Abstract
RNA interference uses small RNAs (sRNA), which target genes for sequence-specific silencing. The parasite Entamoeba histolytica contains an abundant repertoire of 27 nt antisense (AS) sRNA with 5′-polyphosphate termini, but their roles in regulating gene expression have not been well established. We demonstrate that a gene-coding region to which large numbers of AS sRNAs map can serve as a ‘trigger’ and silence the gene fused to it. Silencing is mediated by generation of AS sRNAs with 5′-polyphosphate termini that have sequence specificity to the fused gene. The mechanism of silencing is independent of the placement of the trigger relative to the silenced gene but is dependent on the sRNA concentration to the trigger. Silencing requires transcription of the trigger-gene fusion and is maintained despite loss of the trigger plasmid. We used this approach to silence multiple amebic genes, including an E. histolytica Myb gene, which is upregulated during oxidative stress response. Silencing of the EhMyb gene decreased parasite viability under oxidative stress conditions. Thus, we have developed a new tool for genetic manipulation in E. histolytica with many advantages over currently available technologies. Additionally, these data shed mechanistic insights into a eukaryotic RNA interference pathway with many novel aspects.
INTRODUCTION
RNA interference (RNAi) pathways regulate gene expression in diverse systems ranging from protozoans to humans and can function at the transcriptional or post-transcriptional levels (1–5). Although details of these pathways are known in many eukaryotic model organisms, little is known about the mechanisms in protozoans. Key to the RNAi pathways in all organisms are small RNAs (sRNAs) that associate with Argonaute proteins to mediate sequence-specific gene silencing (6,7). Many classes of sRNAs such as microRNAs, small interfering RNAs (siRNAs) and trans-acting siRNAs are generated by Dicer, an RNAseIII enzyme. Thus, these sRNAs have a 5′-monophosphate and 3′-hydroxyl structure typical of RNAseIII cleavage products. However, some sRNA biogenesis pathways function independent of Dicer processing, including Piwi-interacting sRNAs, which are shown to be specifically expressed in germ-line cells (7,8) and secondary sRNAs in Caenorhabditis elegans and Ascaris (9–11). In nematodes, secondary sRNAs are dependent on RNA-dependent RNA polymerase (RdRP) activity, are based on a mature mRNA template, harbor 5′-triphosphate termini, are of antisense (AS) polarity and associate with a distinct set of Argonaute proteins (9,11–13). The only other system in which these sRNAs with 5′-polyphosphate termini are described is the protozoan parasite Entamoeba histolytica (14–16).
Entamoeba histolytica is an important human pathogen causing diarrheal disease and liver abscesses with ∼50 million people with invasive disease worldwide (17). Conserved elements of the eukaryotic RNAi pathway can be identified in E. histolytica including three genes with Piwi and PAZ domains (characteristic of Argonaute proteins) and two genes with RdRP domains (14,18,19). Endogenous sRNAs have been identified in E. histolytica; an abundant 27-nt sRNA population associates with EhAgo2-2 and harbors properties similar to secondary sRNAs of C. elegans including their 5′-polyphosphate termini, AS nature and bias toward the 5′ ends of genes. Furthermore, the levels of these AS sRNAs correlate inversely with the mRNA expression of their cognate gene, suggesting a role in gene silencing (14). Multiple RNAi-based methods of genetic manipulation have been developed in E. histolytica to achieve gene knockdown (20–23). Unfortunately, although promising, these approaches have difficulties in practical use as (i) the knockdown efficiency varies, (ii) not all genes appear to be amenable to silencing, (iii) the small hairpin RNA (shRNA) approach is labor intensive and (iv) reversal of gene silencing mediated by both double stranded RNA and shRNA has been reported (24) [W. A. Petri Jr. (personal communication)]. Additionally, little is known about the mechanism(s) of how these sRNA approaches target the appropriate mRNA.
In this study, we analyzed the function of the secondary sRNAs in E. histolytica. We show for the first time that AS sRNAs directly mediate gene silencing. Our data demonstrate that a small portion of a gene-coding region to which large numbers of AS sRNAs map is sufficient to ‘trigger’ silencing of a gene fused to it. Gene silencing only occurred in E. histolytica strains in which sRNAs to the coding region trigger existed and was associated with the appearance of 27-nt AS sRNAs to the silenced gene. We adapted this method to trigger gene silencing of chromosomally encoded genes and achieved robust and specific gene knockdown for the highly expressed E. histolytica rhomboid protease (EhROM1) (25), and additionally applied the technique to gain novel insights into the role of a putative Myb transcription factor in response to hydrogen peroxide (H2O2) stress. Analysis of the mechanism of silencing revealed that the newly generated AS sRNAs possess 5′-polyphosphate termini, map to introns indicating that they can be derived from nascent mRNA, are dependent on transcription of the trigger-gene fusion construct and persist after removal of the trigger plasmid, suggesting that an amplification pathway is initiated by the initial silencing plasmid.
We have successfully developed a new approach for robust gene knockdown in E. histolytica, which has many advantages over currently available technologies and will provide a crucial genetic tool for studying the virulence of this important human pathogen. More importantly, our data provide important new insights into novel aspects of a eukaryotic RNAi pathway and contribute to expanding our knowledge of RNAi in eukaryotes.
MATERIALS AND METHODS
Parasite culture, transfection, RNA and DNA isolation
Entamoeba histolytica trophozoites (strains: HM-1:IMSS and Rahman) and Entamoeba invadens IP-1 were grown under standard conditions as previously published (26–28). Parasites were transfected with 20 µg of DNA as described previously (29). All lines used 6 µg/ml G418 unless otherwise stated. As needed, drug selection was removed and loss of plasmid was confirmed by testing the viability of parasites in 3 µg/ml G418. Transfection of E. invadens and luciferase assays were performed using a method published previously (30). The sRNAs were isolated using the mirVana kit (Ambion) as previously published (14). Phenol-chloroform extraction was done as previously described (29), and DNA was isolated using the RNeasy Plant Mini Kit (Qiagen) and concentration was measured on Nanodrop.
Trigger gene selection
The genes used for this study were selected in silico by comparing previously performed microarray analyses (26,27) and two sequenced sRNA libraries from our laboratory, one for the virulent E. histolytica strain HM-1:IMSS and one for the non-virulent E. histolytica strain Rahman (16). The sRNAs mapping to the genes were visualized using the Integrative Genomics Viewer tool (www.broadinstitute.org/igv/). The genes were selected using the criteria (i) AS sRNAs in at least one strain, (ii) not expressed in the strain where AS sRNAs are present under any condition tested in the lab and (iii) active promoters.
Construct cloning
All primers used for cloning in this study are listed in Supplementary Table S2. All constructs used the pKT3M vector as backbone (16), restriction enzymes from New England Biolabs (NEB) were used and the final constructs were sequenced. For the luciferase assays the firefly luciferase gene was cloned into the pKT3M backbone by using the AvrII and XhoI restriction enzymes resulting in the promoter-less pKTluc backbone. The promoters of interest including 21 bp of each gene-coding region (EHI_048600, 480 bp upstream of the ATG; EHI_197520, 310 bp upstream of the ATG; EhSTIRP, 484 bp upstream of the ATG) were then cloned into the pKTluc backbone by using the HindIII and AvrII sites. Each trigger fragment was cloned in between the promoter and luciferase gene by using the AvrII site. The XhoI site was used to clone fragments in between the end (3′) of the luciferase gene and the CS3′ regulatory region. For these constructs, the stop codon was added at the end of the luciferase gene-trigger fragment fusion. For the stable cell line, the EHI_197520 trigger fragment was cloned in between the cysteine synthase (CS) promoter and the gene of interest by using the SmaI and AvrII sites. The Renilla luciferase gene was cloned into the pKTluc backbone using the HindIII and XhoI sites between the CS 5′ and CS 3′ regulatory regions.
