Abstract
The generation of genome-modified animals is a powerful approach to analyze gene functions. The CAS9/guide RNA (gRNA) system is expected to become widely used for the efficient generation of genome-modified animals, but detailed studies on optimum conditions and availability are limited. In the present study, we attempted to generate large-scale genome-modified mice with an optimized CAS9/gRNA system, and confirmed the transmission of these mutations to the next generations. A comparison of different types of gRNA indicated that the target loci of almost all pups were modified successfully by the use of long-type gRNAs with CAS9. We showed that this system has much higher mutation efficiency and much lower off-target effect compared to zinc-finger nuclease. We propose that most of these off-target effects can be avoided by the careful control of CAS9 mRNA concentration and that the genome-modification efficiency depends rather on the gRNA concentration. Under optimized conditions, large-scale (∼10 kb) genome-modified mice can be efficiently generated by modifying two loci on a single chromosome using two gRNAs at once in mouse zygotes. In addition, the normal transmission of these CAS9/gRNA-induced mutations to the next generation was confirmed. These results indicate that CAS9/gRNA system can become a highly effective tool for the generation of genome-modified animals.
INTRODUCTION
Genome-modified mice, in which the specific genes, regulatory elements or other kinds of sequences have been disrupted or displaced are powerful tools for analyzing the roles of target genome sequences and to produce specific phenotypes (1). However, the generation methods for genome-modified mice commonly used today, which are based on homologous recombination in embryonic stem cells, are painstaking, time-consuming and expensive. Especially for a large-scale genome modification, such as the modification of gene clusters, which extend into dozens of kilo bases long, the laboriousness is much increased because of the requirement of repeated homologous recombination steps (2).
Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) comes from the acquired immune system of bacteria and archaea and has a unique function. In this system, exogenous DNA sequences recognized by crRNA are cleaved by a CRISPR-associated (CAS) endonuclease, which is recruited to the target sequences through tracrRNA (3,4). The application of this system to the genome modification of eukaryotic cells; the induction of a DNA double-strand break to a target sequence using guide RNA (gRNA), which is an engineered crRNA + tracrRNA, and a type of CAS protein, CAS9, and subsequent repair-associated mutagenesis have been reported in mammalian culture cells, zebrafish embryos and mouse embryos (5–11). This system is expected to provide an easy, inexpensive and fast method for mammalian genome modification. In addition, improvements in the production efficiency are anticipated because of the reported high success rates of genome modification in culture cells using the CAS9/gRNA system (5).
At present, two contrasting studies have been reported using CAS9/gRNA system in genome modification of mouse zygotes. The successful rate of genome modification in first report was unexpectedly low (10), whereas Wang et al. (11) have recently reported highly efficient genome modification, and, furthermore, double genome mutations have been successful using two gRNAs targeted two different chromosomes at once, in mouse zygotes. These results in the later report indicate the possibility of large-scale genome deletion using CAS9/gRNA system because two breakpoints in the same chromosome have been reported to elicit the deletion of the sequence in between, in culture cells (5). However, reasons for the discrepancy of these two reports, evaluations of efficiency and toxicity of CAS9/gRNA in mouse zygotes, and whether the long deletion is really possible and can be transmitted to the next generation using this system have never been studied.
In the present study, we first synthesized long gRNAs, which have CAS9-associate sequences ∼80 nt long and compared the mutation induction efficiency in mouse embryos against short gRNAs, which are ∼40 nt long and is the same sequence as those used in the former report (10). Then, the off-target effects of CAS9/gRNA system were analyzed, and the toxicity was compared between CAS9/gRNA and zinc-finger nuclease (ZFN) systems in mouse zygotes. After examining the efficient conditions, we injected two gRNAs targeting different loci on the same chromosome into mouse zygotes at once and addressed whether large-scale genome-modified mice could be produced using CAS9/gRNA system in a single step procedure. In addition, we reported for the first time that the mutations generated by CAS9/gRNA system could be transmitted to the next generation.
MATERIALS AND METHODS
Construction of CAS9 expression vector
The human codon-optimized CAS9 was cloned from Plasmid 41815: hCas9 (Addgene: #41815) and SV40 nuclear localization signals (NLS) were added to the N- and C-terminal by PCR using NLS-coding primers. The constructs were inserted into the BglII-Acc65I site of ZFN-platform vector (12) to add the T3 promoter, the FLAG-tag sequence and the Tbpl1 3′UTR with a 95 bp polyadenine tale. Using this construct, the sequence from theT3 promoter to the polyadenine tale was cloned by PCR and inserted into the EcoRI site of pCAGGS vector (referred to as ‘CAS9 vector’). For visualization of CAS9 protein in zygotes, eGFP-tagged CAS9 was constructed by inserting a ΔATG-eGFP sequence between CAS9 and C-terminal NLS of CAS9 vector (referred to as ‘CAS9-eGFP vector’). These vectors were sequenced using a commercial sequencing kit (Applied Biosystems, Foster City, CA, USA) and a DNA sequencer (Applied Biosystems) according to the manufacturer’s instructions. The schematics of these vectors are shown in Supplementary Figure S1A and B, and their sequences are shown in Supplementary Figure S2.
