest cluster of identical elements contains 116 elements. The consensus sequence of Gnome is 209 bp long with TSDs of “TA” dinucleotides, the characteristic feature of elements in the Tc1/mariner superfamily. The TIRs are 28 bp, and the two TIRs are not identical, with the left TIR (TIR-L) and right TIR (TIR-R) differing by one nucleotide at the 14th nucleotide (fig. 1). The high sequence similarity, particularly the large number of identical copies, suggests that Gnome is a newly formed family.
Fig. 1.—
Sequence alignment of representative Gnome MITE sequences. The copy numbers of identical elements are shown to the right of each sequence. Gray arrowheads, TSDs; TIR-L, left TIR; TIR-R, right TIR; blue box, signature base for the left TIR; red box, signature base for the right TIR.
The Autonomous Element of Gnome
The high copy number and intrafamily sequence similarity of Gnome suggest that this element has amplified very recently and may even still be actively transposing. Because MITEs are nonautonomous elements, their amplification requires transposases from their autonomous elements. It is possible that, like observed for the rice Tourist MITE mPing, the transposase is produced from the ancestral element(s) from which Gnome was derived. Alternatively, as observed for the rice Stowaway-35, the transposase can be from an element different from, but related to, the direct ancestral element (Gonzalez and Petrov 2009; Yang et al. 2009). In both cases, the autonomous element is expected to bear TIRs highly similar or identical to those of the Gnome. In the TEfam database, there are 70 Tc1/mariner elements; however, none of them bear the TIR sequence of Gnome. Among the 17 elements in the ITmD37E subgroup, one (TF000892) shares the 6 nt terminal sequences and 10 of them (TF000893–TF000902) share 3–4 nt terminal sequences with Gnome. This suggests that Gnome belongs to the ITmD37E subgroup of the Tc1/mariner superfamily (Shao and Tu 2001; Biedler et al. 2007). As TIRs are critical for recognition by transposase during transposition, it is unlikely that Gnome is mobilized by any of these elements.
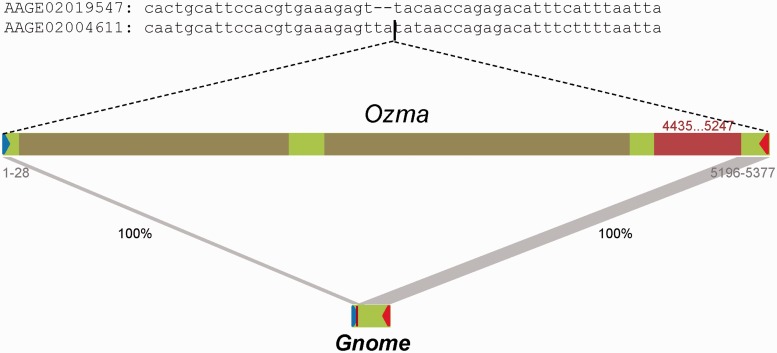
To see whether an autonomous element bearing TIRs of Gnome is present in the genome, even though not present in the TEfam database, Gnome sequence was used as the query sequence for the MAK Anchor function (see Materials and Methods) to retrieve transposase coding sequences bearing the termini of Gnome. Among the retrieved sequences, one element with a size of 5,377 bp was found to bear the left TIR (28 bp) of Gnome on the 5′-end and bear the remaining sequences of Gnome (181 bp) on the 3′-end (fig. 2). Therefore, Gnome is a direct deletion derivative of the long element. This element, here designated as Ozma, contains an intact open reading frame (ORF) of 270 a.a. from position 4435 to 5247 encoding a Tc1/mariner-like transposase. Interestingly, there is only one copy of Ozma in the genome, and it is inserted in the contig AAGE02004611. By using the flanking sequence of Ozma as a query sequence to search against the genome database, a related empty flanking site containing a single copy of the “TA” target site was identified in the contig AAGE02019547. Ozma is inserted at the target site “TA” and generates a duplication of the target site (fig. 2). The size of Ozma is unusual, given the typical sizes of similar elements such as Tc1 and Mariner around 1.5 kb. To see whether the large size of Ozma is caused by insertion of other repetitive sequences, Ozma sequence was used to search against the genomic DNA database. Analyses of the output identified two putative repetitive sequences. The element on the 5′-end bears TIR sequences of “CAGGGTGTCGACT” and is located at positions from 50 to 1,945 bp. Its insertion generated a duplication of the target site “GTTTT.” The element close to the 3′-end does not appear to have TIRs and is located at positions from 2,145 to 4,318 bp. Its insertion appears to have generated a duplication of the target site “AAAA.” This element carries a relic coding region with an ORF of 126 a.a., similar to that of the EEP motif that are commonly found in LINE elements. When the two repetitive sequences were removed, the ORF of Ozma can be extended 201 bp at the 5′-end to result in a protein of 337 a.a. These results suggest that Ozma may have been inactivated by these insertion sequences.
Fig. 2.—
The autonomous element Ozma for Gnome. Sequences on top, the flanking sequence for Ozma element and related empty site. “-” in sequences, gaps in the alignment. Blue and red triangles, left and right TIRs; brown bars, repeats inserted in Ozma; red bar, 270 a.a. ORF; gray stripes, corresponding regions between Gnome and Ozma; percentage, sequence identity; range numbers in red, coordinates for the 270 a.a. ORF; gray number ranges, coordinates of homologous regions with Gnome on Ozma. Element length is to scale.
