Abstract
Unresolved repair of clustered DNA lesions can lead to the formation of deleterious double strand breaks (DSB) or to mutation induction. Here, we investigated the outcome of clusters composed of base lesions for which base excision repair enzymes have different kinetics of excision/incision. We designed multiply damaged sites (MDS) composed of a rapidly excised uracil (U) and two oxidized bases, 5-hydroxyuracil (hU) and 8-oxoguanine (oG), excised more slowly. Plasmids harboring these U-oG/hU MDS-carrying duplexes were introduced into Escherichia coli cells either wild type or deficient for DNA n-glycosylases. Induction of DSB was estimated from plasmid survival and mutagenesis determined by sequencing of surviving clones. We show that a large majority of MDS is converted to DSB, whereas almost all surviving clones are mutated at hU. We demonstrate that mutagenesis at hU is correlated with excision of the U placed on the opposite strand. We propose that excision of U by Ung initiates the loss of U-oG-carrying strand, resulting in enhanced mutagenesis at the lesion present on the opposite strand. Our results highlight the importance of the kinetics of excision by base excision repair DNA n-glycosylases in the processing and fate of MDS and provide evidence for the role of strand loss/replication fork collapse during the processing of MDS on their mutational consequences.
INTRODUCTION
Multiply damaged sites (MDS) or clustered DNA lesions are the most deleterious damages induced by ionizing radiation [for reviews (1,2)]. They are produced by a single radiation track, consist of two or more DNA lesions distributed on both strands within one or two helical turns and comprise various types of lesions, i.e. oxidized bases, abasic sites (AP sites) and strand breaks (3–7). MDS may also be induced by ultraviolet (UV), radiomimetic and alkylating agents (3,8,9). Moreover, MDS constituted by oxidized bases seem to be induced by oxidative stress in human tumor tissues (10).
If single base damage can be efficiently repaired by the base excision repair (BER) when isolated [for review (11)], MDS represent a real challenge for BER. The reparability of MDS is expected to depend not only on the type of lesion but also on their relative positions, including the interlesion spacing. A number of in vitro studies using purified enzymes or cellular extracts showed that this is indeed the case and that most of the BER steps are retarded within a cluster (12–20). For example, an AP site or a single-strand break (SSB) impedes the cleavage at a base damage situated closely on the opposite strand. Those studies also established a hierarchy in the excision/incision of base damage at MDS (15,17). In vivo data have been obtained mainly in Escherichia coli, and also in yeast and human cells, using constructs inserted in plasmids and harboring 7,8-dihydro-8-oxoguanine (oG), 5,6-dihydrothymine (DHT), 5-hydroxyuracil (hU), 5-formyluracil (fU), AP site, uracil (U), SSB or one nucleotide gap (Gap) in various configurations. The results have been summarized in recent reviews (1,2,21). They show that in MDS the BER pathway is compromised compared with isolated lesions, leading to the accumulation of repair intermediates that, by interfering with subsequent repair steps, can result in the persistence of nearby lesions that on replication lead to mutations.
During the course of BER of MDS in plasmids, incision events in opposite strands can lead to the formation of double-strand breaks (DSB) with subsequent loss of plasmid viability. MDS composed of bistranded Us, AP sites or Gap/AP are readily converted to DSB in bacteria and yeast (21–26). Plasmids containing U and thymidine glycol (U/Tg) also lose viability in E. coli (27). In contrast, the processing of MDS containing oG on one strand and either hU, DHT, Tg or U on the other strand results in a limited number of DSB, if any (25–29). In human cells, MDS composed of two opposed U do not seem to induce DSB, unlike the MDS carrying tetrahydrofurans, AP site analogs (30,31). In some cases, DSB formed at MDS in mammalian cells can be repaired by non-homologous end-joining and result in the appearance of large deletions (31). To explain the differences described above, it has been proposed that the kinetics of base damage excision/incision by BER DNA n-glycosylases determine DSB formation (26).
Another consequence of a delay in the repair of lesions within MDS is an enhanced targeted mutagenesis. Results in E. coli showed that targeted mutagenesis at MDS is increased in comparison with single lesions (21,25,27–29,32,33). This seems also to be the case in yeast and human cells (Sage, unpublished results). Additionally, the mutation frequency (MF) at MDS decreases with increase of inter-lesion spacing (32).
Taken together, these results suggest that the sequence in which lesions are processed is critical in defining whether a particular MDS will be efficiently repaired or lead to DSB and death or to mutagenesis. The aim of this work was to investigate how important is the hierarchy of the lesion processing in the biological outcome of MDS. With that purpose, we analyzed in E. coli the processing of MDS composed by three base lesions with very different kinetics of excision by cellular DNA n-glycosylases, U, hU and oG (26). U and oG were placed on the same strand and hU on the other. The role of specific DNA n-glycosylases in the handling of MDS by the cells was analyzed by using mutant strains defective in those glycosylases. This work emphasizes the deleterious biological consequences of U or AP site within oxyclusters and provides an explanation for mutagenesis at MDS.
MATERIALS AND METHODS
Bacterial strains
AM101 [lacZ(Am) CA7020 lacY1 hadR hsdMΔ(ara ABC-leu) 7679 galU galK(Am) galE rpsZ thi] E. coli (34) were provided by Dr. M. Seidman (National Institute of Aging, NIH Baltimore, MD, USA). The mutants AM101Δnth (nth::KanR), AM101ΔnthΔnei (nth::KanR, nei::CamR) and AM101Δung (ung::KanR) were constructed by P1 transduction. BH720 is a Δfpg (fpg::KanR) derivative of AB1157.
Oligonucleotides
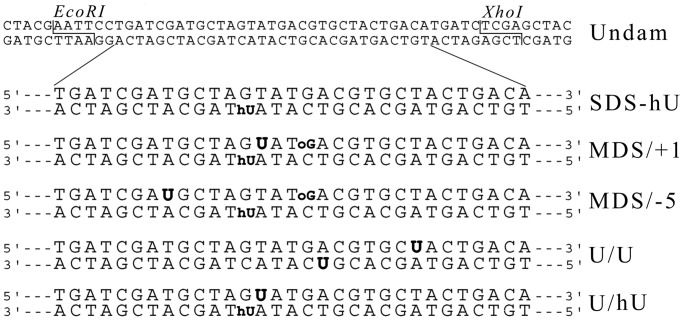
Unmodified oligonucleotides and oligonucleotides carrying U, 8-oxoguanine (oG) and 5-hydroxyuracil (hU) were purchased from Sigma-Aldrich and purified on a 20% denaturing polyacrylamide gel. These oligonucleotides were used to build the various MDS and control constructs indicated in Figure 1. Two restriction sites, EcoRI and XhoI, were placed in 5′ and 3′ ends of the duplexes, respectively, for insertion into the pSP189 plasmid (35).
Figure 1.
Duplexes carrying damaged sites used in this study. Oligonucleotides carrying oG, hU and U. The modified bases are indicated in bold. oG was separated by 3 bp from hU located on the complementary strand in MDS/+1, MDS/−5. Uracil was positioned at +1 position relatively to hU in MDS/+1 and at −5 position in MDS/−5. The control U/U oligonucleotide consists of two bistranded Us separated by 5 bp. Undamaged and MDS-containing 56-mers duplexes harbor EcoRI and XhoI restriction sites at each extremity as indicated.
Annealing of oligonucleotides and duplexes digestion
Equimolar quantities (200 pmol of each) of complementary oligonucleotides were annealed in NEBuffer 3 (New England Biolabs) in a total volume of 20 µl, by heating to 90°C and slowly cooling to 20°C. The obtained DNA duplexes were then simultaneously digested with EcoRI (500 U) and XhoI (500 U) in NEBuffer 3 supplemented with 100 µg/ml BSA, in a total volume of 100 µl, for 20 h at 37°C. The digested duplexes were purified using QIAquick Nucleotide Removal Kit (Qiagen) and quantified by UV absorbance.