Construction of gRNA coding vectors
Target sequences (GGN19−21GG) and gRNAs were designed according to previous work (8), with some modification. The long and short gRNAs each with T3 promoters were synthesized by PCR without the template, using forward and reverse primers containing overlap sequences in 3′-end. The primer sets are shown in Supplementary Table S1. Each PCR product was cloned into a pGEM-T Easy vector and is referred to as ‘gRNA vector’. These vectors were sequenced as described earlier in the text, and the schematics and sequences of inserted gRNA with the T3 promoter are shown in Supplementary Figures S1C and S2, respectively.
In vitro transcription of CAS9, gRNAs and ZFNs
For the in vitro synthesis of CAS9 and CAS9-eGFP mRNAs, each vector was linearized by SphI and transcribed in vitro with T3-RNA-polymerase (Promega) in the presence of m7G(5′)ppp(5′)G to synthesize capped RNA as described previously (12). In the case of gRNAs, each gRNA vector was linearized by DraI and transcribed by the same procedure but without m7G(5′)ppp(5′)G to avoid the formation of cap structure. Preparation of ZFN mRNA for Cdkn1b was performed according to previous protocol (12). The RNA transcripts were precipitated with absolute ethanol, washed and resuspended in RNase-free water. The RNA solutions were stored at −80°C until use.
Microinjection
Following the guidelines for animal experiments at The University of Tokyo, sexually immature female C57BL/6NCr mice (4–5 weeks olds) were superovulated by intraperitoneal injection of 7.5 IU eCG followed by 7.5 IU hCG at an interval of 48 h and mated overnight with C57BL/6NCr male mice that were >12 weeks old. Zygotes were collected after 20 h of hCG injection by oviductal flashing, and pronuclei-formed zygotes were put into the M2 medium. Microinjection was performed using a microinjector (Narishige) equipped microscope. Approximately 4 pl of RNA solution was injected into the cytoplasm of each zygote using continuous pneumatic pressure. After injection, all zygotes were cultured in M16 medium and subjected for following experiments.
Immunoblotting of CAS9
Micro-western blotting was used for the immunoblotting of the zygotes as described in a previous report (13). Eighty zygotes injected with or without 200 µg/ml CAS9 mRNA solution were used in each lane. The antibodies used were anti-Flag M2 monoclonal antibody (F1804, Sigma-Aldrich) and anti-β-actin polyclonal antibody (GTX109639, GeneTex, Inc., CA, USA). To visualize the protein-bound antibodies, horseradish peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) were used for the second layer, respectively, followed by detection procedure using an ECL detection kit (Amersham-Pharmacia) according to the manufacture’s protocol.
Observation of eGFP-tagged CAS9
The zygotes injected with or without 200 µg/ml CAS9-eGFP mRNA solution were fixed in 3.7% formaldehyde in saline at room temperature for 40 min and stained with 45 μg/ml propidium iodide in PBS containing 0.1% PVP (PBS-PVP) for 30 min. The zygotes were then washed in PBS-PVP, mounted on a glass slide and observed using a confocal laser scanning microscope (LSM510-V2.01, Axio-plan MOT; Carl Zeiss, Oberkochen, Germany).
Immunocytechemistry of phospho-Histone H2A
Eight hours after RNA injection, zygotes were fixed in 3.7% paraformaldehyde and permeabilized in 0.2% triton X-100. These zygotes were treated with an anti-phospho-Histone H2A.X (γH2AX) monoclonal antibody (05–636, Millipore) overnight. After washing, the zygotes were incubated in fluorescein-isothiocyanate-conjugated anti-mouse IgG (55494, MP Biomedicals) for 60 min. DNA was visualized by propidium iodide staining as previously mentioned and then examined under a confocal laser scanning microscope (LSM510-V2.01, Axioplan MOT; Carl Zeiss, Oberkochen, Germany).
T7 endonuclease I assay
Genomic DNA was extracted from three to seven blastocysts and subjected to PCR using the primers shown in Supplementary Table S2. The purified PCR products were incubated at 95°C for 10 min and then cooled to 85°C at −2°C/s and to 25°C at −0.5°C/s for annealing of intact and mutated DNA strands. The re-annealed products were incubated with 2 U of T7 endonuclease I (T7EI) at 37°C for 3 h and then subjected to agarose gel electrophoresis. The band intensities were quantified using the Image-J software (NIH), and the amount of mismatch-digestion fragments were calculated according to previous work (5).
