SUMMARY
The NMDA receptor family of glutamate receptor ion channels are formed by obligate heteromeric assemblies of GluN1, GluN2 and GluN3 subunits. GluN1 and GluN3 bind glycine, whereas GluN2 binds glutamate. Crystal structures of the GluN1 and GluN3A ligand-binding domains (LBDs) in their apo states unexpectedly reveal open and closed cleft conformations, respectively, with water molecules filling the binding pockets. Computed conformational free energy landscapes for GluN1, GluN2A, and GluN3A LBDs reveal that the apo state LBDs sample closed cleft conformations, suggesting that their agonists bind via a conformational selection mechanism. By contrast, free energy landscapes for the AMPA receptor GluA2 LBD suggests binding of glutamate via an induced fit mechanism. Principal component analysis reveals a rich spectrum of hinge bending, rocking, twisting, and sweeping motions that are different for the GluN1, GluN2A, GluN3A, and GluA2 LBDs. This variation highlights the structural complexity of signaling by glutamate receptor ion channels.
INTRODUCTION
At most excitatory synapses in the brain, the neurotransmitter glutamate activates a family of ligand-gated ion channels with a unique tetrameric multidomain architecture, different from other receptors and ion channels (Mayer, 2011). Due to their central role in brain function, combined with increasing evidence for dysfunction in neurological and psychiatric diseases, glutamate receptor ion channels (iGluRs) are important eukaryotic membrane protein structural targets. Individual iGluR subunits assemble into three major families, the α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA), kainate, and N-methyl-D-aspartic acid (NMDA) receptors, named after their sensitivity to subtype selective ligands (Watkins and Evans, 1981). Glutamate, or glycine in the case of NMDA receptor GluN1 and GluN3 subunits, is believed to trigger ion channel activation by producing a large conformational change in the bilobed ligand-binding domain (LBD) of iGluRs.
In canonical models for glutamate receptor ion channel activation, in which the four LBDs assemble as a dimer of dimers (Sobolevsky et al., 2009), binding of neurotransmitters results in separation of the lower lobes of LBD dimer assemblies, pulling the ion channel open (Armstrong and Gouaux, 2000). This Venus flytrap or clam shell model for LBD activation was developed on the basis of homology with bacterial periplasmic binding proteins (Mano et al., 1996; Mao et al., 1982), for which structures had been solved for the open-cleft apo and closed-cleft ligand bound states (Quiocho and Ledvina, 1996). Subsequent crystallographic studies, on AMPA receptor GluA2 ligand-binding domains expressed as soluble proteins, revealed a 20° difference in LBD cleft closure for the apo- and glutamate-bound states (Armstrong and Gouaux, 2000), smaller than the 50° closure typical for periplasmic proteins, but sufficient to produce a 20 Å separation of the linkers connecting the LBD to the ion channel α-helical TM segments in the GluA2 tetramer (Sobolevsky et al., 2009). Although more than 200 agonist, partial agonist and antagonist bound crystal structures have since been solved for iGluR LBDs from the AMPA, kainate and NMDA receptor subunit gene families (Mayer, 2011; Pøhlsgaard et al., 2011; Stawski et al., 2010), the apo state has only been crystallized for GluA2 (Armstrong and Gouaux, 2000), and more recently for the orphan receptor GluD2 (Naur et al., 2007), which similar to GluA2 reveals a large difference in domain closure for the apo and ligand (D-serine) bound states.
Central to the Venus flytrap model for iGluR activation is the underlying assumption that the open-cleft apo state is a stable conformational entity distinct from the closed-cleft agonist-bound state. Support for this model was obtained from the calculation of free energy landscapes for the GluA2 LBD, which revealed well-defined, separated basins for the apo and agonist-bound states, the conformations of which agreed well with that for GluA2 LBD crystal structures (Lau and Roux, 2007). The free energy landscapes were also consistent with conformational dynamics observed in single molecule FRET studies (Landes et al., 2011). Although further, indirect support for the flytrap model was provided by work on kainate and NMDA receptor subtype LBDs crystallized with competitive antagonists, which revealed open-cleft conformations thought to resemble the apo state (Furukawa and Gouaux, 2003; Mayer et al., 2006), experimental structures for the apo states of these receptor families have not been determined, and their conformational stability remains unknown.
We have now crystallized the NMDA receptor GluN1 and GluN3A LBDs in their apo states. For GluN1 there is a 25° difference in domain closure compared with the glycine-bound state, but for GluN3A there is only a modest 8° opening, such that the entrance to the binding site is sterically occluded. To explore the mechanism underlying this surprising difference, conformational free energy landscapes were calculated for the apo- and glycine-bound states of both GluN1 and GluN3A, and for the apo- and glutamate-bound states of GluN2A and GluA2. These landscapes reveal that, in contrast to GluA2, the NMDA receptor LBDs can access fully closed-cleft conformations in the absence of agonist. GluN1 and GluN3A can also access open-cleft conformations that are hyperextended compared with GluN1 LBD crystal structures in complex with the competitive antagonist 5,7-dichlorokynurenic acid (DCKA). Low free energy LBD conformers were then studied using principal component analysis to evaluate the characteristic large-scale dynamics inherent to the various LBDs.
RESULTS
Crystal Structure of the GluN1 LBD Apo State
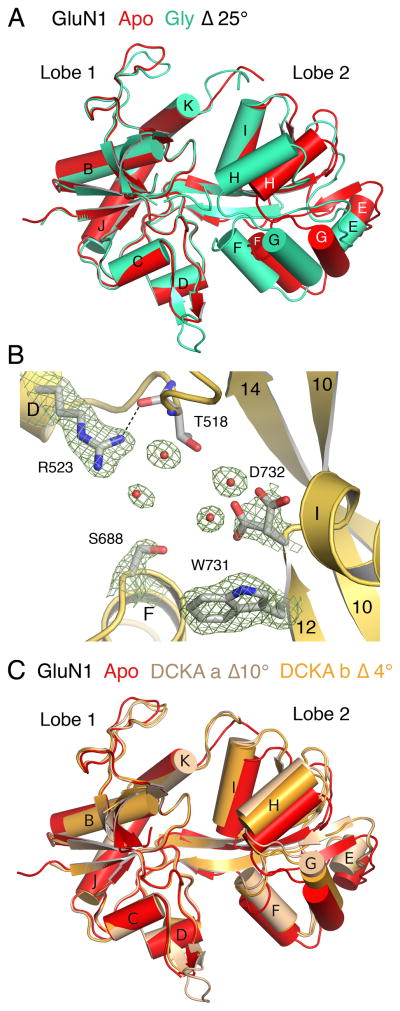
The GluN1 apo state structure was solved to a resolution of 1.9 Å (Table 1) and adopts an open cleft conformation similar to that for the GluA2 and GluD2 LBD apo state crystal structures. To estimate the difference in domain closure in the GluN1 LBD apo crystal structure compared with the GluN1 glycine complex, we performed a least squares superposition using 142 Cα atoms in lobe 1 (RMSD 0.38 Å), and then calculated the rotation angle required for superposition of lobe 2 (RMSD 0.50 Å for 95 Cα atoms). This analysis reveals that the apo structure is open by 25° compared with the glycine complex (Figure 1A). Within the binding pocket, the side chain of Arg523, which interacts with the ligand α-COOH group in the glycine and DCKA complexes, adopts a similar extended conformation in both the presence and absence of ligand, and in the apo state is held in place by a hydrogen bond between the side chain guanidinium group and the main chain carbonyl oxygen of Thr518 (Figure 1B), and by a solvent-mediated hydrogen bond with the Thr518 side chain hydroxyl group. The side chain of Asp732, which binds the glycine α-NH2 group, is disordered in the apo state and is surrounded by a cluster of water molecules (Figure 1B). Due to the change in orientation of lobe 2 in the apo state, Ser688 in α-helix F, which also interacts with the ligand α-COOH group in the glycine complex, moves away from lobe 1 by 4 Å and is exposed to solvent. Inter-lobe interactions, which are extensive in the GluN1 glycine complex, are limited in the apo state to a cluster of van der Waals contacts mediated by Ala734 and Phe738 on α-helix I in lobe 2, which pack against a complimentary hydrophobic surface in lobe 1 formed by Pro407, Phe408, and Phe758. This cluster of contacts is preserved across the apo, DCKA, and glycine complex crystal structures and likely serves as a pivot limiting the relative range of motion of Lobes 1 and 2. An analysis of the dynamics of interactions between Lobes 1 and 2 as well as between water molecules and the binding pocket have previously been described (Kaye et al., 2006).
