Abstract
Comparisons between humans and chimpanzees are essential for understanding traits unique to each species. However, linking important phenotypic differences to underlying molecular changes is often challenging. The ability to generate, differentiate, and profile adult stem cells provides a powerful but underutilized opportunity to investigate the molecular basis for trait differences between species within specific cell types and in a controlled environment. Here, we characterize adipose stromal cells (ASCs) from Clint, the chimpanzee whose genome was first sequenced. Using imaging and RNA-Seq, we compare the chimpanzee ASCs with three comparable human cell lines. Consistent with previous studies on ASCs in humans, the chimpanzee cells have fibroblast-like morphology and express genes encoding components of the extracellular matrix at high levels. Differentially expressed genes are enriched for distinct functional classes between species: immunity and protein processing are higher in chimpanzees, whereas cell cycle and DNA processing are higher in humans. Although hesitant to draw definitive conclusions from these data given the limited sample size, we wish to stress the opportunities that adult stem cells offer for studying primate evolution. In particular, adult stem cells provide a powerful means to investigate the profound disease susceptibilities unique to humans and a promising tool for conservation efforts with nonhuman primates. By allowing for experimental perturbations in relevant cell types, adult stem cells promise to complement classic comparative primate genomics based on in vivo sampling.
Keywords: RNA-Seq, adult stem cell, adipose stromal cell, chimpanzee
The advent of next-generation sequencing has resulted in an explosion of exploratory genomic studies that have identified many candidate genes waiting further examination. For a limited number of species, such as mouse, fruit fly, and zebrafish, these candidates can be directly investigated in the living animal using transgenic technologies. However, for some organisms—including endangered species, animals with husbandry challenges, and those with ethical concerns—investigating the evolution of gene function requires alternative approaches. Adult stem cells allow for ex vivo experiments with multiple cell types, disease states, and physiological conditions, providing the ability to link genomic data and organismal traits. Two species, in particular, that can benefit from this approach are chimpanzees and humans. Sequencing the chimpanzee and human genomes has allowed for new layers of functional data throughout the genome from multiple tissues, including transcript abundance (Enard et al. 2002; Khaitovich et al. 2005), alternative splicing (Blekhman et al. 2010), noncoding transcripts (Babbitt et al. 2010), histone modifications (Cain et al. 2011), methylation (Pai et al. 2011), and chromatin configuration (Shibata et al. 2012). These comparative analyses have provided initial insights into differences in molecular function throughout the genome which may contribute to distinct chimpanzee and human phenotypes. Most of these studies, however, were based on in vivo tissue samples that are composed of multiple cell types, do not control for disparate environmental influences, and are often difficult to obtain.
Moving forward, detailed follow-up studies are necessary to unveil the specific molecular mechanisms that influence trait differences between species. These experiments will need to address the lack of control over environmental variables and the presence of a heterogeneous cellular milieu. Working with cells in culture can overcome both challenges. A pioneering study by Barreiro et al. (2010) investigated immune response to a lipopolysaccharide challenge in human, chimpanzee, and macaque monocytes in cell culture. Currently, this type of investigation is limited to fibroblast and lymphoblast cell lines, because very few other cell lines exist from nonhuman primates. This makes it difficult to study the evolution of organismal phenotypes beyond the restricted set of connective tissue and immune functions carried out by these two cell types.
Adult stem cells offer an efficient approach to overcoming the limited availability of cell lines from nonhuman primates. A single adult stem cell line can differentiate into many different cell types, opening the door to experimental molecular analysis of fundamental questions regarding human origins. In a seminal publication, Cheng et al. (2008) isolated and profiled adult progenitor cells from the dental pulp of an adult female chimpanzee, demonstrating that these cells can differentiate into osteoblasts, adipocytes, and chondrocytes. Another type of adult stem cell, the adipose-derived stromal cell (ASC), is obtained by mechanically and enzymatically processing white adipose tissue (Zuk et al. 2001). In cell culture, ASCs are capable of differentiating into adipocytes, osteoblasts, chondrocytes, hepatocytes, myocytes (smooth, skeletal, and cardiac), endothelial cells, neural cells, epithelial cells, and pancreatic B-cells (Cawthorn et al. 2012). Here, we profile the only publically available population of chimpanzee ASCs and compare it with three human ASC lines. Interestingly, the chimpanzee ASCs were derived from Clint, the first chimpanzee whose genome was sequenced (Chimpanzee Sequencing and Analysis Consortium 2005). To our knowledge, this is the first investigation into the biology of chimpanzee ASCs. Besides providing insights into fundamental differences in functional genomics between humans and chimpanzees, our results demonstrate the need for additional public resources for this kind of research.