Insertion of duplexes into the plasmid pSP189 and E. coli transformation
The pSP189 plasmid (100 µg) was digested with 1000 U of EcoRI and 1000 U of XhoI (New England Biolabs) in NEBuffer 4 in a total volume of 180 µl, for 2 h 30 min at 37°C. The digested plasmid was purified by QIAquick PCR Purification Kit (Qiagen), and after ethanol precipitation, the plasmid was resuspended in H2O. Concentration of linearized plasmid was established by UV absorbance. The efficiency of the digestion was tested by religation of 500 ng of double-digested pSP189, using 400 U of T4 DNA ligase (New England Biolabs) in 10 μl at 16°C overnight, and transformation of E. coli AM101 cells with ligation mixture. Digestion was considered as efficient when <15 transformant colonies were obtained.
Digested duplexes (10.8 pmol) were ligated into linearized pSP189 plasmid (0.9 pmol) using 600 U of T4 DNA ligase (New England Biolabs) in supplied buffer in a total volume of 15 µl at 16°C for 16 h. Electrocompetent AM101 E. coli cells were transformed with 10 ng of ligation mixtures, using Electroporator 2510 apparatus (2.5 kV, 600 Ω, 10 µF) (Eppendorf). Transformants were selected onto LB agar plates containing ampicillin (100 μg/ml).
Relative transformation efficiency (RTE) calculation
Formation of DSB during MDS processing was estimated from the relative transformation efficiency. RTE was calculated as the frequency of transformants with MDS-carrying plasmid divided by the frequency of transformants with plasmid carrying the control undamaged duplex.
Sequence analysis for mutations
To analyze processed pSP189 by sequencing, obtained colonies were amplified in 3 ml of LB with ampicillin (100 μg/ml), and then plasmid was extracted and purified using NucleoSpin kit (Macherey Nagel). Sequencing was performed by Eurofins MWG/operon, using primer 5′-GGCGACACGGAAATGTTGAA-3′ purchased from Sigma-Aldrich.
Statistical analysis of data
For calculation of the statistical significance of obtained results, we used the 2-tailed Fisher’s exact test available online at http://www.langsrud.com/stat/fisher.htm. This test is adapted for comparison of two small samples.
Excision/incision assay by whole-cell extracts
Whole-cell extracts were prepared from transformed human fibroblasts MCR5 VI as previously described (15). Oligonucleotides were 5′-32P-end-labeled by T4 polynucleotide kinase (5). Unincorporated nucleotides were removed using a ProbeQuant G-50 Micro Columns (GE Healthcare). After phenol/chloroform extraction, the radiolabeled oligonucleotides were hybridized with equimolar quantity of radiolabeled complementary strand or 1.25-excess of the non-radiolabeled complementary strand in hybridization buffer [140 mM NaCl, 10 mM Tris-HCl (pH 8), 1 mM ethylenediaminetetraacetic acid (EDTA) (pH 8)] by heating 5 min at 95°C and slow cooling to room temperature. The hybridization efficiency was verified by migration of DNA samples on native 12% polyacrylamide gel [19:1, acrylamide/bisacrylamide (w/w), 100 mM Tris borate and 1 mM EDTA (pH 8)].
The cleavage assay mixtures (14 µl final volume) contained 200 fmoles of radiolabeled double-stranded oligonucleotides and 30 μg of proteins from human whole-cell extract, in the incubation buffer [20 mM Tris-HCl (pH 7.6), 140 mM NaCl, 4 mM EDTA (pH 8) and 8% glycerol]. The reactions were performed at 37°C for various time and stopped by addition of stop-buffer [0.5% sodium dodecyl sulphate (SDS) and 50 mM EDTA (pH 8)] and by incubation with proteinase K (0.8 mg/ml, Eurobio) for 1 h at 37°C. After phenol/chloroform extraction and ethanol precipitation, samples were electrophoresed on a 12% denaturing polyacrylamide gel for 1 h at room temperature at 18.75 V/cm. The reaction products were visualized and quantified using Molecular Dynamics Storm 820 PhosphorImager and ImageQuant 5.2 software (Molecular Dynamics). The cleavage efficiency was expressed as the percentage of the amount of cleaved molecules to the total amount of (cleaved plus uncleaved) molecules.
RESULTS
To analyze the hierarchy in the processing of lesions within MDS and to explore the impact of U or an AP residue on the outcome of such MDS, we built duplex oligonucleotides harboring clustered damage sites containing three lesions displaying different kinetics of excision by DNA n-glycosylases both in vitro and in vivo (17,20,25,26). The MDS-carrying duplexes were then inserted into a plasmid and introduced into E. coli. The formation of DSB as repair intermediate was assessed by determining plasmid survival and mutagenesis at MDS by directly sequencing the propagated plasmid without selection for mutations. The lesions were two oxidized bases, hU and oG, which were placed in opposite strands and spaced by 3 bp, and the fast excised U, which was situated in the oG-carrying strand at positions +1 (MDS/+1) or −5 (MDS/−5) relatively to hU (Figure 1). The U residue is rapidly converted into AP site, a replication blocking DNA lesion, which is particularly cytotoxic if left unrepaired (36,37). Previous work showed that excision/incision at oxidized bases is slower than at U and AP sites, and that cleavage occurs more rapidly at hU than at oG (17,26).
hU is predominantly repaired by endonuclease III in E. coli
Although DNA n-glycosylases involved in BER of oG and U in bacterial cells have been identified, the pathway involved in the in vivo repair of hU, a product of oxidative deamination of cytosine, remains uncertain. To address this point, SDS-hU, carrying hU as single lesion (Figure 1), was inserted into the plasmid pSP189, which was then transformed into E. coli cells. The processed plasmid was recovered from transformants and sequenced. In wild-type (WT) cells, the MF at a single hU was 5% as shown in Table 1. Even though hU has a strong miscoding potential and leads to a MF of 83% when placed in a single-stranded vector (38), our results demonstrate that hU can be efficiently repaired in WT cells when present in double-stranded DNA.
Table 1.
Description of mutations found in studied duplexes within pSP189 plasmid processed in E. coli wild-type strain or strains deficient for various BER DNA n-glycosylases
| Duplex | Strain | Number of sequenced clones | Mutated clones |
|||
|---|---|---|---|---|---|---|
| hUa | oGa | Ua | Non-targeted mutationsa | |||
| Undam | WT | 66 | 12 (18%) | |||
| SDS-hU | WT | 94 | 5 (5%) | 7 (7%) | ||
| Δnth | 92 | 16 (17%) | 3 (3%) | |||
| ΔnthΔnei | 93 | 17 (18%) | 5 (5%) | |||
| Δung | 90 | 3 (3%) | 6 (7%) | |||
| Δfpg | 96 | 1 (1%) | 11 (11%) | |||
| ΔmutS | 94 | 3 (3%) | 10 (11%) | |||
| MDS/+1 | WT | 63 | 61 (97%) | 1 (2%) | 10 (16%) | |
| Δung | 86 | 5 (6%) | 7 (8%) | 3 (3%) | ||
| MDS/−5 | WT | 73 | 61 (84%) | 3 (4%) | 3 (4%) | 4 (6%) |
| Δung | 90 | 2 (2%) | 4 (4%) | 1 (1%) | 8 (9%) | |
aNumber and proportion (%) of clones with mutations targeted at hU, oG, U or non-targeted.
Mutations located at a damaged base or adjacent bases were considered as targeted mutations. One transformant harboring a 7-bp deletion covering U and hU within MDS/+5 in WT cells was counted as targeted mutation at hU and at U. Two transformants harboring simultaneously targeted mutations at hU and oG within MDS/+5 in WT cells were counted as targeted mutations at hU and oG. Clones with targeted mutations may also contain untargeted mutations. One clone of 63 and 9 clones of 73 were found without mutations at MDS/+1 and MDS/−5 in WT cells, respectively.