Embryo transfer
One- or Two-cell embryos injected with CAS9 mRNA and gRNA were transferred into the oviductal ampullas (6–10 embryos per oviduct) of 7–8-week-old female ICR mice mated with vasectomized ICR males at the previous night. After birth, ∼1 mm tail tips were obtained from the 2–4-day-old pups. Genome DNA was extracted from the tail tips and subjected to PCR using the primers shown in Supplementary Table S2. PCR products were purified by agarose gel electrophoresis, and the extracted fragments were directly sequenced as described earlier in the text.
RESULTS
Construction and expression of CAS9 and gRNAs
At first, we constructed a vector that codes a mammalian codon-optimized version of CAS9 with SV40 NLS on both sides of the ORF and the 3′UTR of Tbpl1 with a 95 base polyadenine tale (Supplementary Figures S1 and S2). In vitro synthesized CAS9 mRNAs (Supplementary Figure S3A) were injected into cytoplasm of fertilized mouse eggs. The expression of CAS9 protein was detected at pronuclear stages (Supplementary Figure S3B), and the eGFP-tagged CAS9 protein was observed in the pronuclei, especially in the nucleoli (Supplementary Figure S3C). The localization of NLS-fused CAS9 in nucleoli has been also observed in cultured mammalian cells (5). These results indicated that constructed CAS9 protein could stably exist in the nucleoli both in mammalian somatic cells and in pronuclear embryos.
Comparison of genome-modification efficiencies between long and short gRNAs
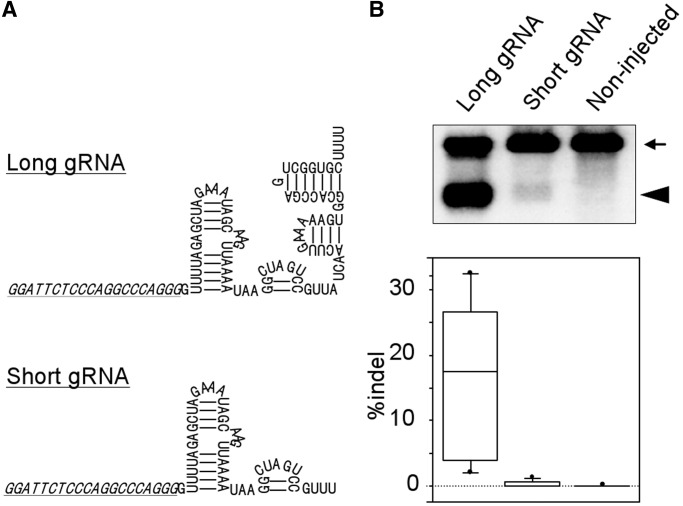
To compare the effects of two gRNA types on the site-directed nuclease activity of CAS9 in mouse embryos, short gRNA and long gRNA were constructed at the same target sequence of Rosa26 intron locus (Figure 1A and Supplementary Figures S1–S3A). One of these gRNAs and CAS9 mRNA were injected into mouse zygotes, and the genome-modification efficiencies were evaluated by a T7EI assay at the blastocyst stage. As a result, much higher efficiency was observed in the genome modification using long gRNA compared with that using short gRNA (Figure 1B). This result indicates that the CAS9-associated sequence of gRNA strongly affects the genome-modification efficiencies in mouse embryos and suggests that the discrepancy between the results of two previous reports was attributed to the difference of the types of gRNA used (10,11).
Figure 1.
Comparison of structure and function between long and short gRNAs. (A) Schematic illustrations of long and short gRNAs for Rosa26 target locus. Target sequence was underlined. (B) Genome modification efficiency of long and short gRNAs evaluated by T7EI assay. Genomic PCR amplicons of blastocysts derived from fertilized eggs injected with CAS9 mRNA (100 µg/ml), and long or short gRNA (10 µg/ml) were subjected for T7EI assay. Experiments were repeated for six times. An example of typical results of T7EI assay was shown in upper part. The arrow and arrowhead indicate PCR amplicons and digested fragments by T7EI, respectively. Calculated efficiencies were shown as boxplot in lower part.