Table 1.
Crystallographic Data collection and refinement statistics
| GluN1 apo | GluN3A apo | |
|---|---|---|
| DATA COLLECTION | ||
| Space group | C2 | P212121 |
| Unit cell dimensions | ||
| a b c (Å) | 87.3 66.4 63.5 | 75.0 83.9 99.1 |
| α β γ (°) | 90 105.2 90 | 90 90 90 |
| Mol per asymmetric unit | 1 | 2 |
| Wavelength (Å) | 1.0000 | 0.9999 |
| Resolution (Å) a | 40 − 1.90 (1.93) | 30 − 1.68 (1.74) |
| Unique observations | 27791 | 72079 |
| Mean redundancy b | 3.9 (3.8) | 7.1 (7.1) |
| Completeness (%) b | 98.7 (98.0) | 98.9 (97.9) |
| Rmergebc | 0.054 (0.59) | 0.059 (0.34) |
| Average I/σ(I) b | 25.3 (2.5) | 27.9 (6.9) |
| REFINEMENT | ||
| Resolution (Å) | 36.9 – 1.89 | 29.5 − 1.68 |
| Protein atoms (alt conf) | 2215 (50) | 4586 (204) |
| Glycerol/PO4 mols | 0/1 | 2/0 |
| Water atoms | 223 | 579 |
| Rwork / Rfree (%) d | 17.2/20.6 | 15.4/18.4 |
| rms deviations | ||
| Bond lengths (Å) | 0.01 | 0.01 |
| Bond angles (degrees) | 1.29 | 1.31 |
| Mean B-Values (Å2) | ||
| Protein overall | 34.6 | 19.4 |
| Main-chain / Side-chain | 30.3/38.8 | 16.5/22.2 |
| Glycerol / PO4 | –/70.7 | 41.7/ – |
| Water | 39.9 | 30.3 |
| Ramachandran % e | 98.5/100 | 98.2/100 |
Values in parenthesis indicate the low resolution limit for the highest-resolution shell of data.
Values in parenthesis indicate statistics for the highest-resolution shell of data.
Rmerge= = (Σ| II − <II>|) / ΣI |II|, where <II> is the mean II over symmetry–equivalent reflections.
Rwork = (Σ ||Fo| − |Fc||) / Σ |Fo|, where Fo and Fc denote observed and calculated structure factors, respectively; 5% of the reflections were set aside for the calculation of the Rfree value.
Preferred/Allowed conformations
Figure 1. Crystal structure of the GluN1 apo state.
(A) Ribbon plot showing a least squares superposition using lobe 1 coordinates of the GluN1 apo and glycine complex crystal structures, illustrating the 25° difference in domain closure. (B) Electron density maps (2mFo-DFc 1.2 sigma) for side chains and water molecules in the GluN1 apo state ligand-binding cleft. (C) Ribbon plot showing a least squares superposition using lobe 1 coordinates of the GluN1 apo and DCKA (chains A and B) complex crystal structures, illustrating 10° and 4° rotations of lobe 2 in the antagonist complexes.
From an analysis of GluN1 LBD structures in complex with the competitive antagonist DCKA, which are 16° to 21° more open than the glycine complex, it was predicted that the GluN1 apo state would have a conformation that is similar to the DCKA complexes (Furukawa and Gouaux, 2003). Least squares superpositions using the GluN1 apo state structure, however, reveals that following a superposition of lobe 1 (resulting in RMSDs of 0.46 Å and 0.50 Å for molecules A and B of the DCKA complex (1PBQ)), rotations of 10° and 4° are required for a superposition of lobe 2 (RMSDs of 0.42 Å and 0.37 Å). Inspection of the superimposed structures reveals that while the GluN1 apo-to-glycine conformational transition appears to result largely from a hinge-bending motion, the apo-to-DCKA transition is instead due to a combined sweeping and twisting motion of lobe 2 (see below), the extent of which is 6° greater for molecule A (Figure 1C).
Crystal Structure of the GluN3A LBD Apo State
The GluN3A apo state structure was solved to a resolution of 1.7 Å (Table 1). The asymmetric unit contains two protomers in similar conformations, arranged as a head-to-tail dimer. Least squares superpositions of the two apo state LBD protomers on the previously published GluN3A glycine complex structure (Yao et al., 2008) gave RMSD values of 0.54 Å and 0.50 Å for lobe 1 (171 Cα atoms). Subsequent superposition of lobe 2 (RMSDs 0.56 Å and 0.66 Å for 92 Cα atoms) required rotations of only 8°, indicating that the GluN3A apo state is substantially more closed than the GluN1 apo state (Figure 2A). Because the GluN3A apo state structures have closed-cleft conformations, numerous inter-lobe contacts in the glycine complex are preserved in the apo state, including hydrogen bonds between Ser633 and Ser801, Asp845 and Tyr873, Glu522 and Thr825, and between the main chain amide of Ala847 with the side chain hydroxyl group of Tyr908. These contacts are formed between lobe 1 residues and α-helices H and I in lobe 2, the tips of which move only 1–2 Å after superposition of lobe 1 (Figure 2A). By contrast, inter-lobe interactions formed between the side chains of Thr614 and Glu799, Lys609 and Glu803, Asn635 and Asp804, and Asn635 and Ser801, which link lobe 1 with helix F in lobe 2, are disrupted in the apo state due to a 3 Å movement of helix F away from lobe 1.
Figure 2. Crystal structure of the GluN3A apo state.
(A) Ribbon plot showing a least squares superposition using lobe 1 coordinates of the GluN3A apo and glycine complex crystal structures, illustrating the 8° difference in domain closure. (B) Electron density omit maps (mFo-DFc, 5σ) for side chains and water molecules in the GluN3a apo state ligand binding cleft, with hydrogen bonds drawn as dashed lines. (C) Molecular surface of the GluN3A LBD drawn with transparent gray shading to illustrate the solvent filled flask shaped cavity (blue) located between Lobes 1 and 2.
Within the ligand-binding pocket of the GluN3A apo state crystal structure, the side chains of both Arg638 and Asp845, which bind the ligand α-carboxy and α-amino groups, adopt similar extended conformations to those in the glycine-bound state (Figure 2B). The Arg638 guanidinium group forms a hydrogen bond with the main chain carbonyl oxygen of Ser633, helping to maintain the extended conformation. Likewise, the extended conformation of Asp845 is stabilized by a hydrogen bond with the Ser801 side chain hydroxyl group. In addition, the side chains of Arg638 and Asp845 are linked by a network of well-ordered water molecules. Omit maps for these solvent molecules are unambiguous and exclude the presence of glycine (Figure 2B). These water molecules lie in a flask shaped cavity, the neck of which has an oval opening of dimensions 2.8 Å × 3.6 Å, too narrow for glycine to enter the ligand binding site (Figure 2C). The constriction at the entrance to the binding site is formed by the side chains of Glu522 and Tyr605 in lobe 1 and by Ser800 and Met844 in lobe 2.
Conformational Free Energy Landscapes
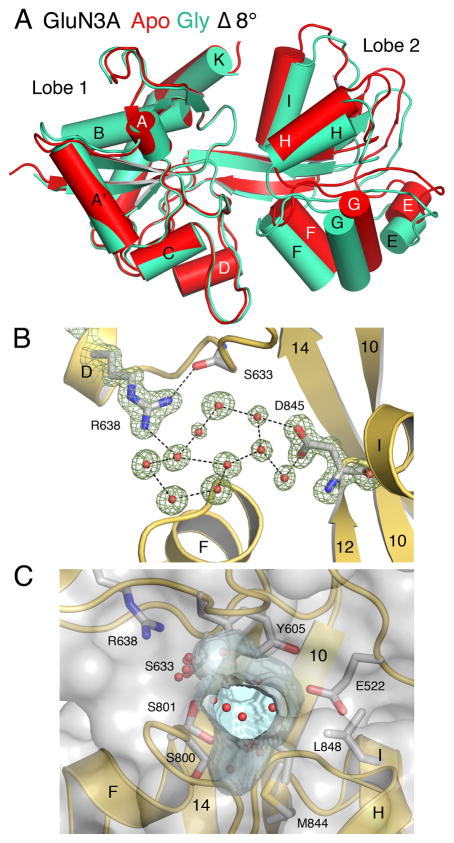
It is not possible to determine from the GluN1 and GluN3A LBD apo state crystal structures whether the large difference in conformation reflects distinct free energy minima between these NMDA receptor subtypes or whether the crystal structures represent conformers belonging to overlapping free energy basins. To address this issue, we calculated free energy landscapes, or potentials of mean force (PMFs), for both the apo and glycine-bound forms of GluN1 and GluN3A. For comparison with a glutamate-binding NMDA receptor subtype, we also calculated PMFs for the GluN2A LBD in both apo and glutamate-bound states. The PMFs were computed using an umbrella sampling molecular dynamics (MD) simulation strategy in which a two-dimensional (2D) order parameter, (ξ1, ξ2), was used to characterize large-scale conformational transitions in the LBD as previously performed for the AMPA receptor GluA2 LBD (Lau and Roux, 2007, 2011). ξ1 and ξ2 are each center of mass (COM) distances between atom selections in lobe 1 and lobe 2 (Figure 3 and Supplemental Information). A calculation of the 2D PMF, W(ξ1, ξ2), enables one to estimate the fraction of the conformational ensemble that populates different conformational states – the probability of observing a conformation (ξ1, ξ2) is proportional to exp[−W(ξ1, ξ2)/kBT], where kB is Boltzmann’s constant, and T is temperature.