The base hU can have the same coding properties as thymine, and thus the hU·G base pair is like a G·T mismatch, the preferential substrate for the mismatch repair (MMR) in E. coli (39,40). Recent work by Zlatanou et al. (41) suggested that tandem lesions carrying hU can be recognized by the MMR proteins in human cells. We thus examined the recognition of a hU·G mismatch by purified E. coli MutS protein using electrophoretic mobility shift assay, and showed that hU·G mismatch in SDS-hU bound MutS protein as efficiently as the G·T mismatch (89 and 91% of DNA-MutS complex, respectively, Supplementary Figure S1 in Supplementary Information). Based on this observation, we reasoned that MMR could be involved in the repair of hU·G mismatch present in the plasmid and therefore analyzed mutagenesis in ΔmutS strain. However, we did not observe any statistically significant change (P = 0.7) in the MF induced by SDS-hU compared with the WT cells (Table 1). Even though hU·G base pair can be recognized by MutS, MMR is not essential for the removal of hU.
We then explored the repair of hU by BER by transforming DNA n-glycosylase mutant strains with the plasmid harboring the SDS-hU (Table 1). Although the MF at hU in Δung or in Δfpg did not statistically differ from that in WT cells (P = 0.7 and 0.1, respectively), that in Δnth or ΔnthΔnei cells was three times higher (P < 0.05). We conclude that EndoIII (Nth) protein, but not EndoVIII (Nei), plays an important role in the repair of hU by BER in E. coli. However, other repair proteins are likely used as backup in the absence of Nth protein, as an MF of 40–50% is expected if hU is not removed at all. In all the strains examined, hU·G→T·A was the predominant type of mutation observed at hU (Supplementary Figure S2).
U-oG/hU MDS are readily converted to DSB
To gain insight into the processing of three lesions (U-oG/hU) MDS, the duplexes shown in Figure 1 were inserted into pSP189 plasmid and the resulting constructs were used to transform WT and DNA n-glycosylases-deficient cells. We first assessed the DSB formation by calculating the relative transformation efficiency (RTE) that compares the transformation efficiency of a plasmid containing damaged duplexes with that of a plasmid containing an undamaged but otherwise identical oligonucleotide. Because the transformation efficiency of a linear plasmid is extremely low in E. coli, RTE for plasmids carrying MDS is a good estimate of DSB formation (42).
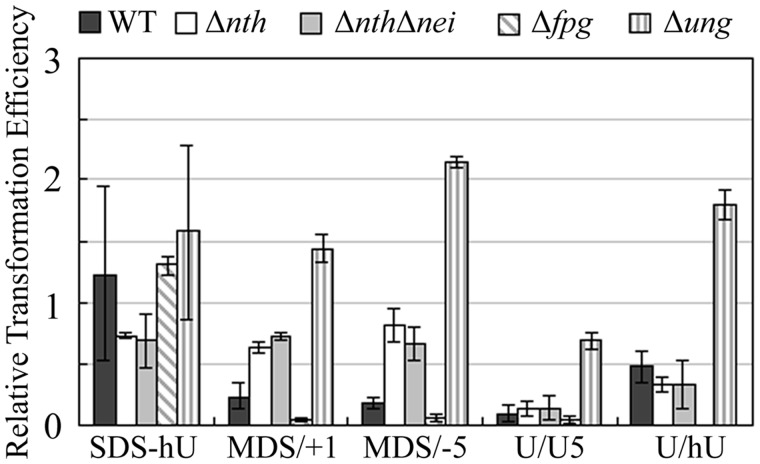
As expected, in WT cells, the RTE for the plasmid containing SDS-hU was close to 1 (Figure 2), whereas that for the plasmid harboring the well-known DSB-inducing U/U duplex was very low (0.09). Interestingly, the RTE for MDS/+1- or MDS/−5-carrying plasmids was relatively low (0.24 and 0.17, respectively), indicating that MDS/+1 and MDS/−5 are predominantly converted into DSB during the repair process. The absence of a significant difference in RTE between MDS/+1 and MDS/−5 indicates that the position of U in the studied MDS has little impact on DSB induction. Unexpectedly, the RTE for U/hU is significantly higher than for U-oG/hU clusters. This implies a role for the presence of oG in DSB formation in the MDS constructs. Thus, unlike U/oG clusters (29), U-oG/hU MDS and U/hU, to a lesser extent, are efficiently converted into DSB.
Figure 2.
RTE of plasmid carrying MDS in WT cells and in cells deficient for various DNA n-glycosylases. Wild type, Δnth, ΔnthΔnei, Δung and Δfpg electrocompetent E. coli cells were transformed with replicative pSP189 plasmid, carrying SDS-hU, MDS/+1, MDS/−5 or U/U. RTE represents the ratio of the number of colonies relative to that with plasmid carrying undamaged oligonucleotide. Error bars represent the standard deviation for at least three independent experiments.
In an attempt to understand the mechanisms underlying the induction of DSB in MDS, we searched for the DNA n-glycosylases involved in the induction of DSB by analyzing plasmid survival in bacterial strains deficient for DNA N-glycosylases EndoIII (Nth), EndoVIII (Nei), Fpg or Ung, which have, as substrates, the lesions present in the MDS. As expected, RTE of plasmid carrying a single hU remained close to 1 in all studied strains (Figure 2). Like in WT cells, MDS carrying two closely located U in opposite strands (U/U) were readily converted into a DSB (RTE in the 0.04–0.15 range) in Δnth, ΔnthΔnei and Δfpg strains. However, DSB induction at U/U is largely diminished in Δung cells lacking U DNA n-glycosylase (RTE = 0.75) (Figure 2). For plasmids carrying MDS/+1 or MDS/−5, the RTE increased in Δnth, ΔnthΔnei and Δung cells in comparison with WT cells, and reached that for SDS-hU, suggesting that initiation of repair at the MDS by those glycosylases contributes to DSB formation. Surprisingly, in Δfpg cells (BH720), plasmids containing MDS/+1, MDS/−5 or U/U displayed an extremely low RTE (below those in WT AM101 cells). This probably reflects a more efficient BER in the AB1157 genetic background. The absence of RTE increase in Δfpg cells demonstrates that Fpg is not directly involved in the DSB formation at MDS/+1 and MDS/−5 in the WT cells. Taken together, these data demonstrate that MDS-containing U-oG/hU are readily converted to DSB, likely due to the fast and simultaneous cleavage at U and hU by Ung and EndoIII, respectively.