Generation efficiency of genome-modified mice and off-target effect of long gRNA and CAS9
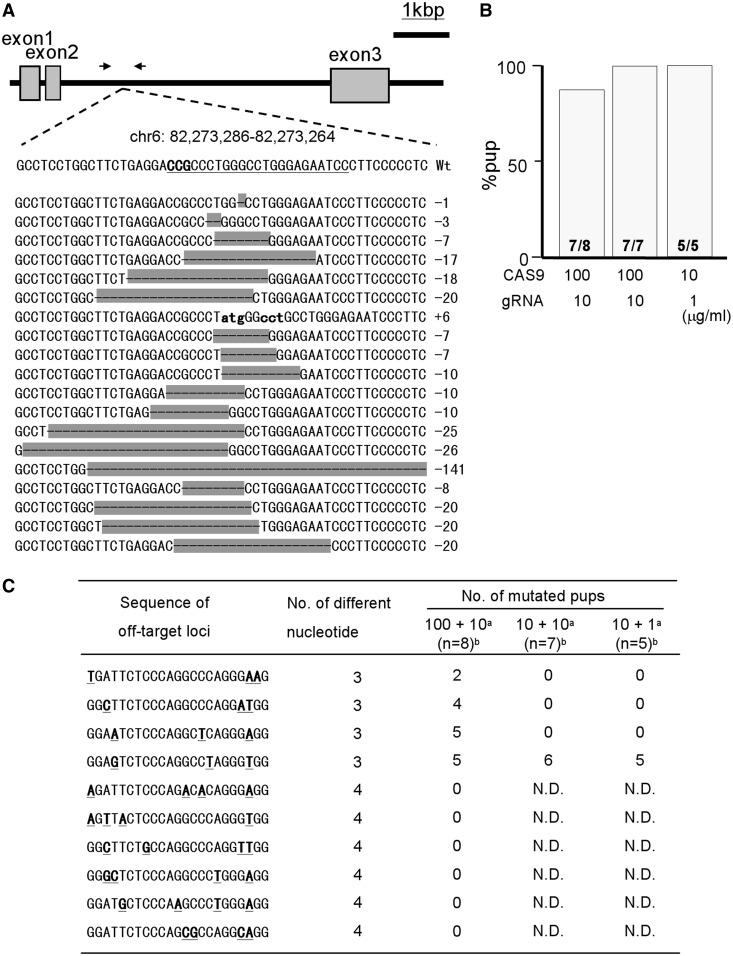
Next, fertilized mouse eggs injected with Rosa26-long gRNA and CAS9 mRNA at various concentrations were transferred into oviducts at 1- or 2-cell stages. The target locus of obtained pups were sequenced, and we found that 96% pups were mutated with a success efficiency ranging from 21.9 to 35% of transferred embryos in all RNA concentrations examined (Figure 2A and B). The DNA waveform data obtained from direct sequencing of genomic PCR products indicated that all of mutated pups had bi-allelic mutations (Supplementary Figure S4). We found some potential off-target sites for Rosa26, in which three or four nucleotides were different from the target sequence and examined the modification of ten off-target sites in the Rosa26-modified-pups. As a result, off-target mutations were detected in three-base-different loci when using a high concentration (100 μg/ml) of CAS9 mRNA (Figure 2C and Supplementary Figure S5), and off-target production effect was much decreased at a low CAS9 mRNA concentration (10 μg/ml) (Figure 2C and Supplementary Figure S5), suggesting that a large proprtion of off-target effects can be avoided by setting the appropriate nuclease concentration in CAS9/gRNA system (Figure 2B and C). No off-target mutations were found in four-base-different loci even at a high CAS9 mRNA concentration (Figure 2C and Supplementary Figure S5).
Figure 2.
Generation of Rosa26-targeted mice mediated by CAS9 and long-gRNA. (A) Schematic illustration of Rosa26 gene structure and sequences of wild-type and mutated alleles around the target locus. Arrows indicate the loci of PCR primers. The targeted locus of gRNA and PAM domain were indicated in the wild-type sequence by underline and bold letters, respectively. Examples of modified-allele sequences obtained from Rosa26 targeted pups are shown below in no particular order. Deleted and inserted nucleotides are indicated by hyphens and small letters, respectively. (B) Comparison of mutation efficiencies of Rosa26 target locus among various concentrations of CAS9 mRNA and gRNA injected into pronuclear-stage embryos. Numbers in the bars indicate the numbers of mutated pups/total pups. (C) Mutation efficiencies ofpotential off-target loci for Rosa26 target sequence. Mismatch nucleotides in examined sequences are shown by bold with underlines. N.D. indicates ‘not determined’. ‘a’ denotes concentrations of CAS9 mRNA + gRNA (µg/ml). ‘b’ denotes number of total pups. The locus information of these sequences are shown in Supplementary Figure S5.
