Abstract
An effective response to DNA damaging agents involves modulating numerous facets of cellular homeostasis in addition to DNA repair and cell-cycle checkpoint pathways. Fluorescence microscopy-based imaging offers the opportunity to simultaneously interrogate changes in both protein level and subcellular localization in response to DNA damaging agents at the single-cell level. We report here results from screening the yeast Green Fluorescent Protein (GFP)-fusion library to investigate global cellular protein reorganization on exposure to the alkylating agent methyl methanesulfonate (MMS). Broad groups of induced, repressed, nucleus- and cytoplasm-enriched proteins were identified. Gene Ontology and interactome analyses revealed the underlying cellular processes. Transcription factor (TF) analysis identified principal regulators of the response, and targets of all major stress-responsive TFs were enriched amongst the induced proteins. An unexpected partitioning of biological function according to the number of TFs targeting individual genes was revealed. Finally, differential modulation of ribosomal proteins depending on methyl methanesulfonate dose was shown to correlate with cell growth and with the translocation of the Sfp1 TF. We conclude that cellular responses can navigate different routes according to the extent of damage, relying on both expression and localization changes of specific proteins.
INTRODUCTION
Several DNA repair and cell-cycle checkpoint pathways have evolved to cope with damage to the genome that can arise from endogenous and exogenous sources (1,2). It is well established that effective cellular responses to DNA damaging agents involve not only modulation of canonical DNA repair and cell-cycle regulation proteins but also modulation of a large number of seemingly unrelated cellular processes (3). Previous studies identified a number of these pathways using ‘transcriptional profiling’ and ‘genomic phenotyping’ in Saccharomyces cerevisiae (4–9). Transcriptional profiling quantified global changes in mRNA levels in response to genotoxic stress using microarrays, whereas genomic phenotyping investigated the sensitivity to DNA damaging agents for almost 6000 S. cerevisiae strains, each deficient in a single gene product. A consensus emerged from these and other studies (9,10) that a general shutdown of protein synthesis occurs under conditions of DNA damage because the transcription of ribosomal protein (RP) genes is repressed under conditions of genotoxic stress. Later studies demonstrated a concomitant preferential translation of specific damage-responsive proteins under conditions of genotoxic stress (11–13), arguing for translational regulation playing a role in the cellular response to DNA damaging agents. Finally, a number of post-transcriptional protein modifications are known to orchestrate the DNA damage response, including phosphorylation, ubiquitylation and sumoylation (1,14).
To probe response pathways at the single-cell level, we developed a quantitative high-throughput fluorescence imaging approach to assess not only changes in protein levels but also changes in nuclear versus cytoplasmic localization in response to the DNA damaging agent methyl methanesulfonate (MMS). This assay was performed for >4000 S. cerevisiae strains expressing individual GFP-tagged fusion-proteins, representing nearly 70% of the yeast proteome (15). Importantly, fusion proteins in this library are expressed from their native promoters, thereby closely reflecting the response of their corresponding genes on MMS exposure.
Previously, this library was used to study genome-wide protein localization using fluorescence microscopy (15) and overall expression levels using flow cytometry (16). Here, we use the library to identify proteins whose expression level is induced or repressed in response to MMS, as well as proteins that concentrate in either the nuclear or cytoplasmic compartment in response to MMS. Analysis of Gene Ontology (GO) and protein–protein interaction networks reveal damage-induced changes in levels and/or localizations for proteins and complexes involved in chromatin remodeling, mRNA processing, RNA polymerase II transcription, proteolysis, ribosome biogenesis, metabolism, lipid synthesis, plus a number of other pathways, in addition to canonical DNA repair and cell-cycle regulation. Further, we characterize the transcription factor (TF) networks linked to changes in protein abundance, revealing a differential regulation of metabolic versus DNA-related processes. Finally, we further investigate the unexpected induction response of RPs, finding a differential response depending on the extent of damage, cellular growth rate and nuclear-to-cytoplasmic translocation of the TF Sfp1 that targets the RP genes (17).
MATERIALS AND METHODS
Cell growth and culture conditions
We used the budding yeast GFP fusion library developed by Huh and colleagues (15). The haploid parent yeast strain was ATCC 201388: MATahis3Δ1leu2Δ0met15Δ0ura3Δ0. This strain is called wild-type (WT) throughout the manuscript. Cells were cultured in minimal SD medium (MP Biomedicals) supplemented with amino acids His, Leu, Ura and Met. Cells were grown at 30°C. Cells were cultured to stationary phase for 3 days and then diluted in fresh medium and allowed to grow overnight in triplicate cultures. Log-phase cultures were incubated in growth medium with or without 0.02% MMS for 3 h. Details of plate preparation for High Content Imaging are presented in the Supplementary Information. For the initial purposes of identifying a better fixation method, either one of two protocols adapted from previous studies was followed (18,19) (Supplementary Information, Supplementary Figures S1–S7). For all subsequent experiments, the second fixation method (called Fix 2) was followed, as it caused lesser loss of GFP fluorescence and also did not introduce any additional autofluorescence (Supplementary Figures S3–S5). Plates for the screen were prepared in triplicate on a Tecan liquid handling robot (Männedorf, Switzerland) running on EVOware software.
Cell processing for flow cytometry and fluorescence microscopy
Cells were treated with 1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI) and 2.5 µg/ml Concanavalin A-Alexa647 (both from Invitrogen Life Technologies) for 30 min to stain DNA and the cell-wall, respectively. Cells used for flow cytometry were resuspended in phosphate buffered saline, whereas cells used for imaging were mounted on concanavalin A-coated 96-well plates to allow adhesion of the cells onto the bottom of the well. Excess cells were washed off and the remaining cells were mounted in 30% glycerol in phosphate buffered saline. Samples were prepared in triplicate for both control untreated and MMS-treated samples.
Flow cytometry and fluorescence microscopy
Flow cytometry was performed on an Accuri C6 Flow Cytometer (Accuri Cytometers Inc., Ann Arbor, MI), unless otherwise mentioned. Imaging was performed on a Cellomics Arrayscan VTi (Thermo Fisher Scientific, Pittsburgh, PA), using the XF93 filter to image GFP, DAPI and Alexa 647 fluorescence. A 40× 0.75 Numerical aperture (N.A.) air objective was used for imaging. High-resolution imaging was performed on an Observer Z1 microscope (Carl Zeiss, Jena, Germany) with a 100× 1.4 N.A. oil immersion objective.
Image analysis
Image analysis was performed using MATLAB (MathWorks Inc., Natick, MA) using custom-written routines for the detection of cellular boundaries from images of the cell-wall stained with Alexa 647 conjugated Concanavalin A. Clusters of cells were eliminated from calculations because nuclear versus cytoplasmic localizations could not be correctly computed for these. DAPI-stained images of DNA were used for computing nuclear positions by thresholding out the nuclei after elimination of cells, which showed uniform DAPI staining by setting cutoff conditions of intensity and area. Masks of nuclei thus obtained were used as a mask on the GFP image to compute GFP fluorescence levels in the nucleus. The rest of the cell was treated as cytoplasm, enabling the computation of nuclear to cytoplasmic ratios. In our approach, the mean GFP intensities are evaluated in the whole cell, nuclear and cytoplasmic masks. This is distinct from a previous work that used an expanded nuclear mask to evaluate cytoplasmic fluorescence (20). All raw images, data analysis programs and single cell level files associated with this study can be accessed at: http://yeastgfpscreen.mit.edu/.
Data analysis
Experiments were performed in triplicate and the mean of each replicate was calculated. The WT control was used to estimate the autofluorescence level, which was subtracted from all the measured strains in that experiment. WT controls were present in every plate. We found that for many low-expressing strains when the measured intensity is close to autofluorescence, there can be spuriously high estimates of fold-changes. Such strains with fluorescence levels close to autofluorescence in the control samples were retained in the analysis because the expression of a protein may be turned on by MMS treatment. To avoid spuriously high fold-changes, we added a constant value, c (equal to the width of the autofluorescence histogram), to every measured value. Although this leads us to underestimate the fold-changes, this ensures that only substantial changes are scored as true responders. Thus, fold-change for an experimental strain is calculated as, f = [IMMS − (IWT − c)]/[IControl − (IWT − c)], where I denotes mean intensity from triplicate samples, and the subscripts denote the conditions and strains. The autofluorescence estimated from the WT strain [as also done in a previous study (21)] is only a ballpark value, and there can be a strain-to-strain variability of true autofluorescence (16). This approach ensures that changes in expression due solely to differences in autofluorescence are not scored as responders. However, because experiments are performed in triplicate, statistical confidence can be attributed even to relatively small changes when they pass the thresholds used. The individual means, obtained from the three experiments, were compared by Student’s t-tests at P ≤ 0.05 and f ≥ 1.5 (for induced proteins) or f ≤ 0.75 (for repressed proteins). For nuclear-to-cytoplasmic ratio (NCR) changes by t-tests, P ≤ 0.05 and IΔNCRI ≥ 0.1 were used. ΔNCR = NCRControl − NCRMMS, and a negative sign indicates nuclear enrichment, whereas a positive sign indicates cytoplasmic enrichment. Typically hundreds to thousands of cells were measured for each strain. If for any particular well the cell count was ≤50, that well was eliminated from all calculations.