Figure 3. The order parameter (ξ1, ξ2) used to describe large-scale conformational transitions in NMDA and AMPA receptor LBDs.
ξ1 and ξ2 each represent the distance (dashed line) between the centers of mass of the atoms shown as spheres. Disulfide bonds are shown in red. The models for GluN2A and GluA2 are based on the PDB entries 2A5S and 1FTO, respectively.
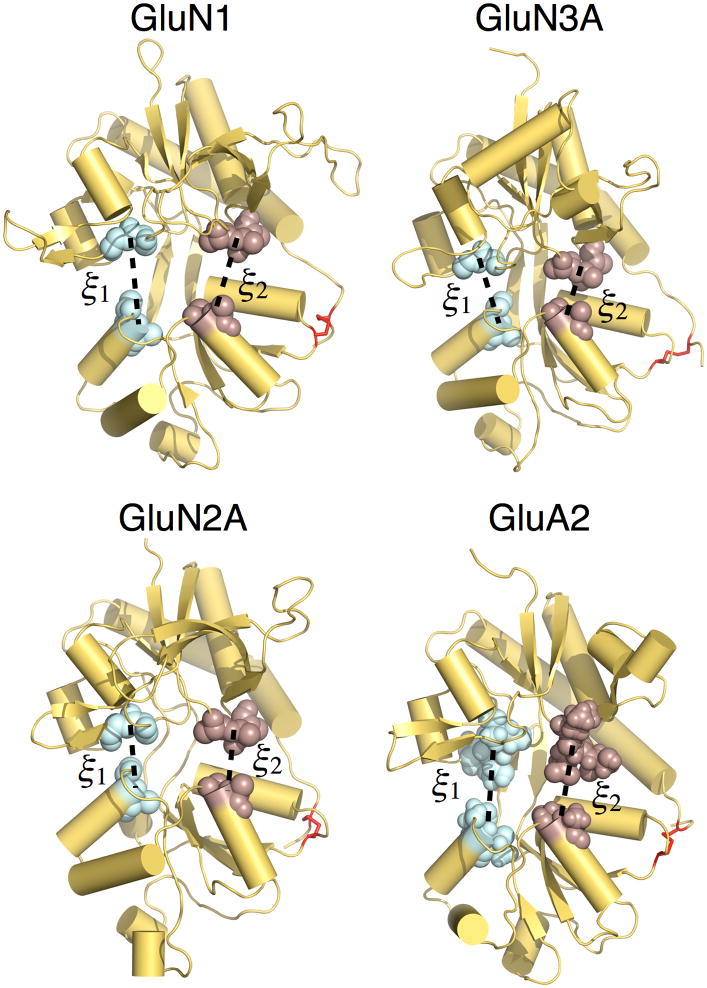
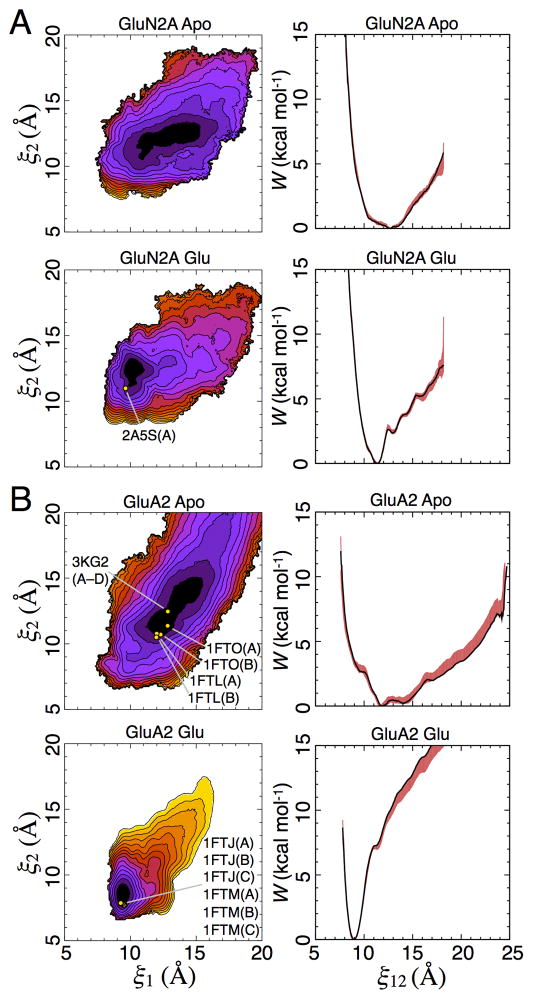
The apo LBD PMFs all feature broad free energy basins (Figures 4, 5, S1, and S2), indicating conformational flexibility in the absence of ligands. The global free energy minima are located as follows: (ξ1, ξ2) = (8.7, 10.3 Å) for GluN1, (9.4, 9.9 Å) for GluN3A, (12.9, 12.5 Å) for GluN2A, and (12.1, 11.5 Å) for GluA2. The populations for W(ξ1, ξ2) < 1 or < 2 kcal/mol are provided in Table S1. These 2D PMFs can be projected onto a single dimension, W(ξ12), where ξ12 = (ξ1+ ξ2)/2 (Figures 4 and 5). This projection results in a simplified conformational PMF that facilitates comparisons among LBD subtypes. The 1D apo PMFs show that GluN1 and GluN3A have double minima at 9.7 and 14.5 Å for GluN1 and 9.5 and 13.1 Å for GluN3A, separated by a ~1 kcal/mol barrier (Figure 4), while GluN2A has a single minimum (Figure 5). As previously reported, the 1D PMF for GluA2 has a broad basin (Lau and Roux, 2007). The populations for W(ξ12) < 1 or < 2 kcal/mol are provided in Table S1. The populations of the GluN1 and GluN3A LBDs in either the closed cleft “left-hand” minima or the open cleft “right-hand” minima are also provided in Table S1. To investigate how these NMDA receptor LBD conformational ensembles compare with those previously computed for the AMPA receptor GluA2 LBD (Lau and Roux, 2007) (Figures 5B and S2B), we examined the shape of the free energy basin by performing a least squares regression fit to ξ2 vs. ξ1 for which W(ξ1, ξ2) < 1 kcal/mol. For the apo GluA2 PMF, this slope has a value of 1.0. By contrast, the same fit to the apo GluN1, GluN3A, and GluN2A PMFs result in slopes of 0.4, 0.5, and 0.3, respectively. This analysis indicates that cleft opening is skewed to be greater along ξ1 than ξ2 for the three NMDA receptor LBDs studied here. Cleft opening for GluA2, on the other hand, is fairly symmetric in the space of (ξ1, ξ2).
Figure 4. Conformational free energy landscapes for GluN1 and GluN3A LBDs.
(A) Free energy landscapes for the GluN1 apo state (top) and glycine complex (bottom) plotted as 2D PMFs (left) and 1D PMFs (right). Contour lines for W(ξ1, ξ2) correspond to a difference of 1 kcal mol−1, with darker colors being lower in free energy. For the 1D PMFs, the red shaded region indicates the range of error estimated by a bootstrap calculation; Figure S1 shows error ranges for the 2D PMFs. The locations of crystal structures of the apo state and the following ligand complexes are indicated by yellow points: apo state (GluN1), cycloleucine (PDB ID code 1Y1M), DCKA (PDB ID code 1PBQ), glycine (PDB ID codes 1PB7 and 2A5T), D-serine (PDB ID code 1PB8), 1-aminocyclobutane-1-carboxylic acid (ACBC) (PDB ID code 1Y1Z), and 1-aminocyclopropane-1-carboxylic acid (ACPC) (PDB ID code 1Y20). (B) Free energy landscapes for the GluN3A apo state (top) and glycine complex (bottom) plotted as 2D PMFs (left) and 1D PMFs (right). The following ligand complexes are indicated: D-serine (2RC8), glycine (2RC7), and ACPC (2RC9). See also Figure S1 and Table S1.