Comparison of toxicity and mutation efficiencies between CAS9/gRNA system and ZFNs
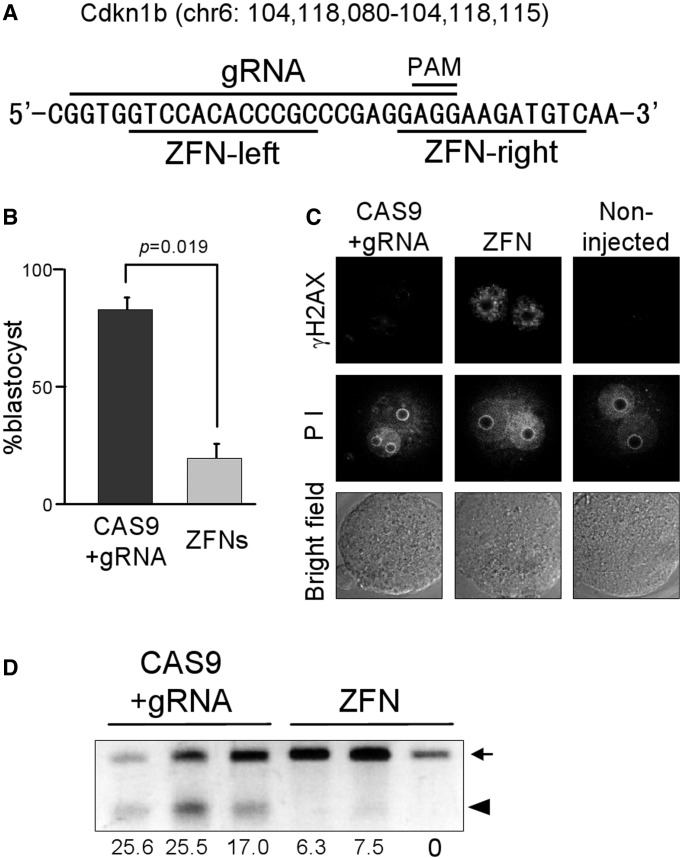
Previously, we studied the use of ZFN for mouse genome modification and observed developmental arrests in the embryos injected with Cdkn1b-targeted ZFN (12). Therefore, we examined the effectiveness of the CAS9/gRNA system on Cdkn1b, at the same target locus as ZFN (Figure 3A) and observed a significantly higher blastocyst rate in the embryos injected with CAS9/gRNA system (100 µg/ml CAS9 and 10 µg/ml gRNA) than those injected with ZFN (5 + 5 µg/ml) (Figure 3B). Immunostaining of γH2AX, which is a marker of DNA double-strand-breaks, showed significantly stronger signals in the pronuclei of ZFN-injected zygotes compared to CAS9/gRNA-injected and non-injected zygotes (Figure 3C). These results indicate that the toxicity, such as off-target effects, is much lower in this system than ZFN. In addition, the mutation efficiency of CAS9/gRNA system was much higher than that of ZFN (Figure 3D).
Figure 3.
Comparison of toxicity and mutation efficiency between CAS9/gRNA system and ZFN on Cdkn1b locus. (A) Cdkn1b target sequences of gRNA and ZFNs. (B) Developmental competence of preimplantation embryos injected with CAS9/gRNA (100 + 10 µg/ml) or ZFNs (5 µg/ml each). Experiments were repeated three times, and the average blastocyst formation rates (mean + SD) are shown. (C) DNA-double-strand breaks of CAS9/gRNA-, ZFN- or non-injected zygotes were visualized by immunocytechemistry of γH2AX (upper panels) at 8 h after microinjection. Pronuclei visualized by propidium iodide (PI) staining (middle panels) and bright field of embryos (lower panels) are also shown. (D) The results of T7EI assay. Genomic PCR amplicons of blastocysts obtained from the previously mentioned experiments were subjected for T7EI assay. Arrow and arrowhead indicate PCR amplicons and digested fragments by T7EI, respectively. The percentage of digestion fragment was calculated and shown under each lane.
Generation of double-mutated or large-scale deletion mice using two long gRNAs
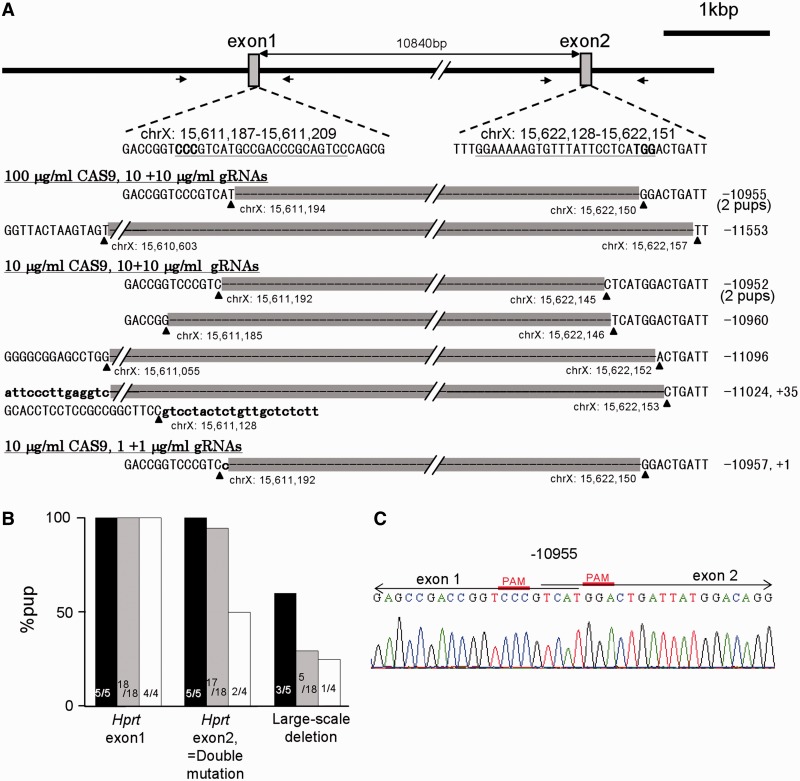
To evaluate whether double mutations and subsequent large-scale deletion could be induced in the mouse genome by the co-injection of two gRNAs targeting different loci in the same chromosome, two gRNAs targeted for Hprt exon1 and exon2 were co-injected with CAS9 mRNA in mouse zygotes (Figure 4A). After the embryo transfer, we confirmed that the exon1 target locus was modified in all newborns and double mutated pups were obtained in 30% of transferred embryos and in 89% of the newborn (Figure 4B). This result shows that the present method can generate double-mutated mice in a single step injection process. Generation rates of double mutants decreased with the dilution of injected gRNAs rather than CAS9 mRNA, and the mutation rates on Hprt exon2 was less effective than on exon1. Furthermore, the DNA waveform data indicated that all female pups (n = 12) had bi-allelic mutations at the exon1 target site, but four pups were mono-allelic at the exon2 target site. These results indicate that a variance in efficiency of the present method depends on the injected gRNA concentration (rather than CAS9 mRNA concentration) and also on the target sequences. Interestingly, total nine of the 27 pups (33%) had deletions that extended across the two target loci (∼10 kb), indicating that the applicability of the present method for chromosome engineering, such as large-scale genome deletion (Figure 4A–C). Off-target effects were examined in 5 loci for exon1 and 7 loci for exon2 in pups injected with a high-concentration (100 μg/ml) of CAS9 mRNA, but no off-target effects were detected in these loci (Supplementary Figure S6).