Protein intensity distributions over cells are often non-Gaussian. However, the means arising from repeated measurements of such distributions are normally distributed, and thus Student’s t-test can be used for comparing the means obtained from such measurements. This assumption of normality does not hold true when comparing the underlying distributions of protein intensities over cells, in which case a non-parametric test is preferable. We used the Kolmogorov–Smirnov (KS) test for determining responders in terms of levels and localization. A strain was considered to be a responder in terms of abundance if KSstat ≥ 0.3 for at least two of the three replicates and f ≥ 1.5 (for induced proteins) or f ≤ 0.75 (for repressed proteins). KSstat ≥ 0.3 for at least two of the three replicates was also used for NCR changes. For KS tests replicates were compared pairwise. For protein levels, no AF correction is performed it is not possible to determine AF for each cell. We also kept track of fold changes and subcellular localization as scored by Huh et al. (15) so that true translocations and NCR changes due to abundance changes can be distinguished.
The Cytoscape program was used for all network analyses (22,23). GO analyses were performed using the ClueGO plugin in Cytoscape (24). The YEASTRACT website was used for TF analyses (http://www.yeastract.com/) (25,26). Further details of statistical methods used can be found in the Supplementary Information File.
RESULTS
Sample preparation and analyses
We set out to monitor expression and localization changes for 4159 GFP-tagged proteins in the same number of S. cerevisiae strains after 3 h of exposure to 0.02% MMS, a relatively non-toxic dose (4,8). In the parental strain, cells showed an expected S-phase arrest and were largely viable at this time point, although the culture showed a ∼40% decrease in colony forming ability (Supplementary Figure S8–S10). To address a specific timepoint across large numbers of samples using multi-spectral fluorescence imaging of distinct cellular compartments, we developed a fixation strategy to minimize the typical attenuation of GFP signal resulting from standard yeast fixation protocols. This was particularly important to detect changes in expression levels for low copy-number proteins that are otherwise obscured by cellular autofluorescence. In a previous study of the 4159 strains expressing the GFP-tagged proteins, only 2700 proteins could be reliably detected using live-cell flow cytometry (16), and of these, the vast majority (85%) of GFP levels resided at the low-end of the expression range (16) (Supplementary Figure S1). We therefore developed an optimized aldehyde-based fixation protocol that minimized intensity loss and, importantly, exhibited a linear relation between fixed and live cell intensities across the entire range of GFP expression so that relative changes upon MMS exposure could be accurately measured (Supplementary Figures S2–S7). Strains were grown in minimal media in triplicate using a robotic liquid handler (see ‘Materials and Methods’ section for details). The importance of replicate measurements for generating greater statistical confidence in results from high-throughput screens has previously been emphasized (27), especially for detecting small but biologically significant responses.
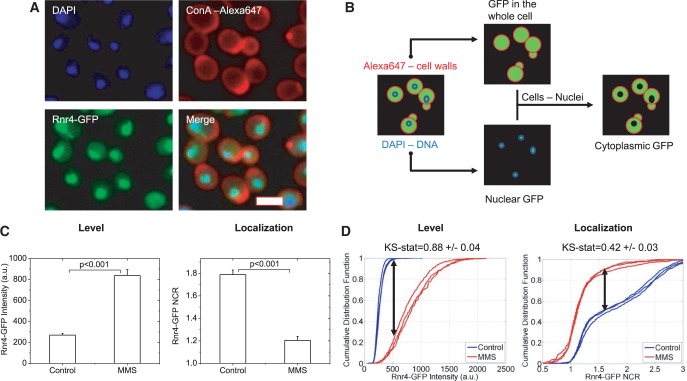
Early log-phase cultures were incubated in control medium or medium containing 0.02% MMS for 3 h. Cell were then fixed, and nuclei and cell-walls were stained. (Figure 1A). Custom-written image analysis programs were used to quantify GFP fluorescence in segmented cytoplasmic and nuclear compartments (Figure 1B, ‘Materials and Methods’ section). The computed intensities from fixed cells in this work compared well with previous studies on live cells (Supplementary Figures S11 and S12). Finally, statistical analyses were performed to identify ‘responders’ defined as strains for which protein levels were significantly induced or repressed, as well as strains for which proteins were enriched in the nucleus or cytoplasm as quantified by their NCR. Significant differences in mean expression or NCR were determined using a Student’s t-test with significance value of P < 0.05 (Figure 1C). However, because the comparison of mean values alone does not reveal differences in the underlying protein distributions themselves, the KS test was also employed (28–31). The KS statistic has been shown to be significantly more sensitive in detecting differences in population responses compared with the comparison of mean responses (29). The KS statistic represents the maximum distance between two cumulative histograms, where values equal to or greater than 0.2 are suggested to denote biological significance in the context of HCS (28,29). In addition to t-tests comparing mean responses over replicates, we used the KS statistic to identify responders in terms of abundance or NCR changes (Figure 1C and D; ‘Materials and Methods’ section). ‘Hit-lists’ for induced and repressed proteins, as well as nucleus- or cytoplasm-enriched proteins, are provided in Supplementary Table S1 as measured by both t-tests and KS tests, with a summary of some major responders presented in Table 1. Also listed is the subcellular localization of each protein, as previously determined (15), and its function as listed in the Saccharomyces Genome Database. Overall, 415 induced proteins, 174 repressed proteins, 133 nuclear-enriched proteins and 10 cytoplasm-enriched proteins were identified. Supplementary Table S2 documents intensity and NCR measurements for every strain measured.
Figure 1.
Overview of sample preparation and analysis. Plates are prepared in triplicate for control and MMS-treated samples (0.02%, 3 h). (A) Samples are fixed and stained with DAPI to mark nuclei and Alexa-647-conjugated Concanavalin A to mark cell walls. Automated imaging is performed on a Cellomics HCSTM fluorescence microscope. A typical raw image is shown for the Rnr4-GFP strain. Scalebar 5 µm. (B) Schematic of image analysis protocol. Raw cell-wall and DNA fluorescence images are used to segment cellular and nuclear boundaries. Mean GFP pixel intensity is evaluated in the full cellular image mask to determine relative protein abundance. The ratio of intensities between the nuclear and cytoplasmic masks is used to determine the NCR on a cell-by-cell basis. (C) Mean response of Rnr4-GFP to DNA damage in terms of both levels and localization. Protein is induced as measure by autofluorescence (AF)-corrected levels and the NCR decreases significantly indicating that the Rnr4-GFP translocates to the cytoplasm on damage. Error bars are standard deviations of the means from the three replicates. P-values are determined using a Student’s t-test. (D) Mean response over cells does not account for the distinct distributions over cell populations. Thus, the same sample as in (C) is evaluated using the KS statistic. The KS statistic is a measure of the maximum distance between the normalized cumulative distribution histograms (indicated by the black double-headed arrows). The KS statistic is independently evaluated for each sample pair. Again, a significantly higher expression and cytoplasmic translocation of Rnr4-GFP is seen with DNA damage by MMS. In all instances, a KS-statistic cutoff of 0.3 is used. At least 200 cells are measured for each curve here.
Table 1.
Some major responders in all investigated categories
 |
The top 20 responders (where applicable) in the induced, repressed, nucleus- and cytoplasm-enriched categories are shown. In the blue rows are significant responders by t-tests, and in the pink rows are the significant responders by KS tests. For the induced and repressed categories, the responders are arranged according to maximal fold change. For the nucleus- or cytoplasm-enriched categories, responders are arranged by maximal NCR change for t-tests and by maximal KS statistic for KS tests. For the complete list of responders, see Supplementary Table S1.