Figure 5. Conformational free energy landscapes for GluN2A and GluA2 LBDs.
(A) Free energy landscapes for the GluN2A apo state (top) and glutamate complex (bottom) plotted as 2D PMFs (left) and 1D PMFs (right). Contour lines for W(ξ1, ξ 2) correspond to a difference of 1 kcal mol−1, with darker colors being lower in free energy; the locations of crystal structures are indicated by yellow points. For the 1D PMFs, the red shaded region indicates the range of error estimated by a bootstrap calculation; Figure S2 shows error ranges for the 2D PMFs. The location of the glutamate complex crystal structure (2A5S) is indicated by a yellow point. (B) Free energy landscapes for the GluA2 apo state (top) and glutamate complex (bottom) plotted as 2D PMFs (left) and 1D PMFs (right) as described above for GluN2A. The locations of crystal structures of the following ligand complexes and apo form are indicated by yellow points: ZK200775 (3KG2), apo (1FTO), DNQX (1FTL), glutamate (1FTJ), and AMPA (1FTM). See also Figure S2 and Table S1.
The agonist-bound NMDA receptor LBD PMFs feature free energy basins that are much more narrow than their apo counterparts, indicating, as expected, a stabilization of closed conformations when the binding site is occupied by either glycine (Figure 4) or glutamate (Figure 5). The global free energy minima are located at the following ξ1, ξ2 values: (9.2, 10.1 Å) for GluN1, (9.0, 10.0 Å) for GluN3A, (10.1, 12.4 Å) for GluN2A, and (9.4, 8.4 Å) for GluA2. For the glycine-binding subunits GluN1 and GluN3A, these minima are within 0.5 Å of the global minima for the apo LBDs. This behavior contrasts with that for the GluN2A glutamate complex, where the minimum shifts 2.8 Å along ξ1, while for the GluA2 LBD glutamate complex, the global free energy minimum shifts 2.7 Å along ξ1 and 3.1 Å along ξ2. Thus glycine appears to preserve the closed-cleft left-hand minimum in the 1D apo PMFs of GluN1 and GluN3A while destabilizing the open cleft right-hand minimum. The result for GluN1 is consistent with previous MD simulations that suggested a stable closed-cleft conformation for the apo LBD (Kaye et al., 2006). Although the position of the minimum shifts for GluN2A, the free energy basin for the apo LBD overlaps with that of the glutamate-bound LBD. For instance, in the 1D PMF, the minimum of the glutamate-bound LBD is at ξ12 = 11.3 Å, which is 0.5 kcal/mol – within 1 kBT – in the PMF of the apo LBD. These observations suggest that the NMDA receptor LBDs bind their agonists via a conformational selection mechanism (Freire, 1998; Ma et al., 1999; Monod et al., 1965). The free energy basin for apo GluA2, however, does not overlap with that of the glutamate-bound LBD. This behavior suggests that GluA2 binds glutamate via an induced fit mechanism (Koshland, 1958). Although we attempt to differentiate the binding mechanisms of the LBDs studied here, we note that the conformational selection and induced fit models are not mutually exclusive, as has been pointed out previously (Bucher et al., 2011). For the GluN1, GluN3A, and GluN2A agonist complexes, the open cleft minima at ξ12= 14.5 Å, 13.1 Å, and 12.7 Å found in the apo state are destabilized by 8.2 kcal/mol, 3.8 kcal/mol, and 2.5 kcal/mol, respectively. For the GluA2 glutamate complex, the apo minimum at ξ12= 11.9 Å is destabilized by 7.4 kcal/mol. The fraction of each conformational ensemble estimated to populate different regions of the PMF is provided in Table S1. For the GluN2A and GluA2 glutamate complexes, the glutamate ligand’s χ1and χ2 sidechain torsion angles primarily occupy a single rotameric state in closed LBD conformations. These torsions, however, explore additional rotamers when the LBD is in conformations where ξ12 > ~12 Å. The χ3 torsion is more dynamic within a closed LBD than χ1and χ 2, but it, too, explores additional configurations when the LBD is open.
Calculating (ξ1, ξ2) for crystal structures of the GluN1 and GluN3A apo states allows us to locate these in the 2D PMFs (Figure 4). The crystal structure of apo GluN1 falls near the minima at larger (ξ1, ξ2), whereas the crystal structure of apo GluN3A falls near the minima at smaller (ξ1, ξ2). Crystal structures of GluN1 bound to the antagonists cycloleucine (1Y1M) and DCKA (1PBQ) lie near minima at smaller (ξ1, ξ2) values than observed for apo GluN1. Antagonist-bound structures for GluN2A have not been reported, but antagonist-bound structures for GluA2 lie within the wide basin of low free energy conformations for the apo state (Figure 5). In contrast to the conformationally diverse apo and antagonist-bound LBD crystal structures, the agonist-bound crystal structures are all tightly clustered near the global free energy minimum for either the glycine- or glutamate-bound proteins (Figures 4 and 5).
Conformational Principal Component Analysis
Hinge-bending motions are the dominant large-scale structural variations observed in crystallographic analyses of GluR LBDs when agonist complexes are compared with the apo state or antagonist complexes (Bjerrum and Biggin, 2008; Mayer, 2011; Pøhlsgaard et al., 2011; Stawski et al., 2010). These motions are highly correlated with the order parameter (ξ1, ξ2). Additional modes of inter-lobe motions orthogonal to (ξ1, ξ2), however, may also contribute to the conformational dynamics of receptor activation (Birdsey-Benson et al., 2010). Such secondary modes are difficult to distinguish from hinge bending using only (ξ1, ξ2) as an indicator. In principle, additional order parameters could be used to differentiate among the various conformational modes, but incorporating them into an umbrella sampling strategy would require substantially more computational time since sampling scales exponentially with the number of order parameters. We thus sought to characterize LBD motions using principal component analysis (PCA), which can determine the large-scale characteristic motions of a protein from an ensemble of protein configurations (García, 1992; Grossfield and Zuckerman, 2009; Levy et al., 1984). A fluctuation correlation matrix constructed from the LBD configurations and then diagonalized yields eigenvectors and eigenvalues. The eigenvectors, or principal components (PCs), represent the characteristic motions observed in the ensemble, and each eigenvalue is the mean square fluctuation associated with the corresponding eigenvector as described in Supplemental Information (Grossfield and Zuckerman, 2009).
We adopted a pseudo-rigid body approach in constructing our ensemble of configurations for PCA. In the initial step of this approach, the configurations from the simulations are superimposed onto the lobe 1 Cα atoms of a reference crystal structure. The lobe 1 Cα atoms of the reference are then joined to the lobe 2 Cα atoms of each simulated configuration to produce the ensemble of configurations. This approach, which is similar to ones used previously (Bjerrum and Biggin, 2008; Wolter et al., 2013), prevents fluctuations in the large loop regions of lobe 1 from entering into the PCA and confounding the examination of inter-lobe dynamics. The loop regions within lobe 2, on the other hand, are small and do not require lobe 2 to be treated as a rigid body. The PCs were separately computed using coordinates from both apo and glycine- or glutamate-bound LBD simulations for which W(ξ1, ξ2) < 2 kcal/mol.