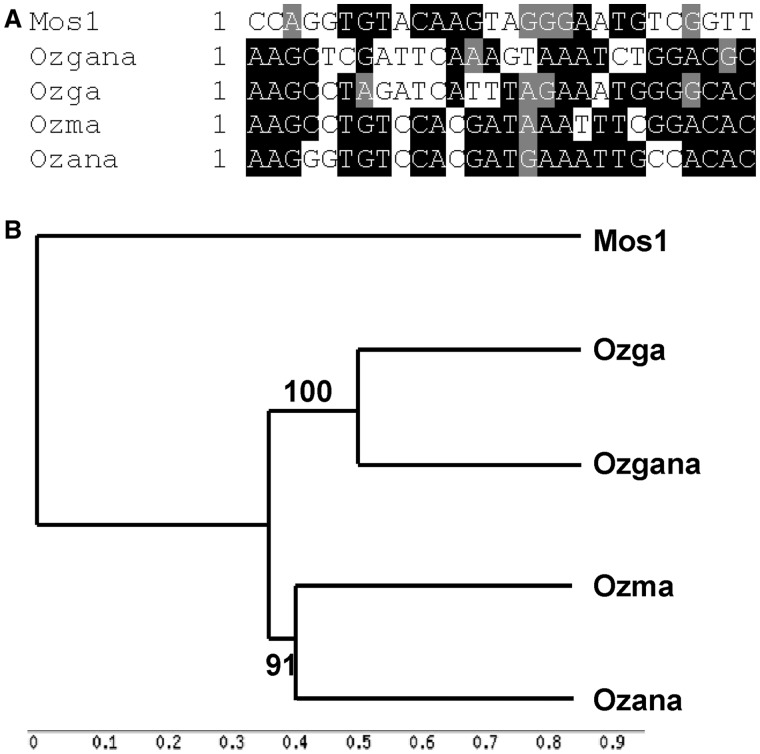
MITEs can be cross-mobilized by transposases encoded by elements different from, but closely related to, their direct ancestral element(s). To identify such related autonomous elements, the Ozma transposase sequence was used with the TpTE function of MAK. Among the elements in the output, three elements (here designated as Ozana, Ozga, and Ozgana) showing the most similar TIRs to that of Ozma were analyzed further. Ozana has 332 copies in the genome and encodes an intact transposase of 336 a.a. Ozga and Ozgana have only 17 and 16 copies, respectively. When the TIRs of these elements were aligned, TIRs of Ozana are the closest to those of Ozma. The TIRs of the other two elements share 6 nt at the 5′-end but differ significantly toward the 3′-end of the TIRs (fig. 3A). When the transposases of the elements were aligned with the mariner element Mos1 (Medhora et al. 1991) and phylogenetic trees were constructed, it is apparent that the transposase of Ozana is the most closely related to Ozma transposase (fig. 3B and supplementary fig. S1, Supplementary Material online).
Fig. 3.—
Autonomous elements related to Ozma. (A) Alignment of the left TIRs of Ozma, Ozana, Ozga, and Ozgana with that of Mos1. (B) Phylogenetic tree of the full-length ORF of the elements. Bootstrap value, 1,000 iterations; see supplementary fig. 1, Supplementary Material online, for alignment. Numbers on branches, percentages of boostrap iterations.
Additional MITE Families Derived from Ozma
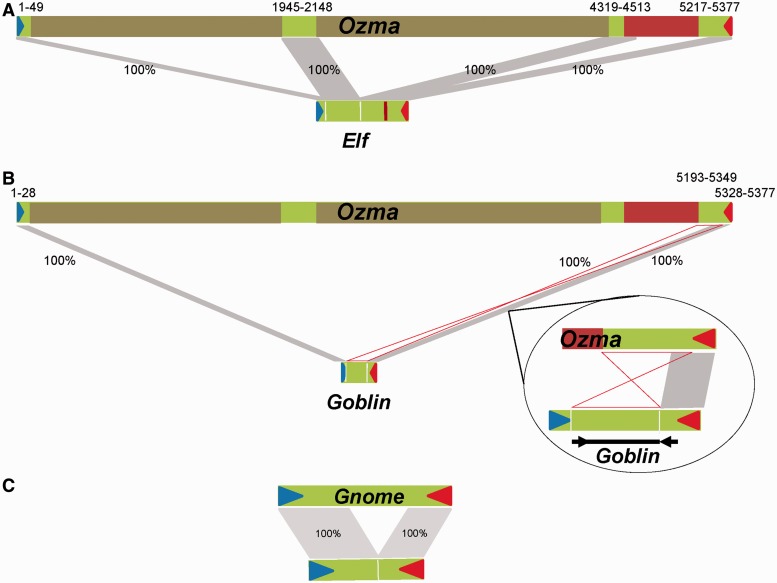
It is believed that an autonomous element often gives rise to one MITE family in a genome. However, among the very few cases where the direct ancestral autonomous elements of MITEs were found, the association of the nematode PIF element with two Tourist MITE families (Cb-mPIF1a and Cb-mPIF1b) suggests the possibility that multiple MITE families may be derived from a single ancestral element (Feschotte et al. 2002). To see whether there are other MITEs derived from Ozma in the genome, sequences bearing the TIRs of Ozma were retrieved. After grouping these elements, in addition to the Gnome family, two other MITE families were identified and designated as Elf and Goblin. Both elements are directly derived from Ozma. Elf is 549 bp long and corresponds to the sequences of four segments of Ozma at following positions: 1–49, 1945–2148, 4319–4513, and 5217–5377 (fig. 4A). The first two junctions (50–1944 and 2149–4318) are at the same positions of the two insertions in Ozma, therefore Elf was likely formed before the insertion of the two repeats in Ozma. The internal deletion of Ozma to form Elf is at the positions between 4514 and 5216. Therefore, Elf contains a small portion (280 bp) of the transposase coding sequence of Ozma. Elf has a copy number of 57 with an overall intrafamily sequence similarity of 97.6%. Goblin is another MITE independently derived from Ozma with a size of 213 bp. Intriguingly, Goblin is not a simple internal deletion of Ozma. The 28 bp on the 5′-end and the 50 bp on the 3′-end of Goblin match the corresponding regions of Ozma. Sequences between the two terminal regions of Goblin match the 135 bp immediately before the 3′ terminal region (50 bp) of Ozma in a reversed orientation (fig. 4B). There are 15 copies of Goblin in the genome with an overall intrafamily sequence similarity of 98.4%. Interestingly, among the deletion derivatives, a single copy of a shortened version of Gnome is present in the genome (fig. 4C).
