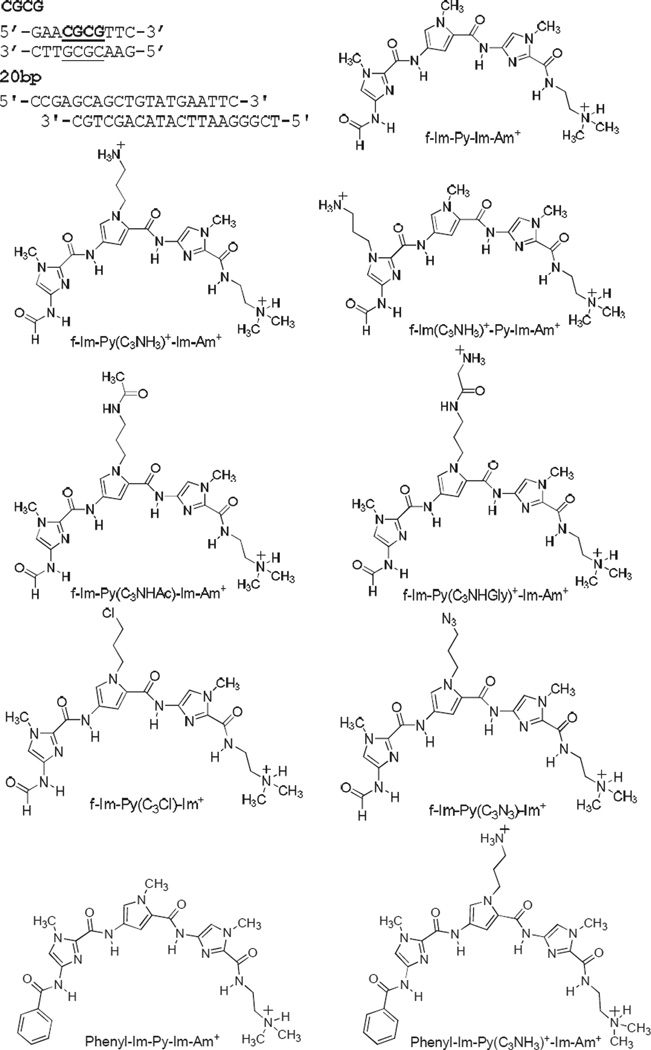
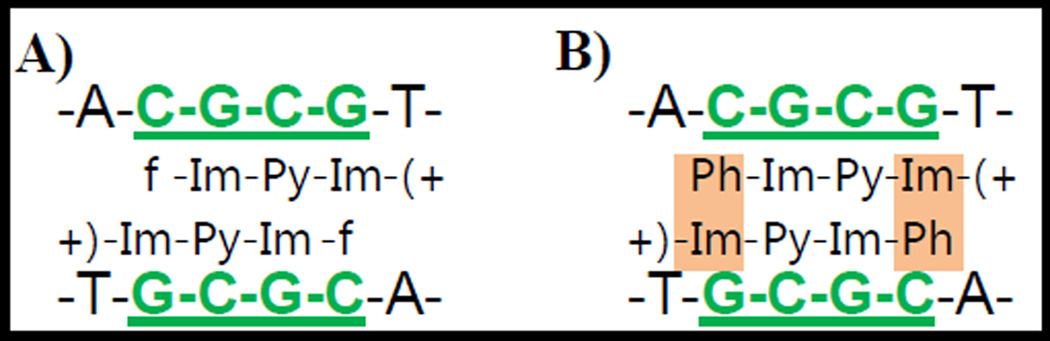Abstract
The DNA sequence 5’-ACGCGT-3’ is in the core site of the Mlu1 cell-cycle box, a transcriptional element in the promoter region of human Dbf4 gene which is highly correlated with a large number of aggressive solid cancers. The polyamide formamido-Imidazole-Pyrrole-Imidazole-Amine+ (f-Im-Py-Im-Am+) can target the minor groove of 5’-ACGCGT-3’ as an antiparallel stacked dimer and has shown good activity in inhibiting transcription factor (TF) binding. Recently, f-Im-Py-Im-Am+ derivatives that involve different orthogonally positioned substituents were synthesized to target the same binding site, and some of them have displayed improved binding and pharmacological properties. In this study, the gel electrophoresis-ligation ladders assay was used to evaluate the conformational effects of f-Im-Py-Im-Am+ and derivatives on the target DNA, an essential factor for establishing the molecular basis of polyamide-DNA complexes and their TF inhibition. The results show that the ACGCGT site in DNA has a relatively wide minor groove and a B-form like overall structure. After binding with f-Im-Py-Im-Am+ derivatives, the DNA conformation is changed as indicated by the different mobilities in the gel. These conformational effects on DNA will at least help to point to the mechanism for the observed Mlu 1 inhibition activity of these polyamides. Therefore, modulating DNA transcription by locking the DNA shape or altering the minor groove geometry to affect the binding affinity of certain TFs is an attractive possible therapeutic mechanism for polyamides. Some of the substituents are charged with electrostatic interactions with DNA phosphate groups and their charge effects on DNA gel mobility have been observed.
Keywords: polyamides, functional side chain, charge effects, conformational changes, DNA rigidity, transcription modulation, Mlu 1
Introduction
From naturally occurring polyamides (PAs), such as distamycin and netropsin, to synthetic pyrrole (Py) and imidazole (Im) containing PAs, remarkable effort has been put into developing PAs for new applications as DNA complexes (Bando and Sugiyama 2006; Chavda et al., 2011; Dervan 2001; Lacy et al., 2003; Satz and Bruice 2002). Starting with the original studies on the distamycin stacked dimer complex (Pelton and Wemmer 1988; Pelton and Wemmer 1989; Pelton and Wemmer 1990), minor groove binding PAs have been designed to recognize a variety of DNA sequences with high affinity (Bando et al., 2002; Buchmueller et al., 2005a; Wemmer 2000). PAs have potential use for development as therapeutics, for example, by inhibiting binding of transcription factors to their DNA recognition sequences (Chenoweth and Dervan 2009; Oyoshi et al., 2003; Raskatov et al., 2012; Wang et al., 2010). This inhibition effect of PAs is highly related to binding affinity and also to the ability to alter the local geometry of DNA upon binding (Chenoweth and Dervan 2010; Wang et al., 2011). Therefore, besides improvement of binding affinity, sequence selectivity, solubility and cellular uptake, the conformational effect of PAs on DNA is an important aspect for the design of therapeutic candidates. A better understanding of PA effects on DNA structure is also essential for establishing the molecular recognition basis of PA-DNA complexes.
In our efforts to develop PAs as potential drugs we have focused on three-ring monomer derivatives that can bind tightly to specific DNA sequences. These PAs also have good pharmacological potential. We have placed specific focus on the formamido-Imidazole-Pyrrole-Imidazole-Amine+ (f-Im-Py-Im-Am+) derivative which has unusually tight binding to the DNA sequence 5’-ACGCGT-3’ (Buchmueller et al., 2005a; Buchmueller et al., 2005b; Buchmueller et al., 2006; Lacy et al., 2002). This sequence is of interest since it is found in the core sequence of the Mlu1 cell-cycle box (MCB), a transcriptional element in the promoter region of the human Dbf4 gene. The level of Dbf4 is critical for the activation of Cdc 7 kinase and has been implicated with a large number of aggressive solid cancers (Hess et al., 1998; Verma et al., 1991; Wu and Lee 2002).