Induced proteins and their interaction networks represent a wide range of cellular processes including DNA repair, proteolysis, chromatin remodeling and ribosome biogenesis
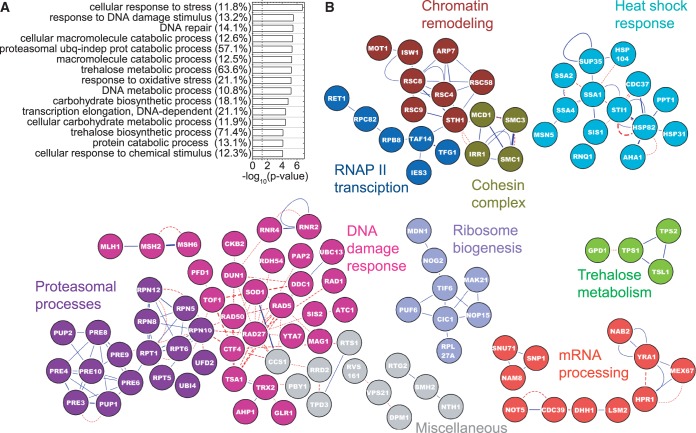
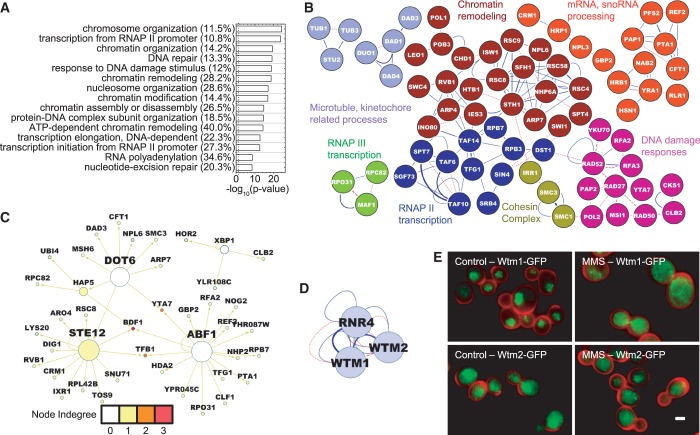
GO functional enrichment analysis for the 415 induced proteins using the ClueGO plugin in Cytoscape (22–24) revealed an expected enrichment of GO-terms related to DNA damage response, DNA repair and cellular stress, as well as other GO-terms related to cellular metabolism, protein degradation, chromatin remodeling, RNA polymerase II transcription, mRNA processing and ribosome biogenesis (Supplementary Table S3 and Figure 2A). Although the absolute number of genes associated with some GO terms can be low, they may still be scored as significant when they represent a large fraction of the total genes associated with that term. For example, 8/14 (57%) proteins associated with the ubiquitin-independent proteasomal machinery, and 5/7 (71%) proteins associated with trehalose metabolism are induced by MMS treatment (Figure 2A). The first indication that proteasome function is enhanced and that trehalose metabolism is affected upon/by MMS exposure came from transcriptional profiling studies (6,7). Subsequent work has now shown that proteasome-mediated responses are directly involved in DNA repair (32), and that trehalose protects cells against different DNA damaging agents (33–35). Thus, although these and the other GO-enriched processes may initially seem disparate, taken together they represent a coordinated response to cellular insult by MMS.
Figure 2.
Functional enrichment and protein–protein interactions of induced proteins. Proteins that showed fold change, f ≥ 1.5 and passed the statistical criteria were used. (A) The top 15 functional categories as determined by a GO analysis using Cytoscape are shown. Negative logarithms to the base 10 of the P-values for the GO terms are plotted. Numbers in parentheses show the percentage of genes associated with a GO term, which are found to be induced. The dashed line shows the position of P = 0.05 in all figures. (B) Induced proteins are projected onto a yeast interactome. Blue lines denote physical protein–protein interactions, and red lines denote genetic interactions. The weakest edges have been removed to parse out isolated modules. Isolated single nodes are not shown. The text on the network is color coded to represent the broad cellular processes represented by the corresponding nodes. Gray nodes do not conform to these categories but are still not isolated single nodes.
Mapping responders onto a previously compiled yeast interactome (36) enabled identification of functional networks of induced proteins. The full interactome annotates physical, genetic and TF interactions. The smaller network that resulted from mapping MMS-induced proteins onto the full interactome had higher connectivity, higher clustering coefficient, and a greater ‘Large Connected Component’ (LCC) than random networks of similar size (Supplementary Figure S13). These general features were previously observed in network analyses of toxicity modulating genes in genomic phenotyping studies (37). Although the protein–protein and genetic interaction maps revealed a number of clusters that were expected from the GO functional enrichment analysis (namely, DNA damage response, ribosome biogenesis, chromatin remodeling, RNAPII transcription and proteolysis), such mapping also highlighted additional functions that were not represented in the GO analysis (Figure 2B). For example, all four proteins known to be associated with the cohesin complex (38) are induced. This group was not represented in the GO analysis because the number of proteins in the cohesin complex is so small that it falls below the threshold used for filtering GO terms. It turns out that previous studies also demonstrated a role for the cohesin complex in DNA damage response beyond its conventional role in sister-chromatid cohesion (39–41). We also identified a distinct cluster of heat shock proteins (Figure 2B), consistent with recent studies that demonstrate links between heat shock and DNA damage responses (42,43). Finally, a group of proteins involved in mRNA processing was present among the subnetworks of induced proteins. Interestingly, a recent genomic phenotyping study involving both essential and non-essential genes also highlighted the importance of mRNA processing and splicing proteins in governing sensitivity to the toxic effects of MMS (8). MMS also induces damage to proteins, RNA and other cellular components, in addition to DNA. The modulation of a large number of RNA processing and ribosomal genes may also be signaled from protein and RNA damage in conjunction with the direct DNA damage response.
Partitioning of biological function among induced proteins according to number of upstream TFs
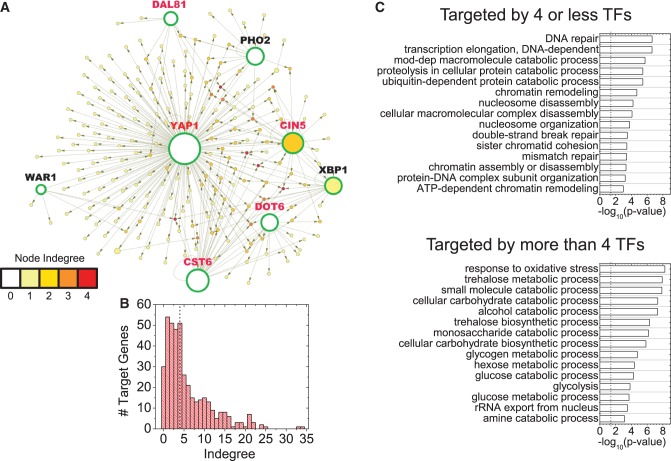
In addition to physical and genetic interactions, we examined TF interactions by mapping induced proteins onto the global TF network constructed from the YEASTRACT database (25,26,36), thereby identifying eight TFs that are themselves induced (Yap1, Cst6, Cin5, Dot6, Xbp1, Pho2, Dal81, War1—circled in green in Figure 3A); these eight TFs collectively have the potential to regulate the expression of 54% of the induced proteins (Figure 3A). Deletion mutants for five of the eight TFs (Yap1, Cst6, Cin5, Dot6, Dal81—see red text in Figure 3A) showed MMS sensitivity in the genomic phenotyping assay (5), and Xbp1 is known to be a stress-responsive TF. In general, 11% of all possible targets for these eight TFs are represented among the induced proteins. Although this is higher than the 6.5% that would be expected for random networks of the same size, it is still a relatively small fraction of all possible targets. Although this may be due in part to the fact that the GFP library represents only 70% of the yeast genome, it more likely indicates a tendency for cells to require combinations of TFs to modulate gene expression in response to environmental challenges, rather than allowing promiscuous non-specific upregulation of all possible targets for a single TF. Although a number of the induced proteins are targeted by more than one of the eight TFs (i.e. have an indegree >1, Figure 3A), these eight MMS-induced TFs by no means represent all of the TFs that are potentially capable of regulating the induced proteins.
Figure 3.