Fig. 4.—
Elf and Goblin MITE families derived from Ozma. (A) Elf element derived from Ozma. (B) Goblin element derived from Ozma. Bubble, close up view of homologous regions between Ozma and Goblin right ends. (C) A deletion derivative of Gnome. Blue and red triangles, left and right TIRs; gray stripes, homologous regions; percentage, sequence similarity; brown bars, repeats inserted in Ozma; red bar, 270 a.a. ORF; number ranges, coordinates of homologous regions with Gnome on Ozma. Hour glass shape, inversed orientation of the region of Ozma on Goblin. Black arrow heads in bubble, inverted sequences of the Ozma subterminal regions on Goblin.
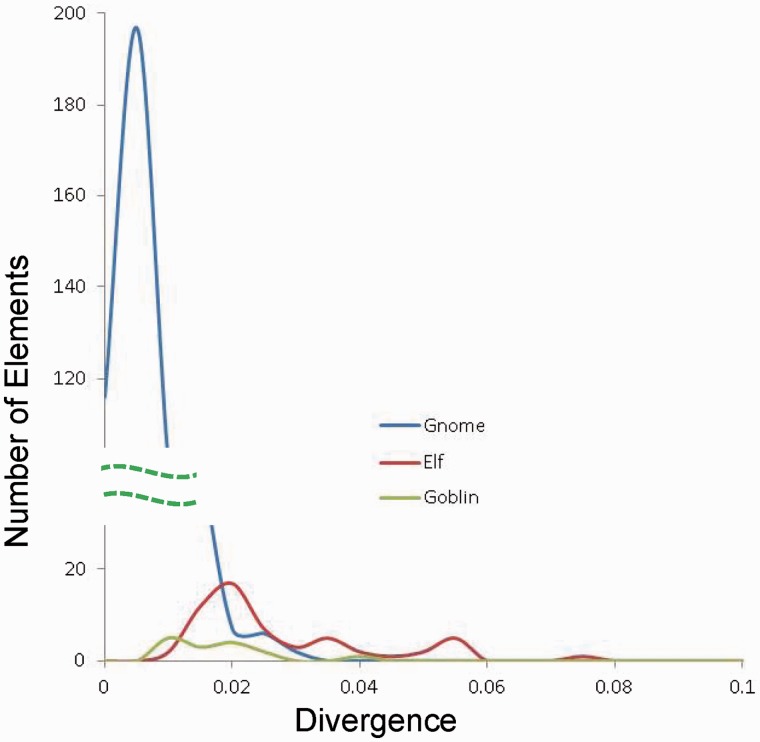
During the amplification of a TE family, mutations in the elements will accumulate. The degree of divergence of the elements from the consensus sequence of a TE family can be used to estimate the relative age of a family (Kapitonov and Jurka 1996). To understand whether Gnome, Elf, and Goblin were generated at the same time during evolution, consensus sequences were generated for each family and the divergence rate of each copy from the consensus was calculated. The average divergence value for the three families, Gnome, Elf, and Goblin, are 0.6%, 2.42%, and 1.64%, respectively. Numbers of copies of an element at a certain range of divergence rate were plotted against the divergence rate (fig. 5). The number of elements peaked at the divergence value of ∼0.5%, ∼2%, and ∼1.5% for Gnome, Elf, and Goblin, respectively, suggesting that the order of appearance for the three families is Elf, Goblin, and Gnome. The highest divergence rates for the three families are 2.82% (Gnome), 7.11% (Elf), and 3.72% (Goblin), in agreement with the order of their appearance. In addition, Gnome has the largest number of identical elements as described earlier, Goblin has five identical copies, and Elf has no identical elements. This observation further supports the order of their appearance during evolution. Based on the rough estimation of the mutation rate for the mosquito genome at 1 × 10−7/base/year (Haag-Liautard et al. 2007; Struchiner et al. 2009), the average time of appearance for these families are estimated to be 60, 164, and 242 thousand years ago. Even though Elf appears to have formed before the insertion of the two repeat elements into Ozma, it is unclear whether Gnome and Goblin arose before or after the insertion events.
Fig. 5.—
Distribution of sequence divergence for Gnome, Elf, and Goblin families. The numbers of elements in a certain range of divergence from the consensus sequences are plotted against the divergence range. Bin size, 0.005. Dashed lines, broken y axis for better view of the three families. x axis, divergence value; y axis, number of elements in a certain range of divergence value.