To improve the DNA binding and pharmacological properties of f-Im-Py-Im-Am+ based derivatives, a variety of modified analogs have been synthesized to target the same ACGCGT binding site (Satam et al., 2012a; Satam et al., 2012b) (Figure 1). Some of the modifications have led to both improved DNA binding and solution properties of f-Im-Py-Im-Am+. The structural influences of PAs on DNA, however, are unclear. To evaluate the conformational effects of the f-Im-Py-Im-Am+ derivatives, we have used gel electrophoresis with ligation ladders. We wish to determine: i) how the binding of f-Im-Py-Im-Am+ stacked dimers affects the DNA structure at the CGCG site, and ii) how the additional functional groups influence the local geometry of minor groove. The results provide a novel mechanism for allosteric modulation of DNA structure, which is complementary to other demonstrated mechanisms for changing DNA groove geometry (Chenoweth and Dervan 2009).
Figure 1.
DNA sequence and compounds used in this study.
Materials and Methods
DNA-Binding PAs
All the PAs in this study were synthesized as described previously (Ramos et al., 2013; Satam et al., 2012a; Satam et al., 2012b), and their structures are shown in Figure 1.
DNA Oligonucleotides and Ligation Ladders
A 10 bp self-complementary duplex with a CGCG site flanking with AT base pairs (Figure 1) was purchased from Integrated DNA Technologies, Inc. (IDT, Coralville, IA) with HPLC purification. Double distilled water was added to the lyophilized DNA oligomers to bring the concentration to approximately 1.0 mM, based on the reported amount of DNA from IDT. The actual concentration of the single strand DNA was then determined using a Cary 300 UV-visible spectrophotometer (Agilent, Santa Clara, CA) at A260 with extinction coefficient calculated by the nearest-neighbor method (Fasman 1975). The self-complementary single strands were annealed in ligation reaction buffer (New England Biolabs, Ipswich, MA) containing 50 mM Tris–HCl, 10 mM MgCl2, 10 mM dithiothreitol and 1 mM ATP. A 20 base-pair ladder (20 bp, Bayou Biolabs, Metairie, LA), which is a commercially available standard sequence with a double intensity band at 100 bp, was used for gel mobility reference.
The ligation procedure is as described previously (Tevis et al., 2009). In brief, annealed duplexes were 5’ phosphorylated using T4 polynucleotide kinase (New England Biolabs) and then ligated with T4 DNA ligase (New England Biolabs) in the buffer provided. Kinasing reactions were incubated for 30 min at 37 °C followed by enzyme deactivation for 20 min at 65 °C. Ligation reactions in 200 µL contained 2 µM DNA with 2400 U of ligase and were done at room temperature overnight. The ligated DNA solutions were stored at 4 °C.
Gel Electrophoresis and Calculation of RL
Ligation ladders were separated by 8% native polyacrylamide gels (1.5 mm thick, 20 cm long) prepared from a 40% acrylamide solution (29:1, acrylamide: bisacrylamide, Acros Organic, NJ). Electrophoresis was carried out at 200 V (10 V/cm) for 170 min at 25 °C in a BioRad Protean II xi gel apparatus with TBE buffer (0.089 M Tris, 0.089 M boric acid, 2.0 mM EDTA, pH 8.3) and a BioRad PowerPac Basic 300. Each sample had a final volume of 20 µL containing 12 µL of 2 µM DNA, 4 µL of 6× load dye (Promega, Madison, WI), and the amount of compounds needed for the desired ratios of compound to binding site. After electrophoresis, gels were stained with SYBR Gold Nucleic Acid Gel Stain (Invitrogen, Carlsbad, CA). The stained gels were imaged using an UltraLum Omega 10 gD Molecular Imaging System (UltraLum, Claremont, CA).
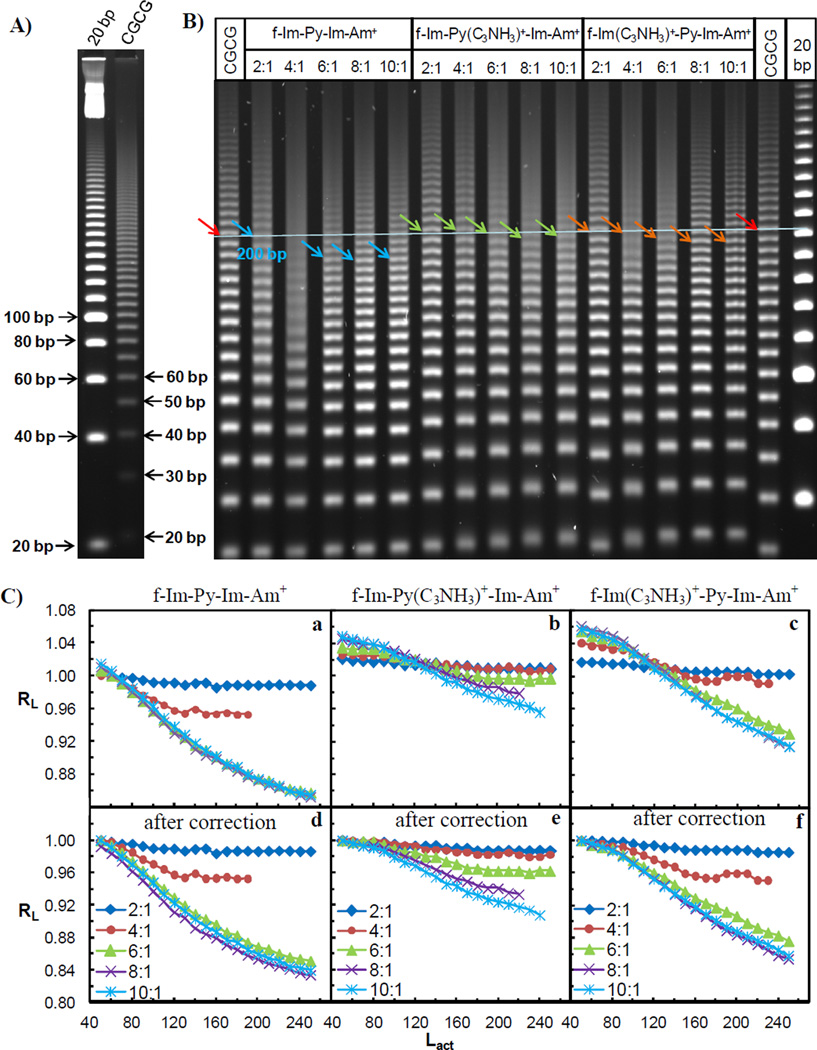
Molecular weight assignments were obtained by adding 10 bp to each sequential gel band starting from the bottom most bands and using the 20 bp ligation ladders as the reference (Figure 2A). The value was in bp and termed as the actual length (Lactual) for each gel band. Using Image Quant TL (GE Healthcare, Piscataway, NJ), the apparent length (Lapparent) of each band was determined based on the distance it migrated in the gel. Relative mobility RL, where RL=Lapparent/Lactual (Ross et al., 1999; Tevis et al., 2009), was calculated for each ligation ladder and plotted as a function of Lactual for each sample. The RL value for each unbound CGCG duplex was normalized to 1.00 in order to compare the mobility changes induced by ligands binding more easily. The relative mobilities were reproducible within ±0.01 experimental error.
Figure 2.
A) 8% (29:1) native PAGE gel of unbound target DNA CGCG and the 20 bp reference sequence. The arrows indicate the molecular weights of the ligation ladders. B) 8% (29:1) native PAGE gel of monocation f-Im-Py-Im-Am+, dications f-Im-Py(C3NH3)+-Im-Am+ and f-Im(C3NH3)+-Py-Im-Am+ with CGCG. Ratios on top refer to compound per binding site of DNA. Free DNA and a 20 bp marker are shown as the migration standards. Colored arrows indicate 200 bp ligated linear multimers. C) Plot of the relative mobility, RL, as a function of Lactual for the CGCG bound with three PAs before (a–c) and after (d–f) the correction of charge effect. Ratios are as in Figure 2B.