Luciferase assays
Luciferase assays were done in triplicate. Parasites were transfected as described previously (29). Cells were harvested 20–22 h and re-suspended in lysis buffer complemented with protease inhibitors (luciferase assay system, Promega E1500, 1x N-acetyl-l-leucyl-l-leucyl-l-argininal, 1x HALT protein inhibitor cocktail, 1x E64) and protein concentration was measured by Bradford assay. Luciferase activity was measured by a luminometer (Monolight 2010). A total of 30 µg of protein was added to luciferase reagent (Promega) and relative light units were obtained. For Renilla luciferase measurements, the assay was performed as described earlier in the text and the Dual-LuciferaseR Reporter Assay System (Promega) according to the manufacturer’s protocol to measure the Renilla luciferase subsequent to the firefly luciferase.
Northern blot analysis, terminator and capping assays
All primers used for northern blotting in this study are listed in Supplementary Table S3. High-resolution northern blot analysis was done according standard protocols; the amount of RNA used is stated in the figure legends (29). Probe for EHI_118130 was as previously published (14). Northern blot analysis was done using standard protocols (29). Terminator and capping assays were done like previously described (14) using 10 µg of sRNA-enriched RNA. For the terminator assay, the sample mixture was treated with terminator enzyme (Epicentre) following the manufacturer’s protocol. For the capping assay, the ScriptCap m7G capping system (Epicentre) was used with a provided alternate cap zero capping protocol. After treatments, samples were resolved on a 12% polyacrylamide gel for sRNA northern blot analysis.
Reverse transcriptase PCR
All primers used for reverse transcriptase polymerase chain reaction (RT-PCR) in this study are listed in Supplementary Table S4. RT-PCR was performed by standard protocol as previously described (29). The negative control was split away as separate reaction after the initial DNAse digestions and treated as the other samples, but ddH2O was added instead of the reverse transcriptase.
Overexpression of EhMyb1 and immunofluorescence assays
The full-length coding region of EHI_197980 was obtained from genomic DNA using PCR (primer Supplementary Table S2). PCR product was cloned into pKT3M, downstream of the cysteine synthase promoter and 3x myc tag (SmaI and XhoI restriction sites) and sequence verified. Overexpression of EHI_197980 was achieved by transfecting E. histolytica HM-1:IMSS as described earlier in the text. Myc-tagged EhMyb1 expressing amebae were imaged as described previously (31). Expression and distribution were determined using α-myc antibody (1:1000) (Cell Signaling) and Alexa 488 α-mouse (1:1000) (Molecular Probes). Slides were prepared using Vectashield mounting medium with DAPI (Vector Laboratories, Inc).
Peroxide stress and viability assay
Parasites in log phase were incubated for 1 h at 37°C with 1 mM fresh H2O2. Cells were then put on ice for 7 min to completely detach from the tube and spun down. Cells were re-suspended in 300 µl of media and mixed 1:1 with Trypan blue immediately before counting on a light microscope. The experiment was performed each time in duplicate on three separate days.
RESULTS
Antisense sRNAs target genes for silencing
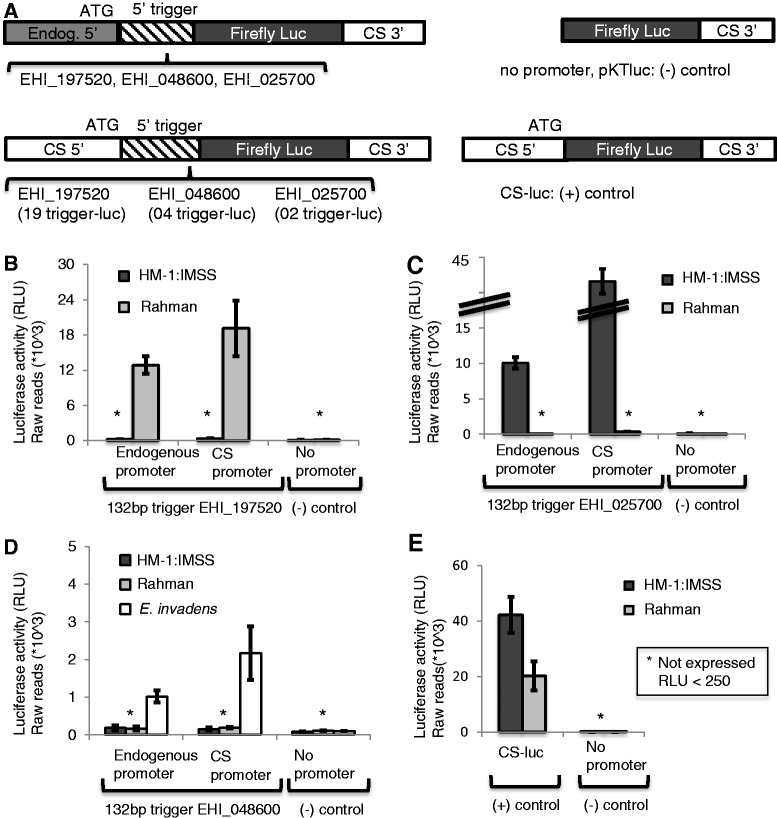
We have recently shown that genes with large numbers of AS sRNAs have low mRNA abundance, suggesting a role for AS sRNAs in gene silencing (14–16). To functionally investigate the role of AS sRNAs in regulation of gene expression, we selected genes that had variable mRNA and sRNA abundance in a strain-specific manner (Supplementary Table S1). This selection was based on microarray analyses (26,27) and two sRNA libraries, one each for the virulent and non-virulent strains E. histolytica HM-1:IMSS and Rahman (16). Genes were selected because they had different patterns of sRNA and gene expression in the E. histolytica strains HM-1:IMSS and Rahman. Specifically, genes selected were not expressed in the strain where AS sRNAs are present under any condition tested in the laboratory. For the E. histolytica HM-1:IMSS strain, extensive transcriptome data sets are available and used to find genes that lacked expression under oxidative or nitrosative stress (32), heat shock (33), histone acetylation (34), DNA methylation (35) or tissue invasion (25,36). EHI_197520 had abundant sRNAs and low gene expression in the HM-1:IMSS strain, but no sRNAs and high gene expression in the Rahman strain. Conversely, EHI_025700 had abundant sRNAs and no gene expression in the Rahman strain but had no sRNAs and high gene expression in the HM-1:IMSS strain. And finally, EHI_048600 had sRNAs and no gene expression in both the HM-1:IMSS and Rahman strains (Supplementary Table S1). To exclude the possibility that the given genes have low mRNA levels due to low promoter activity, we determined the activity of the endogenous promoters by creating luciferase reporter gene constructs, and found that all three promoters are active and share sequence identity in both amebic strains (data not shown). To define the role of the gene-coding region, we investigated whether a ‘trigger’ region (portion of the coding region to which large numbers of AS sRNAs map) could regulate gene expression. We fused the first 132 bp of the EHI_197520 coding sequence, to which large numbers of AS sRNAs map (Supplementary Figure S1), in frame to the 5′ end of a luciferase reporter gene using either the endogenous promoter of the EHI_197520 gene or the highly active promoter of the CS gene (Figure 1A, Supplementary Figure S2A and C). Using either promoter, the fusion of the 132-bp trigger fragment resulted in abrogation of reporter gene expression in the E. histolytica HM-1:IMSS strain, which contains abundant AS sRNAs to the EHI_197520 gene (Figure 1B). As a technical control, we used the E. histolytica Rahman strain, where no AS sRNAs are present to the EHI_197520 gene, and demonstrated that both fusion constructs were highly expressed. Thus, the 132 bp of the EHI_197520 gene serves as trigger to silence the luciferase gene in cell lines that have abundant AS sRNAs to the trigger but cannot induce silencing in the strain that lacks AS sRNAs to the trigger region. Similar analyses were performed for two additional genes: EHI_025700 (or EhSTIRP) (large numbers of AS sRNAs in the Rahman strain but none in HM-1:IMSS) and EHI_048600 (large numbers of AS sRNAs in both Rahman and HM-1:IMSS strains) (Figure 1A, Supplementary Figure S2A and C). Results were consistent with those seen previously: the 132-bp trigger resulted in reporter gene silencing in a strain-specific manner, correlating with the presence of AS sRNAs to the gene in a given strain (Figure 1C and D). The trigger from EHI_048600 resulted in reporter silencing in both HM-1:IMSS and Rahman strains in accordance with the AS sRNAs being present in both strains. Recent RNA sequencing data showed that a conserved homologue to the EHI_048600 gene is expressed in E. invadens (Ehrenkaufer,G., Hall,N. and Singh,U., unpublished data). Therefore, E. invadens was used as a control strain, and both constructs were found to express well, indicating no technical issues with the fusion proteins (Figure 1D). Thus, fusion of a trigger region (to which large numbers of AS sRNAs map) to a reporter gene results in reporter silencing in a promoter-independent manner. Importantly, the three trigger regions used have sequence identity among the different E. histolytica strains (data not shown). Thus, the sequence of the trigger region itself does not provide either the ability or specificity for the trigger-fusion silencing. Instead, silencing is dependent on the presence of sRNAs to the trigger region in a given amebic strain.