TF network of induced proteins. (A) The TF network filtered from the yeast interactome (36) with induced proteins projected onto it. Arrows point away from TFs towards their targets, and the size of nodes relate to their outdegree or number of arrows pointing away from a node. Thus, larger nodes are upstream TFs that target the expression of many of the induced proteins. The eight largest nodes are correspondingly indicated by large font and are circled in green. Deletion mutants of the five TFs marked with red font showed sensitivity in the genomic phenotyping study (5). The color of a node relates to indegree, representing the number of incoming arrows at a node and therefore indicating the number of TFs that target the node. Warmer colored nodes are targeted by multiple TFs. Isolated nodes are not shown. (B) Fifty-nine major TFs, each of which target at least 5% of the induced proteins, were identified. Their targets mapped onto the TF network of induced proteins. As before, node indegree indicates the number of TFs controlling the expression of an induced protein, with node indegree distribution shown here. The black dashed line indicates the median of the distribution at four. (C) GO analyses were performed separately on the 234 proteins with indegree less than or equal to four, representing the relatively sparser nodes and the 176 proteins with indegree greater than four representing targets controlled by many TFs. The first group was enriched in DNA-related processes, whereas the second show a clear enrichment for metabolic processes. The top 15 GO categories are shown.
To obtain a comprehensive picture of all TFs that might govern the expression of induced proteins, irrespective of whether the TFs themselves were induced, we used the updated YEASTRACT database that documents all known major TF interactions in the yeast genome (25,26). This analysis identified 59 TFs, each of which potentially governs the expression of at least 5% of the induced proteins. Most induced genes were governed by more than one of the 59 TFs, with four as the median value of node in-degree, which represents the number of TFs governing the expression of a target protein (Figure 3B).
To test whether the induced protein nodes that can be governed by a large number of TFs serve distinct biological functions compared with more isolated nodes, we filtered out genes that can be targeted by four or fewer of the 59 putative TFs (representing proteins that are regulated by relatively few TFs) from genes that can be targeted by more than four TFs (representing proteins that are regulated by many TFs). GO analysis for these two sets of MMS-induced proteins revealed a clear difference in functional enrichment, with P-values as low as or lower than those for the combined set of all induced proteins shown in Figure 2A (Figure 3C; Supplementary Table S4). Interestingly, induced proteins associated primarily with metabolic processes were found to be targeted by five or more TFs, whereas DNA damage response and chromatin remodeling proteins represent the more isolated nodes that are targeted by four or fewer TFs. In the full transcription network, the median value for indegree is also four, which is similar to the subset of MMS-induced proteins (Supplementary Figure S14). Not surprisingly, when genes are partitioned as mentioned previously in the full network, DNA-related processes do not dominate the more isolated nodes because all cellular functions are now represented. However, metabolic processes continue to be over-represented among the nodes with five or more upstream TFs, indicating that this may be a generic feature of the transcriptional network (Supplementary Figure S14).
Enrichment of lipid biosynthesis and membrane trafficking processes among repressed proteins and networks
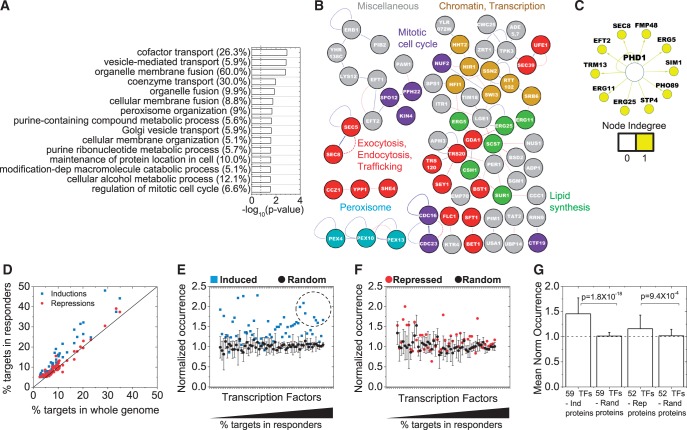
In addition to induced genes, transcriptional profiling studies have revealed many genes whose transcripts are downregulated by MMS (6,7,9). At the protein level, one might expect fewer repression responders due to the longer half-lives of proteins compared with mRNAs, except in the case of targeted protein degradation. In other words, even for a gene that is transcriptionally silenced, its protein products may remain in the cell for some time, particularly under conditions of inhibited growth. In our screen, 174 repressed proteins were identified, even with a lower cut-off threshold of 25% reduction compared with the 415-induced proteins that showed >50% induction. P-values for GO-function enrichment were generally larger for repressed versus induced proteins (Supplementary Table S3 and Figure 4A), and the network of repressed proteins had lower connectivity than the induced proteins as measured by their connectivity that was comparable with random networks of the same size (Supplementary Figure S15). However, cellular membrane organization, secretion, trafficking and peroxisome organization were significantly over-represented among the repressed proteins (Figure 4A and B). Also, the single repressed TF, Phd1, is known to control a number of sterol biosynthesis genes known to be important for lipid and membrane biosynthesis (Figure 4C). Finally, several proteins involved in chromatin remodeling and regulation of the mitotic cell-cycle were also part of the network of repressed proteins (Figure 4B), even though none of these processes was significantly enriched in the GO analysis (Figure 4A).
Figure 4.
Functional enrichment, protein interactions of the repressed proteins and comparison of TF interactions with induction responders. Proteins that showed fold change, f ≤ 0.75 and passed the statistical criteria were used. (A) Top 15 GO categories; (B) Protein–protein interaction networks; and (C) TF interaction networks for the repressed protein responders. The weakest links by edge weight have not been removed in the interaction network in (B), otherwise all representations are similar to Figure 2. Numbers in parentheses illustrate the percentage of genes associated with a GO term. (D) Fifty-two TFs, each of which controls the expression of at least 5% of the repressed proteins, were identified. Major TFs that target the repressed proteins are largely similar to those that target the induced proteins (47/52 TFs). The percentages of targets for TFs in the induction and repression responder lists are plotted against the percentage of targets from the whole genome. Each dot represents a TF, with the solid black line illustrating the 1:1 line expected by random chance. The percentage of targets in the responder lists are divided by the percentage targets in the whole genome for these TFs factors for the (E) Induced and (F) Repressed proteins. This ‘normalized occurrence’ should be one in the absence of specific enrichment or depletion of the targets of a TF in a list of genes. For comparison, we generated five random networks of the same size as the Induced or Repressed protein lists and analyzed them similarly for targets of the chosen major TFs. The means and standard deviations of these random samples are plotted in black. For many TFs, the normalized occurrence in Inductions lies beyond the standard deviation of the normalized occurrence of the random samples. TFs with higher percentage of targets among the induced or repressed proteins are on the right. The dashed circle marks TFs that have both high normalized occurrence and a large number of targets in the genome (Yap1, Met4, Rpn4, Msn2, Msn4, Hsf1 and Pdr3). Many of these are stress-responsive TFs. (G) The mean and standard deviation of normalized occurrence from (E) and (F) for all TFs are plotted. The dashed line shows the expected value for random networks.
We used the YEASTRACT database to identify 52 TFs that can target the expression of 5% or more of the repressed proteins. Most of these (47/52) are found among the TFs that target genes for the induced proteins, presumably because some of these TFs (e.g. Yap1, Sfp1 and Ste12) have high numbers of targets in the genome (Figure 4D). However, although we observed highly significant enrichment of the targets of these TFs for the induced proteins (P = 1.8 × 10−18 overall, Figure 4G), the enrichment was not nearly so high (P = 9.4 × 10−4 overall, Figure 4G) for repressed proteins (Figure 4E–G). The number of targets for each TF in both lists was divided by the number of targets expected by random chance from the whole genome, to assess specific enrichment of the targets for a given TF in each list. This measures the normalized occurrence for each TF (thus a normalized occurrence of 1.5 indicates that 50% more targets are present in the induced or repressed list when compared with random lists of the same size that exhibit a mean normalized occurrence of 1). This analysis revealed the targets of TFs for all three major stress-responsive pathways in yeast [genes with heat shock elements, stress response elements or AP-1 responsive elements (6,9,44–46)] to be particularly enriched among the induced proteins. TFs Yap1, Msn2, Msn4, Rpn4, Hsf1, Met4 all exhibit a high normalized occurrence among the induced proteins (Figure 4E, dotted circle). There were other TFs that showed yet higher normalized occurrence, but the absolute number of targets for these TFs in the genome was low, and in effect, they targeted only several of the induced genes.