Molecular Docking
Molecular docking of PA into the CGCG site was performed by employing the SYBYL-X 1.1 software package (Tripos Inc. St Louis 2010) on a Fedora Core 5 Linux work station. PDB 1B0S, which has an f-Im-Im-Im-Am+ dimer in complex with 5’-GAACCGGTTC-3’ (Yang et al., 1999), was used as a template. The target DNA duplex 5’-GAACGCGTTC-3’ was constructed by editing the 1B0S sequence with the Biopolymer Modules in SYBYL. The target sequence was then minimized by a maximum of 100 iterations using the Conjugate Gradient algorithm and Tripos force field with a termination conjugate gradient of 0.1 kcal/ mol Å (Collar et al., 2010). f-Im-Py(C3NH3)+-Im-Am+ (Figure 1) dimer was assigned Gasteiger-Hückel charges and then minimized for 1000 maximum iterations using the Tripos force field until a terminating conjugate gradient of 0.01 kcal/ mol Å was reached. The f-Im-Im-Im-Am+ dimer in 1B0S was employed as a reference for aligning f-Im-Py(C3NH3)+-Im-Am+ dimer to the DNA sequence and then was removed. f-Im-Py(C3NH3)+-Im-Am+ dimer was moved to a separate molecular area in order to move independently of the DNA in the FlexiDock genetic algorithm (Liu et al., 2012). The FlexiDock module of the SYBYL software package was employed implementing five different random numbers. The docking consisted of 87000 generations, which was calculated by (the number of rotatable bonds + 6) × 500, to ensure that it received the lowest energy conformations. Both DNA and f-Im-Py(C3NH3)+-Im-Am+ dimer were permitted torsional, rotational and translational flexibility in the docking process.
Results
Effects on DNA gel mobility of f-Im-Py-Im-Am+ and analogs with orthogonally positioned charged alkyl amine side chain
Ligation ladders of the target sequence CGCG, with f-Im-Py-Im-Am+ and analogs that contain an additional positively charged alkyl amine side chain that either attaches on the pyrrole moiety or on the formamido-terminus imidazole, f-Im-Py(C3NH3)+-Im-Am+ and f-Im(C3NH3)+-Py-Im-Am+, were compared using gel electrophoresis (Figure 2B). PAs were incubated with ligated sequences at five concentrations to achieve 2:1, 4:1, 6:1, 8:1 and 10:1 ligand to binding site ratios. Higher molar ratios than binding stoichiometries were used to overcome partial dissociation of the positively charged PA dimers during the electrophoresis experiment. As indicated by the colored arrows (Figure 2B), the 200 bp ligated multimers were used to visually compare the migration behaviors of the target sequence CGCG with and without ligands. This length of multimers was selected for two reasons: (1) they migrated far enough in the gel to be fully separated from other ligated multimers and (2) at this length the multimers can display structural features more prominently than shorter ligated oligomers. The gel results show that the free CGCG duplex migrated in the gel at approximately the same rate as the ligated 20 bp random sequence, indicating that this DNA site does not have significant bends. After binding with the monocation f-Im-Py-Im-Am+ dimer, CGCG migrated faster than the free DNA. The mobility changed with an increasing amount of compound until a molar ratio of 6:1. Bands were somewhat smeared at the 4:1 ratio suggesting some exchange between free and bound DNA in the gel experiment. The extent of migration change of CGCG bound with dications f-Im-Py(C3NH3)+-Im-Am+ and f-Im(C3NH3)+-Py-Im-Am+ dimers was less significant and it could only be detected at high molar ratios of ligand to DNA binding sites i.e. at ratios of 8:1 and 10:1.
To compare the DNA mobility changes induced by the binding of PAs, the relative mobility, RL, was determined and is plotted as a function of Lactual (Figure 2C a–c). Since the RL values of free DNA have been normalized to 1.00, a bound sequence with an RL value below 1 indicates that it has a higher mobility than the free DNA, while a value greater than 1 means lower mobility. As shown in Figure 2C a, the dimer formation of monocation f-Im-Py-Im-Am+ in the minor groove increased the DNA mobility. Similar mobility changes have been detected for the two dication complexes but only in longer DNA multimers and are less significant than those induced by the monocation dimer (Figure 2C b–c). The dicationic PAs also had quite divergent behavior depending on the oligomer length: oligomers less than around 80–100 bp had mobilities less than free DNA while larger oligomers had higher mobility. 50 bp ladders were used as the starting bands for the mobility analysis and all the RL values of each sample were subtracted by the difference between the RL value of its 50 bp ligation ladder and 1.00 as shown in Figure 2C d–f. In this way the RL values of each PA-DNA complex are adjusted to start at the same value for direct comparison. This aspect of the analysis is described in more detail in the Discussion Section.
Effects on DNA gel mobility of modification of the side chain structure and charge
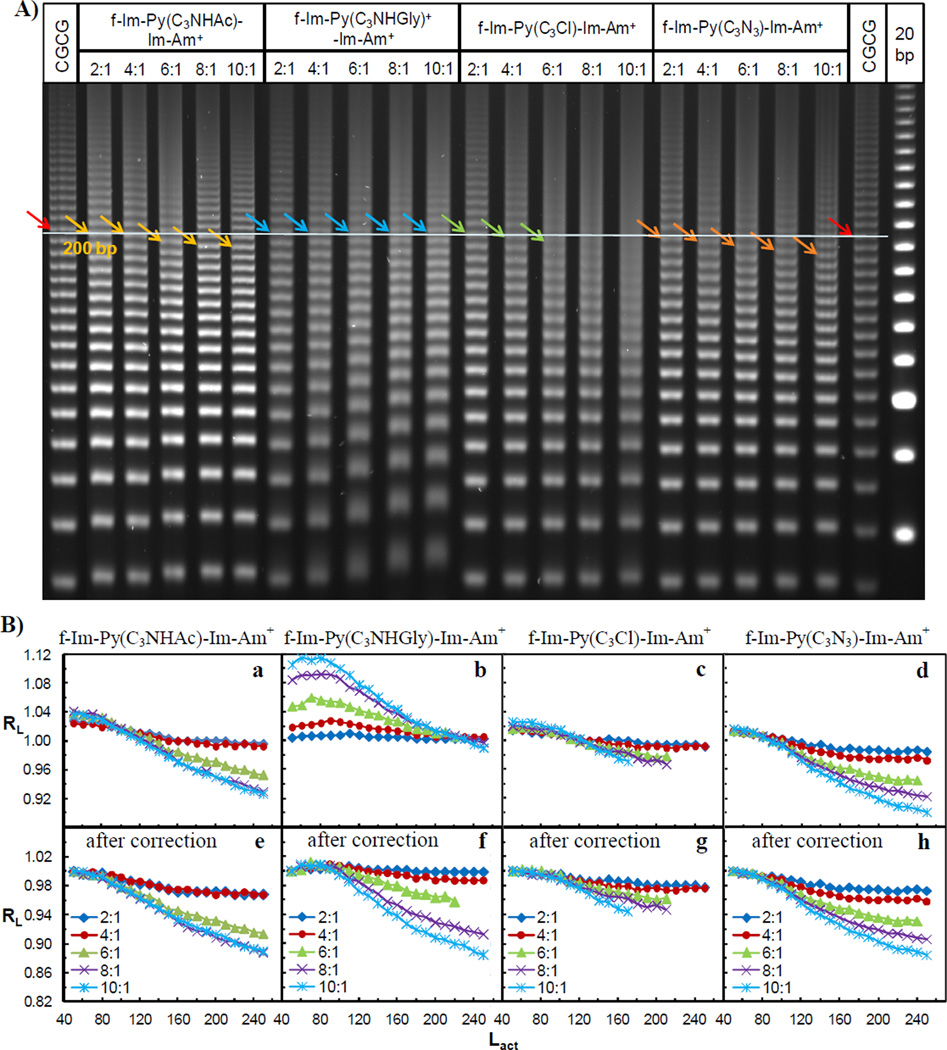
To further test the effects of the addition of a functional side chain on DNA mobility, four more f-Im-Py-Im-Am+ analogs, f-Im-Py(C3NHAc)-Im-Am+, f-Im-Py(C3NHGly)+-Im-Am+, f-Im-Py(C3Cl)-Im-Am+ and f-Im-Py(C3N3)-Im-Am+, were studied using PAGE (Figure 3A). All complexes had small mobility changes. Dimer formation of monocations f-Im-Py(C3NHAc)-Im-Am+, f-Im-Py(C3Cl)-Im-Am+ and f-Im-Py(C3N3)-Im-Am+ slightly increased the DNA mobility but by smaller extents than parent PA f-Im-Py-Im-Am+. In contrast, a slight mobility retardation was detected in the samples containing f-Im-Py(C3NHGly)+-Im-Am+. The amount of retardation decreased with increasing lengths of ligation ladders. RL values were plotted as a function of the length of DNA segment with unbound sequences serving as the reference in Figure 3B a–d. To compare the mobility changes more effectively, the RL values were corrected using the 50 bp ladders as the starting bands as described above (Figure 3B e–h).