Figure 4.
Generation of double-mutated or large-scale deleted mice using two long gRNAs targeted Hprt exon1 and exon2. Different concentrations of CAS9 mRNA and two gRNAs were injected at once into pronuclear-stage embryos, and 1-cell or 2-cell stage embryos were transferred into oviducts. (A) Schematic illustration of Hprt gene structure and sequences around the target loci and obtained large-scale deleted alleles. Arrows in the schema indicate the loci of PCR primers, and the targeted sequences of gRNAs and PAM domain are indicated by underlines and bold letters, respectively. Sequences of all large-scale modified alleles, which were deleted ∼10 kb extended over the two target loci, are shown with respect to each concentration of CAS9 mRNA and gRNAs used. The hyphens and small letters denote deleted and inserted nucleotides, respectively. (B) Mutation efficiencies of Hprt loci. As target-locus of Hprt exon1 in all pups were mutated, the mutation efficiencies of exon2 are identical to the double mutated efficiencies. Results of different RNA concentrations were shown by black bars (100 μg/ml CAS9 and 10 + 10 μg/ml each gRNA), gray bars (10 μg/ml CAS9 and 10 + 10 μg/ml each gRNA) and white bars (10 μg/ml CAS9 and 1 + 1 μg/ml each gRNA). Numbers in the bars indicate the numbers of mutated pups/total pups. (C) An example of sequencing raw data from a pup with large-scale deletion.
Transmission of mutations induced by CAS9/gRNA system to the next generation
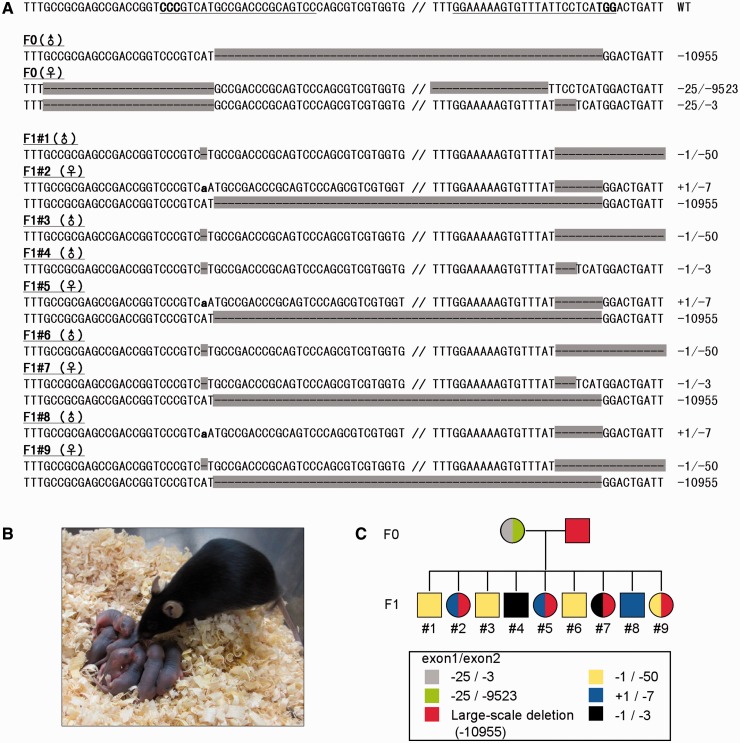
Finally, the transmission of mutations induced by CAS9/gRNA system to the next generation was examined by mating a large-scale deleted male mouse and a double-mutated female mouse. The target loci of obtained F1 pups, five males and four females, were sequenced, and we found that all F1 pups showed transmission of Hprt mutations (Figure 5A–C). A large-scale Hprt deletion was observed in all F1 females, which was the same as F0 male, indicating that the large-scale deletion induced by CAS9/gRNA can be inherited to the next generation (Figure 5A). Moreover, three patterns of exon1–exon2 double mutation, which was most likely inherited from the F0 female, were detected in F1 pups, but these patterns were different from the tail genome of the F0 female (Figure 5A and C). These results show that the F0 female was mosaicism, and that the mutation patterns were different between the tail cells and the germ cells, and they suggest that at least three types of oocytes were present in the ovaries of the F0 female, and that almost all oocytes were double-mutated on the identical allele.