Microhomology-Mediated Transposon Miniaturation
Little is known about mechanisms of origination of MITE families from autonomous elements. The internal deletions of an autonomous element in MITE formation fall in the category of chromosome microdeletion. Different mechanisms responsible for these deletion events may leave their characteristic sequence features at or around break points. To understand what mechanisms may be involved in the generation of these MITE sequences, break point sequences at the junctions were inspected. The break point for Gnome on the left is immediately after the left TIR whereas the break point on the right is 52 bp upstream of the stop codon of the transposase coding sequence. The 8 bp sequence (CGGACACT) after the left break point is very similar to that before the right break point (CGGAACCT) with a mismatch of “CA/AC.” In addition, an information scar of a “T” to “G” transversion is present at the junction of the break points (fig. 6A) (Verdin et al. 2013). The left break point of Elf is 280 bp into the transposase coding sequence and the right break point is 28 bp upstream of the stop codon. Similarly, the 6 bp (GGAAGT) right after the left break point is very similar to that after the right break point (GAAAGT) with a “G/A” mismatch (fig. 6B). Despite the unusual configuration of Goblin as described earlier, break points show a 6 bp (AACTTT) microhomology (fig. 6C). An information scar of a single nucleotide “T” insertion is present at the junction. Though microhomologies of this size range can occur with replication-based mechanisms, mismatches in the microhomologous sites and, particularly, the insertional information scars are hallmark features of MMEJ. Therefore, gap repair of the double-stranded DNA breaks resulted from the excision of Ozma followed by MMEJ repairing was likely to be involved in the generation of Gnome, Elf, and Goblin. The formation of Goblin may also involve template switching during the new strand synthesis as shown in the proposed model (fig. 7). In addition, the miniature element derived from Gnome internal deletion shows microhomology of three nucleotides, suggesting a classical nonhomologous end joining (NHEJ) process (fig. 6D).
Fig. 6.—
Microhomology between break point sequences. Green sequences, left break points; red sequences, right break points; black base letters, aberrant nucleotides introduced; vertical black lines in sequences, junctions; number of bases, length between the two break points; underlined letters, microhomologous sequences.
Fig. 7.—
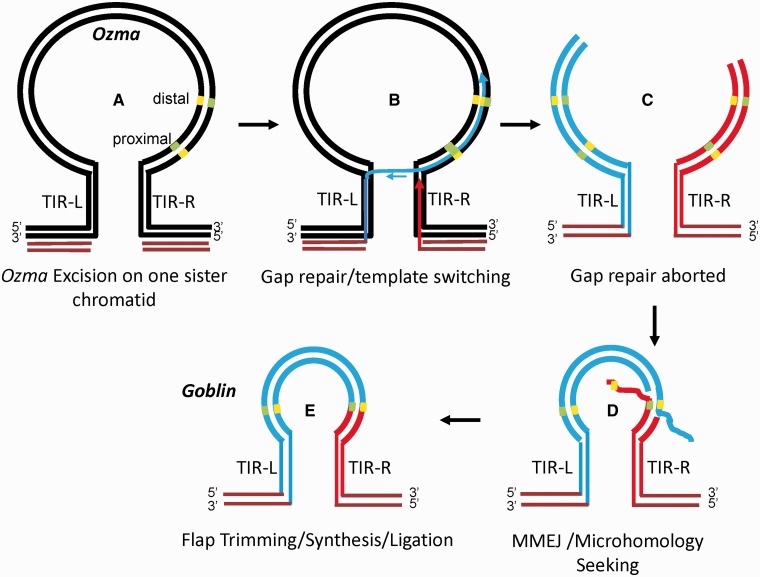
Hypothetical model for the formation of Goblin. The microhomologous sites at the break points are located in the 3′ subterminal region of Ozma element. Yellow and green short bars, complementary microhomologous sites. Ozma is drawn as a loop structure for convenient illustration of template switching. (A) Double-stranded break formed after the excision of Ozma on one of the two sister chromatids. (B) Gap repair initiated and template switching occurred after the replication of the left TIR. When the 3′-end of the top strand of the left TIR is synthesized, it invades the DNA sequences on the right TIR for replication. (C) Gap repair aborted and the newly synthesized strands are released from the template and the lagging strands synthesized. Microhomologous sites on the newly synthesized DNA are in direct repeat orientation of that on the sequences close to the right TIR. (D) Resection occurs to expose the microhomologous sites that anneal to each other, forming single-stranded flaps with the unannealed strands. (E) Flap trimming, synthesis, and ligation, the newly synthesized double-stranded DNA joins the sequences on the right end between the left distal and the right proximal microhomologous sites. Maroon lines, the sequences flanking the excised Ozma; black lines, unexcised Ozma with flanking sequences; blue lines, newly synthesized DNA from the left; red lines, newly synthesized DNA from the right.