Figure 3.
A) 8% (29:1) native PAGE gel of f-Im-Py(C3NHAc)-Im-Am+, f-Im-Py(C3NHGly)+-Im-Am+, f-Im-Py(C3Cl)-Im-Am+ and f-Im-Py(C3N3)-Im-Am+ with target DNA CGCG. Ratios on top are as in Figure 2. Colored arrows indicate 200 bp ligated linear multimers. B) Plot of the relative mobility, RL, as a function of Lactual for the CGCG bound with four PAs before (a–d) and after (e–h) the correction of charge effect. Ratios are as in Figure 2.
Effects on DNA gel mobility of incorporation of an amino terminal benzamide
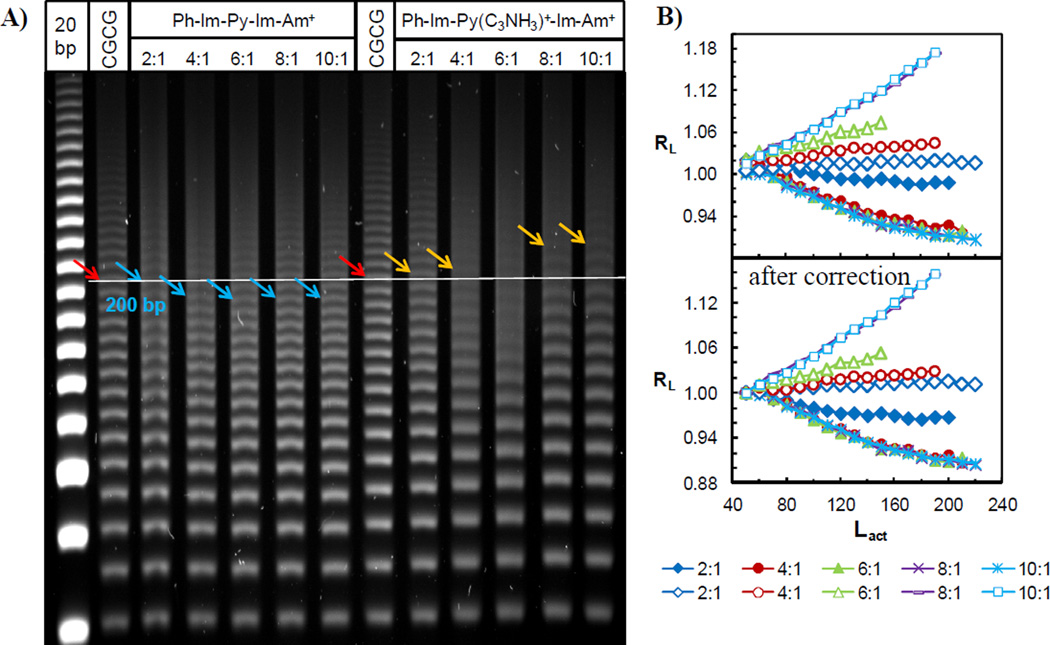
Two more f-Im-Py-Im-Am+ analogs with a benzamide in place of the terminal formamido group, phenyl-Im-Py-Im-Am+ and phenyl-Im-Py(C3NH3)+-Im-Am+ (Figure 1), were incubated with the target CGCG ligated duplex and evaluated by PAGE (Figure 4A). Phenyl-Im-Py-Im-Am+ slightly accelerated the DNA mobility but the dication analog phenyl-Im-Py(C3NH3)+-Im-Am+ significantly retarded the DNA migration. This decreased mobility became more remarkable with longer sequences, and its effects were more pronounced than the retardation induced by f-Im-Py(C3NHGly)+-Im-Am+. Figure 4B shows the RL values as a function of Lactual and obviously displays two different effects on DNA mobility, acceleration and retardation by the phenyl attached monocation and dication, respectively. The corrected RL values using the same method above are shown as well.
Figure 4.
A) 8% (29:1) native PAGE gel of phenyl-Im-Py-Im-Am+ and phenyl-Im-Py(C3NH3)+-Im-Am+ with target DNA CGCG. Ratios on top are as in Figure 2. Colored arrows represent the 200 bp sequences. B) Plot of RL as a function of Lactual for phenyl-Im-Py-Im-Am+ (filled symbols) and phenyl-Im-Py(C3NH3)+-Im-Am+ (empty symbols) before and after the correction of charge effect.