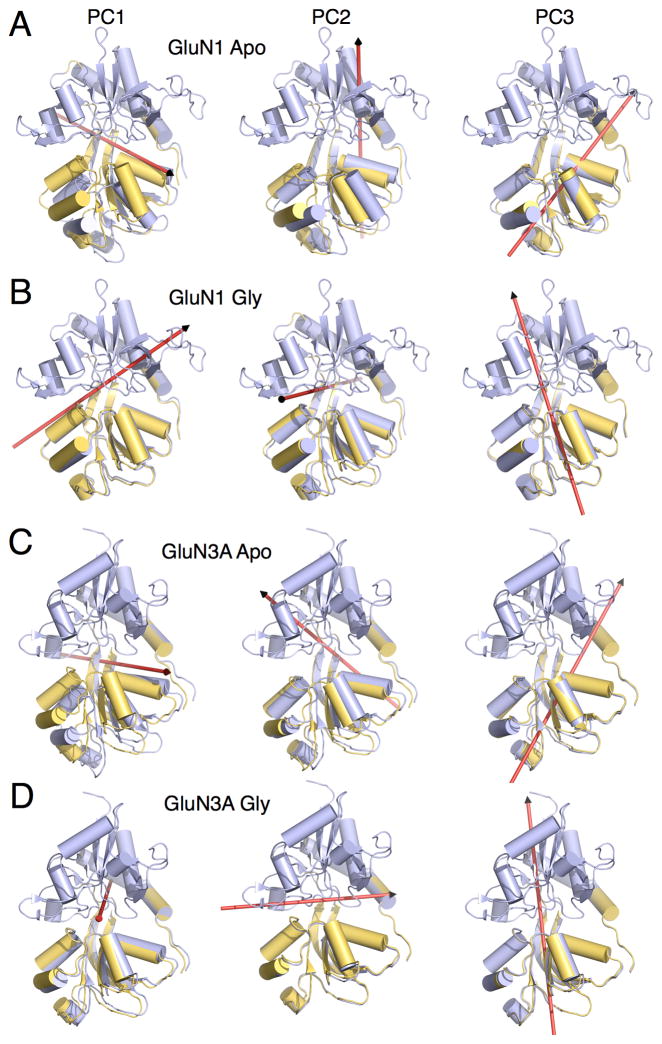
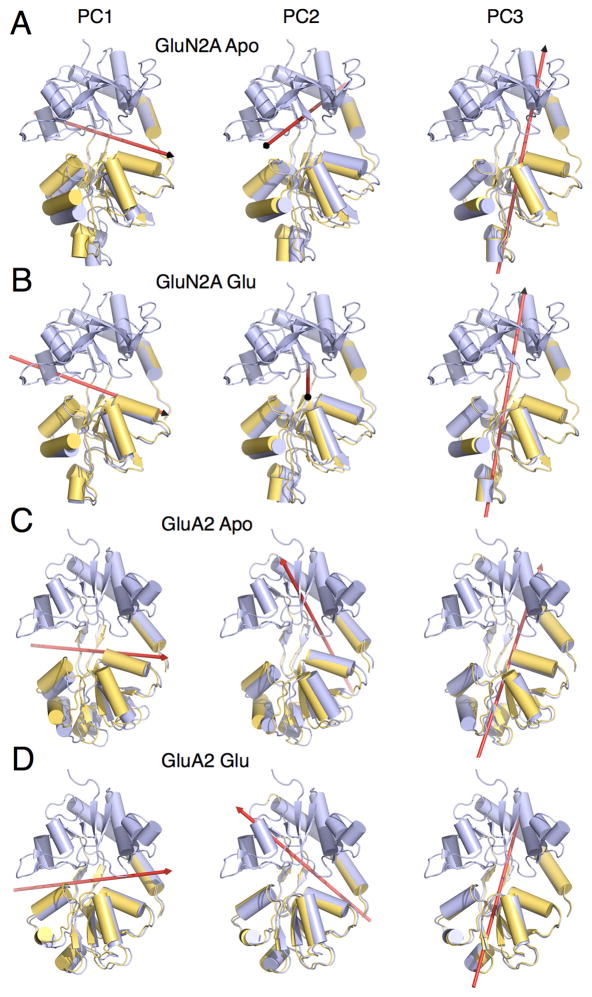
The dominant three PCs that characterize inter-lobe conformational transitions are described for each of the four LBDs (Figures 6 and 7, Tables S2 and S3, Movies S1 and S2). For the apo LBDs, PC1 corresponds, as expected, to a hinge-bending motion (Figures 6A,C and 7A,C, left panel) and accounts for the majority of the proportion of variance in large-scale transitions, with additional eigenvectors accounting for increasingly smaller contributions (Table S2). PC2 corresponds to a sweeping motion for the apo GluN1, GluN3A, and GluA2 LBDs, but a rocking motion for the apo GluN2A LBD (Figures 6A,C and 7A,C, middle panel). PC3 corresponds to a twisting motion for the apo LBDs (Figures 6A,C and 7A,C, right panel). The difference between the sweeping and twisting motions is that the sweeping rotational axis falls “behind” lobe 2, whereas the twisting axis penetrates lobe 2. The first two eigenvectors have previously been computed for the apo GluN1 LBD (Kaye et al., 2006), and they are similar to the PCs described here.
Figure 6. Axis of rotation for GluN1 and GluN3A LBD principal components 1–3.
(A) Representative GluN1 apo LBD conformers for PC1, PC2, and PC3. Red arrows indicate the axis of rotation. PC1, PC2, and PC3 are described as hinge bending, sweeping, and twisting, respectively. (B) GluN1 glycine-bound LBD conformers. PC1, PC2, and PC3 are described as hinge bending, rocking, and twisting (tilted relative to the apo LBD). (C) GluN3A apo LBD conformers. PC1, PC2, and PC3 are described as hinge bending, sweeping, and twisting. (D) GluN3A glycine-bound LBD conformers. PC1, PC2, and PC3 are described as rocking, hinge bending, and twisting (tilted relative to the apo LBD). See also Tables S2 and S3, and Movie S1.
Figure 7. Axis of rotation for GluN2A and GluA2 LBD principal components 1–3.
(A) Representative GluN2A apo LBD conformers for PC1, PC2, and PC3. Red arrows indicate the axis of rotation. PC1, PC2, and PC3 are described as hinge bending, rocking, and twisting, respectively. (B) GluN2A glutamate-bound LBD conformers. PC1, PC2, and PC3 are described as hinge bending, rocking, and twisting. (C) GluA2 apo LBD conformers. PC1, PC2, and PC3 are described as hinge bending, sweeping, and twisting. (D) GluA2 glutamate-bound LBD conformers. PC1, PC2, and PC3 are described as hinge bending, sweeping, and twisting. See also Tables S2 and S3, and Movie S2.
Upon glycine or glutamate binding to the LBD, PC1 remains a hinge-bending motion for all LBDs except GluN3A, which converts to a rocking motion (Figures 6B,D and 7B,D, left panel, Movie S1). Upon glycine binding, PC2 converts from a sweeping to a rocking motion for GluN1 and from a sweeping to a hinge-bending motion for GluN3A (Figure 6B,D, middle panel). By contrast, upon glutamate binding, PC2 remains rocking and sweeping motions for GluN2A and GluA2, respectively (Figure 7B,D, middle panel). PC3 remains a twisting motion for all LBDs upon either glycine or glutamate binding (Figures 6B,D and 7B,D, right panel and Movie S2), although the axis of rotation undergoes a tilt for the glycine-binding LBDs. Note that since the PCs were calculated separately for the apo and glycine/glutamate-bound LBDs, the same eigenvalue does not necessarily correspond to the same conformation. The proportion of variance accounted for by each of the different PCs is provided in Table S2. In comparing the PC motions with (ξ1, ξ2), the hinge-bending motions generally fall along a positive-slope diagonal in (ξ1, ξ2), whereas the other PCs are described in part by configurations off this diagonal. As noted above, the PMFs indicate that cleft opening appears to be skewed to be greater along ξ1 than ξ2 for the three NMDA receptor LBDs compared with GluA2. This skew is reflected in the tilt of the rotational axes corresponding to hinge bending, which is steeper for the glycine binding subunits.
PCA has previously been performed for GluA2 using LBD configurations from crystal structures and MD simulations (Bjerrum and Biggin, 2008; Wolter et al., 2013). The dominant three PCs, determined using the crystal structures, are hinge-bending (or clamshell closure), twisting, and rocking motions, listed in order of decreasing contribution to the proportion of variance. The fact that the eigenvectors and eigenvalues in these studies are not identical may be due to the different ensemble of configurations used in the PCA (Balsera et al., 1996). Alternatively, the umbrella sampling approach used here may have introduced some degree of conformational bias into the ensemble. Umbrella sampling, however, is not expected to obviate access to low free energy conformations associated with large-scale motions. The most energetically relevant conformations are therefore expected to be included in the structural ensembles used in the PCA.