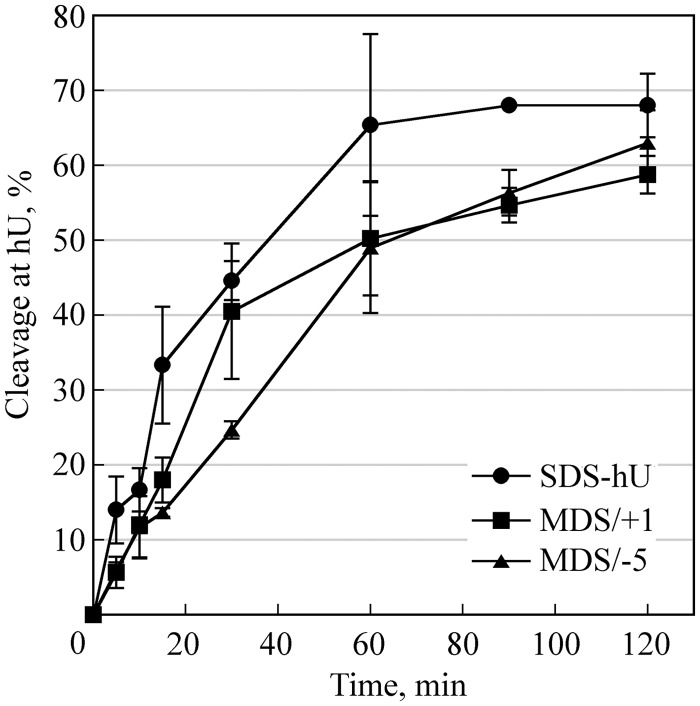
We examined the cleavage efficiency at hU and U in MDS/+1 and MDS/−5 using human whole-cell extract. Figure 3 shows that hU is efficiently cleaved, although at a slightly slower rate in comparison with single hU. The excision/incision at U in MDS/+1 and MDS/−5 is complete and fast (Supplementary Figure S3 in Supplementary Information) and, as expected, the rate of cleavage is higher than that at hU and similar to that of U/U by yeast extracts (26). Because the BER repair is well conserved from bacteria to mammalian cells, the high cleavage efficiency observed here at U and hU is consistent with the elevated induction of DSB at U-oG/hU in E. coli.
Figure 3.
Cleavage efficiency at hU by human cell extracts. 32P-labeled duplexes SDS-hU, MDS/+1 or MDS/−5 were incubated with 30 µg of proteins from whole-cell extract at 37°C for various periods and separated on 12% denaturing polyacrylamide gels. Data represent the means of at least three independent experiments. Preparation of whole-cell extracts and analysis of cleavage activity were performed as described in (15).
Surviving U-oG/hU MDS-carrying plasmids display an elevated mutagenesis targeted at hU
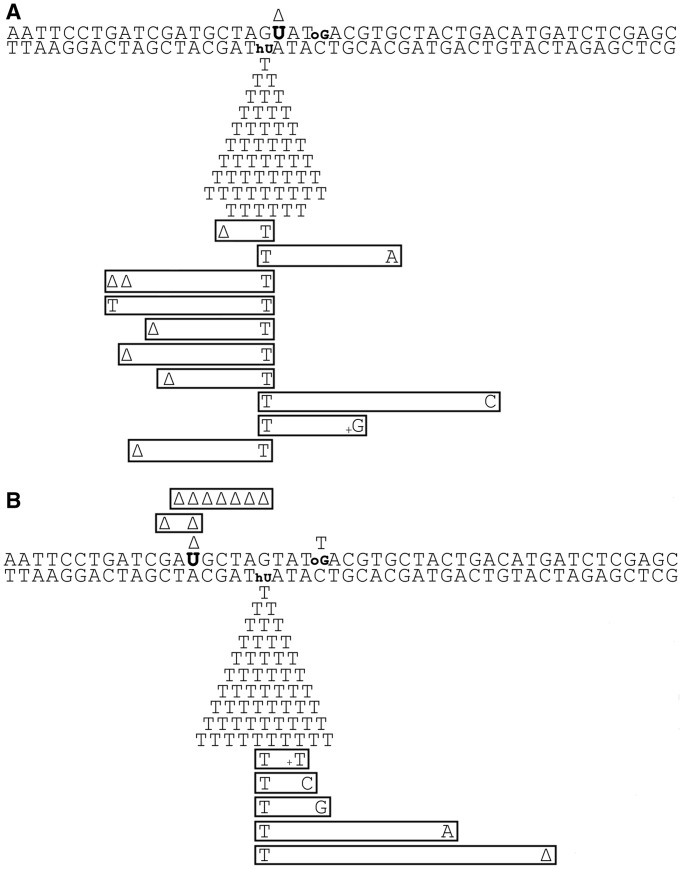
While in the WT cells, MDS/+1 and MDS/−5 are predominantly converted to a DSB, a fraction (<20%) of MDS-carrying plasmids escaped DSB formation and replicated. Sequencing of the surviving plasmids showed that in all cases a relatively high level of non-targeted mutations, i.e. which could not be associated with the modified bases, was detected (Table 1). Because the levels of non-targeted mutations were similar in the control undamaged duplex (Supplementary Figure S4), with no statistically significant difference (P > 0.05) among duplexes, or strains (Table 1), these base substitutions could be due to the presence of modifications on the oligonucleotides possibly generated during chemical synthesis. More interestingly, the table reveals that almost all the plasmids recovered from the transfection with plasmids containing MDS/+1 or MDS/−5 carried mutations at the site of the lesions within the MDS (98 and 88%, respectively). Moreover, the mutation spectra at MDS/+1 and MDS/−5 displayed in Figure 4A and B, show that a large majority of mutations (97 and 84% of the mutated clones, respectively) were point mutations targeted at the original hU position (Table 1). This unexpectedly high mutagenesis contrasts with the 5% MF found for the single hU. In fact, only one G·C→T·A transversion targeted at oG, within MDS/−5, and 1 bp deletion associated with U in both MDS were recovered. The low number of mutations at these lesions’ positions is consistent with the fact that the MF at single oG in WT E. coli cells is expected to be 1–2% (29,32,33,43,44) and that U is paired with adenine in MDS/+1 and MDS/−5, and therefore not expected to lead to mutations (45). Among the mutations at hU in MDS, most were hU·G→T·A substitutions (Figure 4), consistent with the mutation spectra obtained for SDS-hU in various strains (Supplementary Figure S2 and Table 1). Moreover, excision of hU is almost as efficient in MDS/+1 and MDS/−5 as in SDS-hU (Figure 3), suggesting that induction of mutagenesis at SDS-hU and at the studied MDS follows different mechanisms.
Figure 4.
Mutation spectra induced at MDS/+1 (A) and at MDS/−5 (B) in E. coli WT cells. DNA lesions are indicated in bold. Induced mutations are indicated below and above the duplex sequence. Each symbol corresponds to a base change in one mutated clone. Multiple mutations in the same clone are boxed. Delta represents 1 bp deletion.
The mutation frequencies at the hU site in MDS/+1 and MDS/−5 in WT cells, which were in the range of 84 and 97%, raise the question of their origin. Such mutation level cannot be related to the MF at hU in SDS-hU in either Δnth or ΔnthΔnei cells (17 and 18%, respectively), but rather to the 85% observed at single hU in a single-stranded vector (38). Most importantly, the fact that the mutation frequencies at hU found in MDS were higher than 50%, and that there was an almost complete absence of mutations derived from lesions on the opposite strand, indicates that most surviving clones come from the replication of hU-carrying strand. Therefore, this result strongly suggests the loss of U-oG-carrying strand during MDS processing.