Morphological and Molecular Characterization of Chimpanzee ASCs
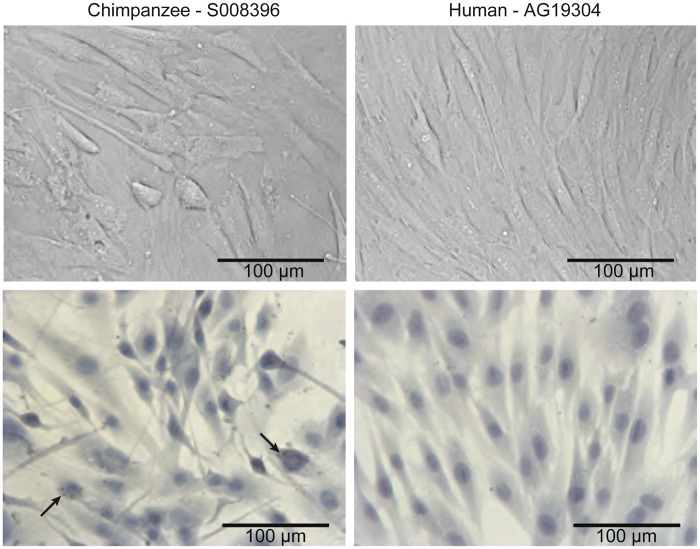
We visualized cultured chimpanzee ASCs using several staining protocols (fig. 1). Most chimpanzee ASCs adopt a fibroblast-like phenotype in vitro (fig. 1), consistent with previous reports of human ASCs (Zuk et al. 2001) and with the human ASCs profiled in this study (fig. 2). In comparison with human ASCs, Clint’s ASCs are not uniform, displaying a range of sizes and shapes (fig. 2). When grown to confluence, the chimpanzee ASCs migrate on top of one another and appear to exhibit lower levels of contact inhibition than the human ASCs (fig. 2). Decreased contact inhibition has been noted in other stem cell populations, such as embryonic stem cells (Burdon et al. 2002). Interestingly, small lipid droplets are present in some of the chimpanzee ASCs but were not seen in any of the human stromal cells profiled in this study (fig. 2, arrows).
Fig. 1.—
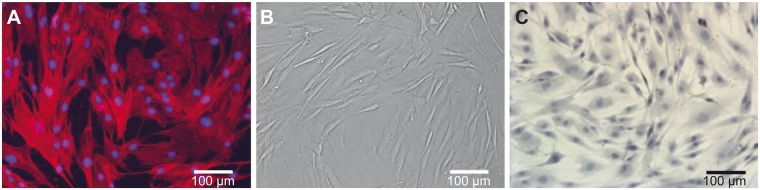
Chimpanzee ASCs in culture. (A) Fluorescence image depicting the nucleus (blue: DAPI) and the actin filaments (red: phalloidin). (B) Brightfield image of ASCs at confluence before collection. (C) Brightfield image depicting the nucleus (blue: Mayer’s hematoxylin) and lipid droplets (red: Oil Red O).
Fig. 2.—
Comparison of chimpanzee and human ASCs in culture. The top panel represents brightfield images of ASC at confluence before collection. The bottom panel contains brightfield images depicting the nucleus (blue: Mayer’s hematoxylin) and lipid droplets (red: Oil Red O). Arrows indicate examples of chimpanzee ASCs that contain small lipid droplets. No lipid droplets were seen in any of the human ASC lines.
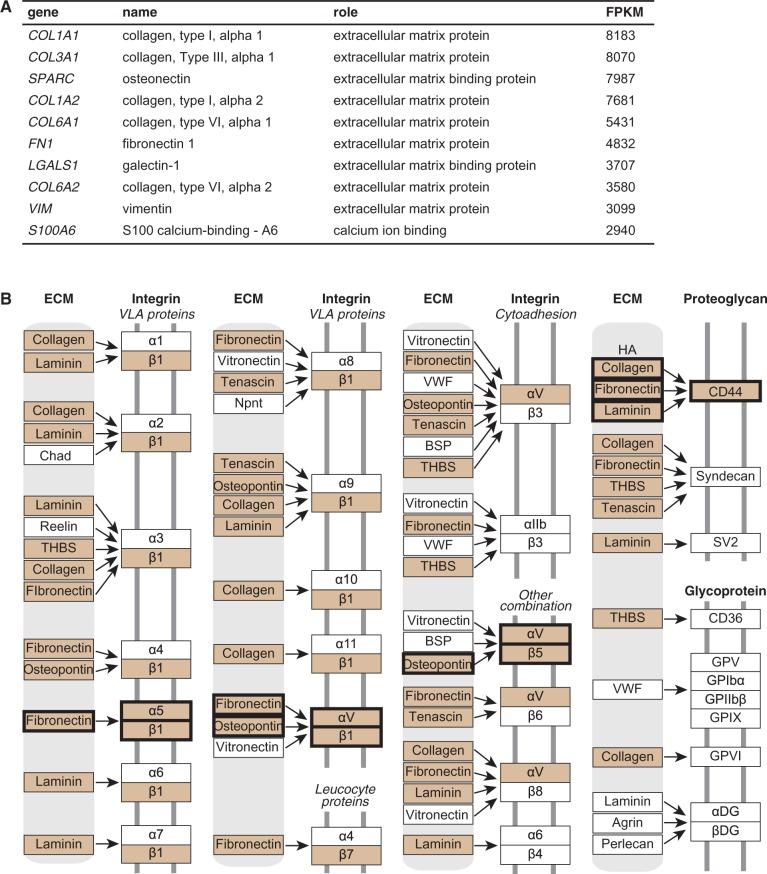
To uncover fundamental properties of the chimpanzee ASC transcriptome, RNA extracted from the confluent chimpanzee stromal cells (figs. 1B and 2A) was made into Illumina TruSeq libraries for RNA-Seq. Approximately 48 million reads were mapped (∼94% of the total) to the panTro3 chimpanzee genome assembly. The highest expressed genes in chimpanzee ASC overwhelmingly encode extracellular matrix (ECM) components (fig. 3A). This makes sense, as cells of the connective tissue produce, organize, and degrade the ECM. In turn, the ECM provides organization, strength, and signaling mechanisms for cells of the connective tissue. The dominance of ECM gene expression we observe in chimpanzee ASCs is consistent with a previous study of human ASCs (Katz et al. 2005). We also observed that the 10 highest expressed genes in chimpanzee were represented within the top 14 highest expressed genes in humans (fig. 3A). It is not surprising that genes encoding collagen, the most abundant family of proteins in mammals and primary source of strength and structure in the ECM, represent five of the top ten highest expressed genes (fig. 3A) (Alberts et al. 2008).