Figure 1.
Fusion of the trigger to the luciferase coding region results in reporter gene silencing. Evaluation of the fusion constructs (outlined in Figure 1A and C) was performed by transient transfection and luciferase assays. Experiments were done in triplicate; average and standard errors are shown. For all experiments, the negative control has no luciferase expression. (A) Schematic of constructs used in this assay. The trigger was fused upstream of the luciferase-coding region and driven by either the endogenous promoter or the CS promoter. (B) The 132-bp trigger of EHI_197520 results in luciferase reporter silencing specifically in the E. histolytica HM-1:IMSS strain, where AS sRNAs to the trigger are present regardless of whether expression is driven by the endogenous or the CS promoter. The constructs are expressed well in the Rahman strain, where no AS sRNAs are present. (C) The 132-bp trigger of EHI_025700 (EHI_STIRP) results in luciferase reporter silencing specifically in the E. histolytica Rahman strain, where AS sRNAs to the trigger are present regardless of whether expression is driven by the endogenous or the CS promoter. The constructs are expressed well in the HM-1:IMSS strain, where no AS sRNAs are present. (D) The 132-bp trigger of EHI_048600 results in luciferase reporter silencing in both the E. histolytica HM-1:IMSS and Rahman strains, where AS sRNAs to the trigger are present regardless of whether expression is driven by the endogenous or the CS promoter. The constructs are expressed well in E. invadens. (E) An additional control the CS-luc control construct was transfected in HM-1:IMSS and Rahman, and expression levels were monitored.
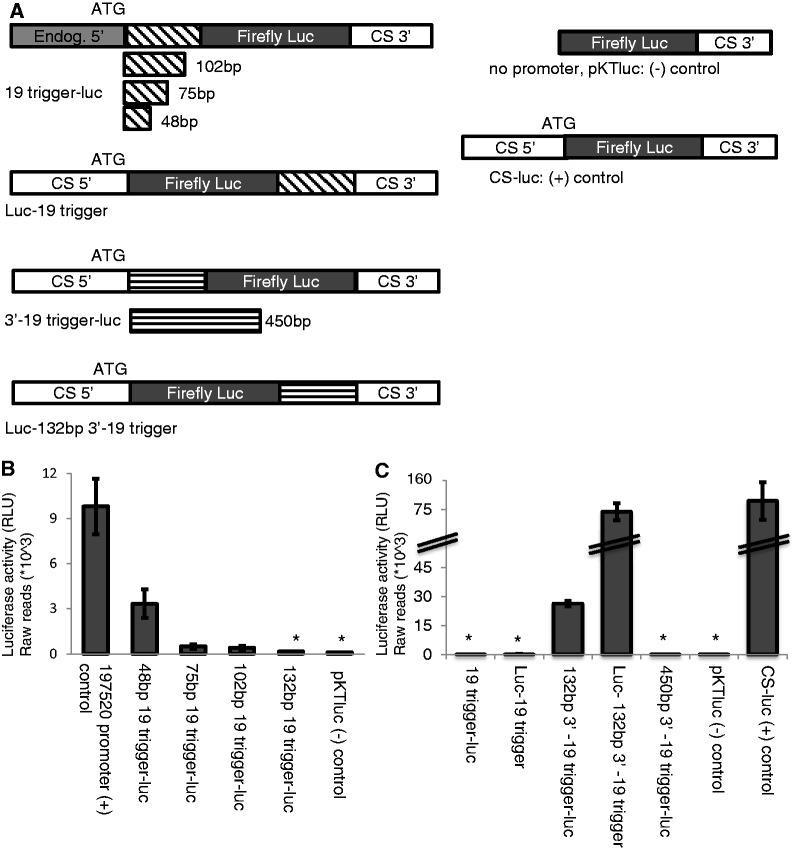
Reporter silencing occurs independent of position of trigger fragment but is dependent on the amount of AS sRNAs to the trigger region
To determine whether location or origin of the trigger were important for gene silencing, we tested placement of trigger (at the 5′ or 3′ end of genes) and the region of the gene from which the trigger is derived (from the 5′ or 3′ end of coding regions). Fusion of the EHI_197520 trigger to the 3′ end of the luciferase gene resulted in reporter silencing, indicating that this mechanism of gene silencing is independent of the position of the trigger fragment (Figure 2A, Supplementary Figure S2C). To determine specificity of the trigger region, we used a 132-bp portion from the 3′ end of the EHI_197520 gene-coding region (Figure 2A, Supplementary Figures S1 and S2D). Transient transfection and luciferase reporter gene analysis revealed that this 3′ fragment (fused either to the 5′ or 3′ end of the luciferase reporter gene) could not induce gene silencing (Figure 2C). However, given that only few sRNAs map to this region, this lack of silencing could be because of low sRNA abundance. Thus, we extended the size of the trigger from the 3′ end of EHI_197520 to 450 bp (a region to which larger numbers of AS sRNAs map) (Figure 2A, Supplementary Figures S1 and S2D); luciferase reporter analysis revealed that this fragment could induce gene silencing (Figure 2C). This indicates that the amount of AS sRNAs to a trigger may be a crucial element in the induction of gene silencing. To confirm the dependence of gene silencing on amount of AS sRNAs to the trigger region, we cloned a series of constructs, where we attached the first 48, 75, 102 or 132 bp of the EHI_197520 trigger region to its endogenous promoter (Figure 2A, Supplementary Figure S2B). We observed that increasing the size of the trigger reduced reporter gene expression (Figure 2B). All constructs expressed well in the Rahman strain where no AS sRNAs to the trigger fragments are present (data not shown). Taken together, the data suggest that (i) the trigger region can mediate gene silencing when placed at either the 5′ or 3′ end of a coding region, (ii) triggers from the 5′ or 3′ end of a gene are equally functional and (iii) the amount of sRNAs mapping to the trigger region is crucial in mediating gene silencing.
Figure 2.