The fast excision of U in U-oG/hU MDS induces strand loss and leads to high mutagenesis at hU
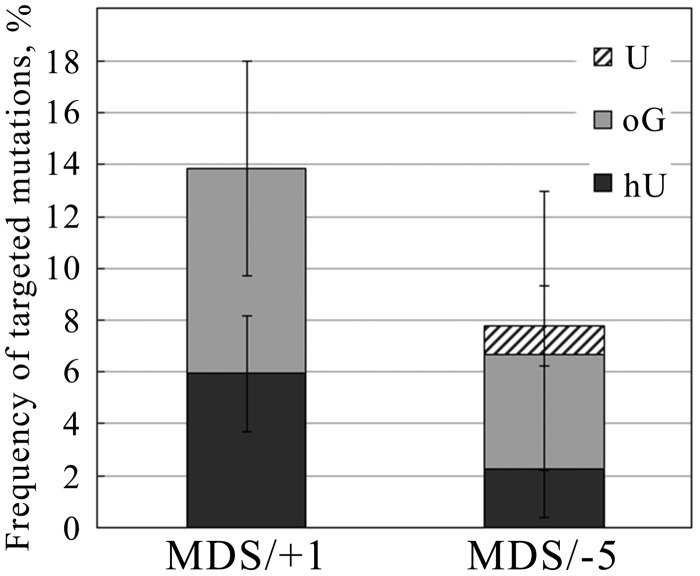
It has been reported that excision of U kinetics is faster than that of oG (26,46,47). Therefore, excision of U is likely to be at the origin of the loss of the U-oG-carrying strand. To test this hypothesis, we analyzed mutation frequencies and spectra at MDS/+1 and MDS/−5 in a Δung strain. Interestingly, in Δung cells, the MF at MDS/+1 and MDS/−5 went down to 14 and 8% (Figure 5), respectively, in comparison with 98 and 88% in WT cells (Table 1). Moreover, in Δung cells, mutation frequencies at hU within MDS/+1 and MDS/−5 were 6 and 2%, respectively, the level observed at single hU in WT cells (5%). In addition, mutation frequencies at oG were 8 and 4%, respectively, thus higher than that in WT cells and that expected for single oG. Therefore, in Δung cells, hU in MDS/+1 and MDS/−5 is repaired as efficiently as the single lesion, but repair at oG is somehow reduced. The decrease to <50% of the MF at hU within MDS in the absence of U excision and the occurrence of mutagenesis at oG indicate that both strands can be replicated in a Δung strain, in contrast with what happened in WT cells. Conversely, in WT cells, the strand carrying U-oG is lost. This is the first demonstration of strand loss during the processing of MDS in WT cells. Altogether, these data suggest that in WT cells the fast excision of U in MDS/+1 and MDS/−5 is a key step leading to the loss of the strand carrying U and oG, and consequently to exclusive and elevated mutagenesis at hU.
Figure 5.
Targeted mutation frequencies at MDS/+1 and MDS/−5 in Δung strain. Processed plasmid carrying MDS/+1 or MDS/−5 was recovered from surviving bacterial clones and sequenced. Clones with mutations targeted at modified bases were considered as mutated clones. Non-targeted mutations were not considered in the calculation of MF at MDS/+1 and MDS/−5.
DISCUSSION
We have previously reported a strong hierarchy in excision/incision by cell extracts of oxidized bases within 2–5-lesion clustered DNA damage (15,17). These results were consistent with DSB induction at MDS in yeast (26). In the present study, we investigated in E. coli the processing of MDS composed of U-oG/hU, three lesions presenting rather different kinetics of excision by cellular DNA n-glycosylases (46,47). Uracil is rapidly converted to an AP site, which is itself a frequent radiolesion in MDS (6). We found that the processing of U-oG/hU MDS largely leads to DSB formation (either directly or through a replication block) as seen from plasmid inactivation, whereas the low amount of surviving plasmids are almost all mutated at hU position, with virtually no occurrence of error-free repair. These results emphasize the importance of the sequence of repair events in the fate of MDS. Moreover, they provide the first demonstration of strand loss of a plasmid triggered by the processing of MDS containing U or AP site and consequently the first indication of such a deleterious role of U or AP site within oxyclusters.
Three-lesion clusters comprising U (AP site) and oxybases are more readily converted into DSB than a related two-lesion cluster
It has been observed that in E. coli a DSB is efficiently formed at U/U, AP/AP, U/AP, F/F (clusters composed by tetrahydrofurans), Gap/U or Gap/AP bistranded clusters (20,21,23,24). Albeit to a lower extent, a DSB is also formed when U, AP site or a gap are associated with DHT, another modified pyrimidine, on the opposite strand (21). In contrast, DSB formation was not significantly detected for clusters composed of multiple oG, oG/DHT and oG/U in E. coli (21,23,28,29) or of oxidized pyrimidines/oG in yeast (26). Here, we show that DSB is formed at high frequency (in ∼80% of the cases) in three-lesion oxyclusters containing a single U (AP site), and that it results from the cleavage at U and the oxidized pyrimidine (hU). These results are in agreement with the fact that oxidized pyrimidine-specific DNA n-glycosylases are the first to cleave at MDS composed of mixed oxidized bases (17,25,26). They are also consistent with the fact that repair intermediates such as a SSB or a gap generated nearby opposite oG retard the repair of the oxidized guanine both in eukaryotes and in prokaryotes (15,17,48–51). Although DSB formation at our U-oG/hU MDS was not totally unexpected, as hU is efficiently cleaved by human cell extracts within MDS/+1 and MDS/−5 or when placed nearby an opposite gap (17), surprisingly, DSB are not as efficiently produced in bistranded U/hU cluster. Because U is excised much more rapidly than hU or oG (26), we suspect that in the U-oG/hU MDS, excision/incision at U occurs first, generating an SSB or a gap, the repair of which is not completed before excision/incision at hU. However, even though our results do not indicate any cleavage at oG in MDS/+1 and MDS/−5 (Figure 2), the presence of oG retards or prevents gap filling that occurs to some extent at U/hU. These observations are in full agreement with Cunniffe et al. (52) who found that in tandem clusters, the efficiency of rejoining at an incised AP site by mammalian cell extract is reduced at least 10-fold by the presence of oG. In Δnth or ΔnthΔnei cells, a back-up repair pathway may operate at hU, for instance Nfo protein through nucleotide incision repair (NIR) (53), and generate a toxic intermediate, reflected by the lower RTE for SDS-hU and U/hU compared with that in WT cells. Altogether, our data show that delayed or impaired repair at three-lesion clusters does not necessarily prevent the DSB formation and further underline the role of the hierarchy in the processing of individual lesions within MDS on biological consequences of these complex lesions.
Uracil (AP site) initiates strand loss/replication fork collapse in three-lesion cluster
An unexpected result arose from the analysis of mutagenesis in the fraction of clones that survived after transfection with MDS/+1- or MDS/−5-carrying plasmid. The MF in WT cells was extremely high and almost exclusively targeted on one strand, at hU (MF ∼90%). Even though repair of individual lesions at MDS can be strongly impaired, the targeted mutation frequencies have never been reported yet to exceed 50%, even in BER-deficient cells (20,25,27–29,33). Furthermore, MDS containing U (AP site) and thymidine glycol, a replication-blocking lesion, have been shown to lead to the loss of colonies after transfection but not to mutagenesis (27). In any case, neither U, hU nor oG is known as replication blocking lesions. Moreover, mutations at MDS can be recovered on both strands (33; Kozmin and Sage, unpublished results), consistently with the fact that in normal conditions both strands serve as template for DNA replication. Therefore, the exclusive and elevated mutagenesis observed in this study at hU shows that most surviving colonies come from replication of the hU-carrying strand. As discussed in Shikazono et al. (25), an MF >50% (90% in our case) indicates the loss of the other strand.
Strand loss has been observed for plasmids carrying unrepaired bulky lesion (54). It was also reported that in Ung-proficient cells, the plasmid strand carrying multiple Us is rapidly hydrolyzed, leading to strand loss (55). Additionally, Nogushi et al. (33) observed an almost exclusive targeting of mutations at oG located on the strand carrying the single lesion at a Gap-oG/oG cluster in ΔfpgΔmutY strain, and suggested strand loss. Like Cunniffe et al.(52), they also proposed that the very low MF observed at tandem GAP/oG in a ΔfpgΔmutY strain is owing to strand loss. Meanwhile, the MF at tandem AP/oG in WT cells is at least 10 times higher than at single oG, which would not occur in case of strand loss (52). In support of a mutation pathway involving strand loss at our U-oG/hU clusters in WT cells, we show here that the MF at hU within U-oG/hU MDS in a strain deficient for U excision (Δung) is drastically reduced to values <10%, whereas in WT cells, >80% of the recovered clones were mutated at hU (Figure 5). Moreover, in the Ung-deficient strain, mutations are recovered on both strands, indicating that in the absence of U-excision within U-oG/hU MDS, both strands were replicated. These observations confirmed our hypothesis that in WT cells, the U-oG-carrying strand in our U-oG/hU MDS is lost and that this loss strictly depends on the cleavage at U. Repair synthesis starting at the incision left by the U excision could operate in conjunction with strand displacement. However, the same MF is observed whether U is placed upstream or downstream of hU. The more likely explanation for the loss of U-oG-carrying strand is that in the MDS context, once U is excised and the resulting AP site incised, the next steps of BER are impaired by the proximity of other lesions, i.e. hU on the opposite strand and oG on the same strand, as shown for tandem AP-oG clusters (52). In favor of this mechanism, in the rare surviving colonies in Δfpg cells that were sequenced, 7 of the 13 relevant transformants were mutated and exhibited 3 hU to T mutations, 2 G to T mutations and 2 deletions at oG. In WT cells, the strand loss probably does not rely on cleavage at oG by Fpg, but rather by stalling of Fpg at oG. The replication machinery may collide with the complex formed by repair proteins recruited at the repair intermediate SSB and at unrepaired hU and oG, causing the collapse of the replication fork. Similarly, at the chromosome level in prokaryotic or eukaryotic cells, a replication fork may collapse or stall if it encounters a repair complex blocked at a repair intermediate.