Discussion
Most of TEs in an eukaryotic genome are nonautonomous, and a major portion of them are internally deleted versions of autonomous elements with MITEs being exemplary cases. Mechanisms involved in such chromosome microdeletions can be 1) homologous recombination based such as non-allelic homologous recombination (NAHR) (Stankiewicz and Lupski 2002; Sen et al. 2006; Han et al. 2008); 2) replication-based such as fork stalling and template switching (FoSTeS) (Lee et al. 2007), replication slippage (RS) (Streisinger et al. 1966; Niel et al. 2004; Tancredi et al. 2004), and serial replication slippage (SRS) (Chen et al. 2005); 3) DNA break-and-repair based such as NHEJ (Lieber 2008), MMEJ (McVey and Lee 2008), and single-strand annealing (SSA) (Sugawara et al. 2000); 4) combined mechanisms such as break-induced SRS (BISRS) and microhomology-mediated break-induced replication (MMBIR) (Sheen et al. 2007; Hastings et al. 2009). In synthesis-dependent strand annealing (SDSA), 3′ overhangs produced by resection of the double-strand break invade DNA duplex containing homologous sequences to form displacement loops that translocate during strand extension (Resnick 1976; Nassif et al. 1994). Interruption of SDSA can lead to AGR: while premature ending of SDSA during the repairing of a double-stranded DNA break (DSB) results in internal deletions of the template sequence, template switching during SDSA may result in the capture of stuff sequences from other genomic loci (Rubin and Levy 1997). As MITEs are end products of microdeletion events, break points and junction sequences are the only reminders of such deletion events and can serve as clues to uncover the underlying mechanisms.
The observed microhomology at the break points of Gnome, Elf, and Goblin excluded the NAHR mechanism which requires relatively long stretches of homologous sequences between the two sites (Stankiewicz and Lupski 2002). Microhomology at break points can be produced by several deletion generation mechanisms including the replication-based mechanisms such as FoSTeS, RS, and MMBIR or DNA break-and-repair based mechanisms such as NHEJ, MMEJ, and SSA. Microhomolgy is optional for NHEJ, and the size of microhomology involved is 1–4 nt (Lieber 2008). NHEJ often results in small deletions and insertions (1–4 nt). The microhomology required by SSA is >30 bp, and nucleotide insertion at the junction sites were never observed. Therefore, these deletion events in the formation of the three MITE families are not likely to have resulted from classical NHEJ and SSA events. In the replication-based mechanisms (FoSTeS, RS, and MMBIR), microhomologous sites are used for priming in replication, and mismatches in these sites are rare. Particularly, nucleotide insertion at the junction site is not expected.
Break repair based mechanisms start with the generation of a DSB. Such a break in AGR is caused by the excision of a transposon; therefore, the deletion derivatives of the element are newly synthesized. The released free ends may undergo end joining processes. Alternatively, in cases where transposons are resistant to gap repairing (Dooner and Martinez-Ferez 1997; Yamashita et al. 1999), a DSB occurring inside of an element independent of transposition may lead to deletion derivatives. Direct DSBs can also be caused by endonucleases and ionizing irradiation (e.g., UV and radioisotopes) (Goettel and Messing 2009). The major source of endogeneous DSBs is single-strand DNA lesions (SSLs) resulted from factors such as thermofluctations, hydrolysis, apurinic/apyrimidinic sites, topoisomerases, reactive oxygen species, 8-oxoG, thymine glycol, and 3-methyladenine (Vilenchik and Knudson 2003). Eukaryotic nuclear DNA is subjected to SSLs at a frequency of about 1 × 10−6 per base per S phase. A small portion (1%) of SSLs that escape repair cause the collapse of replication forks during S phase and result in endogeneous DSBs at about 10−8 per base per cell cycle. The majority (>95%) of these DSBs are repaired. Assuming the number of mitotic cell divisions before spermitogenesis in yellow fever mosquito is similar to that of the fruit fly with 25 cell divisions and the number of generations in a year is 10, around 10,000 DSBs would be expected in one year with a population size of a million for a DNA fragment the size of Ozma (1,871 bp). The generation of deletion derivatives from the repair of a DSB occurring inside an element does not require active transposition.
Although the generation of Gnome and Elf can be explained with a typical MMEJ repairing of a DSB, the configuration of Goblin is intriguing in that the subterminal sequences on both sides of the junction seem to have derived from the right subterminal region of Ozma with different lengths (fig. 4B). Unlike a typical MMEJ event that uses direct repeats of microhomologous sites on Ozma, the two microhomologous sites that led to the junction in Goblin are in inverted orientation on Ozma (fig. 6). In this case, template switching during SDSA of the newly synthesized left TIR to the right TIR on the template and subsequent extension of the right subterminal region may explain the inversion of the right subterminal sequences. The released ends may have then been repaired in an end joining process. As a number of MITE families bear long TIRs or a whole element is a hairpin even though the related autonomous elements do not bear long TIRs, it is possible that these MITE families arose in a similar process.
To see whether a similar break repair process may also be involved in the formation of other MITEs, the junctions of several MITEs with identified ancestral autonomous element were inspected. The rice MITE mPing is a deletion derivative of the element Ping. There are four subtypes of mPing, each having different break points though all of the break points are located in a narrow region on Ping (Jiang et al. 2003). The break points in subtypes A and B do not show microhomology or nucleotide insertions. The break points in subtype C show a single nucleotide “C” without any nucleotide insertion. The break points in subtype D shows a 2 bp “CT” microhomology with a 6 bp insertion at the junction. These break points and junction features favor the classical NHEJ mechanism. The human MITE Made1 is a deletion derivative of the human mariner-like element Hsmar1. Similar to Gnome, Elf, and Goblin, 6 bp (TGAAAT) of microhomology can be identified with an insertion of 6 bp at the junction, features fitting those of the MMEJ. These observations indicate that although MMEJ appears to be common in MITE formation, the formation of different MITE families may involve different mechanisms for internal deletions of ancestral autonomous elements. These miniaturization processes may have also led to the non-MITE miniature versions TEs that are much more abundant than autonomous elements such as the Ac/Ds elements in maize (Du et al. 2011).