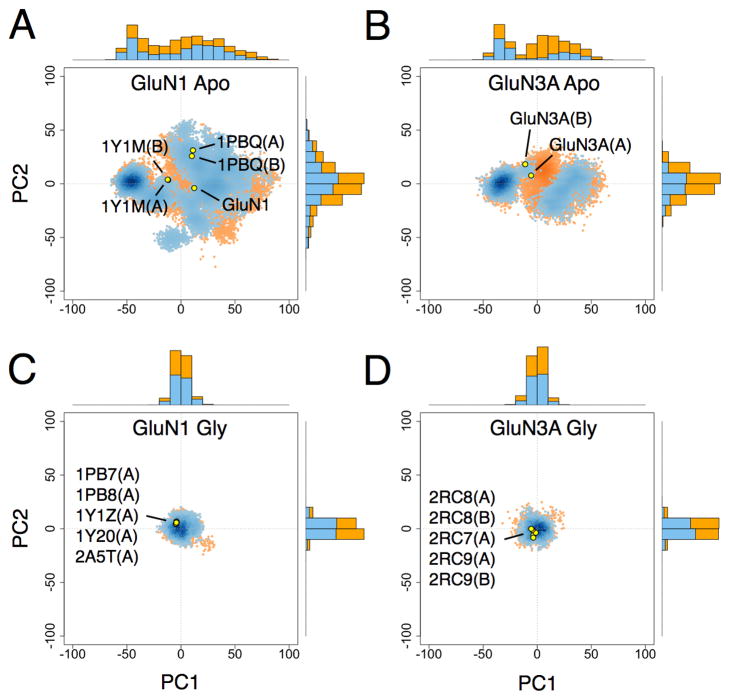
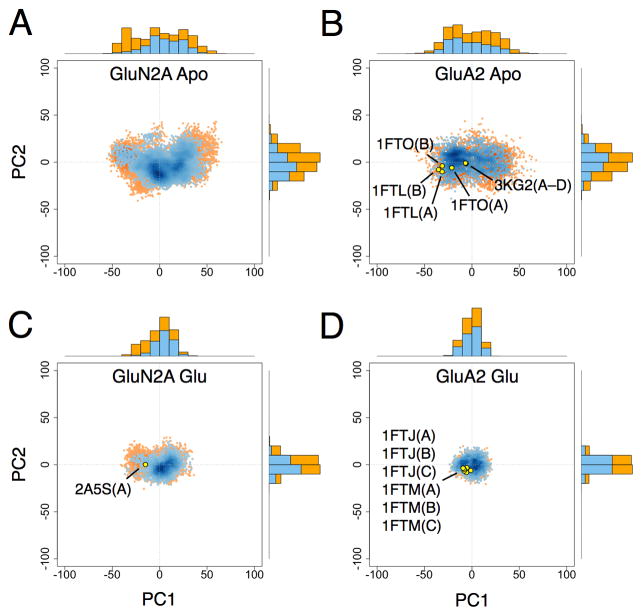
Scatter plots of the PCs for NMDA and AMPA receptor LBDs are shown in Figures 8, 9, S3, and S4. Conformers in either the apo or glycine/glutamate-bound state with W(ξ1, ξ2) < 1 kcal/mol and W(ξ1, ξ2) < 2 kcal/mol are plotted separately in order to show how the distribution of PCs vary with conformational free energy. For the apo LBDs, the PC1 distributions appear bimodal for GluN1 and GluN3A and unimodal for GluN2A and GluA2 (see the 1D PC1 projections in Figures 8 and 9). These modalities mirror the distribution of minima in the apo LBD 1D PMFs. All other PC distributions, whether for apo or glycine/glutamate-bound states, appear unimodal.
Figure 8. Principal component distributions for GluN1 and GluN3A.
(A) 2D projection of apo GluN1 LBD conformers generated from umbrella sampling simulations onto the PC1–PC2 plane with W(ξ1, ξ2) < 1 and < 2 kcal mol−1 shown in blue and orange, respectively; 1D projections onto either PC1 or PC2 are represented by marginal histograms; the 2D projections of selected crystal structures are shown as yellow points. (B) Projection of apo GluN3A conformers. (C) Projection of GluN1-glycine conformers. (D) Projection of GluN3A-glycine conformers. See also Figure S3.
Figure 9. Principal component distributions for GluN2A and GluA2.
(A) 2D projection of apo GluN2A LBD conformers generated from umbrella sampling simulations onto the PC1–PC2 plane with W(ξ1, ξ2) < 1 and < 2 kcal mol−1 shown in blue and orange, respectively; 1D projections onto either PC1 or PC2 are represented by marginal histograms. (B) Projection of apo GluA2 conformers; the 2D projections of selected crystal structures are shown as yellow points. (C) Projection of GluN2A-glutamate conformers. (D) Projection of GluA2-glutamate conformers. See also Figure S4.
The crystal structures that are localized in the 2D PMFs are also indicated in the PC scatter plots. For GluN1, the apo structure differs in conformation from the cycloleucine-bound structures (1Y1M) primarily in PC1. However, the differences between the apo structure and the DCKA-bound structures (1PBQ) are primarily in PC2 and PC3. For GluN3A, the apo structures (chains A and B) differ mainly by a small change in PC2. For GluA2, the apo and antagonist-bound structures differ primarily in PC1 with only small changes in PC2 and PC3.
DISCUSSION
Crystal structures of glutamate receptor ligand-binding domains provide a detailed but static picture of a highly dynamic segment of the receptor. The Venus flytrap model for large-scale rearrangements in response to the binding of ligands has been extensively studied in other proteins, especially maltose-binding protein (MBP). On the basis of paramagnetic relaxation enhancement data, it has been established that in the apo state, MBP is a rapidly exchanging mixture of a predominantly (~95%) open form, similar to the apo state crystal structure, and a minor (~5%) partially closed species that differs from the maltose complex (Tang et al., 2007). In the present analysis we show that the NMDA receptor subunits differ substantially from GluA2 in two ways. First, the apo state for GluN1, GluN3A, and GluN2A appears able to adopt the same fully closed conformation stabilized by agonist, suggesting a conformational selection mechanism for agonist binding. This behavior contrasts with that observed for the GluA2 LBD, for which the agonist complex adopts a more closed conformation than the apo state can easily access, suggesting an induced fit mechanism. Second, the proportion of the LBD population in the closed-cleft apo state, roughly 11%, 28%, and 15% for GluN1, GluN3A, and GluN2A, respectively (Table S1), is substantially greater than the minor closed conformation observed for MBP.
Our analysis reveals that iGluR apo LBDs can sample a large region of conformational space. The GluN1 and GluN3A subunits exhibit double conformational free energy minima in the 1D PMFs, where the minima are separated by a barrier of ~1 kcal/mol, whereas GluN2A and GluA2 are better described by a single basin. At present, it is unknown whether these PMFs would change in the context of LBD dimer and tetramer assemblies, and if so, how. Indeed, PMFs computed using higher-order LBD assemblies as well as full-length receptor subunits, while requiring considerable computational resources, would provide important insights into the structural thermodynamics of intact iGluRs.
Comparing crystal structures for iGluR LBD apo states and antagonist complexes with the free energy landscapes reveals several features. First, the GluN1 and GluN3A apo state crystal structures fall outside both open and closed cleft free energy minima, although they are still in low free energy regions, suggesting modest conformational rearrangement by the crystal lattice. Of note, GluN1 apo-state LBD crystals, which grew in similar conditions to those reported previously for the GluN1 DCKA antagonist complex (Furukawa and Gouaux, 2003), crystallized in the same C2 space group, but with a shortening by one-half of the c axis length. Second, while there is a large region of conformational space available for the binding of antagonists, the crystal structures solved to date exploit only a small region, suggesting that antagonists with different structures could be designed to trap more open cleft conformations.
The dominant three principal components for all four LBDs may be classified as hinge bending, sweeping, rocking, or twisting motions. PC analysis reveals that the apo state and DCKA crystal structures have similar extents of hinge bending (PC1) but different conformations that arise from a combination of sweeping (PC2) and twisting (PC3) transitions. For the apo LBDs, PC1 corresponds to hinge bending, and PC3 corresponds to twisting. PC2 corresponds to sweeping for all apo LBDs except GluN2A, which is rocking. The characteristic motion for PC2 therefore differentiates neither between NMDA and AMPA receptors nor between glycine- and glutamate-binding LBDs. Upon glutamate binding to GluN2A and GluA2, the characteristic motions for PC 1–3 remain the same. By contrast, many of these motions change upon glycine binding to GluN1 and GluN3A: sweeping converts to rocking for GluN1 (PC2), and for GluN3A, hinge bending goes to rocking (PC1), and sweeping to hinge bending (PC2). Additionally, the rotational axis for twisting undergoes a tilt (PC3). The different behaviors for the glycine- and glutamate-binding LBDs may result from the different binding site architectures and mechanics required to selectively bind glycine versus glutamate.