The mutagenic potential of hU was previously shown in vitro (56) and in vivo (38). Using a single-stranded vector, where hU cannot be removed by DNA n-glycosylases, Kreutzer and Essigmann reported an MF of 83% at this lesion (38). Replication through hU mostly yielded hU to T mutations. Our results confirm the high mutagenic capacity of hU and give some hints on the mechanisms for its repair in E. coli. We showed (Table 1) that the presence of Nth protein protects partially from mutation at single hU in double-stranded vector, suggesting the existence of other repair pathways able to act on this lesion. Fpg (Sage, unpublished results), MMR or NIR could serve as backup for Nth. In addition, our results show that there is no repair pathway that removes the misincorporated base opposite hU, unlike MutY for the misincorporated A opposite oG.
Role of U or AP site in the biological consequences of MDS
Our data show that all the events that occur at U-oG/hU MDS, i.e. extensive formation of DSB, collapse of replication fork, high MF on one strand, can be attributed to the initiation of repair at the U. In the absence of U excision (Δung strain), DSBs were not detected, and the MF at hU was only 2–6%, which is about that observed at single hU in WT cells. This showed that in this context, hU is as efficiently repaired as a single hU. In contrast, inactivation of ung results in mutation frequencies at oG significantly higher than those expected for single oG (4–8% versus 1–2%). This indicates that when the loss of U-oG-carrying strand does not occur, hU and oG are repaired sequentially, and that the excision of hU leads to retardation of oG repair, as reported previously (12,15,17,20,50). Thus, the processing of U or AP site at MDS is the crucial and initial event that triggers DSB formation and DNA strand loss/replication fork collapse, with mutational consequences.
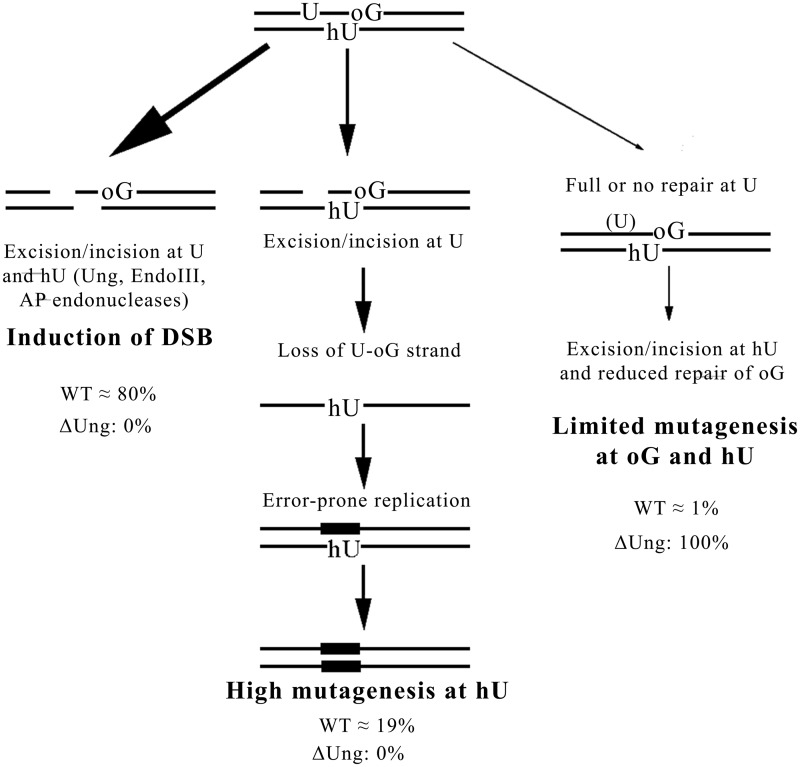
Such data lead us to propose a model for the processing of MDS composed of oxidized purines and pyrimidines and one U or AP site and in which U and an oxidized pyrimidine are on opposite strands (Figure 6). According to this model, in WT cells, the majority of MDS is converted to deleterious DSB. In this pathway, the activities of the AP endonuclease following that of Ung and that of EndoIII (or oxidized pyrimidine DNA n-glycosylases) on their cognate lesions trigger DSB formation. The presence of two Us/AP sites on opposite strands would increase the extent of DSB. Conversely, this pathway is inhibited in cells lacking DNA n-glycosylases or AP endonucleases. However, in WT cells a small fraction of MDS escapes conversion to the DSB, and the processing of lesions within MDS leads to strand loss. As most DNA n-glycosylases are unable to excise base damage on single-stranded DNA, this pathway is a major actor in mutagenesis. Nevertheless, the mutagenesis rate will depend on the capacity of the lesion left unrepaired on the remaining strand to be bypassed in error-free or error-prone manner. It may range from 90% for hU to no mutation for DHT, or to ∼4% of mutagenesis at oG on single-stranded DNA (57). Finally, a small fraction of MDS (<1%) is processed without DSB formation or mutagenesis induction. This implies sequential repair of each of the comprised lesions. In this last process, the interlesion spacing should play a role. However, in a strain deficient for Ung, the inductions of DSB and of strand loss do not occur, unveiling a third pathway potentially involved in the processing of such MDS.
Figure 6.
The model proposed for processing of MDS composed by two oxidized bases and U. We propose that U-oG/hU MDS are processed by three pathways. A major pathway (∼80%) converts MDS into DSB in WT cells owing to the excision/incision at U and hU by Ung, EndoIII (Nth), AP endonucleases, whereas it does not occur in Δung cells. This was determined from the fraction of viable colonies (∼20%) after transformation with MDS-containing DNA. For ∼19% of the events in WT cells, the fast excision/incision at U produces an SSB, which, in the presence of surrounding lesions, is not further repaired and leads to the loss of the U-oG-strand. Error-prone replication of hU-carrying strand thus occurs and leads to elevated mutagenesis at hU (reaching almost 100% of surviving clones). This pathway is not activated in Δung strain. In rare cases (∼1% of total events in WT cell), when the full repair of U is extremely fast (WT) or does not occur (in Δung), the hU is repaired before oG and the excision/incision at hU partly inhibits or retards repair of oG. Consequently, the mutagenesis rate at hU within MDS is as at single hU (in WT cells), and mutagenesis at oG within studied MDS is slightly increased in comparison with that at single oG. This third pathway represents the only possible pathway in Δung cells. It should be slightly more represented in the processing of MDS/−5 than for MDS/+1 because of greater interlesion spacing.