The newly formed MITE Gnome and its related elements in yellow fever mosquito genome provided a unique opportunity to look into the formation of MITE families. The analyses of these elements revealed features in MITE origin and amplification including 1) one autonomous element gives rise to multiple MITE families bearing different internal sequences of the ancestral element; 2) the internal deletion of autonomous element may be mediated by microhomology at the break points; 3) MITEs with longer TIRs can be generated during internal deletion of the autonomous element. The identification of the direct ancestral element of the three MITE families opens the possibility to use these MITEs as vectors for gene transfer. The two insertions in the Ozma element may result in the inactivation of this element. However, the Gnome family may still be actively transposing in mosquito populations if intact copies of Ozma are still present. Alternatively, Gnome may be cross-mobilized by transposases other than Ozma in the mosquito genome. Although the presence of such cross-mobilizing transposase sources has not yet been demonstrated, the Ozma transposase, which can be easily reconstructed from the inactivated element, is an obvious choice for establishing an in vivo or in vitro transposition system for studies of MITE transposition mechanisms and to test for its potential utility as a gene driver in mosquito genetic control applications.
Supplementary Material
Supplementary file and figure S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr Cedric Feschotte and the reviewers for insightful comments. This work was supported by Natural Sciences and Engineering Research Council (RGPIN371565 to G.Y.), Canadian Foundation for Innovation (24456 to G.Y.), Ontario Research Fund (24456 to G.Y.), and University of Toronto.
Literature Cited
- Ade J, Belzile FJ. Hairpin elements, the first family of foldback transposons (FTs) in Arabidopsis thaliana. Plant J. 1999;19:591–597. doi: 10.1046/j.1365-313x.1999.00567.x. [DOI] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Leebens-Mack J, Bennetzen JL. Natural selection on gene function drives the evolution of LTR retrotransposon families in the rice genome. Genome Res. 2009;19:243–254. doi: 10.1101/gr.083360.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Mukabayire O, Bedell JA, Lusz H. Pegasus, a small terminal inverted repeat transposable element found in the white gene of Anopheles gambiae. Genetica. 1996;98:119–129. doi: 10.1007/BF00121360. [DOI] [PubMed] [Google Scholar]
- Biedler JK, Shao H, Tu Z. Evolution and horizontal transfer of a DD37E DNA transposon in mosquitoes. Genetics. 2007;177:2553–2558. doi: 10.1534/genetics.107.081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR. Stowaway—a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell. 1994;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier B, et al. Bigfoot: a new family of MITE elements characterized from the Medicago genus. Plant J. 1999;18:431–441. doi: 10.1111/j.1365-313x.1999.00469.x. [DOI] [PubMed] [Google Scholar]
- Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Complex gene rearrangements caused by serial replication slippage. Hum Mutat. 2005;26:125–134. doi: 10.1002/humu.20202. [DOI] [PubMed] [Google Scholar]
- Chen M, et al. Microcolinearity in sh2-homologous regions of the maize, rice, and sorghum genomes. Proc Natl Acad Sci U S A. 1997;94:3431–3435. doi: 10.1073/pnas.94.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Egan JB. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK, Martinez-Ferez IM. Germinal excisions of the maize transposon activator do not stimulate meiotic recombination or homology-dependent repair at the bz locus. Genetics. 1997;147:1923–1932. doi: 10.1093/genetics/147.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H, Weil C. Molecular genetics and epigenetics of plant transposons. Hoboken (NJ): John Wiley & Sons; 2013. Transposons and gene creation; pp. 143–167. [Google Scholar]
- Doseff A, Martienssen R, Sundaresan V. Somatic excision of the Mu1 transposable element of maize. Nucleic Acids Res. 1991;19:579–584. doi: 10.1093/nar/19.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus DH, Emmons SW. A transposon-related palindromic repetitive sequence from C. elegans. Nucleic Acids Res. 1991;19:1871–1877. doi: 10.1093/nar/19.8.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Hoffman A, He L, Caronna J, Dooner HK. The complete Ac/Ds transposon family of maize. BMC Genomics. 2011;12:588. doi: 10.1186/1471-2164-12-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels WR, Johnson-Schlitz DM, Eggleston WB, Sved J. High-frequency P element loss in Drosophila is homolog dependent. Cell. 1990;62:515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- Fattash I, et al. Miniature inverted-repeat transposable elements (MITEs): discovery, distribution and activity. Genome. 2013 doi: 10.1139/gen-2012-0174. Advance Access published March 8, 2013, doi: 10.1139/gen-2012-0174. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Swamy L, Wessler SR. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs) Genetics. 2003;163:747–758. doi: 10.1093/genetics/163.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Zhang X, Wessler SR. 2002. Miniature inverted-repeat transposable elements (MITEs) and their relationship with established DNA transposons. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington (DC): American Society of Microbiology Press. p. 1147–1158.