Different lengths and position of the trigger to the luciferase-coding region results in reporter gene silencing. (A) Schematic of constructs used in this assay. (B) Different lengths of the EHI_197520 trigger were fused upstream of the luciferase-coding region. A 48-bp trigger does not silence luciferase, the 102-bp and 75-bp triggers significantly decreased luciferase expression and the 132-bp trigger resulted in complete silencing. (C) Evaluation of the fusion constructs where a trigger was fused either upstream or downstream of the luciferase-coding region. The first 132 bp of EHI_197520 trigger results in luciferase reporter silencing regardless of whether the trigger is fused up- or downstream of the luciferase gene. The last 132 bp of EHI_197520 trigger does not result in luciferase reporter silencing, regardless of whether the trigger is fused up- or downstream of the luciferase gene. The last 450 bp of EHI_197520 trigger results in luciferase reporter silencing when the trigger is fused upstream of the luciferase gene. All constructs with trigger luciferase are expressed well in the Rahman strain, where no AS sRNAs are present (data not shown).
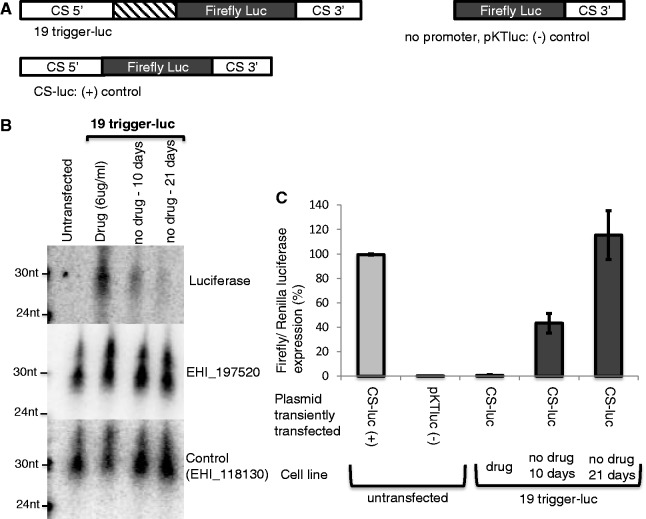
Luciferase silencing is associated with the biogenesis of AS sRNAs to the reporter gene, which do not persist after plasmid removal
To investigate the mechanism of gene silencing more thoroughly, we generated a stable cell line in which the 132-bp trigger region of EHI_197520 was fused to the luciferase gene (cell line: 19-trigger-luc) (Figure 3A, Supplementary Figure S2C). In these parasites, the plasmid would be maintained episomally under G418 drug selection. Despite raising drug selection to 60 µg/ml G418, a concentration typically used to overexpress genes in Entamoeba, no luciferase expression could be detected (data not shown). To determine whether gene silencing was linked to sRNAs, we performed northern blot analysis and detected ∼27-nt AS sRNAs to the luciferase gene (Figure 3B).
Figure 3.
Fusion of the 132-bp trigger to the luciferase reporter gene results in generation of functional luciferase AS sRNAs. The 19-trigger-luc cell line is E. histolytica HM-1:IMSS parasites stably transfected with the plasmid containing the EHI_197520 trigger fused to luciferase. (A) Schematic of constructs used in this assay. (B) High-resolution northern blot of the 19-trigger-luc and untransfected (HM-1:IMSS) cell lines probed for luciferase AS sRNAs (90 µg of RNA enriched for sRNAs). AS sRNAs to the luciferase reporter gene were detected in abundance, which decreased substantially over time with drug removal. An abundant population of AS sRNA to the EHI_197520 trigger was detected, which increased in the 19-trigger-luc cell line. Loading control: sRNAs to EHI_118130. (C) Transient transfection of the 19-trigger-luc cell line with a luciferase reporter construct (CS-luc). The construct expresses well in the untransfected cell line but does not express in the 19-trigger-luc cell line. Ten days after drug release the CS-luc construct can be partially expressed, and 21 days after drug release a full recovery of CS-luc expression is observed. The values were normalized to a CS-Renilla-luc expression. Experiments were done in triplicate; average and standard errors are shown.
To test if the luciferase sRNAs could target an additional luciferase template for silencing, we transiently transfected the cell line with sRNAs to luciferase with a construct that expresses firefly luciferase (Figure 3A, Supplementary Figure S2F). The CS-luc construct was silenced in the cell line where luciferase AS sRNAs were present, but was highly expressed in the wild-type cell line (Figure 3C). To test the stability of the silencing, we released the 19-trigger-luc cell line from drug selection, which resulted in loss of the trigger plasmid. There was increased expression of CS-luc in the cell lines after 10 days with no drug selection, which increased to full recovery of luciferase expression after 21 days of drug removal (Figure 3C). Concomitant with recovery of luciferase expression, northern blot analysis revealed substantial loss of AS sRNAs to the luciferase gene at 10 days and 21 days after drug removal (Figure 3B). These findings demonstrate that the sRNAs generated to the luciferase gene are functional and can silence genes both in cis and in trans, and that loss of the trigger plasmid results in loss of the sRNAs.
Biogenesis of functional AS sRNAs to an amebic gene
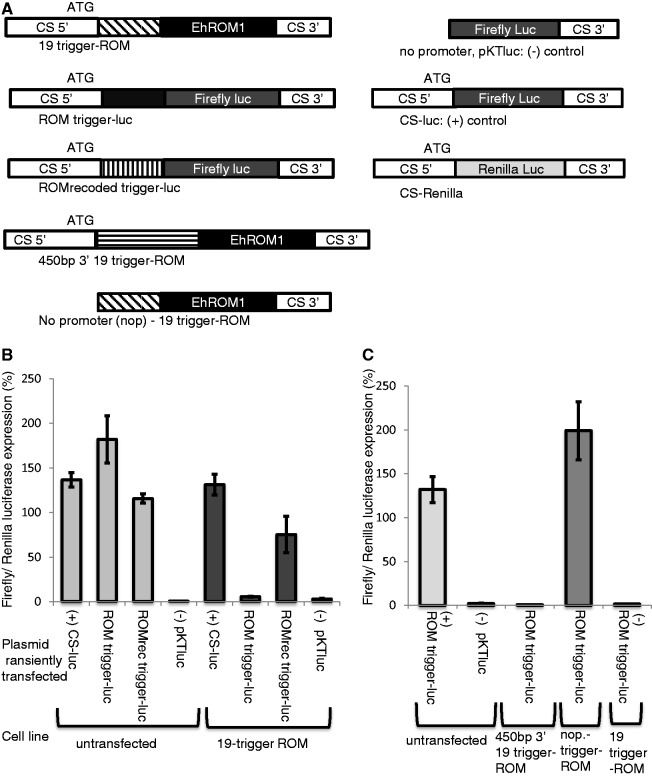
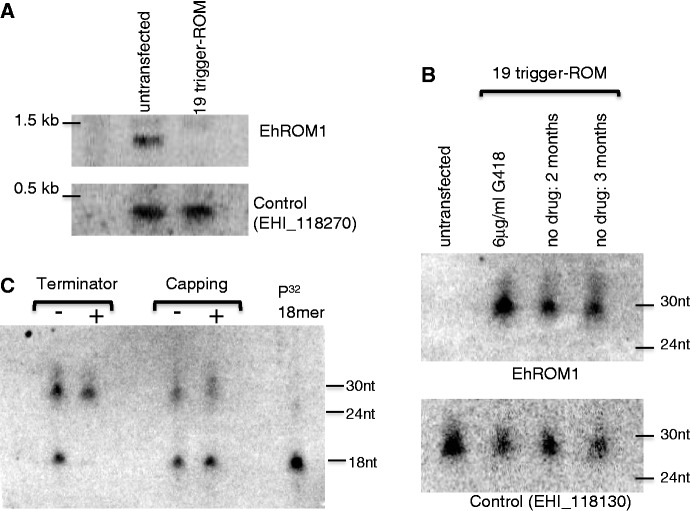
As we were successful in generating functional sRNAs and silencing a reporter gene, we proceeded to determine whether we could achieve gene knockdown of chromosomally encoded amebic genes. We chose EhROM1, a highly expressed single copy gene that is a known virulence factor involved in host–parasite interactions (25,29). We fused the full-length coding region of EhROM1 to the EHI_197520 132-bp trigger fragment and generated a stable cell line (cell line: 19-trigger-ROM, Figure 4A). We confirmed presence of an ∼27-nt population of sRNAs to EhROM1 (Figure 5B). To determine if sense sRNAs to EhROM1 were also generated, we probed for these but could not detect any by northern blot analysis (data not shown). This result suggests that fusion to a trigger results in biogenesis of only AS sRNAs. Although deep sequencing would be needed to definitively make this statement, this result is consistent with and parallels patterns seen for the endogenous sRNA population.