In summary, the analyses presented here reveal a rich spectrum of ligand and subtype conformational dynamics in iGluR LBDs that are not apparent from analyses of apo state crystal structures nor agonist and antagonist complexes. This spectrum likely has multiple functional consequences within an intact receptor related to the following: the action of full and partial agonists, the requirement in NMDA but not AMPA receptors for activation by different ligands, and the role of structurally distinct proximal and distal iGluR subunits in ion channel gating.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The GluN1 and GluN3A LBD S1S2 constructs were expressed as soluble proteins in OrigamiB (DE3) Escherichia coli (Novagen) and purified to homogeneity using Ni-NTA affinity, size exclusion, and SP Sepharose ion-exchange chromatography as described previously (Yao and Mayer, 2006). The GluN1 construct contains residues M394-K544 and R663-S800 joined by a GT dipeptide linker; the GluN3A construct contains residues N511-R660 and E776-K915 joined by a GT dipeptide linker. To prepare apo proteins the following changes were made to the published protocol; low affinity ligands, 30 mM L-serine for GluN1 and 1 mM L-glutamate for GluN3A, were added to all chromatography buffers to displace endogenous bound glycine, followed by exhaustive dialysis over 3–4 days against ligand free buffer, with a total volume change of 1015 prior to crystallization; for GluN1 an additional size exclusion chromatography step in crystallization buffer was performed prior to setting up drops, as described in detail in Supplemental Information,
Structure Determination and Refinement
X-ray diffraction data was collected from single crystals at 100 K at beamline ID22 at the Advanced Photon Source (APS). Datasets were indexed, scaled and merged using DENZO and SCALEPACK from the HKL2000 suite (Otwinowski and Minor, 1997). None of the datasets showed twinning as analyzed by Phenix xtriage (Adams et al., 2010). The GluN1 apo structure was solved by molecular replacement with Phaser using the glycine complex (PDB 1PB7) broken down into separate search probes for lobe 1 (RFZ1 = 21.8, TFZ1 = 21.6) and lobe 2 (RFZ1 = 14.6, TFZ1 = 35.5). The GluN3A apo structure contained 2 protomers in the asymmetric unit; using Lobes 1 and 2 of the glycine complex (PDB 2RC7) as search probes for molecular replacement, the following solution was obtained (RFZD1a = 21.8, TFZD1a = 21.6l; RFZD1b = 21.8, TFZD1b = 21.6; RFZD2a = 21.8, TFZD2a = 21.6; RFZD2b = 21.8, TFZD2b = 21.6). Alternate cycles of crystallographic refinement with PHENIX (Adams et al., 2010) coupled with rebuilding and real space refinement with COOT (Emsley et al., 2010), using TLS groups determined by motion determination analysis for GluN1 and GluN3A (Painter and Merritt, 2006) and at the final stages of refinement, individual anisotropic displacement parameters for the GluN1 glycine complex, was performed until Rfree converged to a stable value and mFo-DFc maps had no interpretable features. The final models (Table 1) were validated with MOLPROBITY (Davis et al., 2004); clash scores and Molprobity scores, with values in parenthesis for the percentile compared to structures of comparable resolution, with 100 equal the best, were GluN1 apo 1.36 (100) and 0.87 (100); GluN3A apo 1.55 (99) and 0.90 (100), respectively. Figures were prepared using PyMol (Schrödinger, LLC).
Free energy landscapes, W(ξ1,ξ2)
The protein conformational free energy landscapes, or potentials of mean force (PMFs), were computed using umbrella sampling simulations. A two-dimensional order parameter (ξ1, ξ2) is used to describe large-scale conformational transitions in each LBD. ξ1 and ξ2 each indicate the distance between the center of mass (COM) of an atom selection in lobe 1 and the COM of an atom selection in lobe 2. The atom selections are detailed in Supplemental Information.
Coordinates for the umbrella sampling windows consist of LBD conformations positioned in 1 Å increments along ξ1 and ξ2. These coordinates were obtained via targeted (biased-potential) MD simulations using CHARMM (Brooks et al., 2009) initiated from the following crystal structures: 2A5T (glycine-bound GluN1 LBD), 4KCC (apo GluN1 LBD), 2RC7 (glycine-bound GluN3A LBD), 4KCD (apo GluN3A LBD), 2A5S (glutamate-bound GluN2A LBD), 1FTJ (glutamate-bound GluA2 LBD) and 1FTO (apo GluA2 LBD). Chain A and residue isomer AC1 were used from the entries where multiple options exist. Missing residues were built using the MODLOOP server (Fiser and Sali, 2003), and missing residue sidechains were built using SCWRL4 (Krivov et al., 2009). During the targeted MD simulations, RMSD restraints were applied separately to each lobe to minimize intra-lobe structural distortions, and the glycine and glutamate ligands were restrained to remain docked to lobe 1.
93 umbrella sampling windows were used to compute each of the GluN1, GluN2A, and GluN3A PMFs. Computation of the GluA2 PMFs have been described previously (Lau and Roux, 2007). Sampling for the GluA2-glutamate PMF was extended by ~12 ns from the original 200 ns. All simulations were performed using CHARMM with explicit solvent at 300 K. The all-atom potential-energy function PARAM27 for proteins (MacKerell, et al., 1998; Mackerell et al., 2004) and the TIP3P potential energy function for water (Jorgensen et al., 1983) were used. The total simulation time for each of the GluN PMFs is ~70 ns. A timestep of 2 fs was used. The number of atoms in each simulation system is ~47,000 for GluN1, ~44,000 for GluN2A, and ~35,000 for GluN3A. Crystallographic waters in each ligand-binding cleft were included in our models. Na+ and Cl− ions were added in the bulk solution to give 150 mM NaCl and an electrically neutral system. Periodic boundary conditions were used with an orthorhombic cell with approximate dimensions 88 Å × 78 Å × 72 Å for GluN1, 94 Å × 72 Å × 68 Å for GluN2A, and 88 Å × 64 Å × 64 Å for GluN3A. Harmonic biasing potentials with a force constant of 2 kcal mol 1 Å 2 centered on (ξ1, ξ2) were used. Each PMF, W(ξ1, ξ2), was computed using the weighted histogram analysis method (WHAM) (Kumar et al., 1992; Souaille and Roux, 2001) to unbias and recombine the sampled distribution functions in (ξ1, ξ2) from all windows. To prevent ligand dissociation in open LBD conformations, an asymmetric harmonic potential (force constant of 20 kcal mol−1 Å−2) was applied to the distance between the α-carboxylate oxygen atoms of the ligand and the guanidinium nitrogen atoms of the arginine in lobe 1 to which it docks (Arg485 for GluA2, Arg523 for GluN1, Arg518 for GluN2A, and Arg638 for GluN3A) for a subset of window trajectories (6% of sampling for GluA2, 27% for GluN1, 19% for GluN2A, and 4% for GluN3A). The restraint was active only when this distance exceeded 2.8 Å. This restraint is not expected to adversely affect conformational sampling because dissociation only occurred in open LBD conformations where interactions between the ligand and lobe 2 were weak. The majority of simulations did not involve the restraint; sampling associated with trajectories where the ligand dissociated from the LBD was discarded.
The range of variation in each PMF was estimated using a bootstrapping approach (Wehrens et al., 2000). (ξ1, ξ2) for each GluN umbrella window was resampled using a window of 300 ps. The resampling window was 200 ps for GluA2. On average, correlated fluctuations in (ξ1, ξ2) decay reasonably close to zero within these windows, as assessed from a calculation of the autocorrelation function (Box et al., 2008):
where N is the total number of time steps, k is the number of time steps corresponding to a given lag time, ξ is either ξ1 or ξ2, and the overbar indicates an average.