Docking of f-Im-Py(C3NH3)+-Im-Am+ to cognate DNA
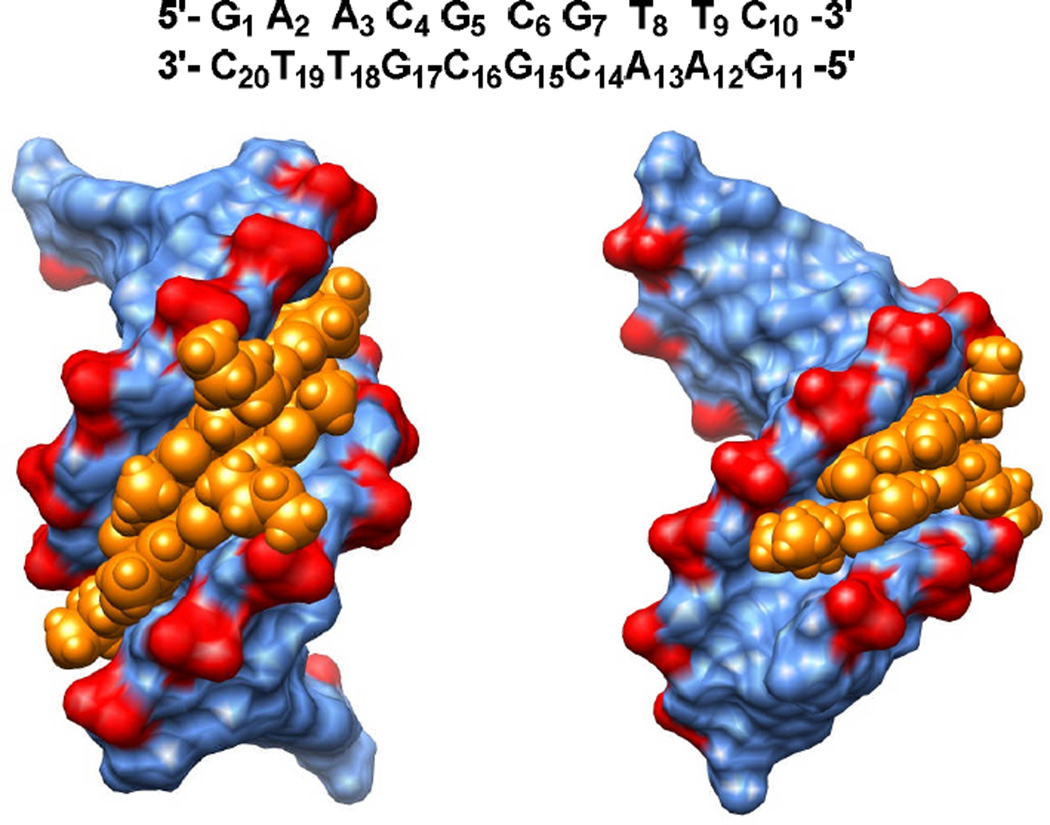
To provide a better understanding of the binding mode of PAs in this work, the f-Im-Py(C3NH3)+-Im-Am+ dimer was docked into the 5′-GAACGCGTTC-3′ target sequence (Figure 5). The left view in Figure 5 is directly into the minor groove while the right view is rotated 90 degrees. Two f-Im-Py(C3NH3)+-Im-Am+ molecules form an antiparallel dimer in the minor groove within the central CGCG region and the Im-Py-Im heterocycles form H-bonds with the CGCG sequence at the floor of the groove. The positively charged alkyl amine (C3NH3)+ side chains extend out in opposite directions and interact with the phosphate groups of G7 and G17. This interaction helps explain the extra stabilization of the dimer complex that is provided by the alkyl amine substituents. It was also found that the distance between the two alkyl amine side chains and the neighboring phosphate groups of G7 and G17 are too large to form H-bonds. The f-Im-Py(C3NH3)+-Im-Am+ dimer also has intermolecular H-bonds between the charged tail amide NH and formamido O groups. Since the DNA sequence is symmetric, there are similar interactions between the two molecules and the bases on complementary strands.
Figure 5.
Docking of f-Im-Py(C3NH3)+-Im-Am+ to its cognate DNA sequence 5′-GAACGCGTTC-3′. Front (left) and side (right) views of the docked model in the minor groove of CGCG site were built with Chimera. The PA is shown in golden and DNA sequence is colored as blue with the highlighted phosphate groups in red. The full sequence with bases numbered is shown at the top of the figure.
Discussion
1. Factors that determine DNA gel mobility
a. DNA bending and minor groove geometry
The ligation ladder-PAGE assay has been validated as a sensitive and qualitative tool to analyze global DNA curvature and the local variation in DNA minor groove geometry through comparison of DNA mobilities (Crothers and Drak 1992; Diekmann et al., 1992; Ross et al., 1999). For example, DNA sequences with a narrow minor groove, such as phased A tracts (Digabriele et al., 1989), have a locally bent structure that yields a curved helix axis and anomalously migration in gels (Koo and Crothers 1988; Lu and Stellwagen 2008; Maki et al., 2004). Sequences possessing a wider, more B-form like, minor groove, like alternating AT sequences (Yoon et al., 1988), are less curved and display higher gel mobility (Crothers and Drak 1992; Tevis et al., 2009; Wang et al., 2011). This ligation ladder-PAGE method has been extended to detect DNA conformational changes induced by the binding of small molecules in the minor groove (Hunt et al., 2011; Wilson et al., 2008). DNA mobility changes observed upon ligand binding demonstrate that compounds, such as netropsin (Tevis et al., 2009) and distamycin (Wang et al., 2011), which target AT rich minor groove sites can constrict or widen the minor groove and thus cause bending or straightening of DNA structures.
In this study, a DNA sequence, which contains in phase CGCG sites, has been investigated with minor groove binding PAs by the ligation ladder PAGE method for the first time. Unlike the relatively slow migration for sequences containing A tracts, the target DNA CGCG displayed a mobility in the gel that is similar to the reference sequence, a random 20 bp ladder, lacking A/T or G/C binding sites for the PAs in Figure 1. This observation indicates that the target CGCG DNA has a relatively straight B-form structure with a wider minor groove binding site than with A tracts. This result is in good agreement with crystallographic analysis that the introduction of G/C base pairs widens the minor groove (Goodsell et al., 1993; Heineman and Alings 1989) relative to that for AT rich sites.
b. Ligation ladder length and the ligand charge effects
Experimental and modeling results (Allison et al., 2002; Diekmann et al., 1992) illustrate that, in general, DNA ligation ladders that are shorter than 50 bp are too short to display gel mobility differences in 8% polyacrylamide gel (Tevis et al., 2009; Wilson et al., 2008). Therefore, the relative mobility RL values for any 50 bp or shorter DNA sequence are similar to the migration standard, around 1.0 after normalization under the gel conditions used in the experiments reported here. With increasing length of DNA, the helix axis and global curvature become more pronounced, and their effects on mobility can be detected. Another factor that can affect the DNA mobility is the positive charges of the bound ligands. Cationic ligands result in less negatively charged complexes that can migrate slower than the free DNAs under the same conditions. The local influence of single or double positive charges from a ligand on DNA mobility has not had a significant effect during the gel electrophoresis in compounds evaluated to date, however, perhaps due to counterion release from DNA on binding of the cationic ligands (Chaires 1997; Record et al., 1978).
The nine PAs in this study have the same basic Im-Py-Im heterocyclic structure but with various substituents. This set allows evaluation of any steric or the structural effects on DNA of compound modifications but also provides the possibility to detect differential charge effects on mobility. All of the PAs caused at least some changes of the DNA mobility, either acceleration or retardation, but both the magnitude and direction of the effects were dependent on the PAs substituents. The relative mobilities RL values for the 50 bp complexes with monocation derivatives are quite similar to that for the unbound DNA due to the lack of PAGE sensitivity for curvature of this length of DNA, as discussed above. However, an unusual phenomenon was observed with the RL values of the dimer complexes involving dication derivatives, f-Im-Py(C3NH3)+-Im-Am+, f-Im(C3NH3)+-Py-Im-Am+ and f-Im-Py(C3NHGly)+-Im-Am+. The complexes formed with short ligation ladder migrated slower than the unbound DNA, while the ones with longer sequence moved faster relative to the unbound DNA. This inconsistent DNA length dependent mobility change can be explained by functional group charge effects. The PAs bind in the minor groove as stacked dimers and when each PA is a dication, this gives a tetracationic minor groove complex that could exceed the local ability of counterion release. The decreased DNA motilities resulting from charge effects are observed at short DNA segments, up to around 100 bp in length (Figures 2C b–c). As the target CGCG sequence gets longer, the structural influence by PA binding accumulates and becomes larger than charge effects.
Among the dications, f-Im-Py(C3NHGly)+-Im-Am+ showed the most remarkable charge effects (Figures 2C b–c, 3B b). This may be due to its longer positively charged side chain relative to the alkyl amide of the other two derivatives (Figure S1). This longer and more extended side chain, C3NHGly+, could interact with two phosphate groups on the DNA backbone rather than one, as the shorter C3NH3+ does (Figure 5) and this may account for its more significant charge effects on the DNA gel mobility.