- Goettel W, Messing J. Change of gene structure and function by non-homologous end-joining, homologous recombination, and transposition of DNA. PLoS Genet. 2009;5:e1000516. doi: 10.1371/journal.pgen.1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J, Macpherson JM, Petrov DA. A recent adaptive transposable element insertion near highly conserved developmental loci in Drosophila melanogaster. Mol Biol Evol. 2009;26:1949–1961. doi: 10.1093/molbev/msp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J, Petrov D. MITEs—the ultimate parasites. Science. 2009;325:1352–1353. doi: 10.1126/science.1179556. [DOI] [PubMed] [Google Scholar]
- Haag-Liautard C, et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445:82–85. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- Han K, et al. L1 recombination-associated deletions generate human genomic variation. Proc Natl Acad Sci U S A. 2008;105:19366–19371. doi: 10.1073/pnas.0807866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MJ, et al. Burst expansion, distribution and diversification of MITEs in the silkworm genome. BMC Genomics. 2010;11:520. doi: 10.1186/1471-2164-11-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka A, Kawahara A. Lineage-specific tandem repeats riding on a transposable element of MITE in Xenopus evolution: a new mechanism for creating simple sequence repeats. J Mol Evol. 2004;59:738–746. doi: 10.1007/s00239-004-2664-1. [DOI] [PubMed] [Google Scholar]
- Hollister JD, et al. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci U S A. 2011;108:2322–2327. doi: 10.1073/pnas.1018222108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia AP, Schnable PS. DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon, MuDR. Genetics. 1996;142:603–618. doi: 10.1093/genetics/142.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8(2):e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki M, Rooke R, Yang G. Bioinformatics and genomic analysis of transposable elements in eukaryotic genomes. Chromosome Res. 2011;19:787–808. doi: 10.1007/s10577-011-9230-7. [DOI] [PubMed] [Google Scholar]
- Jiang N, Ferguson AA, Slotkin RK, Lisch D. Pack-Mutator-like transposable elements (Pack-MULEs) induce directional modification of genes through biased insertion and DNA acquisition. Proc Natl Acad Sci U S A. 2011;108:1537–1542. doi: 10.1073/pnas.1010814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Feschotte C, Zhang XY, Wessler SR. Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs) Curr Opin Plant Biol. 2004;7:115–119. doi: 10.1016/j.pbi.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Jiang N, et al. An active DNA transposon family in rice. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kapitonov V, Jurka J. The age of Alu subfamilies. J Mol Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- Kurkulos M, Weinberg JM, Roy D, Mount SM. P element-mediated in vivo deletion analysis of white-apricot: deletions between direct repeats are strongly favored. Genetics. 1994;136:1001–1011. doi: 10.1093/genetics/136.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Lepetit D, Pasquet S, Olive M, Theze N, Thiebaud P. Glider and Vision: two new families of miniature inverted-repeat transposable elements in Xenopus laevis genome. Genetica. 2000;108:163–169. doi: 10.1023/a:1004173315419. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- Lin R, et al. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318:1866. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D, Chomet P, Freeling M. Genetic characterization of the Mutator system in maize: behavior and regulation of Mu transposons in a minimal line. Genetics. 1995;139:1777–1796. doi: 10.1093/genetics/139.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas J, Koblizkova A, Neumann P. Characterization of Stowaway MITEs in pea (Pisum sativum L.) and identification of their potential master elements. Genome. 2005;48:831–839. doi: 10.1139/g05-047. [DOI] [PubMed] [Google Scholar]
- Marzo M, Bello X, Puig M, Maside X, Ruiz A. Striking structural dynamism and nucleotide sequence variation of the transposon Galileo in the genome of Drosophila mojavensis. Mob DNA. 2013;4:6. doi: 10.1186/1759-8753-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medhora M, Maruyama K, Hartl DL. Molecular and functional analysis of the mariner mutator element Mos1 in Drosophila. Genetics. 1991;128:311–318. doi: 10.1093/genetics/128.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niel F, et al. Rapid detection of CFTR gene rearrangements impacts on genetic counselling in cystic fibrosis. J Med Genet. 2004;41:e118. doi: 10.1136/jmg.2004.022400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosumi T, Garlick B, Belknap WR. Identification and characterization of putative transposable DNA elements in solanaceous plants and Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:8886–8890. doi: 10.1073/pnas.92.19.8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne PW, Luke GN, Holland PW, Ferrier DE. Identification and characterisation of five novel miniature inverted-repeat transposable elements (MITEs) in amphioxus (Branchiostoma floridae) Int J Biol Sci. 2006;2:54–60. doi: 10.7150/ijbs.2.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Fiston-Lavier AS, Lipatov M, Lenkov K, Gonzalez J. Population genomics of transposable elements in Drosophila melanogaster. Mol Biol Evol. 2011;28:1633–1644. doi: 10.1093/molbev/msq337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS One. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurek O, Nishihara H, Okada N. The evolution of two partner LINE/SINE families and a full-length chromodomain-containing Ty3/Gypsy LTR element in the first reptilian genome of Anolis carolinensis. Gene. 2009;441:111–118. doi: 10.1016/j.gene.2008.11.030. [DOI] [PubMed] [Google Scholar]