Signatures of translational regulation of induced proteins
Cells use both transcription and translation to regulate gene expression. Transcriptional responses to DNA damaging agents are well documented (6,9), and recent studies have unveiled a translational component to such responses (12,13). The study by Begley et al. (2007) computationally identified a set of 425 genes with a skewed codon usage pattern such that their translation would be promoted by the Trm9 tRNA methyltransferase that catalyzes specific tRNA modifications that change codon-anticodon affinity. Such modifications affect the efficiency of translation for a subset of transcripts rich in specific codons, especially under conditions of DNA damage (13). Ribosomal, metabolism and stress response genes were enriched in this group of 425 potential preferentially translated (PPT) genes. However, belonging to the PPT group does not ensure induction under conditions of damage, for transcriptional components can offset translational responses. Despite this, we find 57 PPT proteins among the MMS-induced proteins, approximately twice the number that is expected by random chance (Supplementary Figure S16A). These 57 proteins form a close network with several RP genes; GO analysis reveals enrichment of processes related to carbohydrate and trehalose metabolism, ribosome biogenesis, oxidative stress and deoxyribonucleotide production (Supplementary Figure S16B and C).
Nucleus and cytoplasm enriched proteins and networks
The list of proteins that become enriched in the nucleus in response to MMS includes two categories: (i) nuclear proteins whose relative expression increases in response to MMS and (ii) proteins that translocate from the cytoplasm to the nucleus. Although not all induced nuclear proteins are represented in this list, instances where the NCR increases simply reflects the induction of a protein were retained because they captured subtle expression increases in the nucleus not represented in the total list of induced proteins (Supplementary Table S1). For example, Ubi4, yeast ubiquitin, is scored as both an induction and a nuclear enrichment responder, but Rxt2, a subunit of the histone deacetylase Rpd3 complex implicated in the activation of DNA damage induced genes (47) is found only in the nuclear enrichment list. Remarkably, GO analysis of this relatively small list of proteins (133) produced extraordinarily high-significance values for functional enrichments, and perhaps not surprisingly, most of the nuclear-enriched functional categories pertained to DNA and RNAPII-related processes, with a notable absence of the metabolic, proteasomal and ribosomal processes seen in the total list of inductions. P-values for functional enrichment (Supplementary Table S3 and Figure 5A) were substantially lower even than those for the induced proteins as a whole (Figure 2A), and the network of nuclear-enriched proteins exhibited connectivity far greater than what would be expected for similarly sized random networks, reflecting the close functional interactions among these proteins (Supplementary Figure S17). For several GO terms associated with the nuclear-enriched proteins, >25% of all associated proteins were represented (Figure 5A). The physical and genetic protein interaction networks were dominated by components of chromatin remodeling, RNAP II-dependent transcription, plus mRNA and snoRNA processing proteins. Components of the cohesin complex were also present among the nuclear-enriched proteins.
Figure 5.
Functional enrichment and protein–protein and TF interactions of the nuclear or cytoplasmic enrichment responders. (A) The nuclear enriched protein list exhibits very significant enrichment of DNA-related terms, despite being smaller in size than both induction and repression responder lists. Top 15 GO terms are shown. The numbers in parentheses show the percentage of genes associated with a GO term. (B) The protein–protein interaction networks too show functionally clustered modules. The weakest links have been removed like the induced proteins. In addition to DNA damage responsive proteins, a large number of chromatin remodelers, transcription components and mRNA processing proteins make up this network. (C) The TF network identifies two additional TFs, Ste12 and Abf1. (D) Rnr4, Wtm1, Wtm2 make a close connected network in the few cytoplasm-enriched proteins. (E) The cytoplasmic translocation of Wtm1-GFP or Wtm2-GFP shown with high resolution 100 × 1.4 N.A. images. The cell wall is shown in red, whereas the GFP-tagged protein is shown in green. Many cells show cytoplasmic protein upon MMS treatment, whereas some still have nuclear protein. The scalebar is 2 µm. Representations in (B), (C) and (D) are similar to Figure 2.
TF network analysis identified three nuclear-enriched TFs (Ste12, Abf1, Dot6) that regulate a number of proteins in the list of nuclear-enriched proteins. Although other TFs such as Ixr1 previously implicated in the DNA damage response (48,49) were not part of this network, they were present in the list of nuclear enrichments (Supplementary Table S1). These are instances of subtle inductions that were only seen in the nucleus but missed in the total list of induced proteins. Conversely, two proteins that are a part of the induction list and known to be nuclear-enriched upon damage (Ubc13, Yap1) were not found in this study. Although Yap1 with a KS-statistic value of 0.28 lay just below the cutoff of 0.3 for nuclear enrichment, it should be noted that in previous work, more than a 10-fold higher MMS dose was used to induce its nuclear translocation, compared with the present study (50). Thus, nuclear enrichment of a protein can be dose-dependent.
Evidence from several distinct lines of work have demonstrated connections between chromatin remodeling and the DNA damage response in eukaryotes (4,6,7,9,47,51–53). Our data (Figures 2B and 5B) also indicate extensive changes in proteins for the remodeling of chromatin and for nucleosome disassembly that are mobilized on exposure to MMS, presumably to allow repair machinery access to sites of DNA lesions.
Surprisingly, few cytoplasmic enrichments (10 proteins) were identified in our screen (Supplementary Table S1). This may in part be due to the fact that the cytoplasmic volume is significantly larger than the nuclear volume, rendering it difficult to detect translocation of low-expressed proteins. However, even amongst this small number of proteins, interesting features stand out. For example, we find that the Wtm1 and Wtm2 proteins translocate out of the nucleus into the cytoplasm in response to MMS, as does Rnr4 (Figure 5D and E). Wtm proteins are involved in nuclear anchoring of the RNR small-subunits (54–57), with one study previously implicating the Wtm proteins in the control of RNR transcription (58). It appears that controlling the subcellular localization of the Wtm proteins may provide an additional mode of RNR regulation.
RP response to MMS is dose dependent
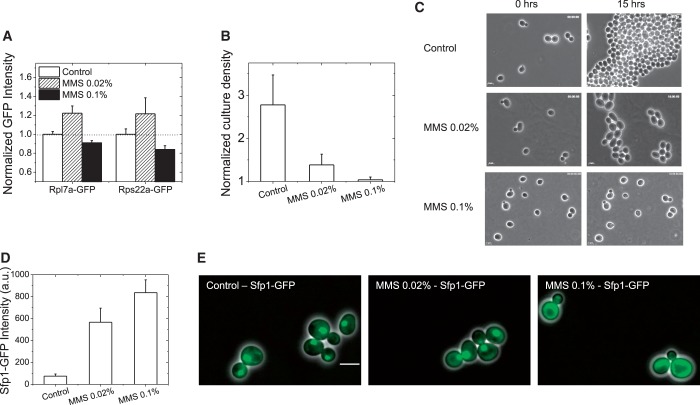
A surprising GO category for induced proteins was ribosome biogenesis and components of the ribosomal machinery because previous studies from several groups have shown that ribosomal genes are generally transcriptionally repressed under conditions of DNA damage (6,7,9,10). The regulation of ribosomal genes is thought to be primarily at the level of transcription in yeast (59), but almost 90% of all RP genes are also found in the list of PPT genes (13), suggesting cells use both transcription and translation to tune ribosome numbers (also see Supplementary Figure S16). A recent study similar to ours (discussed in more detail later in the text) also identified this group among induced proteins (20). To investigate this apparent discrepancy, we examined two such induced RPs (Rpl7a-GFP and Rps22a-GFP) using live-cell flow cytometry. We found that at moderate doses of MMS (0.02%, 2 h), both proteins were induced, whereas at higher MMS dose (0.1%, 2 h), there was a small but significant repression of the proteins (Figure 6A). The 2-h time-point was chosen because it was intermediate between the 3-h time-point at which the present screen is performed, and the 1-h time-point of the first transcriptional profiling study that revealed transcriptional repression of the ribosomal genes at 0.1% MMS (7). This suggests, not surprisingly, that cellular response differs according to the extent of damage, presumably depending on whether the cell can repair damage and proceed through the cell-cycle, or whether growth halts completely. A similar pattern of induction was seen in Tandem Affinity Purification (TAP)-tagged strains of Rpl7a and Rps22a (Supplementary Figure S18). Because ribosome biogenesis is intimately linked with cell growth, we investigated how cell growth characteristics differ at the two MMS doses.
Figure 6.