2. A novel mechanism for allosteric modulation of DNA transcription
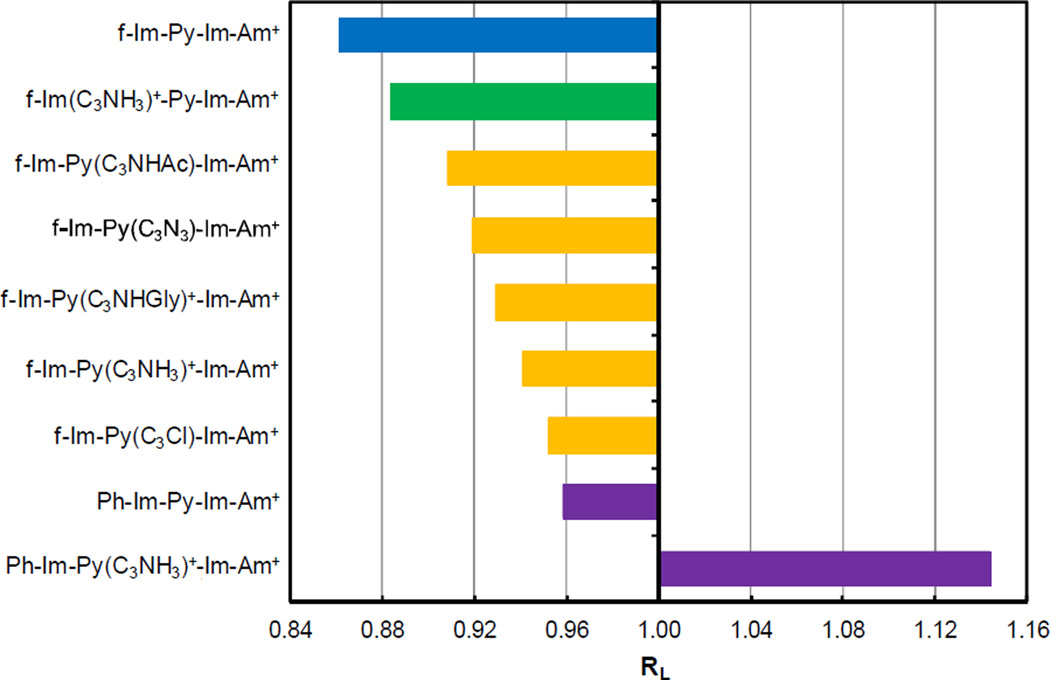
For the better comparison of the structural effect of all derivatives, 50 bp ladders were chosen as the starting multimers since they rarely display conformational effects. In this case charge effects can be eliminated by subtracting the difference between the values of 50 bp bound DNA and the normalized free CGCG from each of the RL. In this way, the RL values of the nine 200 bp complexes were corrected to 1.0 at 50 bp (Figure 6). The RL values with charge effect corrections show us that the mobility of the target sequence CGCG has been enhanced to some degree by dimer formation of all the PAs tested in this study except phenyl-f-Im-Py(C3NH3)+-Im-Am+ (Figure 6). The increased migration indicates the DNA is less curved or the flexibility is reduced by the PA binding. When the rigidity is enhanced, the DNA migrates through the gel pores faster with less resistance than with similar but more flexible duplexes (Anderson 1986; Ohyama et al., 1998; Stellwagen 2009). Some compounds targeting the narrow and curved minor groove of A-tract sites have shown the ability to increase the DNA mobility by straightening the DNA (Tevis et al., 2009). For the target sequence CGCG in this work, the overall structure of free DNA is essentially straight, relative to the control sequence, and the minor groove is relatively wide as discussed above. It seems likely that increased rigidity of the duplex structure allows the DNA to move more easily through the gel. The wide minor groove is an ideal location for the dimer formation, and this excellent space fitting by the dimer complex increases the energetic cost for the DNA to be flexible and thus can rigidify the DNA structure. The reduced flexibility effectively straightens the average structure of the DNA helix axis.
Figure 6.
Plot of RL values of 200 bp ladders as a function of Lactual after charge effect correction. The target sequence CGCG is shown with f-Im-Py-Im-Am+ and other eight derivatives in an 8:1 compound: DNA molar ratio. Compounds are classified by colors and the structure is indicated at the left.
Besides accelerating DNA mobility, the rigidifying effect might provide new insight on how PAs could act as allosteric modulators of DNA transcription. By locking the DNA structure with regard to the groove width and overall curvature, PAs can inhibit the binding of certain transcription factors that modify DNA structure on binding. This is a novel possible allosteric modulation mechanism that could be involved in inhibition of the binding of Mlu 1 to the MCB site (5’-ACGCGT-3’) of the human Dbf4 gene by f-Im-Py-Im-Am+. This inhibition mechanism is based on an increased energy barrier of DNA conformational changes and it is complementary to changing DNA groove geometry by PAs as found, for example, by Chenoweth and Dervan (Chenoweth and Dervan 2009; Chenoweth and Dervan 2010). Bruice and coworkers (reviewed in Satz and Bruice 2002) synthesized and studied positively charged tripyrrole PAs with several different cationic groups on the central pyrrole. Their compounds are all similar to f-Im-Py(C3NH3)+-Im-Am+ but have only pyrroles. They found that their compounds bind very strongly to AT-rich DNA sequences. Some of their PAs can bend AT-rich DNAs and change the DNA conformation based on the NMR and AFM results.
3. Effects of the modified side chains on DNA structure
Figure 6 shows that PAs affect the target DNA in the following order: f-Im-Py-Im-Am+ > f-Im(C3NH3)+-Py-Im-Am+ > f-Im-Py(C3NHAc)-Im-Am+ > f-Im-Py(C3N3)-Im-Am+ > f-Im-Py(C3NHGly)+-Im-Am+ > f-Im-Py(C3NH3)+-Im-Am+ > f-Im-Py(C3Cl)-Im-Am+> phenyl-Im-Py-Im-Am+ >phenyl-Im-Py(C3NH3)+-Im-Am+. Based on the classification of the various modifications on these compounds, the order above is: compound without side chain > compounds with side chain on Im ring > compounds with side chain on Py ring > compound with an additional phenyl ring > compound with both a phenyl ring and a side chain on Py. This observation suggests that the side chains on the Py ring, no matter what their charge states, have weaker effects on DNA structure relative to modification on the formamido-terminal Im ring. The Py rings of two ligands forming the antiparallel dimer are closer in structure over the two formamido-terminal Im rings (Collar et al., 2010) and thus the side chains attaching on Py have stronger steric and electrostatic repulsion if the side chain is positively charged. For the derivatives with an uncharged side chain, there is no electrostatic interaction between the neutral group and the DNA backbone, which results in more unfavorable steric clash between the side chains on the dimer complex. These repulsion forces or steric effects between side chains will reduce the stacking intensity between the Im and Py rings, and the flexibility of the PA-DNA complexes will be increased as a consequence.
Phenyl rings
The migration properties of phenyl-containing PAs on CGCG multimers were significantly influenced by the cationic side chain. The binding of monocation phenyl-Im-Py-Im-Am+ increased the DNA mobility in a smaller extent than all the non-phenyl analogs. The dication, phenyl-Im-Py(C3NH3)+-Im-Am+, possesses both a side chain on Py and a terminal benzamide, and it significantly decreased the DNA migration (Figure 4). This PA, in contrast to the monocation, causes DNA bending. It could be due to the fact that the minor groove with a wide CGCG site has been narrowed by the dimer formation of phenyl-Im-Py(C3NH3)+-Im-Am+. To obtain a better understanding of how the phenyl ring may play a role in structural influence, both the binding modes of f-Im-Py-Im-Am+ and those with terminal benzamide are illustrated in Figure 7. It is clear that there are two pairs of stacked rings, Im/Py and Py/Im, in the dimer involving ligands without the phenyl (Figure 7A). With the addition of terminal phenyl rings, the imidazole ring in the charge terminus in one ligand can be stacked together with the phenyl ring in the other ligand through the π-π interaction. This means that the introduction of phenyl rings brings two more pairs of stacked rings into the dimer unit (Figure 7B). Once the positively charged alkyl amide side chains are attached on the central Py ring, they will attract the DNA backbones to fold towards the minor groove for a special compensation through the electrostatic interaction. Therefore, the CGCG minor groove would become narrower and DNA structure would become curved with decreased gel mobilities.