Fig. 3.—
Transcriptomic insights into chimpanzee ASCs. (A) Highest expressed genes using FPKM. (B) KEGG pathway: ECM–receptor interaction. Highlighted genes have FPKM ≥100 and the connections that contain a complete ECM-ligand and receptor pair are shown in bold. Immunoglobin superfamily is not shown.
We next sought to determine what is unique about the collection of highest expressed chimpanzee ASC genes (Fragments Per Kilobase of transcript per Million counted reads [FPKM] ≥ 100, n = 614). Using Database for Annotation, Visualization and Integrated Discovery (DAVID), we identified the ECM–receptor interaction KEGG pathway as the most enriched pathway when compared with all ASC-expressed genes (P = 2.9E−10, corrected P = 4.1E−8) and the second most when compared with all of the genes in the genome (P = 4.4E−11, corrected P = 3.1E−9) (Huang et al. 2009a, 2009b). The hallmark of the ECM–receptor interaction pathway is the relationship between structural proteins, including collagen, and α/β integrins (fig. 3B). Integrins are responsible for mediating a physical and chemical connection between the internal ASC cytoskeleton (actin shown in fig. 1B) and the external matrix environment. This connection is necessary for a variety of critical cell behaviors including proliferation, migration, adhesion, differentiation, and apoptosis (Alberts et al. 2008). The particular α/β heterodimer dictates which ECM ligand(s) the integrin interacts with (fig. 3B). In our analysis, four complete integrin pairings were represented among the highest expressed chimpanzee genes: fibronectin-α5β1, fibronectin-αVβ1, osteopontin-αVβ1, and osteopontin-α5β5 (fig. 3B). Fibronectin, one of the top ten expressed chimpanzee ASC genes (fig. 3A), binds to other ECM proteins, including collagens (Alberts et al. 2008). As cells of the connective tissue, much of the structure and function of ASCs are mediated by ECM–receptor interactions.
Chimpanzee ASC Pluripotency and Differentiation Status
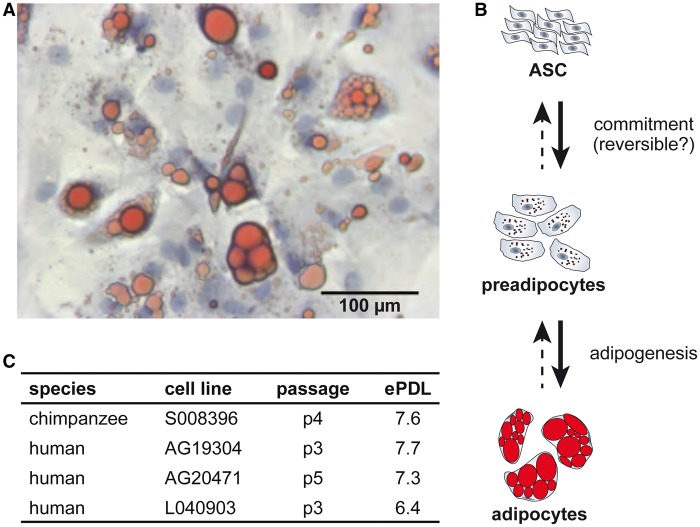
A hallmark of stem cells is pluripotency, the ability to differentiate into cell types of the three germ layers. Although the ability of ASCs to self-renew and differentiate into ectodermal lineages in vivo has not been definitively established (Cawthorn et al. 2012), these cells can be used to investigate many different cell types in culture. We successfully differentiated Clint’s ASCs into mature adipocytes in vitro using a cocktail of adipocyte differentiation media. These cells contain prominent lipid droplets (fig. 4A), a marker of mature adipocytes, here visualized by Oil Red O staining. Although this result does not confirm pluripotency for chimpanzee ASCs, it does demonstrate their ability to differentiate into mesodermal lineages.
Fig. 4.—
Pluripotency insights. (A) Brightfield image of adipocytes after 14 days of differentiation depicting the nucleus (blue: Mayer’s hematoxylin) and lipid droplets (red: Oil Red O). (B) Schematic of ASC differentiation into adipocytes, modified from Cawthorn et al. (2012). (C) Relative age of cell lines in this study measured by passage number and estimated population doubling level (ePDL).
ASCs and preadipocytes (a slightly more differentiated state) are members of the white adipose tissue expansion continuum and share many of the same cell surface markers (fig. 4B) (Katz et al. 2005; Cawthorn et al. 2012). As mentioned earlier, Clint’s ASCs show evidence of lipid accumulations that were not detected in the human cells we profiled (fig. 2) nor reported in the literature (Zuk et al. 2001). These lipid droplets are substantially smaller then those found in Clint’s differentiated adipocytes (fig. 4A). Three possibilities may explain the presence of lipid droplets in chimpanzee but not human ASCs: 1) the chimpanzee cells are further differentiated than the human cells, 2) an artifact was introduced during collection prior to receipt of the cell lines, or 3) these lipids represent a species-specific difference in ASCs.