Differential response of RPs to different levels of MMS damage. The response of two candidate induced RPs (Rpl7a-GFP, Rps22a-GFP) to MMS damage was investigated with live-cell flow cytometry, at 2 h. (A) At 0.02% MMS, the RPs are induced, whereas at the higher dose (0.1%), they are repressed. (B) Parental BY4741 cells were subjected to similar treatments, and culture densities (events/µl) were determined by flow cytometry (optical density measurements are expected to be not accurate at these early time points, as MMS arrest can cause an increase in cell-size). For three early log-phase cultures, initial densities were measured, and a third of each culture was subjected to 0, 0.02%, 0.1% MMS treatment. All densities at 2 h are normalized by the density of the initial cultures. 0.02% MMS significantly retards growth, but does not abrogate it altogether, whereas there is almost no growth in the presence of 0.1% MMS. (C) In a different assay from liquid cultures, even for cells growing on agar-pads with or without MMS, there is slow growth with 0.02% MMS, whereas no growth is seen at 0.1% MMS. Images of the same fields at 0 and 15 h are shown. (D) The GFP-tagged TF Sfp1, which controls RP gene expression, is induced at both doses of MMS as determined by live-cell flow cytometry. AF-corrected intensities are plotted; however, the expression of the protein is low in the absence of damage, making estimations of fold change difficult. (E) At 0.02% MMS, the protein is still nuclear, whereas at 0.1% MMS, it is clearly cytoplasmic, concomitant with the induction and repression of the RP proteins. Live cells were imaged with a 100× oil-immersion objective, given the low expression of the protein. Overlays of GFP and phase images are presented. The scalebar is 5 µm.
Using both liquid and agar assays, we found that at 0.02%, MMS cells proceed through the cell-cycle, albeit slowly, whereas at 0.1%, MMS cell growth is abrogated entirely (Figure 6B and C, Supplementary Figure S19). Conditions of general cellular stress inhibit growth, and at the same time, ribosome biogenesis in a Target of Rapamycin (TOR)-dependent manner (17,59). The TOR pathway has also been shown to control DNA damage responses by controlling dNTP production (60), and the TOR pathway effectors Sch9 and Sfp1 are known to be involved in both ribosome biogenesis and stress responses (17,61,62). Sfp1 is a nuclear-localized TF that regulates RP expression and translocates to the cytoplasm in response to various stresses including DNA damage by MMS (0.1%), thus turning off RP gene expression (17). Sfp1 itself is also induced by MMS damage, which may appear to conflict with the aim of shutting down ribosome biogenesis (63). We therefore investigated the levels and localization of Sfp1-GFP in response to moderate and high doses of MMS. At both doses, Sfp1-GFP was induced as assessed by live-cell flow cytometry (Figure 6D). Indeed, although Sfp1-GFP showed only a 27% increase in the original screen, and was not scored as induced (only proteins with >50% increase in expression were considered to be responders), this small increase was significant (P = 0.003). Expression of Sfp1-GFP in the absence of damage is low, making estimations of fold change difficult. Our conservative estimates of fold change in the global screen are systematically underestimated when the fluorescence signal is close to cellular autofluorescence (see ‘Materials and Methods’ section), causing Sfp1-GFP to be absent from the list of induced proteins. However, when observed with higher resolution microscopy, at the moderate MMS dose (0.02%), Sfp1-GFP was induced and clearly nuclear, thus being available to upregulate the expression of RP genes. At the higher dose (0.1% MMS), Sfp1-GFP, although still present at induced levels, became cytoplasmic, concomitant with the repression of ribosomal genes. This provides a clear example where tuning both the abundance and the nuclear-versus-cytoplasmic localization of a protein can be used by cells to effect induction and repression of gene expression.
DISCUSSION
Identification of repressed proteins
The present study provides a comprehensive single-cell-level view of an orchestrated cellular response to damage induced by MMS. Previous studies using the yeast GFP fusion library to interrogate protein level changes in response to MMS, by imaging (20) or flow cytometry (21) used a single replicate of live cells. Here, fixation of cells was essential to ensure a specific and consistent exposure time to the damaging agent, as well as unambiguous identification of the cell nucleus and cell-wall without expression of an additional fluorescent protein as a nuclear marker. In previous studies (20,21), no proteins met the cutoff set for downregulation, even though many genes are known to be transcriptionally repressed in response to MMS (6,7,9). As discussed, protein level repressions may manifest as small changes at early time points after damage exposure, and hence replicate measurements may be necessary to identify them reliably. Similar doses and time points were used in all these studies: Lee et al. used 0.02% MMS for 4 h; Tkach et al. used 0.03% MMS for 2 h, whereas we use 0.02% MMS for 3 h. Here, triplicate measurements enabled the identification of a number of proteins whose expressions are reduced on DNA damage. Processes of membrane-trafficking, lipid synthesis and peroxisome function were enriched among the repressed proteins. In breast cancer models, mutations in the critical tumor suppressor protein p53 have been associated with expression of sterol biosynthesis genes (64). In mouse liver, activation of the protein Nrf2 by oxidative stress has been shown to be associated with downregulation of lipid biosynthesis, to allow the scavenging of reactive oxygen species (ROS) by reduced nicotinamide adenine dinucleotide phosphate (NADPH) that functions both in lipid metabolism pathways and in ROS scavenging (65,66). Peroxisomes may act as a source of ROS that affect nucleic acids, proteins and lipid, and they are kept under tight control in yeast (67); indeed, peroxisome proliferation causes oxidative DNA damage in rat livers and plays a role in hepatocarcinogensis (68). Although these findings span a number of distinct model systems, they provide plausible explanations for the genes repressed by MMS treatment, as seen in this study, and indicate that these may be general features of regulation under conditions of damage.
Comparisons with other studies
Despite the attenuation of GFP signals on fixation, the intensities measured in fixed cells in this work compared well with previous studies on live cells performed with both flow cytometry and imaging methods (16,20) (Supplementary Figures S11 and S12). Further, MMS-induced protein groups are largely similar between this study and that by Tkach et al. (20). In terms of protein localization, manual annotation of protein localization in the prior study allowed finer discrimination of subcellular localization into organelles, but at the cost of replicate measurements. In comparison, here, we measure broad nuclear versus cytoplasmic localization in a fully automated manner and identify several highly nuclear-enriched DNA-related processes, as well as abundance changes that span a wide variety of cellular responses. Although we lose detail in the granularity of subcellular organelles, we detect GFP intensity automatically in the full nuclear and cytoplasmic masks, combined with greater statistical confidence from replicate measurements.
The number of responders identified in this study is substantially fewer than the transcriptional profiling studies. However, comparison with an early transcriptional profiling study from our laboratory (7) reveals that 62% of all the induced proteins are also transcriptionally induced in response to MMS. In contrast, only 6.3% of the repressed proteins were found to be transcriptionally repressed. Indeed, although correlations between transcript and protein levels in unperturbed cell populations are poor (69,70), it has recently been shown that under conditions of environmental stress, transcript induction correlates well with protein induction, but transcript reduction produces negligible change in protein levels (71). Protein level repression under conditions of DNA damage presumably occurs due to targeted degradation (72) instead of transcriptional repression. Several studies from our laboratory, including the present one, have indicated a key link between the proteasomal machinery and response to MMS-induced damage (6,73).
TF network analysis
Extensive TF analyses identified Yap1 as a major induced TF. Yap1 has many targets in the yeast genome, and its role in stress responses is well-established (45,50,74). In addition to causing direct alkylation damage, MMS can also elicit an oxidative stress response in cells, and Yap1 was recently shown to be a major regulator of this response (74). We also found specific enrichment of the targets of several other major stress-responsive TFs (Rpn4, Msn2, Msn4, Hsf1 and Met4) among the induced proteins. Combinations of a large number of TFs were upstream of the observed metabolic response genes, whereas fewer TFs directly targeted genes for DNA-related processes.
It should be borne in mind that the network analyses performed here projects responders onto static protein–protein, genetic and TF interaction networks. It is thought that genetic interactions may shift on DNA damage, whereas physical interactions remain largely unchanged (75). TF combinations affecting a gene may also shift on DNA damage (49), and the periodically updated YEASTRACT database documents most known TF interactions under various conditions from different sources, based on both bioinformatic and experimental data. However, under any one condition, only a subset of the targets is likely to be active for a given TF.
DNA-related processes in nuclear-enriched proteins
Interestingly, nuclear-enriched proteins are found to form a closely connected network, with a corresponding significant enrichment of DNA-related processes that include transcription, chromatin remodeling and DNA repair, and a notable absence of proteins of the metabolic, ribosomal and proteolytic machinery. Thus, the list of nuclear-enriched proteins clearly segregates the central DNA level responses from the other cellular processes that together orchestrate the total cellular response to MMS insult.