Figure 7.
Binding modes of f-Im-Py-Im-Am+ and N-benzamide analogs.
Conclusions
In conclusion, the dimer formation of PA in minor groove has the ability to stiffen the overall DNA structure without grossly affecting DNA conformation. The PAs can also bend DNA through inducing a narrower minor groove with ligand modifications. These various conformational effects might allow these PAs to be allosteric modulators of DNA transcription by locking the DNA shape or altering the minor groove width to reduce or enhance the binding affinity of DNA transcription factors that bind in the major groove of the same sequence.
Supplementary Material
Acknowledgements
The authors thank NIH NIAID AI 064200 to W. D. W, NSF CHE 0809162 to W. D. W and M. L, and the Center for Diagnostic and Therapeutic for a fellowship to S. W. The authors thank Carol W. Wilson for manuscript proofreading.
Abbreviations
- PAGE
polyacrylamide gel electrophoresis
- bp
base pair
- f
formamido
- Im
imidazole
- Py
pyrrole
- Am
amine
References
- Allison SA, Li ZY, Reed D, Stellwagen NC. Modeling the gel electrophoresis of short duplex DNA by Brownian dynamics: Cubic gel lattice with direct interaction. Electrophoresis. 2002;23:2678–2689. doi: 10.1002/1522-2683(200208)23:16<2678::AID-ELPS2678>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Anderson JN. Detection, sequence patterns and function of unusual DNA structures. Nucleic Acids Res. 1986;14:8513–8533. doi: 10.1093/nar/14.21.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando T, Sugiyama H. Synthesis and biological properties of sequence-specific DNA-alkylating pyrrole-imidazole polyamides. Acc. Chem. Res. 2006;39:935–944. doi: 10.1021/ar030287f. [DOI] [PubMed] [Google Scholar]
- Bando T, Narita A, Saito I, Sugiyama H. Molecular design of a pyrrole-imidazole hairpin polyamides for effective DNA alkylation. Chem. Eur. J. 2002;8:4781–4790. doi: 10.1002/1521-3765(20021018)8:20<4781::AID-CHEM4781>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Buchmueller KL, Staples AM, Uthe PB, Howard CM, Pacheco KAO, Cox KK, Henry JA, Bailey SL, Horick SM, Nguyen B, Wilson WD, Lee M. Molecular recognition of DNA base pairs by the formamido/pyrrole and formamido/imidazole pairings in stacked polyamides. Nucleic Acids Res. 2005a;33:912–921. doi: 10.1093/nar/gki238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmueller KL, Staples AM, Howard CM, Horick SM, Uthe PB, Le NM, Cox KK, Nguyen B, Pacheco KAO, Wilson WD, Lee M. Extending the language of DNA molecular recognition by polyamides: Unexpected influence of imidazole and pyrrole arrangment on binding affinity and specificity. J. Am. Chem. Soc. 2005b;127:742–750. doi: 10.1021/ja044359p. [DOI] [PubMed] [Google Scholar]
- Buchmueller KL, Bailey SL, Matthews DA, Taherbhai ZT, Register JK, Davis ZS, Bruce CD, O'Hare C, Hartley JA, Lee M. Physical and structural basis for the strong interactions of the -ImPy- central pairing motif in the polyamide f-ImPyIm. Biochemistry. 2006;45:13551–13565. doi: 10.1021/bi061245c. [DOI] [PubMed] [Google Scholar]
- Chaires JB. Energetics of drug-DNA interactions. Biopolymers. 1997;44:201–215. doi: 10.1002/(SICI)1097-0282(1997)44:3<201::AID-BIP2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Chavda S, Liu Y, Babu B, Davis R, Sielaff A, Ruprich J, Westrate L, Tronrud C, Ferguson A, Franks A, Tzou S, Adkins C, Rice T, Mackay H, Kluza J, Tahir SA, Lin SC, Kiakos K, Bruce CD, Wilson WD, Hartley JA, Lee M. Hx, a Novel Fluorescent, Minor Groove and Sequence Specific Recognition Element: Design, Synthesis, and DNA Binding Properties of p-Anisylbenzimidazole-imidazole/pyrrole-Containing Polyamides. Biochemistry. 2011;50:3127–3136. doi: 10.1021/bi102028a. [DOI] [PubMed] [Google Scholar]
- Chenoweth DM, Dervan PB. Allosteric modulation of DNA by small molecules. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13175–13179. doi: 10.1073/pnas.0906532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenoweth DM, Dervan PB. Structural basis for cyclic Py-Im polyamide allosteric inhibition of nuclear receptor binding. J. Am. Chem. Soc. 2010;132:14521–14529. doi: 10.1021/ja105068b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collar CJ, Lee M, Wilson WD. Setting Anchor in the Minor Groove: in Silico Investigation into Formamido N-Methylpyrrole and N-Methylimidazole Polyamides Bound by Cognate DNA Sequences. J. Chem Inf. Model. 2010;50:1611–1622. doi: 10.1021/ci100191a. [DOI] [PubMed] [Google Scholar]
- Crothers DM, Drak J. Global features of DNA-structure by comparative gel-electrophoresis. Methods Enzymol. 1992;212:46–71. doi: 10.1016/0076-6879(92)12005-b. [DOI] [PubMed] [Google Scholar]
- Dervan PB. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- Diekmann S, David MJL, James ED. Analyzing DNA curvature in polyacrylamide gels. Analyzing DNA curvature in polyacrylamide gels. Methods Enzymol. 1992;212:30–46. doi: 10.1016/0076-6879(92)12004-a. [DOI] [PubMed] [Google Scholar]
- Digabriele AD, Sanderson MR, Steitz TA. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc. Natl. Acad. Sci. U. S. A. 1989;86:1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasman GD. Handbook of Biochemistry and Molecular Biology, Nucleic Acids. Vol. I. Ohio: CRC Press; 1975. p. 589. [Google Scholar]
- Goodsell DS, Kopka ML, Cascio D, Dickerson RE. Crystal structure of CATGGCCATG and its implications for A-tract bending models. Proc. Natl. Acad. Sci. U. S. A. 1993;90:2930–2934. doi: 10.1073/pnas.90.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineman U, Alings C. Crystallorgraphic study of one ture of G/C-rich B DNA. J. Mol. Biol. 1989;210:369–382. doi: 10.1016/0022-2836(89)90337-9. [DOI] [PubMed] [Google Scholar]
- Hess GF, Drong RF, Weiland KL, Slightom JL, Sclafani RA, Hollingsworth RE. A human homolog of the yeast CDC7 gene is overexpressed in some tumors and transformed cell lines. Gene. 1998;211:133–140. doi: 10.1016/s0378-1119(98)00094-8. [DOI] [PubMed] [Google Scholar]
- Hunt RA, Munde M, Kumar A, Ismail MA, Farahat AA, Arafa RK, Say M, Batista-Parra A, Tevis D, Boykin DW, Wilson WD. Induced topological changes in DNA complexes: influence of DNA sequences and small molecule structures. Nucleic Acids Res. 2011;39:4265–4274. doi: 10.1093/nar/gkq1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo HS, Crothers DM. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc. Natl. Acad. Sci. U. S. A. 1988;85:1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy ER, Le NM, Price CA, Lee M, Wilson WD. Influence of a terminal formamido group on the sequence recognition of DNA by polyamides. J. Am. Chem. Soc. 2002;124:2153–2163. doi: 10.1021/ja016154b. [DOI] [PubMed] [Google Scholar]