To investigate the differentiation state of these cell lines, we examined the expression of two transcriptional regulators that mark committed preadipocytes in white adipose tissue, PPARγ and Zfp467 (Cawthorn et al. 2012). We found that PPARγ is expressed at higher level in chimpanzee ASCs (false discovery rate [FDR]-adjusted P = 0.0316, log2 fold change = 1.43), whereas Zfp467 is not expressed in either chimpanzee or human ASCs. The significantly higher expression levels of PPARγ in chimpanzee ASCs indicates that perhaps this population of cells is more differentiated than human ASCs. It is also consistent with the small lipid droplets present in the chimpanzee ASCs, as this transcription factor regulates lipid processing (Neve et al. 2000; Alaynick 2008). However, the absence of Zfp467 expression in both species suggests that the story is more complicated. We also examined a third gene, MMP3, that encodes a metalloprotease produced by committed preadipocytes (Cawthorn et al. 2012). This gene is expressed at much higher levels in human than chimpanzee ASCs and shows the second greatest fold difference between the two species genome-wide (FDR-adjusted P = 5E−09, log2 fold change = 7.51). Based on these three incongruent expression markers for a limited selection of differentiation markers, it remains unclear where specifically the chimpanzee and human cells lie on the white adipose tissue expansion continuum (fig. 4B).
Additional information about the degree of differentiation comes from the estimated population doubling level and passage number, which act as a proxy for cell age. These are relevant, as the length of time ASCs are in culture changes their immunophenotypes (Mitchell et al. 2006), increases senescence (Gruber et al. 2012), and is inversely related to their pluripotency (Katz et al. 2005; Wall et al. 2007). No substantial species differences in passage number or estimated population doubling level distinguish the cells used in this study (fig. 4C), suggesting that these factors are unlikely to account for the phenotypic difference between species.
Finally, a species-specific collection bias is also improbable, as the ASCs were collected from at least three different institutions and harvested by different investigators. Despite widely known differences in ASC processing strategies, several groups have commented on the consistency in immunophenotype and molecular profiles of ASCs across studies (Katz et al. 2005; Gimble et al. 2007). Therefore, it is unlikely that the approach employed by the scientists and veterinarians who harvested Clint’s cells was so radically different that they induced a phenotypic change in the ASC line.
Several differences have been documented in adipose tissue derived mesenchymal stem cells from humans and the nonhuman primate Macaca mulatta. Izadpanah et al. (2006) found that human ASCs retained their adipogenic potential longer than those from macaque. Although these results do not directly speak to differences in lipid droplet formation between human and chimpanzee ASCs (no droplets were detected in any of the undifferentiated macaque ASCs), they do demonstrate that phenotypic differences exist in ASCs among primate species (Izadpanah et al. 2006). The lipid droplet difference in ASCs reported in the current study could be indicative of biological differences between humans and chimpanzee ASCs (fig. 2), but without more chimpanzee adult stem cell resources, the nature of these differences remains unclear.
Transcriptomic Differences between Chimpanzee and Human ASCs
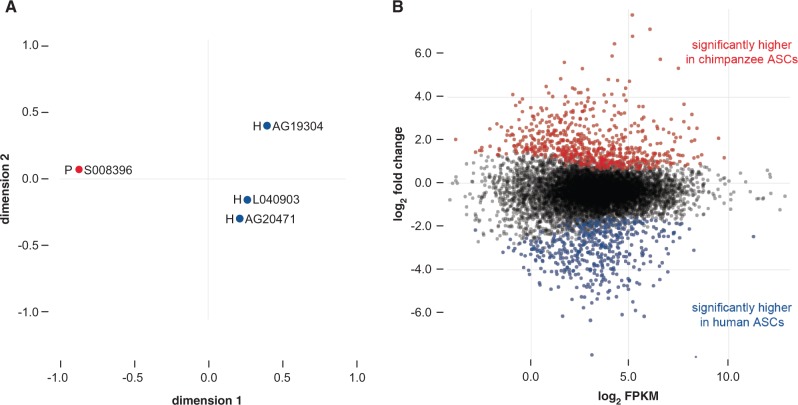
In order to find genes that are differentially regulated between human and chimpanzee, we next compared Clint’s ASC transcriptome with three human ASC transcriptomes. The human ASC samples yielded on average 44 million reads mapped to hg19 (∼95% of total reads). Based on these reads and the chimpanzee reads discussed earlier, we were able to compare 10,021 orthologous genes between species. As expected, the three human ASCs are more similar in expression to each other than any of them are to the chimpanzee ASC, with the major axis in a MDS plot clearly separating the species and explaining 69.12% of the distance between samples (fig. 5A). Next, we sought to determine which genes distinguish Clint’s ASCs from the human ASC samples. At a 5% FDR (Benjamini and Hochberg 1995), 679 genes are expressed at significantly higher levels in chimpanzee ASCs (fig. 5B, red) and 486 genes are expressed at significantly higher levels in human ASCs (fig. 5B, blue).
Fig. 5.—
Visualizing the normalized ASC transcriptomes. (A) Multidimensional scaling plot of Euclidean distances among the four transcriptomes investigated in this study. The first and second dimensions explain 69.12% and 18.73% of the distance between samples. (B) MA-plot where each dot represents a gene and those that are significantly differentially expressed at a FDR-adjusted P <0.05 are red (up in chimpanzee) or blue (up in human).