Figure 4.
Triggering functional AS sRNAs to an amebic gene. E. histolytica HM-1:IMSS parasites stably transfected with the plasmid containing the EHI_197520 trigger fused to the full-length EhROM1 gene (cell line: 19-trigger-ROM). (A) Schematic of constructs used in this assay. (B) Untransfected and 19-trigger-ROM cell lines were transfected with a number of constructs including CS-luc (positive control) and pKT-luc (negative control). ROM trigger-luc and ROM-recoded trigger-luc constructs express well in the wild-type untransfected cell line. In the 19-trigger-ROM cell line, the ROM trigger-luc vector does not express, whereas the ROM-recoded trigger-luc vector expresses well. The values were normalized to a CS-Renilla-luc expression. Experiments were done in triplicate; average and standard errors are shown. (C) A wild-type untransfected cell line transfected with a ROM trigger-luc and pKTluc constructs worked as expected (a positive and negative control, respectively). A cell line stably transfected with 450-bp 3′19-trigger ROM did not express luciferase when transfected with ROM trigger-luc, whereas a no-promoter (nop) trigger-ROM cell line transfected with the same plasmid expressed well. The 19-trigger-ROM cell line when transfected with the ROM trigger-luc did not express luciferase. The values were normalized to a CS-Renilla-luc expression. Experiments were done in triplicate; average and standard errors are shown.
Figure 5.
Probing for functional AS sRNAs to the amebic gene. (A) Northern blot (10 µg of total RNA) probed for EhROM1 mRNA reveals absence of EHROM1 mRNA in the 19-trigger-ROM cell line. Control (EHI_118270) indicates equivalent mRNA in both lanes. (B) High-resolution northern blot analysis of 19-trigger-ROM and untransfected cell lines probed for EhROM1 AS sRNA (20 µg of enriched sRNA). An abundant population of AS sRNAs to the EhROM1 gene were detected, which persisted 2 and 3 months after drug release. Control (EHI_118130) indicates equivalent sRNA in all lanes. (C) Biochemical analysis of the EhROM1 AS sRNAs reveals that they are resistant to treatment with terminator enzyme and increase in size when treated with capping enzyme; both features are indicative of a 5′-polyphosphate structure. As a control a radioactively labeled 5′-monophosphate 18-mer (included in each lane) is degraded by terminator enzyme and does not increase in size when treated with capping enzyme (10 µg of enriched sRNA).
To address the functionality and specificity of the sRNAs to EhROM1, we designed two constructs: the first in which we used a portion of the EhROM1 coding region as a trigger fused to luciferase (ROM1-trigger luc); the second in which we used a portion of a recoded EhROM1 gene (with 66% DNA sequence identity, but unchanged amino acid sequence) as trigger fused to luciferase (ROM1 recoded trigger-luc) (Figure 4A, Supplementary Figure S2E). We transiently transfected these constructs into the cell line 19-trigger-ROM, which contains sRNAs to EhROM1. We observed that the ROM1 trigger-luc construct was silenced by the EhROM1 sRNAs, confirming the functionality of the AS EhROM1 sRNAs (Figure 4B). In contrast, the ROM1 recoded trigger-luc construct was not silenced in the cell line with EhROM1 sRNAs, indicating that the specificity of the EhROM1 sRNAs being dependent on the nucleotide sequence but independent of the encoded amino acid sequence.
To investigate the mechanism of sRNA generation, we aimed to determine whether a trigger derived from the 3′ end of the EHI_197520 gene would induce generation of sRNAs. We fused a 450-bp portion of the 3′ region that was successful in transient luciferase reporter silencing (Figure 4A, Supplementary Figure S2C and D), to EhROM1 and generated a stable cell line (Figure 4A, Supplementary Figure S2E). Transient transfection with the ROM1 trigger-luc construct revealed that functional EhROM1 sRNAs were generated in the 450-bp 3′ 19-trigger-ROM cell line (Figure 4C). This indicates that any portion of a gene that has abundant AS sRNAs can function as a trigger to induce the generation of AS sRNAs and subsequent gene silencing.
To determine whether transcription is needed for the trigger-induced silencing method, we generated a promoter-less construct (nop-19-trigger-ROM) and made stable cell lines expressing this construct. Transient transfection with the ROM1 trigger-luc construct revealed that luciferase was expressed and thus no EhROM1 AS sRNAs were generated from the nop-19-trigger-ROM cell line (Figure 4A and C, Supplementary Figure S2E). This result suggests that the presence of the trigger DNA is not sufficient to induce sRNAs but that transcription of the trigger-fusion construct is needed for the generation of sRNAs and subsequent gene silencing. It has previously been observed that sRNAs map to open reading frames but are largely absent in surrounding intergenic regions (16). The combined data indicate that sRNA generation occurs on an mRNA template, the presence of which limits the extent of sRNAs to transcribed regions and prevents spreading to adjacent genomic regions. In C. elegans, the secondary sRNAs are generated by an amplified RdRP dependent pathway where mRNA is targeted for amplification (37); a similar pathway might be present in E. histolytica.
Robust knockdown of amebic genes by trigger-derived AS sRNAs
To determine whether the functional sRNAs to EhROM1 could silence the endogenous copy of the gene, we determined the expression level of EhROM1 in the 19-trigger-ROM cell line. Using northern blot analysis, as well as RT-PCR, we found that the EhROM1 mRNA was undetectable and that robust gene knockdown was achieved (Figure 5A, Supplementary Figure S3). In addition, we performed RT-PCR for EhROM2, which has 56% sequence identity to EhROM1 and found that its mRNA level was unaffected, confirming that the gene silencing by the sRNAs is sequence specific. In the E. histolytica G3 cell line, coincidental gene silencing was linked to generation of sRNAs (15), but the mechanism that triggered the silencing or generation of sRNAs was not apparent.
To determine whether silencing persists after loss of the trigger plasmid, we removed the EhROM1 knockdown cell line from drug selection. After 2 months, the parasites were susceptible to 3 µg/ml G418, indicating that the drug resistance plasmid was no longer present. However, despite loss of the trigger plasmid, EhROM1 mRNA silencing persisted (Supplementary Figure S3A and B). In addition, EhROM1 sRNAs were maintained after 2 and 3 months (Figure 5B). This result is in contrast with what we observed with the luciferase gene, where loss of the plasmid resulted in loss of sRNAs and loss of silencing. The observation that despite loss of the trigger plasmid that sRNAs and silencing is maintained, indicates that the chromosomal copy of the EhROM1 gene may serve as a template for an amplification pathway in which EhROM1 sRNAs are continuously generated. We have continuously maintained a cell line which initially harbored the trigger-EhROM1 construct for up to 18 months without drug selection and monitored the continuous presence of sRNA to EhROM1 and silencing of EhROM1. This observation is similar to data from the G3 cell line, where silencing of genes and maintenance of sRNAs is stable despite loss of the initial plasmid (15), indicating that similar mechanisms may be at play for long-term control of silencing.