Principal component analysis
PCA calculations involve the diagonalization of the covariance matrix Cij of positional deviations among an ensemble of protein structures (García, 1992; Grossfield and Zuckerman, 2009; Levy et al., 1984):
where xi represents a specific Cα coordinate, and both the overbar and 〈·〉 indicate average values. The ensemble, generated separately for each iGluR LBD in either the apo or glycine-/glutamate-bound state, consists of protein conformers from all umbrella sampling windows in which W(ξ1, ξ2) < 2 kcal/mol. This subset includes the vast majority of conformations that are expected to be observed in a Boltzmann-weighted ensemble while excluding conformations that are energetically difficult to access. PCs were also computed using cutoffs of 1 and 3 kcal/mol, and the results were similar. Only Cα coordinates were used in the calculations. All conformers were first superimposed onto lobe 1 of a reference structure using a selection of Cα atoms that excludes flexible loops. The references for GluN1, GluN2A, GluN3A, and GluA2 are 4KCC (Figure 1), 2A5S, 4KCD(A) (Figure 2), and 1FTO(A), respectively. The residue selections are detailed in Supplemental Information. Next, the lobe 1 Cα atoms of the reference structure were joined to the lobe 2 Cα atoms of each simulated conformer to construct an ensemble of pseudo-rigid body structures. Again, the residue selections are detailed in Supplemental Information. The ensembles for apo and glycine-bound GluN1 include 22262 and 7007 conformers, respectively. The ensembles for apo and glutamate-bound GluN2A include 12685 and 7436 conformers. The ensembles for apo and glycine-bound GluN3A include 11917 and 4709 conformers. The ensembles for apo and glutamate-bound GluA2 include 3869 and 2970 conformers. PCA calculations were performed using Bio3D (Grant et al., 2006). The PCs characterizing the LBDs in which W(ξ1, ξ2) < 1 kcal/mol were obtained by projection onto the PCs calculated for the LBDs in which W(ξ1, ξ2) < 2 kcal/mol.
The axes of rotation characterizing PC1–3 were calculated using the DomainSelect method provided by the DynDom server (Hayward and Berendsen, 1998). Endpoint coordinates were generated using Bio3D. Lobe 1 is the “fixed” domain, and lobe 2 is the “moving” domain. For GluN1, lobe 1 is defined as residues 399–534 and 758–786, and lobe 2 is defined as residues 537–754. For GluN2A, lobe 1 is defined as residues 407–529 and 761–789, and lobe 2 is defined as residues 532–757. For GluN3A, lobe 1 is defined as residues 515–649 and 873–901, and lobe 2 is defined as residues 652–869. For GluA2, lobe 1 is defined as residues 395–496 and 732–760, and lobe 2 is defined as residues 499–728.
Supplementary Material
Acknowledgments
We thank Barry Grant for helpful discussions on using the Bio3D package. Data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (NIH) and resources provided by the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant OCI-1053575. This work was supported by the Intramural Research Program of the National Institute of Child Health and Human Development, NIH, Department of Health and Human Services (M.L.M.) and NIH grant GM094495 (A.Y.L.).
Footnotes
ACCESSION NUMBERS
Protein Data Bank accession codes for atomic coordinates and structure factors are 4KCC and 4KCD for GluN1 and GluN3A, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Balsera MA, Wriggers W, Oono Y, Schulten K. Principal Component Analysis and Long Time Protein Dynamics. The Journal of Physical Chemistry. 1996;100:2567–2572. [Google Scholar]
- Birdsey-Benson A, Gill A, Henderson LP, Madden DR. Enhanced efficacy without further cleft closure: reevaluating twist as a source of agonist efficacy in AMPA receptors. J Neurosci. 2010;30:1463–1470. doi: 10.1523/JNEUROSCI.4558-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerrum EJ, Biggin PC. Rigid body essential X-ray crystallography: distinguishing the bend and twist of glutamate receptor ligand binding domains. Proteins. 2008;72:434–446. doi: 10.1002/prot.21941. [DOI] [PubMed] [Google Scholar]
- Box GEP, Jenkins GM, Reinsel GC. Time series analysis: forecasting and control. Hoboken, N.J: John Wiley; 2008. [Google Scholar]
- Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher D, Grant BJ, McCammon JA. Induced fit or conformational selection? The role of the semi-closed state in the maltose binding protein. Biochemistry. 2011;50:10530–10539. doi: 10.1021/bi201481a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- Freire E. Statistical thermodynamic linkage between conformational and binding equilibria. Adv Protein Chem. 1998;51:255–279. doi: 10.1016/s0065-3233(08)60654-3. [DOI] [PubMed] [Google Scholar]
- Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García AE. Large-amplitude nonlinear motions in proteins. Phys Rev Lett. 1992;68:2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- Grossfield A, Zuckerman DM. Quantifying uncertainty and sampling quality in biomolecular simulations. Annu Rep Comput Chem. 2009;5:23–48. doi: 10.1016/S1574-1400(09)00502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79:926. [Google Scholar]
- Kaye SL, Sansom MSP, Biggin PC. Molecular dynamics simulations of the ligand-binding domain of an N-methyl-D-aspartate receptor. J Biol Chem. 2006;281:12736–12742. doi: 10.1074/jbc.M512728200. [DOI] [PubMed] [Google Scholar]
- Koshland DE. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc Natl Acad Sci USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivov GG, Shapovalov MV, Dunbrack RL., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. The weighted histogram analysis method for free-energy calculations on biomolecules. I The method. Journal of Computational Chemistry. 1992;13:1011–1021. [Google Scholar]
- Landes CF, Rambhadran A, Taylor JN, Salatan F, Jayaraman V. Structural landscape of isolated agonist-binding domains from single AMPA receptors. Nat Chem Biol. 2011;7:168–173. doi: 10.1038/nchembio.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AY, Roux B. The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure. 2007;15:1203–1214. doi: 10.1016/j.str.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AY, Roux B. The hidden energetics of ligand binding and activation in a glutamate receptor. Nat Struct Mol Biol. 2011;18:283–287. doi: 10.1038/nsmb.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy RM, Srinivasan AR, Olson WK, McCammon JA. Quasi-harmonic method for studying very low frequency modes in proteins. Biopolymers. 1984;23:1099–1112. doi: 10.1002/bip.360230610. [DOI] [PubMed] [Google Scholar]
- Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. The Journal of Physical Chemistry B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Mano I, Lamed Y, Teichberg VI. A venus flytrap mechanism for activation and desensitization of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors. J Biol Chem. 1996;271:15299–15302. doi: 10.1074/jbc.271.26.15299. [DOI] [PubMed] [Google Scholar]
- Mao B, Pear MR, McCammon JA, Quiocho FA. Hinge-bending in L-arabinose-binding protein. The “Venus’s-flytrap” model. J Biol Chem. 1982;257:1131–1133. [PubMed] [Google Scholar]
- Mayer ML. Emerging models of glutamate receptor ion channel structure and function. Structure. 2011;19:1370–1380. doi: 10.1016/j.str.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Ghosal A, Dolman NP, Jane DE. Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. J Neurosci. 2006;26:2852–2861. doi: 10.1523/JNEUROSCI.0123-06.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Naur P, Hansen KB, Kristensen AS, Dravid SM, Pickering DS, Olsen L, Vestergaard B, Egebjerg J, Gajhede M, Traynelis SF, et al. Ionotropic glutamate-like receptor delta2 binds D-serine and glycine. Proc Natl Acad Sci USA. 2007;104:14116–14121. doi: 10.1073/pnas.0703718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Methods in Enzymology. Elsevier; 1997. [20] Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Pøhlsgaard J, Frydenvang K, Madsen U, Kastrup JS. Lessons from more than 80 structures of the GluA2 ligand-binding domain in complex with agonists, antagonists and allosteric modulators. Neuropharmacology. 2011;60:135–150. doi: 10.1016/j.neuropharm.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souaille M, Roux B. Extension to the weighted histogram analysis method: combining umbrella sampling with free energy calculations. Computer Physics Communications. 2001;135:40–57. [Google Scholar]
- Stawski P, Janovjak H, Trauner D. Pharmacology of ionotropic glutamate receptors: A structural perspective. Bioorg Med Chem. 2010;18:7759–7772. doi: 10.1016/j.bmc.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- Watkins JC, Evans RH. Excitatory amino acid transmitters. Annu Rev Pharmacol Toxicol. 1981;21:165–204. doi: 10.1146/annurev.pa.21.040181.001121. [DOI] [PubMed] [Google Scholar]
- Wehrens R, Putter H, Buydens LMC. The bootstrap: a tutorial. Chemometrics and Intelligent Laboratory Systems. 2000;54:35–52. [Google Scholar]
- Wolter T, Steinbrecher T, Elstner M. Computational study of synthetic agonist ligands of ionotropic glutamate receptors. PLoS ONE. 2013;8:e58774. doi: 10.1371/journal.pone.0058774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Mayer ML. Characterization of a soluble ligand binding domain of the NMDA receptor regulatory subunit NR3A. J Neurosci. 2006;26:4559–4566. doi: 10.1523/JNEUROSCI.0560-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML. Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. EMBO J. 2008;27:2158–2170. doi: 10.1038/emboj.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.