Differential response of RPs according to MMS dose
Finally, differing responses for several RPs were observed depending on the MMS dose. These responses correlated with altered cell growth and the cytoplasmic translocation of the Sfp1 TF that results in shutting off RP gene expression. Although Sfp1 is a central player for RP gene expression, other factors also affect decisions to either stall growth or to repair damage and proceed through the cell-cycle. Recently, proteins of the cohesin complex have been shown to be involved in RP gene regulation (76), in addition to direct DNA damage responses (39,40). The cohesin complex proteins are upregulated under the conditions of damage used here, which could in turn feed back onto RP expression. Future work will explore whether the upregulation of specific RPs is for the replacement of damaged ribosomal components (77,78) or to make functionally specialized stress-specific ribosomes (79,80) or perhaps both.
Outlook
Taken together, our work presents a global systems-level proteomic view of the cellular response to MMS damage. Transcription may represent the first level of regulation, but now, we reveal the protein responders in terms of levels and broad subcellular localization. Although focused studies investigate a few genes in isolation, intricate connections within the yeast proteome suggest that no response can be isolated from cascading effects within the network. Yet, despite the seemingly daunting complexity, the overall picture that emerges reveals a coordinated response in terms of DNA repair, chromatin remodeling, proteolysis and cell growth, indicating that the system is tuned to buffer a fairly large range of genotoxic challenges.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [81–95].
FUNDING
NCI [R01-CA055042] [now NIEHS R01-ES022872]; CEHS center grant [NIEHS P30-ES002109]; KI center grant [NCI U54-CA112967]; Cancer Center Support (core) [NCI P30-CA14051]. Funding for open access charge: NIEHS [R01-ES022872].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors also acknowledge the MIT Faculty Start-up Funds and the Samuel A. Goldblith Career Development Professorship to M.B., and the CSBi Merck-MIT postdoctoral fellowship to A.M. L.D.S. is an American Cancer Society Research Professor. They thank Professor Gerald Fink for the gift of the TAP-tagged yeast strains, and Professor Wendy Gilbert and Dr Sara Gosline for useful discussions.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, D.C: American Society for Microbiology Press; 2006. [Google Scholar]
- 2.Langerak P, Russell P. Regulatory networks integrating cell cycle control with DNA damage checkpoints and double-strand break repair. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:3562–3571. doi: 10.1098/rstb.2011.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fry RC, Begley TJ, Samson LD. Genome-wide responses to DNA-damaging agents. Annu. Rev. Microbiol. 2005;59:357–377. doi: 10.1146/annurev.micro.59.031805.133658. [DOI] [PubMed] [Google Scholar]
- 4.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol. Cancer. Res. 2002;1:103–112. [PubMed] [Google Scholar]
- 5.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol. Cell. 2004;16:117–125. doi: 10.1016/j.molcel.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Jelinsky SA, Estep P, Church GM, Samson LD. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl Acad. Sci. USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svensson JP, Pesudo LQ, Fry RC, Adeleye YA, Carmichael P, Samson LD. Genomic phenotyping of the essential and non-essential yeast genome detects novel pathways for alkylation resistance. BMC Syst. Biol. 2011;5:157. doi: 10.1186/1752-0509-5-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powley IR, Kondrashov A, Young LA, Dobbyn HC, Hill K, Cannell IG, Stoneley M, Kong YW, Cotes JA, Smith GC, et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 2009;23:1207–1220. doi: 10.1101/gad.516509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol. Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 13.Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S, Zhao X. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol. Cell. 2012;45:422–432. doi: 10.1016/j.molcel.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 16.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 17.Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O’Shea EK. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl Acad. Sci. USA. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossanese OW, Soderholm J, Bevis BJ, Sears IB, O’Connor J, Williamson EK, Glick BS. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J. Cell. Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C, et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell. Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee MW, Kim BJ, Choi HK, Ryu MJ, Kim SB, Kang KM, Cho EJ, Youn HD, Huh WK, Kim ST. Global protein expression profiling of budding yeast in response to DNA damage. Yeast. 2007;24:145–154. doi: 10.1002/yea.1446. [DOI] [PubMed] [Google Scholar]
- 22.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdulrehman D, Monteiro PT, Teixeira MC, Mira NP, Lourenco AB, dos Santos SC, Cabrito TR, Francisco AP, Madeira SC, Aires RS, et al. YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res. 2011;39:D136–D140. doi: 10.1093/nar/gkq964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, Alenquer M, Freitas AT, Oliveira AL, Sa-Correia I. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat. Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 28.Giuliano KA, Chen YT, Taylor DL. High-content screening with siRNA optimizes a cell biological approach to drug discovery: defining the role of P53 activation in the cellular response to anticancer drugs. J. Biomol. Screen. 2004;9:557–568. doi: 10.1177/1087057104265387. [DOI] [PubMed] [Google Scholar]
- 29.Giuliano KA, Johnston PA, Gough A, Taylor DL. Systems cell biology based on high-content screening. Methods Enzymol. 2006;414:601–619. doi: 10.1016/S0076-6879(06)14031-8. [DOI] [PubMed] [Google Scholar]
- 30.Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 31.Wilson CJ, Si Y, Thompsons CM, Smellie A, Ashwell MA, Liu JF, Ye P, Yohannes D, Ng SC. Identification of a small molecule that induces mitotic arrest using a simplified high-content screening assay and data analysis method. J. Biomol. Screen. 2006;11:21–28. doi: 10.1177/1087057105280726. [DOI] [PubMed] [Google Scholar]
- 32.Ben-Aroya S, Agmon N, Yuen K, Kwok T, McManus K, Kupiec M, Hieter P. Proteasome nuclear activity affects chromosome stability by controlling the turnover of Mms22, a protein important for DNA repair. PLoS Genet. 2010;6:e1000852. doi: 10.1371/journal.pgen.1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benaroudj N, Lee DH, Goldberg AL. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 2001;276:24261–24267. doi: 10.1074/jbc.M101487200. [DOI] [PubMed] [Google Scholar]
- 34.Todorova T, Pesheva M, Stamenova R, Dimitrov M, Venkov P. Mutagenic effect of freezing on nuclear DNA of Saccharomyces cerevisiae. Yeast. 2012;29:191–199. doi: 10.1002/yea.2901. [DOI] [PubMed] [Google Scholar]
- 35.Yoshinaga K, Yoshioka H, Kurosaki H, Hirasawa M, Uritani M, Hasegawa K. Protection by trehalose of DNA from radiation damage. Biosci. Biotechnol. Biochem. 1997;61:160–161. doi: 10.1271/bbb.61.160. [DOI] [PubMed] [Google Scholar]
- 36.Stuart GR, Copeland WC, Strand MK. Construction and application of a protein and genetic interaction network (yeast interactome) Nucleic Acids Res. 2009;37:e54. doi: 10.1093/nar/gkp140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Said MR, Begley TJ, Oppenheim AV, Lauffenburger DA, Samson LD. Global network analysis of phenotypic effects: protein networks and toxicity modulation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2004;101:18006–18011. doi: 10.1073/pnas.0405996101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ball AR, Jr, Yokomori K. Damage-induced reactivation of cohesin in postreplicative DNA repair. Bioessays. 2008;30:5–9. doi: 10.1002/bies.20691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watrin E, Peters JM. Cohesin and DNA damage repair. Exp. Cell. Res. 2006;312:2687–2693. doi: 10.1016/j.yexcr.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 41.Tittel-Elmer M, Lengronne A, Davidson MB, Bacal J, Francois P, Hohl M, Petrini JH, Pasero P, Cobb JA. Cohesin association to replication sites depends on rad50 and promotes fork restart. Mol. Cell. 2012;48:98–108. doi: 10.1016/j.molcel.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadin S, Ciocca D. In: DNA Damage Repair, Repair Mechanisms and Aging. Thomas A, editor. Hauppauge, NY: Nova Science Publishers; 2010. [Google Scholar]