Biochemical analysis of the EhROM1 AS sRNAs with terminator enzyme (a 5′ to 3′ exonuclease, which specifically cleaves 5′-monophosphate species) as well as capping enzyme (only caps di or tri-phosphate termini RNA) was performed to determine the 5′ structure of these sRNAs (14). The EhROM1 AS sRNAs were not degraded by terminator enzyme and an increase in size was observed with the capping enzyme, whereas the control 5′-monophosphate 18-mer sample was appropriately degraded by terminator and did not increase in size with capping enzyme (Figure 5C). Thus, the EhROM1 AS sRNAs are 5′-polyphosphate species and thus similar to the 27-nt endogenous sRNA population in E. histolytica (14). Based on the 5′ structural features, both the endogenous sRNAs to the trigger as well as the sRNAs generated as a result of trigger fusion, appear to be generated via a Dicer-independent pathway (9,14).
Taken together, we successfully managed to silence the virulence factor EhROM1 in E. histolytica using a novel approach, which works through the generation of AS sRNAs to the target gene. The AS sRNAs have sequence specificity, persist after plasmid removal and have 5′-features indicative of a Dicer-independent generation mechanism. Some features of our new silencing approach are similar to that developed in the amebic G3 strain; however, our method offers some significant advantages: (i) the triggering event and trigger are known (in G3 the specific nucleotide sequence that induces the silencing is not absolutely clear), (ii) gene silencing can be initiated in a wild-type virulent strain (in contrast to the non-virulent G3 strain in which multiple genes are already knocked down) and (iii) the silencing method can be adapted for use in other Entamoeba strains and species. These features significantly advance the ability of genetic manipulation in ameba.
Knockdown of EhMyb1 reveals a role in protection against hydrogen peroxide stress
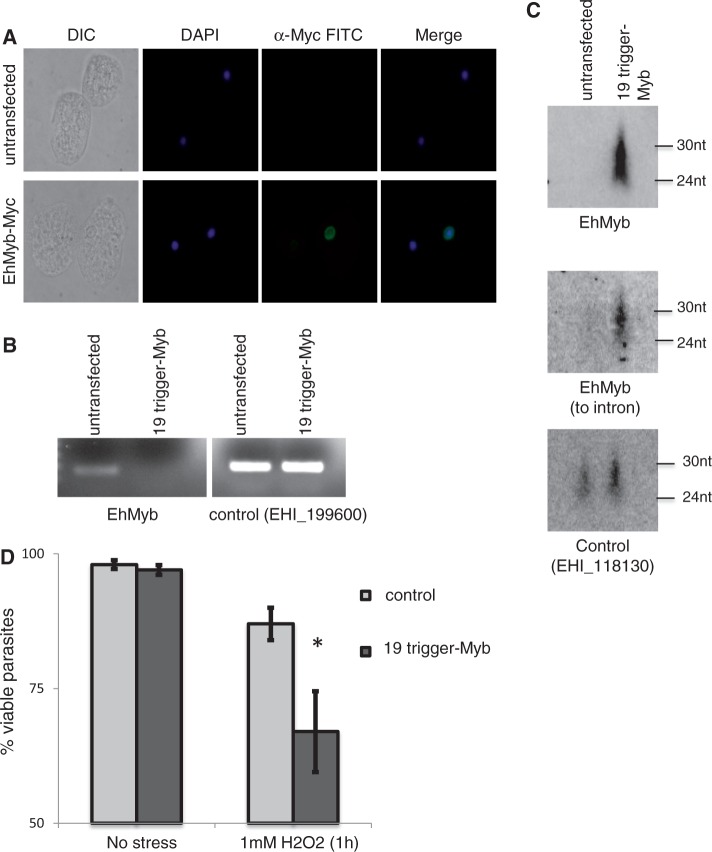
To characterize EhMyb1 (EHI_197980), a putative transcription factor that was upregulated during H2O2 stress (32), we tagged the protein with an N-terminal Myc tag. Protein localization to the parasite nucleus supports the hypothesis that EhMyb1 is a transcription factor (Figure 6A). We designed a 19-trigger-EhMyb1 construct and achieved robust gene knockdown in the E. histolytica HM-1:IMSS strain; silencing was associated with generation of an abundant population of sRNAs to the EhMyb1 gene (Figure 6B and C). As the EhMyb1 gene has an intron, we probed for AS sRNAs to the intronic region and found that they could be detected, indicating that the AS sRNAs can be generated from nascent transcript. This is similar to data from sequencing endogenous sRNA populations, where sRNAs were detected to introns as well as exon–exon junctions (16,38). To determine the relative abundance of AS sRNAs along the gene, we probed for AS sRNAs at discrete locations and observed that AS sRNAs were most abundant at the 5′ portion of the gene, with a decrease in the 3′ portion of the gene (data not shown). We also probed for sense sRNAs but could not detect them (data not shown). Overall, the sRNAs generated by the trigger-gene fusion (abundant AS sRNAs biased to the 5′ end of a gene with 5′-polyphosphate termini and no detectable sense sRNAs) are highly similar to the endogenous 27-nt sRNA population in E. histolytica (14).
Figure 6.
Gene knockdown of EhMyb1 reveals that AS sRNAs are generated to nascent transcript. (A) N-terminal Myc-tagged EhMyb has nuclear localization. (B) E. histolytica HM-1:IMSS parasites stably transfected with the plasmid containing the EHI_197520 trigger fused to the EhMyb1 gene (cell line: 19-trigger-Myb). RT-PCR for EhMyb1 in the 19-trigger-Myb cell line demonstrates the absence of EhMyb mRNA in the 19-trigger-Myb cell line. Control (EHI_199600) indicates equivalent mRNA in both lanes. (C) Northern blot of the 19-trigger-Myb cell line probed for EhMyb1 AS sRNAs reveals an abundant population of AS sRNA to the EhMyb1 gene. AS sRNAs that map to the intron of EhMyb1 were detected. Control (EHI_118130) indicates equivalent sRNA in both lanes (75 µg of enriched sRNA). (D) Viability assay of stress-treated parasites (1 h, 1 mM H2O2) revealed that EhMyb1 knockdown parasites had significantly decreased viability in response to stress.
Given that EhMyb1 was regulated by oxidative stress (32), we further analyzed the EhMyb1 knockdown cell line for a phenotype in response to exposure to H2O2. A viability assay revealed a significantly reduced ability of the EhMyb1-silenced parasites to survive under H2O2 stress, indicating that EhMyb1 indeed has an important role in parasite stress response (Figure 6D). Given the nuclear localization of EhMyb1 and its role in promoting parasite survival under stress, further studies to more fully elucidate its role in amebic biology are warranted.
DISCUSSION
The mechanisms of RNAi-mediated silencing are not well understood in non-model organisms. We describe a novel application of RNAi-based gene silencing in the single-celled eukaryote E. histolytica. We observe that Dicer-independent sRNA biogenesis is triggered by mRNA transcription of DNA fragments that have large numbers of endogenous sRNAs that map to them and that the sRNAs persist through an amplification mechanism independent of the initiating plasmid. Based on these insights, we have harnessed the robust silencing mediated by endogenous AS sRNAs to develop a new tool for genetic manipulation in E. histolytica. This new tool has many advantages to current methodologies in the system, including that it can be used in a virulent strain of E. histolytica and thus be used to study virulence-associated phenotypes, it does not require high levels of drug selection for initiation and it can be maintained long-term despite removal of drug pressure.
We have previously observed an inverse correlation of AS sRNA abundance and gene expression (14,15). This finding indicated that AS sRNAs may be involved in gene silencing in E. histolytica, but no direct causal data were available. We now demonstrate that the use of a 132-bp trigger (to which endogenous AS sRNAs map) and fusion of this trigger to a gene, leads to gene silencing. Importantly, this gene silencing occurs only in amebic strains where AS sRNAs to the trigger are present and fusion of a gene to the trigger leads to generation of AS sRNAs to the fused gene.