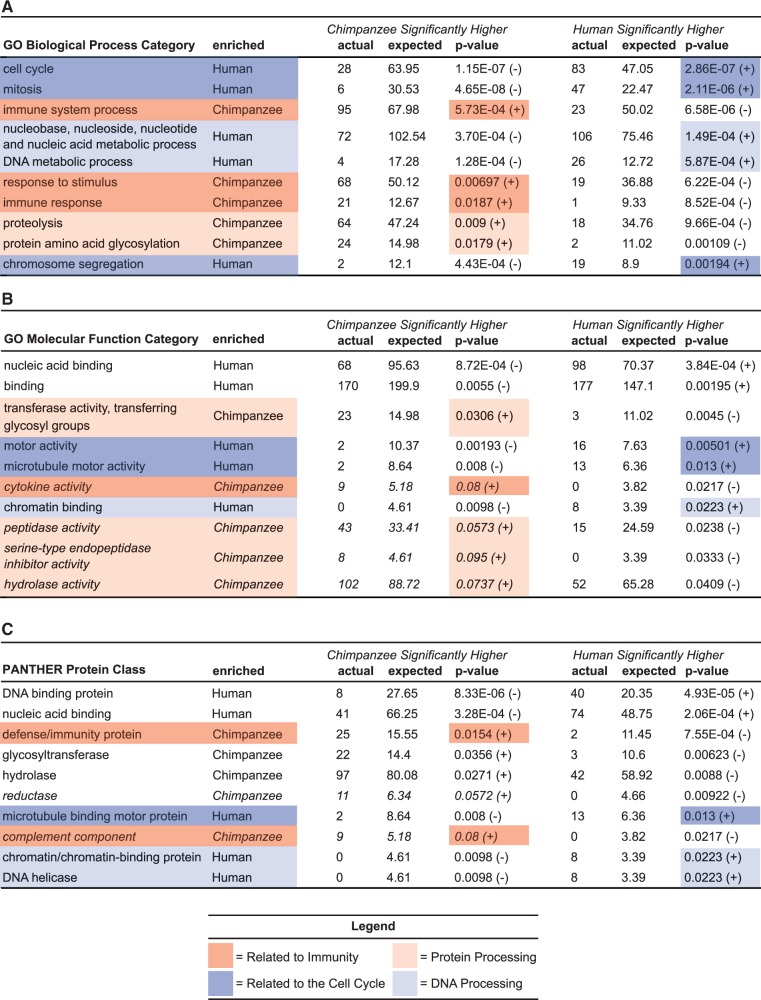
To uncover functional differences between chimpanzee and human ASCs, we interrogated the red (chimpanzee higher) and blue (human higher) genes in figure 5B using the PANTHER tools database (Mi et al. 2005). These standard categorical enrichments include gene ontology (GO) biological processes, GO molecular functions, and PANTHER protein classes. Chimpanzee ASCs have higher expression for genes involved in immunity (dark red) and protein processing (light red), whereas human ASCs have higher expression for genes involved in the cell cycle (dark blue) and DNA processing (light blue) (fig. 6). Strikingly, every one of the broader highlighted categories is distributed perfectly onto either the human or chimpanzee branch (for instance, all six cell cycle subcategories are enriched on the human branch). The most significant enrichments for the chimpanzee are processes involved in the development and functioning of the immune system, which responds to potential invasive or internal threats (fig. 6) (Gene Ontology Consortium 2000). A previous study also found that ASCs are enriched for immune-related expression when compared with other stem cells populations (Jansen et al. 2010). Another complementary category that is enriched in the chimpanzee and significantly depauperate in humans is cytokine activity (fig. 6). The chemokines, one class of cytokines, elicit homing behavior in bone marrow stem cells by sensing tissue injury and migrating to the site of damage (Shyu et al. 2006). Higher expression of genes involved in immunity and cytokine activity is consistent with anecdotal evidence that both captive and wild chimpanzees have faster epidermal wound healing abilities compared with humans (Hedlund et al. 2007). These results provide a glimpse into the molecular differences underlying the human and chimpanzee condition.
Fig. 6.—
PANTHER gene function categories enriched for differential expression by species. The queried genes include those significantly higher in the chimpanzee ASCs (red in fig. 5b) and significantly higher in the human ASCs (blue in fig. 5b). These were assessed against the background set of all significant ASC genes in this study. Shown are the top five most significantly enriched categories for both human and chimpanzee; italicized categories are not statistically significant for the species they are enriched in. The sign next to the nominal P value indicates whether the category is enriched (+) or depauperate (−) for the given species. (A) GO biological process enrichments, (B) GO molecular function enrichments, and (C) PANTHER protein class enrichments.
Stem Cells Can Greatly Expand the Number of Ex Vivo Models for Comparative Primate Genomics
Recently, the National Research Council (National Academy of Sciences, USA) reaffirmed the important role of comparative genomic research involving chimpanzees, highlighting numerous insights that have and will likely continue to come from these data (Altevogt et al. 2011). This report set forth two criteria for research involving chimpanzees: 1) “the studies provide otherwise unattainable insight” and 2) “all experiments are performed on acquiescent animals in a manner that minimizes distress” (Altevogt et al. 2011). The majority of comparative genomic research that would use chimpanzee adult stem cells not only meets both of these criteria but also offers the opportunity to significantly expand the number of available approaches for fruitful inquiry. Moving forward, the use of adult stem cells from chimpanzees can complement existing data on the in vivo state of an evolutionarily relevant tissue by providing access to a single cell type from that tissue, where experiments can be carried out in a controlled ex vivo setting. Combined in vivo and ex vivo comparative functional genomic analyses can provide a unique perspective with the potential to uncover novel results that would not otherwise be accessible.