- 43.Misri S, Pandita S, Pandita TK. Detecting ATM-dependent chromatin modification in DNA damage and heat shock response. Methods Mol. Biol. 2009;523:395–410. doi: 10.1007/978-1-59745-190-1_26. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 45.Ruis H, Schuller C. Stress signaling in yeast. Bioessays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- 46.Menant A, Baudouin-Cornu P, Peyraud C, Tyers M, Thomas D. Determinants of the ubiquitin-mediated degradation of the Met4 transcription factor. J. Biol. Chem. 2006;281:11744–11754. doi: 10.1074/jbc.M600037200. [DOI] [PubMed] [Google Scholar]
- 47.Sharma VM, Tomar RS, Dempsey AE, Reese JC. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol. Cell. Biol. 2007;27:3199–3210. doi: 10.1128/MCB.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsaponina O, Barsoum E, Astrom SU, Chabes A. Ixr1 is required for the expression of the ribonucleotide reductase Rnr1 and maintenance of dNTP pools. PLoS Genet. 2011;7:e1002061. doi: 10.1371/journal.pgen.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowe LA, Degtyareva N, Doetsch PW. DNA damage-induced reactive oxygen species (ROS) stress response in Saccharomyces cerevisiae. Free Radic. Biol. Med. 2008;45:1167–1177. doi: 10.1016/j.freeradbiomed.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharp JA, Rizki G, Kaufman PD. Regulation of histone deposition proteins Asf1/Hir1 by multiple DNA damage checkpoint kinases in Saccharomyces cerevisiae. Genetics. 2005;171:885–899. doi: 10.1534/genetics.105.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end-resection and checkpoint deactivation. Mol. Cell. Biol. 2012;32:4727–4740. doi: 10.1128/MCB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Begley TJ, Jelinsky SA, Samson LD. Complex transcriptional responses to macromolecular damaging agents: regulatory responses specific for SN2 alkylation and the MAG1 gene. Cold Spring Harb. Symp. Quant. Biol. 2000;65:383–393. doi: 10.1101/sqb.2000.65.383. [DOI] [PubMed] [Google Scholar]
- 54.Lee YD, Elledge SJ. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev. 2006;20:334–344. doi: 10.1101/gad.1380506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z, An X, Yang K, Perlstein DL, Hicks L, Kelleher N, Stubbe J, Huang M. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc. Natl Acad. Sci. USA. 2006;103:1422–1427. doi: 10.1073/pnas.0510516103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee YD, Wang J, Stubbe J, Elledge SJ. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol. Cell. 2008;32:70–80. doi: 10.1016/j.molcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mazumder A, Tummler K, Bathe M, Samson LD. Single-cell analysis of ribonucleotide reductase transcriptional and translational response to DNA damage. Mol. Cell. Biol. 2013;33:635–642. doi: 10.1128/MCB.01020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tringe SG, Willis J, Liberatore KL, Ruby SW. The WTM genes in budding yeast amplify expression of the stress-inducible gene RNR3. Genetics. 2006;174:1215–1228. doi: 10.1534/genetics.106.062042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 60.Shen C, Lancaster CS, Shi B, Guo H, Thimmaiah P, Bjornsti MA. TOR signaling is a determinant of cell survival in response to DNA damage. Mol. Cell. Biol. 2007;27:7007–7017. doi: 10.1128/MCB.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu Z, Norris D. The SFP1 gene product of Saccharomyces cerevisiae regulates G2/M transitions during the mitotic cell cycle and DNA-damage response. Genetics. 1998;150:1419–1428. doi: 10.1093/genetics/150.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, Barsotti A, Chicas A, Li W, Polotskaia A, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu KC, Cui JY, Klaassen CD. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol. Sci. 2011;123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu KC, Liu J, Klaassen CD. Role of Nrf2 in preventing ethanol-induced oxidative stress and lipid accumulation. Toxicol. Appl. Pharmacol. 2012;262:321–329. doi: 10.1016/j.taap.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 67.Aksam EB, de Vries B, van der Klei IJ, Kiel JA. Preserving organelle vitality: peroxisomal quality control mechanisms in yeast. FEMS Yeast Res. 2009;9:808–820. doi: 10.1111/j.1567-1364.2009.00534.x. [DOI] [PubMed] [Google Scholar]
- 68.Reddy JK, Rao MS. Oxidative DNA damage caused by persistent peroxisome proliferation: its role in hepatocarcinogenesis. Mutat. Res. 1989;214:63–68. doi: 10.1016/0027-5107(89)90198-x. [DOI] [PubMed] [Google Scholar]
- 69.de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 70.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee MV, Topper SE, Hubler SL, Hose J, Wenger CD, Coon JJ, Gasch AP. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 2011;7:514. doi: 10.1038/msb.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL. CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell. 2008;29:477–487. doi: 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 73.Burgis NE, Samson LD. The protein degradation response of Saccharomyces cerevisiae to classical DNA-damaging agents. Chem. Res. Toxicol. 2007;20:1843–1853. doi: 10.1021/tx700126e. [DOI] [PubMed] [Google Scholar]
- 74.Rowe LA, Degtyareva N, Doetsch PW. Yap1: a DNA damage responder in Saccharomyces cerevisiae. Mech. Ageing. Dev. 2012;133:147–156. doi: 10.1016/j.mad.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bose T, Lee KK, Lu S, Xu B, Harris B, Slaughter B, Unruh J, Garrett A, McDowell W, Box A, et al. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet. 2012;8:e1002749. doi: 10.1371/journal.pgen.1002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujii K, Kitabatake M, Sakata T, Miyata A, Ohno M. A role for ubiquitin in the clearance of nonfunctional rRNAs. Genes Dev. 2009;23:963–974. doi: 10.1101/gad.1775609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hinnebusch AG. Active destruction of defective ribosomes by a ubiquitin ligase involved in DNA repair. Genes Dev. 2009;23:891–895. doi: 10.1101/gad.1800509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gilbert WV. Functional specialization of ribosomes? Trends Biochem. Sci. 2011;36:127–132. doi: 10.1016/j.tibs.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chalfie M, Kain S, editors. Green Fluorescent Protein: Properties, Applications, and Protocols. 2006. 2 ed. John Wiley & Sons, Inc, Malden, MA. [Google Scholar]
- 82.Brock R, Hamelers IH, Jovin TM. Comparison of fixation protocols for adherent cultured cells applied to a GFP fusion protein of the epidermal growth factor receptor. Cytometry. 1999;35:353–362. doi: 10.1002/(sici)1097-0320(19990401)35:4<353::aid-cyto8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 83.Haase SB, Reed SI. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1:132–136. [PubMed] [Google Scholar]
- 84.Agne B, Infanger S, Wang F, Hofstetter V, Rahim G, Martin M, Lee DW, Hwang I, Schnell D, Kessler F. A toc159 import receptor mutant, defective in hydrolysis of GTP, supports preprotein import into chloroplasts. J. Biol. Chem. 2009;284:8670–8679. doi: 10.1074/jbc.M804235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baumstark-Khan C, Palm M, Wehner J, Okabe M, Ikawa M, Horneck G. Green fluorescent protein (GFP) as a marker for cell viability after UV-irradiation. J. Fluorescence. 1999;9:37–43. [Google Scholar]
- 86.Bence NF, Bennett EJ, Kopito RR. Application and analysis of the GFPu family of ubiquitin-proteasome system reporters. Methods Enzymol. 2005;399:481–490. doi: 10.1016/S0076-6879(05)99033-2. [DOI] [PubMed] [Google Scholar]
- 87.Fu D, Collins K. Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev. 2006;20:531–536. doi: 10.1101/gad.1390306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Z, Meray RK, Grammatopoulos TN, Fredenburg RA, Cookson MR, Liu Y, Logan T, Lansbury PT., Jr Membrane-associated farnesylated UCH-L1 promotes alpha-synuclein neurotoxicity and is a therapeutic target for Parkinson’s disease. Proc. Natl Acad. Sci. USA. 2009;106:4635–4640. doi: 10.1073/pnas.0806474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ward WW, Cody CW, Hart RC, Cormier MJ. Spectrophotometric identity of the energy transfer chromophores in Renilla and Aequorea green–fluorescent proteins. Photochem. Photobiol. 1980;31:611–615. [Google Scholar]
- 90.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 91.Giuliano KA, Cheung WS, Curran DP, Day BW, Kassick AJ, Lazo JS, Nelson SG, Shin Y, Taylor DL. Systems cell biology knowledge created from high content screening. Assay Drug Dev. Technol. 2005;3:501–514. doi: 10.1089/adt.2005.3.501. [DOI] [PubMed] [Google Scholar]
- 92.Saville DJ. Multiple Comparison Procedures: The Practical Solution. Am. Stat. 1990;44:174–180. [Google Scholar]
- 93.Sheskin DJ. Handbook of Parametric and Nonparametric Statistical Procedures. 2nd Edn. Boca Raton: Chapman & Hall/CRC; 2000. [Google Scholar]
- 94.Pearson ES, Hartley HO. Cambridge: Cambridge University Press; 1972. Biometrika Tables for Statisticians. [Google Scholar]
- 95.Gorenstein J, Zack B, Marszalek JR, Bagchi A, Subramaniam S, Carroll P, Elbi C. Reducing the multidimensionality of high-content screening into versatile powerful descriptors. Biotechniques. 2010;49:663–665. doi: 10.2144/000113492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.