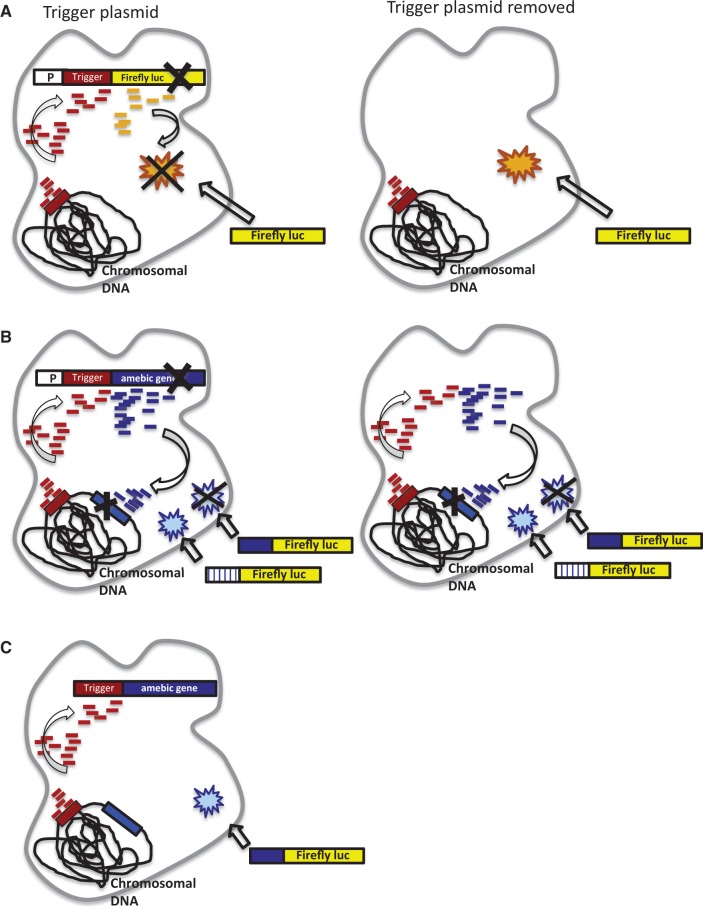
Importantly, the trigger-mediated silencing was dependent on the presence of large numbers of AS sRNAs to the trigger region, but independent of the placement of the trigger to a fused gene (5′ or 3′) or to the portion of the gene from which the initial trigger was derived. Interestingly, the same DNA sequence could serve as a trigger in one E. histolytica strain, but not in another strain (if no endogenous sRNAs were present to the trigger sequence). A number of additional important insights into the mechanism of silencing were obtained (outlined in Figure 7). The sRNAs generated by this mechanism have 5′-polyphosphate termini, are 5′ biased, can be generated on nascent transcript, have sequence specificity, require transcription of the trigger-gene fusion and are maintained despite loss of the initial trigger-gene plasmid. Importantly, the generation of functional AS sRNAs occurs regardless of whether the silenced gene has a chromosomal copy (i.e. sRNAs are generated to both luciferase and EhROM1 and both genes are silenced using this mechanism). However, the maintenance of sRNAs after loss of the episomal plasmid is strikingly different: genes with a chromosomal copy have long-term persistence of sRNAs and gene silencing, whereas for genes without a second chromosomal copy (i.e. luciferase) the sRNAs rapidly disappear after removal of the trigger plasmid. The persistence of sRNAs and gene silencing despite loss of the initiating plasmid suggests that the chromosomal locus is able to maintain sRNA production and amplification. Whether this amplified silencing process can be initiated for all genes or can occur from a second episomal copy is not currently known.
Figure 7.
Schematic of siRNA trigger-mediated silencing in E. histolytica. (A) Functional sRNAs are generated to the reporter gene fused to the trigger and the construct is silenced by these sRNAs. sRNAs are lost after plasmid removal. (B) Functional sRNAs are generated to an amebic gene fused to the trigger, which silence the endogenous gene and exogenous constructs with sequence identity to the silenced gene. The sRNAs and gene silencing remain after plasmid removal. (C) No functional sRNAs are generated if a promoter does not drive the trigger fusion construct.
One question is what determines the boundaries of sRNAs and prevents the amplification pathway from being rampant in the genome. We have previously identified that sRNAs in E. histolytica are enriched on coding regions, rather than intergenic regions and that they are likely templated on both nascent and mature transcript (16). Structural features indicate that these AS sRNAs are generated independent of Dicer processing, and instead are likely from an RdRP-dependent pathway similar to the secondary siRNA pathway in C. elegans (9,12,37,39). In C. elegans, the generation of secondary sRNAs is RdRP dependent and depends on the presence of mRNA and the precedent generation of Dicer-derived primary siRNAs (37). The factors that limit the extent of amplified gene silencing were recently defined in C. elegans with the demonstration that the primary siRNAs can act as triggers but not as templates, which may attenuate RdRP-dependent amplification to avoid indefinitely amplified silencing (37). It is unknown if the E. histolytica pathway also depends on the precedent generation of primary siRNAs because despite extensive efforts, no canonical Dicer has been identified (Pompey,J.M. and Singh,U., unpublished data) and our sequencing efforts have so far failed to detect a clear population of primary sRNAs with typical Dicer-derived features.
The effects of the sRNAs on the silenced genomic loci are not known and dissecting the mechanism(s) is an important area of future investigation. In the epigenetic transcriptional gene silencing mechanism that has been described in the E. histolytica G3 strain (40), gene silencing is dependent on a significant change in the level of histone methylation (H3K4) at the silenced gene loci and associated with an abundant presence of AS sRNAs to the silenced genes (15). The persistence of gene silencing despite removal of the silencing plasmid, noted in both the G3 strain as well as in our trigger silencing approach, suggests that changes in genomic loci might play a crucial role in the ability of sRNAs to silence specific genomic loci in E. histolytica. The phenomenon is also reminiscent of the RNA-directed DNA methylation pathways in plants where DNA methylation through DNA methyltransferase depends on the biogenesis of 24-nt siRNA by RdRP and leads to de novo cytosine DNA methylation (41–43). Studies in organisms ranging from the fission yeast Schizosaccharomyces pombe to Drosophila melanogaster and Tetrahymena thermophila have found that non-coding RNAs function in the assembly and function of heterochromatin (44). In the fission yeast S. pombe, it was shown that RNA-induced initiation of transcriptional gene silencing contains Dicer-generated siRNAs, which are also required for heterochromatic silencing. The deletion of RNAi-related genes leads to loss of heterochromatic gene silencing, highly reduced H3K9 methylation at centromeric repeats and accumulation of non-coding RNAs (44,45). Another study in S. pombe shows that RNA-induced initiation of transcriptional gene silencing tethering to silenced chromosomal domains is essential for transcriptional and post-transcriptional silencing and for the generation of additional siRNAs for heterochromatin assembly. Also, Rdp1 was shown to be an essential component of this self-enforcing RNAi loop and was also given a critical role for its RdRP activity in siRNA production necessary for heterochromatin formation (46,47). Thus, studies to determine whether there is a direct link between sRNAs and DNA modification are of high interest in E. histolytica. Going forward, it will also be interesting to determine whether some genes and genomic regions are resistant to the sRNA-mediated silencing approach we have developed and if so, the mechanism(s) of this resistance.
We describe a novel gene knockdown technique in E. histolytica as well as present mechanistic insights into a eukaryotic RNAi pathway that shows distinct features from RNAi pathways in model systems. We observed that the E. histolytica relies on an amplified sRNA pathway that offers a reliable system for sRNA-dependent gene knockdown. Taken together, this study opens important avenues for further investigation of a novel eukaryotic RNAi pathway, presents a simple technique on how to apply this RNAi pathway in E. histolytica genetic manipulation and overall sheds insight into the broad diversity of RNAi-based gene silencing in eukaryotes.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online, including [48].