Moving forward, adult stem cells promise to transform comparative primate genomics. Here, we profiled just one type of adult stem cell, the ASC. The primary nature of ASCs makes them especially attractive for experimental and medical applications (Gimble et al. 2012). Outside of in vivo strategies, primary cells are the closest representative of a cell type, as they have been taken directly from the living organism and have not been genetically transformed or reprogrammed. Another type of adult stem cell, the induced pluripotent stem cell (iPSC), offers different advantages for comparative functional studies. iPSCs are artificially derived through genomic reprogramming of fibroblasts (Takahashi et al. 2007). Romero et al. (2012) recently commented on the potential of iPSCs for evolutionary genomics approaches, and indeed they may be the most promising source of adult stem cells from chimpanzees. There are established methods for dedifferentiating fibroblasts into iPSCs, and several companies have developed kits specifically for this purpose (Takahashi et al. 2007). Importantly, a large catalog of chimpanzee fibroblast lines is currently available: the Coriell Institute alone has ∼50, whereas just one chimpanzee ASC line exists to our knowledge. The available chimpanzee fibroblasts are derived from both sexes, providing a window into the effects of biological variation, something the current study was unable to examine. In addition, iPSCs can be passaged many times, providing a steady supply of material. In contrast, ASCs can only be cultured for a few passages before their ability to differentiate is diminished (Wall et al. 2007). Obtaining the amount of cells one needs for a complete analysis with ASCs is difficult when working with the chimpanzee, an endangered animal with minimal body fat. This limitation makes follow-up experiments a challenge when relatively large numbers of cells are needed, as in DNase-Seq experiments (Shibata et al. 2012) or when carrying out physiological challenge experiments in vitro.
The toolkit of experimental manipulations available for ex vivo studies is vast and includes physiological and hormonal challenges, targeted gene knock-downs, co-culturing multiple cells, and a variety of environmental manipulations. Responses to these experiments can be assayed through numerous cellular phenotypes, including proliferation and apoptosis rates, migration ability, cell size and shape, organelle content, import and export of specific compounds, and detailed metabolic profiles. To date, only a tiny fraction of the vast array of informative experimental manipulations and phenotypic assays that are possible using culture systems has been exploited. Interesting potential follow-ups to our preliminary observations (fig. 6) include eliciting an immune response by challenging cells with immunomodulating chemokines and carrying out classic in vitro scratch migration assays. These experiments could provide molecular insights into the presumed wound healing differences between human and chimpanzees (Hedlund et al. 2007).
The utility of adult stem cells extends to conservation efforts as well. Chimpanzees are listed by the U.S. Fish and Wildlife Service (2012) as threatened in captivity and endangered in the wild, whereas the International Union for Conservation of Nature considers them as endangered with a declining population (Oates et al. 2008). ASCs can assist in chimpanzee conservation efforts by protecting their genetic diversity. The idea that cells can be used in this manner is becoming more widely recognized as the utility of “frozen zoos” is gaining credibility (Ben-Nun et al. 2011). Recently, Ben-Nun et al. (2011) created iPSC from two highly endangered species, the northern white rhinoceros and the drill monkey. The authors expressed the hope that these resources could “facilitate the reintroduction of genetic material into the population” in the future—a prospect that seems increasingly practical with the development of methods for reprogramming adult stem cells into haploid spermatogenic cells (Equizabal et al. 2011; Easley et al. 2012). Using a transcriptomic approach, a recent study found that an endangered primate population contained considerable genetic variation, which could be capitalized on for conservation efforts (Perry et al. 2012).
An important opportunity in primate comparative genomics is using adult stem cells to carry out controlled experiments aimed at investigating molecular differences between humans and our closest living relatives. In vivo approaches based on tissue samples have proved valuable and will continue to provide useful information. However, the ability to work with cell culture systems provides opportunities for functional studies that are otherwise impossible for practical or ethical reasons. These ex vivo approaches provide a powerful complementary set of experimental tools that will likely become an increasingly important component of primate evolutionary genomics.
Materials and Methods
Culturing and Differentiating Stromal Cells
Adult male ASCs from two different species are investigated in this study: one chimpanzee (S008396 from the Coriell Institute for Biomedical Research) and three humans (AG19304 and AG20471 from the Coriell Institute for Biomedical Research and L040903 from Zen-Bio). The stromal cells were recovered from cryofreeze in MesenPro RS Medium (Invitrogen 12746-012) supplemented with 200 mM l-glutamine (Invitrogen 25030-081) and 1% penicillin-streptomycin solution (Invitrogen 15140-122). These cells were allowed to expand to 70% confluency and then were removed using TryPLE (Invitrogen 12604-021) and plated at 40,625 cells/cm2 in 6-well plates (Corningstar 3516). ASCs were cultured for 24–48 h until confluent and were then differentiated into adipocytes using ZenBio’s Adipocyte Differentiation Medium (DM-2-PRF) and Adipocyte Maintenance Medium (AM-1-PRF) supplemented with 250 µm of linoleic acid (Sigma L9530) following the differentiation and maintenance protocol (ZBM0001.03).