In conclusion, this work further highlights the importance of the first event occurring at a complex MDS and of the hierarchy in the processing of lesions at MDS, on the fate of the MDS, DSB formation or mutation induction. It provides evidence for replication fork collapse being a relatively general process and a major mechanism for mutagenesis at complex MDS. In eukaryotes, a collapsed or stalled replication fork is mainly resolved by homologous recombination, which can cause chromosome rearrangements and translocations (58). Alternatively, inability to replicate part of the genome may lead to cell death. Our data also reveal that, depending on the base lesions that constitute MDS, mutation induction can be extremely high. The consequences of such three damaged bases MDS generated in a mammalian chromosome, would be, with high likelihood, large deletions, insertions, translocations and point mutations, if not cell death.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [59].
FUNDING
Centre National de la Recherche Scientifique (CNRS) (to E.S., J.P.R. and Y.S.); Institut Curie (to E.S. and Y.S.); Electricité de France (EDF) (to E.S. and J.P.R.); Agence Nationale de la Recherche [ANR-09-PIRI-0022-01 to E.S. and J.P.R.]. Funding for open access charge: Institut Curie.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. M. Seidman for providing us AM101 E. coli strain and Dr. A. El Marjou for BL21 (λDE3 Plyss) E. coli strain. We would also like to thank Dr. J.-M. Malinge for providing us the pTX412 expression vector.
REFERENCES
- 1.Sage E, Harrison L. Clustered DNA lesion repair in eukaryotes: relevance to mutagenesis and cell survival. Mutat. Res. 2011;711:123–133. doi: 10.1016/j.mrfmmm.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eccles LJ, O’Neill P, Lomax ME. Delayed repair of radiation induced clustered DNA damage: friend or foe? Mutat. Res. 2011;711:134–141. doi: 10.1016/j.mrfmmm.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat. Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 4.Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int. J. Radiat. Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 5.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered damages and total lesions induced in DNA by ionizing radiation: oxidized bases and strand breaks. Biochemistry. 2000;39:8026–8031. doi: 10.1021/bi9927989. [DOI] [PubMed] [Google Scholar]
- 6.Georgakilas AG, Bennett PV, Sutherland BM. High efficiency detection of bi-stranded abasic clusters in gamma-irradiated DNA by putrescine. Nucleic Acids Res. 2002;30:2800–2808. doi: 10.1093/nar/gkf393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gulston M, Fulford J, Jenner T, de Lara C, O’Neill P. Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res. 2002;30:3464–3472. doi: 10.1093/nar/gkf467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song JM, Milligan JR, Sutherland BM. Bistranded oxidized purine damage clusters: induced in DNA by long-wavelength ultraviolet (290-400 nm) radiation? Biochemistry. 2002;41:8683–8688. doi: 10.1021/bi020137x. [DOI] [PubMed] [Google Scholar]
- 9.Ho EL, Parent M, Satoh MS. Induction of base damages representing a high risk site for double strand break formation in genomic DNA by exposure of cells to DNA damaging agents. J. Biol. Chem. 2007;282:21913–21923. doi: 10.1074/jbc.M610651200. [DOI] [PubMed] [Google Scholar]
- 10.Nowsheen S, Wukovich RL, Aziz K, Kalogerinis PT, Richardson CC, Panayiotidis MI, Bonner WM, Sedelnikova OA, Georgakilas AG. Accumulation of oxidatively induced clustered DNA lesions in human tumor tissues. Mutat. Res. 2009;674:131–136. doi: 10.1016/j.mrgentox.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace SS. Enzymatic processing of radiation-induced free radical damage in DNA. Radiat. Res. 1998;150:S60–S79. [PubMed] [Google Scholar]
- 12.Chaudhry MA, Weinfeld M. Reactivity of human apurinic/apyrimidinic endonuclease and Escherichia coli exonuclease III with bistranded abasic sites in DNA. J. Biol. Chem. 1997;272:15650–15655. doi: 10.1074/jbc.272.25.15650. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto M, Imhoff B, Ali MM, Kow YW. HU protein of Escherichia coli has a role in the repair of closely opposed lesions in DNA. J. Biol. Chem. 2003;278:28501–28507. doi: 10.1074/jbc.M303970200. [DOI] [PubMed] [Google Scholar]
- 14.Lomax ME, Cunniffe S, O’Neill P. 8-OxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single-strand break when present within a clustered DNA damage site. DNA Repair. 2004;3:289–299. doi: 10.1016/j.dnarep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Eot-Houllier G, Eon-Marchais S, Gasparutto D, Sage E. Processing of a complex multiply damaged DNA site by human cell extracts and purified repair proteins. Nucleic Acids Res. 2005;33:260–271. doi: 10.1093/nar/gki165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mourgues S, Lomax ME, O’Neill P. Base excision repair processing of abasic site/single-strand break lesions within clustered damage sites associated with XRCC1 deficiency. Nucleic Acids Res. 2007;35:7676–7687. doi: 10.1093/nar/gkm947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eot-Houllier G, Gonera M, Gasparutto D, Giustranti C, Sage E. Interplay between DNA N-glycosylases/AP lyases at multiply damaged sites and biological consequences. Nucleic Acids Res. 2007;35:3355–3366. doi: 10.1093/nar/gkm190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paap B, Wilson DM, III, Sutherland BM. Human abasic endonuclease action on multilesion abasic clusters: implications for radiation-induced biological damage. Nucleic Acids Res. 2008;36:2717–2727. doi: 10.1093/nar/gkn118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byrne S, Cunniffe S, O’Neill P, Lomax ME. 5,6-Dihydrothymine impairs the base excision repair pathway of a closely opposed AP site or single-strand break. Radiat. Res. 2009;172:537–549. doi: 10.1667/RR1830.1. [DOI] [PubMed] [Google Scholar]
- 20.Eccles LJ, Lomax ME, O’Neill P. Hierarchy of lesion processing governs the repair, double (strand break formation and mutability of three-lesion clustered DNA damage. Nucleic Acids Res. 2010;38:1123–1134. doi: 10.1093/nar/gkp1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shikazono N, O’Neill P. Biological consequences of potential repair intermediates of clustered base damage site in Escherichia coli. Mutat. Res. 2009;669:162–168. doi: 10.1016/j.mrfmmm.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Dianov GL, Timchenko TV, Sinitsina OI, Kuzminov AV, Medvedev OA, Salganik RI. Repair of uracil residues closely spaced on the opposite strands of plasmid DNA results in double-strand break and deletion formation. Mol. Gen. Genet. 1991;225:448–452. doi: 10.1007/BF00261686. [DOI] [PubMed] [Google Scholar]
- 23.D’Souza DI, Harrison L. Repair of clustered uracil DNA damages in Escherichia coli. Nucleic Acids Res. 2003;31:4573–4581. doi: 10.1093/nar/gkg493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison L, Brame KL, Geltz LE, Landry AM. Closely opposed apurinic/apyrimidinic sites are converted to double strand breaks in Escherichia coli even in the absence of exonuclease III, endonuclease IV, nucleotide excision repair and AP lyase cleavage. DNA Repair. 2006;5:324–335. doi: 10.1016/j.dnarep.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shikazono N, Pearson C, O'Neill P, Thacker J. The roles of specific glycosylases in determining the mutagenic consequences of clustered DNA base damage. Nucleic Acids Res. 2006;34:3722–3730. doi: 10.1093/nar/gkl503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozmin SG, Sedletska Y, Reynaud-Angelin A, Gasparutto D, Sage E. The formation of double-strand breaks at multiply damaged sites is driven by the kinetics of excision/incision at base damage in eukaryotic cells. Nucleic Acids Res. 2009;37:1767–1777. doi: 10.1093/nar/gkp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellon S, Shikazono N, Cunniffe S, Lomax M, O'Neill P. Processing of thymine glycol in a clustered DNA damage site: mutagenic or cytotoxic. Nucleic Acids Res. 2009;37:4430–4440. doi: 10.1093/nar/gkp422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malyarchuk S, Youngblood R, Landry AM, Quillin E, Harrison L. The mutation frequency of 8-oxo-7,8-dihydroguanine (8-oxodG) situated in a multiply damaged site: comparison of a single and two closely opposed 8-oxodG in Escherichia coli. DNA Repair. 2003;2:695–705. doi: 10.1016/s1568-7864(03)00040-5. [DOI] [PubMed] [Google Scholar]