- Lacy ER, Madsen EM, Lee M, Wilson WD. Polyamide dimer stacking in the DNA minor groove and recognition of TG mismatched base pairs in DNA. In: Demeunynck M, Bailly C, Wilson WD, editors. DNA and RNA Binders, From Small Molecules to Drugs. Weinheim: WILEY-VCH Verlag GmbH & Co.; 2003. pp. 384–413. [Google Scholar]
- Liu Y, Chai Y, Kumar A, Tidwell RR, Boykin DW, Wilson WD. Designed Compounds for Recognition of 10 Base Pairs of DNA with Two AT Binding Sites. J. Am. Chem. Soc. 2012;134:5290–5299. doi: 10.1021/ja211628j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YJ, Stellwagen NC. Monovalent cation binding by curved DNA molecules containing variable numbers of A-tracts. Biophys. J. 2008;94:1719–1725. doi: 10.1529/biophysj.107.121236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki AS, Kim TW, Kool ET. Direct comparison of A- and T-strand minor groove interactions in DNA curvature at A tracts. Biochemistry. 2004;43:1102–1110. doi: 10.1021/bi035340m. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Tsujibayashi H, Tagashira H, Inano K, Ueda T, Hirota Y, Hashimoto K. Suppression of electrophoretic anomaly of bent DNA segments by the structural property that causes rapid migration. Nucleic Acids Res. 1998;26:4811–4817. doi: 10.1093/nar/26.21.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyoshi T, Kawakami W, Narita A, Bando T, Sugiyama H. Inhibition of transcription at a coding sequence by alkylating polyamide. J. Am. Chem. Soc. 2003;125:4752–4754. doi: 10.1021/ja029196o. [DOI] [PubMed] [Google Scholar]
- Pelton JG, Wemmer DE. Structural modeling of the distamycin A-d(CGCGAATTCGCG)2 complex using 2D NMR and molecular mechanics. Biochemistry. 1988;27:8088–8096. doi: 10.1021/bi00421a018. [DOI] [PubMed] [Google Scholar]
- Pelton JG, Wemmer DE. Structual characterization of a 2-1 distamcyin A-d(CGCAAATTGGC) complex by two-dimensional NMR. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5723–5727. doi: 10.1073/pnas.86.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton JG, Wemmer DE. Binding modes of distamcyin A with d(CGCAAATTTGCG)2 determined by 2-dimensional NMR. J. Am. Chem. Soc. 1990;11:1393–1399. [Google Scholar]
- Ramos JP, Babu B, Chavda S, Liu Y, Plaunt A, Ferguson A, Savagian M, Lee M, Tzou S, Lin SC, Kiakos K, Wang S, Lee M, Hartley JA, Wilson WD. Affinity and kinetic modulation of polyamide-DNA interactions by N-modification of the heterocycles. Biopolymers. 2013 doi: 10.1002/bip.22205. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskatov JA, Meier JL, Puckett JW, Yang F, Ramakrishnan P, Dervan PB. Modulation of NF-kappa B-dependent gene transcription using programmable DNA minor groove binders. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1023–1028. doi: 10.1073/pnas.1118506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record MT, Jr, Anderson CF, Lohman TM. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Quart. Rev. Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- Ross ED, Den RB, Hardwidge PR, Maher LJ. Improved quantitation of DNA curvature using ligation ladders. Nucleic Acids Res. 1999;27:4135–4142. doi: 10.1093/nar/27.21.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satam V, Babu B, Porte A, Savagian M, Lee M, Smeltzer T, Liu Y, Ramos J, Wilson WD, Lin S, Kiakos K, Hartley JA, Lee M. Synthesis and DNA binding properties of 1-(3-aminopropyl)-imidazole-containing triamide f-Im*PyIm: a novel diamino polyamide designed to target 5'-ACGCGT-3'. Bioorg. Med. Chem. Lett. 2012a;22:5898–5902. doi: 10.1016/j.bmcl.2012.07.071. [DOI] [PubMed] [Google Scholar]
- Satam V, Babu B, Chavda S, Savagian M, Sjoholm R, Tzou S, Liu Y, Kiakos K, Lin SC, Wilson WD, Hartley JA, Lee M. Novel diamino imidazole and pyrrole-containing polyamides: Synthesis and DNA binding studies of mono- and diamino-phenyl-ImPy*Im polyamides designed to target 5 '-ACGCGT-3 '. Bioorg. Med. Chem. 2012b;20:693–701. doi: 10.1016/j.bmc.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Satz AL, Bruice TC. Recognition in the minor groove of double-stranded DNA by microgonotropens. Acc. Chem. Res. 2002;35:86–95. doi: 10.1021/ar0101032. [DOI] [PubMed] [Google Scholar]
- Stellwagen NC. Electrophoresis of DNA in agarose gels, polyacrylamide gels and in free solution. Electrophoresis. 2009;30:S188–S195. doi: 10.1002/elps.200900052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevis DS, Kumar A, Stephens CE, Boykin DW, Wilson WD. Large, sequence-dependent effects on DNA conformation by minor groove binding compounds. Nucleic Acids Res. 2009;37:5550–5558. doi: 10.1093/nar/gkp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripos Inc. St Louis M. SYBYL Molecular Modeling Software. Version X1.2. 2010 [Google Scholar]
- Verma R, Patapoutian A, Gordon CB, Campbell JL. Idetification and purification of a factor that binds to the Mlu 1 cell cycle box of yeast DNA replication genes. Proc. Natl. Acad. Sci. U. S. A. 1991;88:7155–7159. doi: 10.1073/pnas.88.16.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Munde M, Wang SM, Wilson WD. Minor groove to major groove, an unusual DNA sequence-dependent change in bend directionality by a distamycin dimer. Biochemistry. 2011;50:7674–7683. doi: 10.1021/bi201010g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Nagase H, Watanabe T, Nobusue H, Suzuki T, Asami Y, Shinojima Y, Kawashima H, Takagi K, Mishra R, Igarashi J, Kimura M, Takayama T, Fukuda N, Sugiyama H. Inhibition of MMP-9 transcription and suppression of tumor metastasis by pyrrole-imidazole polyamide. Cancer Sci. 2010;101:759–766. doi: 10.1111/j.1349-7006.2009.01435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmer DE. Designed sequence-specific minor groove ligands. Annu. Rev. Bioph. Biom. 2000;29:439–461. doi: 10.1146/annurev.biophys.29.1.439. [DOI] [PubMed] [Google Scholar]
- Wilson WD, Tanious FA, Mathis A, Tevis D, Hall JE, Boykin DW. Antiparasitic compounds that target DNA. Biochimie. 2008;90:999–1014. doi: 10.1016/j.biochi.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Lee H. Human Dbf4/ASK promoter is activated through the Sp1 and MluI cell-cycle box (MCB) transcription elements. Oncogene. 2002;21:7786–7796. doi: 10.1038/sj.onc.1205914. [DOI] [PubMed] [Google Scholar]
- Yang XL, Kaenzig C, Lee M, Wang AHJ. Binding of AR-1-144, a tri-imidazole DNA minor groove binder, to CCGG sequence analyzed by NMR spectroscopy. Eur. J. Biochem. 1999;263:646–655. doi: 10.1046/j.1432-1327.1999.00515.x. [DOI] [PubMed] [Google Scholar]
- Yoon C, Prive GG, Goodsell DS, Dickerson RE. Structure of an alternating B DNA helic and its relationship to A-tract DNA. Proc. Natl. Acad. Sci. U. S. A. 1988;85:6332–6336. doi: 10.1073/pnas.85.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.