Stromal Cell Transcriptomics
Stromal cells were collected at confluency and RNA isolated using QIAzol (Qiagen 79306) followed by miRNeasy Mini extraction kit (Qiagen 217004), followed by DNase I treatment. RNA quality was verified using the Agilent Bioanalyzer 2100 (minimum RIN = 10). Illumina TruSeq SBS libraries were constructed with 1 µg of RNA and put through cluster generation. We used 50 bp, paired-end Illumina HiSeq Sequencing, with all four libraries multiplexed in one lane. Sequencing took place at Duke Institute for Genome Sciences & Policy’s Genome Sequencing & Analysis Core Resource. The CASAVA-trimmed reads were mapped to hg19 and panTro3 with TopHat v1.4.1 using default settings (Trapnell et al. 2009). The mapped reads were counted with htseq-count 0.5.1p1 using the settings union and strandedness (http://www-huber.embl.de/users/anders/HTSeq/doc/count.html, last accessed October 18, 2013). Gene models for each species were constructed using Primate Exon Orthology Database version 2 (http://giladlab.uchicago.edu/orthoExon/, last accessed October 18, 2013). These models were further filtered using the Ensembl database (http://useast.ensembl.org/index.html, last accessed October 18, 2013). To remove genes with unclear homologies, we eliminated Human–Chimpanzee homology types one2many and many2many as well as the ribosomal families RPL, RPS MRPL, and MRPS. We also removed genes where the original chromosome assignment did not match the Ensembl chromosome assignment and where multiple Ensembl gene IDs were assigned to the same HGNC gene name (http://www.genenames.org, last accessed October 18, 2013). Genes with less then five counts per 10 million fragments were removed from every library. Counts were normalized by estimating the tagwise dispersion, and significance was calculated used the program edgeR 1.6.0 (Robinson et al. 2010) in R. FDR corrections for multiple comparisons were calculated using the Benjamini–Hochberg method (Benjamini and Hochberg 1995). Expression level was calculated as FPKM. These data are available from the investigators upon request in any standard configuration (.bam files, raw counts, normalized counts, etc.).
To interrogate the highly expressed chimpanzee ASC genes (FPKM ≥ 100, n = 614), we used DAVID v6.7 KEGG Pathway tool (Huang et al. 2009a, 2009b). The P values were corrected using the Benjamini–Hochberg method (Benjamini and Hochberg 1995). As a background list, both the 10,021 ASC genes in this study as well as the entire genome were assessed. When comparing the chimpanzee and human transcriptomes, categorical gene enrichments were calculated with the PANTHER tools’ gene expression data analysis feature using the compare gene lists function (http://www.pantherdb.org/tools/, last accessed October 18, 2013). The queried genes are those expressed at a significantly higher level in the chimpanzee ASC and those expressed at a significantly higher level in the human ASC at an FDR-adjusted P <0.05. These were assessed against the background set of all significant ASC-expressed genes in this study.
Staining and Imaging
For the florescence imagining, 22 × 22 mm glass coverslips were coated with FNC mix (AthenaES), and cells were plated at 5,263 cells/cm2 in 6-well plates. Cells were cultured for 24 h, fixed for 10 min with 4% paraformaldehyde (Electron Microscopy Sciences), washed with 1× phosphate-buffered saline (PBS), and stained with TRITC-phalloidin (Sigma) and DAPI (Sigma), washed with 1× PBS and mounted on slides. The florescence and black and white images were taken with the Zeiss Axio Observer A1 inverted stand microscope with a Zeiss HBO arc lamp and power supply using a Hamamatsu Orca ER digital camera in the Light Microscopy Core Facility at Duke University. These images were obtained using the MetaMorph software (v7.6.5). ASC and adipocytes were stained for lipid content using the Oil Red O Stain Kit and protocol (ScyTek ORK-1) and imaged prior to confluence and on day 14 of differentiation. The color images were taken with a Leica DM IRB microscope using a Zeiss AxioCam ICc1 digital camera. These images were obtained using the AxioVision software.
Acknowledgments
The authors thank Courtney Babbitt, Adam Pfefferle, Dan Runcie, and Jennifer Wygoda for comments. They also thank the Duke Institute for Genome Sciences & Policy’s Genome Sequencing & Analysis Core Resource, especially Olivier Fedrigo and Fangfei Ye, for library preparation and sequencing assistance; the Light Microscopy Core Facility at Duke University, especially Sam Johnson and Yasheng Gao, for microscopy training and imaging support; the team at ZenBio for advice on adipocyte differentiation and a human ASC line, particularly Ben Buehrer, Jim Nicoll, and Renee Lea-Currie; and the Coriell Institute for Medical Research, especially Bernie Goldstein, for assistance with ordering chimpanzee and human ASC lines. This work was supported by Duke Primate Genomics Initiative Fellowship to L.W.P.; a Wenner Gren Dissertation Fieldwork Grant to L.W.P.; and HOMINID Grant BCS-08-27552 from the NSF to G.A.W.