FUNDING
Swiss National Science Foundation for funding [PBZHP3-130988 and PBZHP3-141513 to L.M.]; National Institute of Health [AI085178 and AI053724 to U.S.]; Training Grant in Microbiology [T32 AI07328 to R.P.]. Funding for open access charge: NIH [AI085178 and AI053724].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge all members of the Singh lab for invaluable input, especially Hanbang Zhang, Gretchen Ehrenkaufer and Ruchie Bhardwaj.
REFERENCES
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batista TM, Marques JT. RNAi pathways in parasitic protists and worms. J. Proteomics. 2011;74:1504–1514. doi: 10.1016/j.jprot.2011.02.032. [DOI] [PubMed] [Google Scholar]
- 3.Vance V, Vaucheret H. RNA silencing in plants—defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- 4.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 5.Ketting RF. The many faces of RNAi. Dev. Cell. 2011;20:148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 6.Ender C, Meister G. Argonaute proteins at a glance. J. Cell Sci. 2010;123:1819–1823. doi: 10.1242/jcs.055210. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell. Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 8.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 9.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 10.Faehnle CR, Joshua-Tor L. Argonaute MID domain takes centre stage. EMBO Rep. 2010;11:564–565. doi: 10.1038/embor.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Czech B, Crunk A, Wallace A, Mitreva M, Hannon GJ, Davis RE. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011;21:1462–1477. doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 13.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Ehrenkaufer GM, Pompey JM, Hackney JA, Singh U. Small RNAs with 5′-polyphosphate termini associate with a Piwi-related protein and regulate gene expression in the single-celled eukaryote Entamoeba histolytica. PLoS Pathog. 2008;4:e1000219. doi: 10.1371/journal.ppat.1000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Alramini H, Tran V, Singh U. Nucleus-localized antisense small RNAs with 5′-polyphosphate termini regulate long term transcriptional gene silencing in Entamoeba histolytica G3 strain. J. Biol. Chem. 2011;286:44467–44479. doi: 10.1074/jbc.M111.278184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito-Nakano Y, Yasuda T, Nakada-Tsukui K, Leippe M, Nozaki T. Rab5-associated vacuoles play a unique role in phagocytosis of the enteric protozoan parasite Entamoeba histolytica. J. Biol. Chem. 2004;279:49497–49507. doi: 10.1074/jbc.M403987200. [DOI] [PubMed] [Google Scholar]
- 17.WHO. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico City, Mexico 28–29 January, 1997. Epidemiol. Bull. 1997;18:13–14. [PubMed] [Google Scholar]
- 18.Zhang H, Pompey JM, Singh U. RNA interference in Entamoeba histolytica: implications for parasite biology and gene silencing. Future Microbiol. 2011;6:103–117. doi: 10.2217/fmb.10.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 20.Kaur G, Lohia A. Inhibition of gene expression with double strand RNA interference in Entamoeba histolytica. Biochem. Biophys. Res. Commun. 2004;320:1118–1122. doi: 10.1016/j.bbrc.2004.06.064. [DOI] [PubMed] [Google Scholar]
- 21.Linford AS, Moreno H, Good KR, Zhang H, Singh U, Petri WA., Jr Short hairpin RNA-mediated knockdown of protein expression in Entamoeba histolytica. BMC Microbiol. 2009;9:38. doi: 10.1186/1471-2180-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacFarlane RC, Singh U. Identification of an Entamoeba histolytica serine-, threonine-, and isoleucine-rich protein with roles in adhesion and cytotoxicity. Eukaryot. Cell. 2007;6:2139–2146. doi: 10.1128/EC.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solis CF, Santi-Rocca J, Perdomo D, Weber C, Guillen N. Use of bacterially expressed dsRNA to downregulate Entamoeba histolytica gene expression. PLoS One. 2009;4:e8424. doi: 10.1371/journal.pone.0008424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacFarlane RC, Singh U. Loss of dsRNA-based gene silencing in Entamoeba histolytica: implications for approaches to genetic analysis. Exp. Parasitol. 2008;119:296–300. doi: 10.1016/j.exppara.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baxt LA, Baker RP, Singh U, Urban S. An Entamoeba histolytica rhomboid protease with atypical specificity cleaves a surface lectin involved in phagocytosis and immune evasion. Genes Dev. 2008;22:1636–1646. doi: 10.1101/gad.1667708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell. Microbiol. 2007;9:1426–1444. doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- 27.MacFarlane RC, Singh U. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis. Infect. Immun. 2006;74:340–351. doi: 10.1128/IAI.74.1.340-351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 1978;72:431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- 29.Baxt LA, Rastew E, Bracha R, Mirelman D, Singh U. Downregulation of an Entamoeba histolytica rhomboid protease reveals roles in regulating parasite adhesion and phagocytosis. Eukaryot. Cell. 2010;9:1283–1293. doi: 10.1128/EC.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrenkaufer GM, Singh U. Transient and stable transfection in the protozoan parasite Entamoeba invadens. Mol. Biochem. Parasitol. 2012;184:59–62. doi: 10.1016/j.molbiopara.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrenkaufer GM, Hackney JA, Singh U. A developmentally regulated Myb domain protein regulates expression of a subset of stage-specific genes in Entamoeba histolytica. Cell. Microbiol. 2009;11:898–910. doi: 10.1111/j.1462-5822.2009.01300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vicente JB, Ehrenkaufer GM, Saraiva LM, Teixeira M, Singh U. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stresses: implications for amebic pathogenesis. Cell. Microbiol. 2009;11:51–69. doi: 10.1111/j.1462-5822.2008.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacFarlane RC, Shah PH, Singh U. Transcriptional profiling of Entamoeba histolytica trophozoites. Int. J. Parasitol. 2005;35:533–542. doi: 10.1016/j.ijpara.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Ehrenkaufer GM, Eichinger DJ, Singh U. Trichostatin A effects on gene expression in the protozoan parasite Entamoeba histolytica. BMC Genomics. 2007;8:216. doi: 10.1186/1471-2164-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali IK, Ehrenkaufer GM, Hackney JA, Singh U. Growth of the protozoan parasite Entamoeba histolytica in 5-azacytidine has limited effects on parasite gene expression. BMC Genomics. 2007;8:7. doi: 10.1186/1471-2164-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rastew E, Vicente JB, Singh U. Oxidative stress resistance genes contribute to the pathogenic potential of the anaerobic protozoan parasite, Entamoeba histolytica. Int. J. Parasitol. 2012;42:1007–1015. doi: 10.1016/j.ijpara.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pak J, Maniar JM, Mello CC, Fire A. Protection from feed-forward amplification in an amplified RNAi mechanism. Cell. 2012;151:885–899. doi: 10.1016/j.cell.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samuelson J, Robbins P. A simple fibril and lectin model for cyst walls of Entamoeba and perhaps Giardia. Trends Parasitol. 2011;27:17–22. doi: 10.1016/j.pt.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meneses E, Cardenas H, Zarate S, Brieba LG, Orozco E, Lopez-Camarillo C, Azuara-Liceaga E. The R2R3 Myb protein family in Entamoeba histolytica. Gene. 2010;455:32–42. doi: 10.1016/j.gene.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Mirelman D, Anbar M, Bracha R. Epigenetic transcriptional gene silencing in Entamoeba histolytica. IUBMB Life. 2008;60:598–604. doi: 10.1002/iub.96. [DOI] [PubMed] [Google Scholar]
- 41.Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 43.Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 44.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 46.Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 47.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl Acad. Sci. USA. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Ehrenkaufer GM, Hall N, Singh U. Small RNA pyrosequencing in the protozoan parasite Entamoeba histolytica reveals strain-specific small RNAs that target virulence genes. BMC Genomics. 2013;14:53. doi: 10.1186/1471-2164-14-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.