- Plasterk RH. The origin of footprints of the Tc1 transposon of Caenorhabditis elegans. EMBO J. 1991;10:1919–1925. doi: 10.1002/j.1460-2075.1991.tb07718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham EJ. Transposable elements and factors influencing their success in eukaryotes. J Hered. 2009;100:648–655. doi: 10.1093/jhered/esp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesneville H, Nouaud D, Anxolabehere D. P elements and MITE relatives in the whole genome sequence of Anopheles gambiae. BMC Genomics. 2006;7:214. doi: 10.1186/1471-2164-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, et al. Chompy: an infestation of MITE-like repetitive elements in the crocodilian genome. Gene. 2005;362:1–10. doi: 10.1016/j.gene.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- Remigereau MS, et al. Tuareg, a novel miniature-inverted repeat family of pearl millet (Pennisetum glaucum) related to the PIF superfamily of maize. Genetica. 2006;128:205–216. doi: 10.1007/s10709-005-5703-y. [DOI] [PubMed] [Google Scholar]
- Resnick MA. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Rubin E, Levy AA. Abortive gap repair: underlying mechanism for Ds element formation. Mol Cell Biol. 1997;17:6294–6302. doi: 10.1128/mcb.17.11.6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Sen SK, et al. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Tu Z. Expanding the diversity of the IS630-Tc1-mariner superfamily: discovery of a unique DD37E transposon and reclassification of the DD37D and DD39D transposons. Genetics. 2001;159:1103–1115. doi: 10.1093/genetics/159.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen CR, et al. Double complex mutations involving F8 and FUNDC2 caused by distinct break-induced replication. Hum Mutat. 2007;28:1198–1206. doi: 10.1002/humu.20591. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Streisinger G, et al. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Struchiner CJ, Massad E, Tu Z, Ribeiro JM. The tempo and mode of evolution of transposable elements as revealed by molecular phylogenies reconstructed from mosquito genomes. Evolution. 2009;63:3136–3146. doi: 10.1111/j.1558-5646.2009.00788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol Cell Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surzycki SA, Belknap WR. Characterization of repetitive DNA elements in Arabidopsis. J Mol Evol. 1999;48:684–691. doi: 10.1007/pl00006512. [DOI] [PubMed] [Google Scholar]
- Tancredi M, et al. Haplotype analysis of BRCA1 gene reveals a new gene rearrangement: characterization of a 19.9 KBP deletion. Eur J Hum Genet. 2004;12:775–777. doi: 10.1038/sj.ejhg.5201223. [DOI] [PubMed] [Google Scholar]
- Tu Z. Genomic and evolutionary analysis of Feilai, a diverse family of highly reiterated SINEs in the yellow fever mosquito, Aedes aegypti. Mol Biol Evol. 1999;16:760–772. doi: 10.1093/oxfordjournals.molbev.a026161. [DOI] [PubMed] [Google Scholar]
- Tu Z. Molecular and evolutionary analysis of two divergent subfamilies of a novel miniature inverted repeat transposable element in the yellow fever mosquito, Aedes aegypti. Mol Biol Evol. 2000;17:1313–1325. doi: 10.1093/oxfordjournals.molbev.a026415. [DOI] [PubMed] [Google Scholar]
- Tu Z. Eight novel families of miniature inverted repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci U S A. 2001;98:1699. doi: 10.1073/pnas.041593198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsal K, Morgan GT. A novel group of families of short interspersed repetitive elements (SINEs) in Xenopus: evidence of a specific target site for DNA-mediated transposition of inverted-repeat SINEs. J Mol Biol. 1995;248:812–823. doi: 10.1006/jmbi.1995.0262. [DOI] [PubMed] [Google Scholar]
- Verdin H, et al. Microhomology-mediated mechanisms underlie non-recurrent disease-causing microdeletions of the FOXL2 gene or its regulatory domain. PLoS Genet. 2013;9:e1003358. doi: 10.1371/journal.pgen.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci U S A. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang L, Meyer E, Matz MV. Characterization of a group of MITEs with unusual features from two coral genomes. PLoS One. 2010;5:e10700. doi: 10.1371/journal.pone.0010700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaakov B, Ceylan E, Domb K, Kashkush K. Marker utility of miniature inverted-repeat transposable elements for wheat biodiversity and evolution. Theor Appl Genet. 2012;124:1365–1373. doi: 10.1007/s00122-012-1793-y. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Takano-Shimizu T, Kitamura K, Mikami T, Kishima Y. Resistance to gap repair of the transposon Tam3 in Antirrhinum majus: a role of the end regions. Genetics. 1999;153:1899–1908. doi: 10.1093/genetics/153.4.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Dong J, Chandrasekharan MB, Hall TC. Kiddo, a new transposable element family closely associated with rice genes. Mol Genet Genomics. 2001;266:417–424. doi: 10.1007/s004380100530. [DOI] [PubMed] [Google Scholar]
- Yang G, Hall T. MAK, a computational tool kit for automated MITE analysis. Nucleic Acids Res. 2003a;31:3659. doi: 10.1093/nar/gkg531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Hall TC. MDM-1 and MDM-2: two mutator-derived MITE families in rice. J Mol Evol. 2003b;56:255–264. doi: 10.1007/s00239-002-2397-y. [DOI] [PubMed] [Google Scholar]
- Yang G, Nagel DH, Feschotte C, Hancock CN, Wessler SR. Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science. 2009;325:1391–1394. doi: 10.1126/science.1175688. [DOI] [PubMed] [Google Scholar]
- Yang G, Wong A, Rooke R. ATon, abundant novel nonautonomous mobile genetic elements in yellow fever mosquito (Aedes aegypti) BMC Genomics. 2012;13:283. doi: 10.1186/1471-2164-13-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, et al. P instability factor: an active maize transposon system associated with the amplification of Tourist-like MITEs and a new superfamily of transposases. Proc Natl Acad Sci U S A. 2001;98:12572–12577. doi: 10.1073/pnas.211442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LQ, et al. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.