- 29.Malyarchuk S, Brame KL, Youngblood R, Shi R, Harrison L. Two clustered 8-oxo-7,8-dihydroguanine (8-oxodG) lesions increase the point mutation frequency of 8-oxodG, but do not result in double strand breaks or deletions in Escherichia coli. Nucleic Acids Res. 2004;32:5721–5731. doi: 10.1093/nar/gkh911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malyarchuk S, Harrison L. DNA repair of clustered uracils in HeLa cells. J. Mol. Biol. 2005;345:731–743. doi: 10.1016/j.jmb.2004.10.079. [DOI] [PubMed] [Google Scholar]
- 31.Malyarchuk S, Castore R, Harrison L. DNA repair of clustered lesions in mammalian cells: involvement of non-homologous end-joining. Nucleic Acids Res. 2008;31:822–826. doi: 10.1093/nar/gkn450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson CG, Shikazono N, Thacker J, O'Neill P. Enhanced mutagenic potential of 8-oxo-7,8-dihydroguanine when present within a clustered DNA damage site. Nucleic Acids Res. 2004;32:263–270. doi: 10.1093/nar/gkh150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noguchi M, Urushibara A, Yokoya A, O’Neill P, Shikazono N. The mutagenic potential of 8-oxoG/single strand break-containing clusters depends on their relative positions. Mutat. Res. 2012;732:34–42. doi: 10.1016/j.mrfmmm.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Radany EH, Dornfeld KJ, Sanderson RJ, Savage MK, Majumdar A, Seidman MM, Mosbaugh DW. Increased spontaneous mutation frequency in human cells expressing the phage PBS2-encoded inhibitor of uracil-DNA glycosylase. Mutat. Res. 2000;461:41–58. doi: 10.1016/s0921-8777(00)00040-9. [DOI] [PubMed] [Google Scholar]
- 35.Parris CN, Seidman MM. A signature element distinguishes sibling and independent mutations in a shuttle vector plasmid. Gene. 1992;117:1–5. doi: 10.1016/0378-1119(92)90482-5. [DOI] [PubMed] [Google Scholar]
- 36.Guillet M, Boiteux S. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J. 2002;21:2833–2841. doi: 10.1093/emboj/21.11.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boiteux S, Guillet M. Abasic sites in DNA:repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Kreutzer DA, Essigmann JM. Oxidized, deaminated cytosines are a source of C –> T transitions in vivo. Proc. Natl Acad. Sci. USA. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su S, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc. Natl Acad. Sci. USA. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown J, Brown T, Fox KR. Affinity of mismatch-binding protein MutS for heteroduplexes containing different mismatches. Biochem. J. 2001;354:627–633. doi: 10.1042/0264-6021:3540627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zlatanou A, Despras E, Braz-Petta T, Boubakour-Azzouz I, Pouvelle C, Stewart GS, Nakajima S, Yasui A, Ishchenko AA, Kannouche PL. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol η in response to oxidative DNA damage in human cells. Mol. Cell. 2011;43:649–662. doi: 10.1016/j.molcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 42.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 43.Moriya M, Ou C, Bodepudi V, Johnson F, Takeshita M, Grollman AP. Site-specific mutagenesis using a gapped duplex vector: a study of translesion synthesis past 8-oxodeoxyguanosine in E. coli. Mutat. Res. 1991;254:281–288. doi: 10.1016/0921-8777(91)90067-y. [DOI] [PubMed] [Google Scholar]
- 44.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G—T and A—C substitutions. J. Biol. Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 45.Pytel D, Słupianek A, Ksiazek D, Skórski T, Błasiak J. Uracil-DNA glycosylases [in Polish] Postepy Biochem. 2008;54:362–370. [PubMed] [Google Scholar]
- 46.Cappelli E, Degan P, Frosina G. Comparative repair of the endogenous lesions 8-oxo-7,8-dihydroguanine (8-oxoG), uracil and abasic site by mammalian cell extracts: 8-oxoG is poorly repaired by human cell extracts. Carcinogenesis. 2000;21:1135–1141. [PubMed] [Google Scholar]
- 47.Cappelli E, Hazra T, Hill JW, Slupphaug G, Bogliolo M, Frosina G. Rates of base excision repair are not solely dependent on level of initiating enzymes. Carcinogenesis. 2001;22:387–393. doi: 10.1093/carcin/22.3.387. [DOI] [PubMed] [Google Scholar]
- 48.Harrison L, Hatahet Z, Wallace SS. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 49.David-Cordonnier MH, Laval J, O'Neill P. Recognition and kinetics for excision of a base lesion within clustered DNA damage by the Escherichia coli proteins Fpg and Nth. Biochemistry. 2001;40:5738–5746. doi: 10.1021/bi002605d. [DOI] [PubMed] [Google Scholar]
- 50.Lomax ME, Cunniffe S, O'Neill P. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry. 2004;43:11017–11026. doi: 10.1021/bi049560r. [DOI] [PubMed] [Google Scholar]
- 51.Lomax ME, Salje H, Cunniffe S, O'Neill P. 8-OxoA inhibits the incision of an AP site by the DNA glycosylases Fpg, Nth and the AP endonuclease HAP1. Radiat. Res. 2005;163:79–84. doi: 10.1667/rr3284. [DOI] [PubMed] [Google Scholar]
- 52.Cunniffe SM, Lomax ME, O’Neill P. An AP site can protect against the mutagenic potential of 8-oxoG when present within a tandem clustered site in E. coli. DNA Repair. 2007;6:1839–1849. doi: 10.1016/j.dnarep.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Ischenko AA, Saparbaev MK. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature. 2002;415:183–187. doi: 10.1038/415183a. [DOI] [PubMed] [Google Scholar]
- 54.Koffel-Schwartz N, Maenhaut-Michel G, Fuchs RP. Specific strand loss in N-2-acetylaminofluorene-modified DNA. J. Mol. Biol. 1987;193:651–659. doi: 10.1016/0022-2836(87)90348-2. [DOI] [PubMed] [Google Scholar]
- 55.Feig DI, Sowers LC, Loeb LA. Reverse chemical mutagenesis: identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc. Natl Acad. USA. 1994;91:6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purmal AA, Lampman GW, Kow YW, Wallace SS. The sequence context-dependent mispairing of 5-hydroxycytosine and 5-hydroxyuridine in vitro. Ann. N. Y. Acad. Sci. 1994;726:361–363. doi: 10.1111/j.1749-6632.1994.tb52852.x. [DOI] [PubMed] [Google Scholar]
- 57.Moriya M, Grollman AP. Mutations in the mutY gene of Escherichia coli enhance the frequency of targeted G:C–>T:A transversions induced by a single 8-oxoguanine residue in single-stranded DNA. Mol. Gen. Genet. 1993;239:72–76. doi: 10.1007/BF00281603. [DOI] [PubMed] [Google Scholar]
- 58.Iraqui I, Chekkal Y, Jmari N, Pietrobon V, Fréon K, Costes A, Lambert SA. Recovery of arrested replication forks by homologous recombination is error-prone. PLoS Genet. 2012;8:e1002976. doi: 10.1371/journal.pgen.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sedletska Y, Fourrier L, Malinge JM. Modulation of MutS ATP-dependent functional activities by DNA containing a cisplatin compound lesion (base damage and mismatch) J. Mol. Biol. 2007;369:27–40. doi: 10.1016/j.jmb.2007.02.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.