Figure 5.
Transmission of mutations generated by CAS9/gRNA system. (A) Target sequences obtained form tail-tip DNA of F0 male and female mated, and F1 pups are shown with wild-type sequence. The sequences on the same line, which are connected by double slash, are obtained from same alleles. The hyphens and small letters denote deleted and inserted nucleotides, respectively. (B) The F0 female and obtained F1 pups used for the sequence analyses. (C) A pedigree obtained from analyses in (A). The identical alleles are shown by the same colors. The large-scale deletion of F0 male is wholly transmitted to the female pups. F0 female is genetically mosaic, and the target sequences of germ cells and tale-tip are different.
DISCUSSION
In the present study, we examined the experimental conditions suitable for the efficient genome modification using CAS9/gRNA system, and succeeded in the generation of double-mutated mice and subsequent large-scale deleted mice in a high efficiency by the use of two gRNAs targeting different loci in a same chromosome. A deletion of the inner sequence located between two target loci has been reported using a CAS9/gRNA system in somatic cells, but the distance of two target loci was small, and the deletion size was only 118 bp in this previous report (5). Our present results showed that CAS9/gRNA system can be used for efficient modification of large-scale chromosome engineering. In addition, we confirmed the transmission of the large-scale deletion and double mutations to the next generation by mating the Hprt mutated male and female mice and indicated the normal inheritance of CAS9/gRNA-generated mutations for the first time.
At present, the induction of genome reduction by double cutting in a fertilized egg has been studied in silkworm using two pairs of Transcription activator-like effector nuclease (TALEN), and the generation of a 795 bp deletion has been reported.(14). In the case of TALEN and ZFN, however, two constructs containing endonuclease activity have to be designed against both 3′ and 5′ sides of one target sequence, and their genome modification function was exerted after dimerization on the target locus (15). Therefore, targeting multiple loci at once by TALEN or ZFN results in an increase of nuclease concentration and subsequently may increase the risk of off-target cutting. In addition, painstakingly created ZFN and TALEN do not always satisfyingly function on target modifications. The mutation-creating rates of ZFN and TALEN in early embryos are usually only 10–20%, and it is difficult to prepare two efficient pairs for two target loci at one time (12,16–19). In contrast, our results suggested that CAS9/gRNA system could induce double mutations in almost pups under the suitable conditions, and that the low toxicity of this system was demonstrated by the absence of developmental arrest of the embryos mutated by CAS9/gRNA system unlike in the case of ZFN. These results indicate the availability of CAS9/gRNA system for the large-scale deletion and imply that the CAS9/gRNA system can contribute largely to genomics studies with its applicability for chromosome engineering without homologous recombination.
In the present study, we examined several conditions that affect the efficiency of the CAS9/gRNA system. Comparison of mutation inducing efficiency between long and short gRNAs, which targeted the same Rosa26 locus, revealed much higher efficiency in long gRNA than short gRNA. Because the additional sequence present in long gRNA but not in short gRNA derived from 3′ terminal of tracrRNA (8), which bound and recruit CAS9, the conformation of this part in tracrRNA may be important for the CAS9 recognition and may have had a critical effect on genome modification efficiency. This result strongly suggests that the difference of mutation inducing efficiency between previously reported two CAS9/gRNA systems is attributed to the different gRNA types they used (10,11). Comparison of several concentrations of gRNA and CAS9 mRNA revealed that the mutation rates of Rosa26 and Hprt exon1 loci were not affected by the concentration used in either gRNA or CAS9 mRNA, whereas the rate mutation on Hprt exon2 was generally lower than the two other loci and was affected by the gRNA concentration, but not with the CAS9 mRNA concentration. Because of the low GC content in Hprt exon2 target locus comparing with other two loci, the DNA–RNA-binding strength between gRNA and Hprt exon2 target locus may possibly be lower than those of other two loci. Low GC contents in CAS9/gRNA-ineffective target loci were observed also in a previous report in Zebrafish embryo (8). There results suggest that the binding efficiency of gRNA to a target locus is affected by the GC content of the target sequence, indicating that the consideration of GC content for the selection of the target locus of gRNA is required.