Literature Cited
- Alaynick WA. Nuclear receptors, mitochondria and lipid metabolism. Mitochondrion. 2008;8:329–337. doi: 10.1016/j.mito.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, et al. Molecular biology of the cell. New York: Garland Science, Taylor & Francis Group, LLC; 2008. [Google Scholar]
- Altevogt BM, Pankevich DE, Shelton-Davenport MK, Kahn JP. Chimpanzees in biomedical and behavioral research: assessing the necessity. Washington (DC): The National Academies Press; 2011. [PubMed] [Google Scholar]
- Babbitt CC, et al. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol Evol. 2010;2010:67–79. doi: 10.1093/gbe/evq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro LB, Marioni JC, Blekhman R, Stephens M, Gilad Y. Functional comparison of innate immune signaling pathways in primates. PLoS Genet. 2010;6:e1001249. doi: 10.1371/journal.pgen.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Nun IF, et al. Induced pluripotent stem cells from highly endangered species. Nat Methods. 2011;8:829–831. doi: 10.1038/nmeth.1706. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:1–11. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- Cain CE, Blekhman R, Marioni JC, Gilad Y. Gene expression differences among primates are associated with changes in a histone epigenetic modification. Genetics. 2011;187:1225–1234. doi: 10.1534/genetics.110.126177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthorn WP, Scheller EL, MacDougald OA. Adipose tissue stem cells meet preadipocyte commitment: going back to the future. J Lipid Res. 2012;53:227–246. doi: 10.1194/jlr.R021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng PH, et al. Postnatal stem/progenitor cells derived from the dental pulp of adult chimpanzee. BMC Cell Biol. 2008;9:1–11. doi: 10.1186/1471-2121-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Easley CA, et al. Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep. 2012;2:440–446. doi: 10.1016/j.celrep.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- Equizabal C, et al. Complete meiosis from human induced pluripotent stem cells. Stem Cells. 2011;29:1186–1195. doi: 10.1002/stem.672. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble JM, Bunnell BA, Guilak F. Human adipose-derived cells: an update on the transition to clinical translation. Regen Med. 2012;7:225–235. doi: 10.2217/rme.11.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble JM, Katz AJ, Bunnell BA. Adipose-derived stem cells for regenerative medicine. Circ Res. 2007;100:1249–1260. doi: 10.1161/01.RES.0000265074.83288.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber HE, et al. Human adipose-derived mesenchymal stem cells: serial passaging, doubling time and cell senescence. Biotech Histochem. 2012;87:303–311. doi: 10.3109/10520295.2011.649785. [DOI] [PubMed] [Google Scholar]
- Hedlund M, et al. N-glycolylneuraminic acid deficiency in mice: Implications for human biology and evolution. Mol Cell Biol. 2007;27:4340–4346. doi: 10.1128/MCB.00379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Izadpanah R, et al. Biologic properties of mesenchymal stem cells derived from bone marrow and adipose tissue. J Cell Biochem. 2006;99:1285–1297. doi: 10.1002/jcb.20904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen BJH, et al. Functional differences between mesenchymal stem cell populations are reflected by their transcriptome. Stem Cells Dev. 2010;19:481–489. doi: 10.1089/scd.2009.0288. [DOI] [PubMed] [Google Scholar]
- Katz AJ, Tholpady A, Tholpady SS, Shang HL, Ogle RC. Cell surface and transcriptional characterization of human adipose-derived adherent stromal (hADAS) cells. Stem Cells. 2005;23:412–423. doi: 10.1634/stemcells.2004-0021. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:6. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Mi H, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:5. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JB, et al. Immunophenotype of human adipose-derived cells: temporal changes in stromal-associated and stem cell-associated markers. Stem Cells. 2006;24:376–385. doi: 10.1634/stemcells.2005-0234. [DOI] [PubMed] [Google Scholar]
- Neve BP, et al. Role of the peroxisom proliferator-activated receptors (PPAR) in atherosclerosis. Biochem Pharmacol. 2000;60:1245–1250. doi: 10.1016/s0006-2952(00)00430-5. [DOI] [PubMed] [Google Scholar]
- Oates JF, et al. IUCN red list of threatened species: Version 2011.2. 2008. International Union for Conservation of Nature and Natural Resources. [cited 2013 Oct 18]. Available from: www.iucnredlist.org. [Google Scholar]
- Pai AA, Bell JT, Marioni JC, Pritchard JK, Gilad Y. A genome-wide study of DNA methylation patterns and gene expression levels in multiple human and chimpanzee tissues. PLoS Genet. 2011;7:e1001316. doi: 10.1371/journal.pgen.1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, et al. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 2012;22:602–610. doi: 10.1101/gr.130468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero IG, Ruvinsky I, Gilad Y. Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet. 2012;13:505–516. doi: 10.1038/nrg3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, et al. Extensive evolutionary changes in regulatory element activity during human origins are associated with altered gene expression and positive selection. PLoS Genet. 2012;8:e1002789. doi: 10.1371/journal.pgen.1002789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu WC, Lee YJ, Liu DD, Lin SZ, Li H. Homing genes, cell therapy and stroke. Front Biosci. 2006;11:899–907. doi: 10.2741/1846. [DOI] [PubMed] [Google Scholar]
- Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Fish and Wildlife Service. Chimpanzee. 2012 (Pan troglodytes). [cited 2013 Oct 18]. Available from: http://ecos.fws.gov/speciesProfile/profile/speciesProfile.action?spcode=A06C. [Google Scholar]
- Wall ME, Bernacki SH, Loboa EG. Effects of serial passaging on the adipogenic and osteogenic differentiation potential of adipose-derived human mesenchymal stem cells. Tissue Eng. 2007;13:1291–1298. doi: 10.1089/ten.2006.0275. [DOI] [PubMed] [Google Scholar]
- Zuk PA, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]