Off-target effects of the CAS9/gRNA system have not been detected in mouse genome modification in previous reports (10,11). Although it was once reported that a single-base mismatch at up to 11 bp into the 5′ direction from the PAM domain completely abolished off-target effects of the CAS9/gRNA system in somatic cells (5), the significant off-target potential of the CAS9/gRNA system in such cases has recently become clear thorough studies using human cell lines (20,21). Furthermore, off-target effects are detected at the loci that vary in five nucleotides to the target sequence (20). In the present study, the presence of off-target effect in CAS9/gRNA system was also shown in mouse embryos for the first time at a high CAS9 concentration. Interestingly, one of these off-target sequences, off-target 1, does not have the PAM domain but an NNG sequence at its 3′-end, indicating that the CAS9/gRNA system has potential to target such sequences in mouse zygote. Although these results support the recent studies showing high off-target effects of the CAS9/gRNA system, this effect should be much lower in mouse zygotes than in somatic cells because no off-target effects were detected in four-base-different off-target loci for Rosa26 and all off-target loci we found for Hprt exon1 and exon2. In addition, the disappearance of most of off-target effects at low CAS9 concentrations indicates the importance of concentration adjustment of CAS9 mRNA for the avoidance of off-target effects of the CAS9/gRNA system.
In the present study, we confirmed the normal transmission of the CAS9/gRNA-induced mutation to the next generation for the first time in mice. Although the paternal large-scale deletion was transmitted in all female pups without any changes, three mutation patterns were inherited from the maternal genome, and all of these were different forms of the modification pattern detected from the maternal tale-tip DNA, indicating the mosaicism of the maternal genome. The presence of mosaicism in the tail genome has also been reported in a previous study of mouse genome modification by CAS9/gRNA system (10). In other report, the single-step generation of double mutated mice was postulated by the information of only the tail-tip genome (11). Considering the generation of mosaicism in the tail genome by CAS9/gRNA system that was exhibited in this study, confirmations of a single-cell genome or of the transmission to the next generation should be necessary for the verification of multi-loci mutations. Our present results, which showed a large-scale deletion on the X chromosome of male mouse and double mutations in males in the next generation, is the first report showing in a convincing way that CAS9/gRNA system can generate a double mutation on an identical allele by a single step operation.
In summary, our results indicate that the CAS9/gRNA system may provide an easy, inexpensive and short-term method for mammalian genome modification, and that the large-scale genome-modification could be produced using multiple gRNAs together. In addition, we showed that off-target effects of this system were much lower than ZFN, and we propose that the off-target effect could be regulated by the CAS9 mRNA concentration, whereas the genome-modification efficiency depends rather on the gRNA concentration. Furthermore, we confirmed the transmission of the CAS9/gRNA system-induced mutation to the next generation. The present CAS9/gRNA system will contribute to the future expansion of functional genomics studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for JSPS Fellows [11J02886 to W.F.]; Grant-in-Aid for Scientific Research [22380147 and 25252056 to K.N.]; Grant-in-Aid for Challenging Exploratory Research [24658232 to K.N.] from Japan Society for the Promotion of Science. Funding for open access charge: Grant-in-Aid for Scientific Research [25252056] from the Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 2.Wallace HA, Marques-Kranc F, Richardson M, Luna-Crespo F, Sharpe JA, Hughes J, Wood WG, Higgs DR, Smith AJ. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell. 2007;128:197–209. doi: 10.1016/j.cell.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 3.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 4.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 8.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a -Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii W, Kano K, Sugiura K, Naito K. Repeatable construction method for engineered zinc finger nuclease based on overlap extension PCR and TA-cloning. PLoS One. 2013;8:e59801. doi: 10.1371/journal.pone.0059801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naito K, Kagii H, Iwamori N, Sugiura K, Yamanouchi K, Tojo H. Establishment of a small-scale western blotting system named as “Micro-Western Blotting” for mammalian ova analysis. J. Mamm. Ova Res. 1999;16:154–157. [Google Scholar]
- 14.Ma S, Zhang S, Wang F, Liu Y, Liu Y, Xu H, Liu C, Lin Y, Zhao P, Xia Q. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS One. 2012;7:e45035. doi: 10.1371/journal.pone.0045035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer M, de Angelis MH, Wurst W, Kühn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc. Natl Acad. Sci. USA. 2010;107:15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermann M, Maeder ML, Rector K, Ruiz J, Becher B, Bürki K, Khayter C, Aguzzi A, Joung JK, Buch T, et al. Evaluation of OPEN zinc finger nucleases for direct gene targeting of the ROSA26 locus in mouse embryos. PLoS One. 2012;7:e41796. doi: 10.1371/journal.pone.0041796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW. Knockout mice created by TALEN-mediated gene targeting. Nat. Biotechnol. 2013;31:23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- 19.Wefers B, Meyer M, Ortiz O, Hrabé de Angelis M, Hansen J, Wurst W, Kühn R. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc. Natl Acad. Sci. USA. 2013;110:3782–3787. doi: 10.1073/pnas.1218721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.