Abstract
In the yeast Saccharomyces cerevisiae, the Edc3 protein was previously reported to participate in the auto-regulatory feedback loop controlling the level of the RPS28B messenger RNA (mRNA). We show here that Edc3 binds directly and tightly to the globular core of Rps28 ribosomal protein. This binding occurs through a motif that is present exclusively in Edc3 proteins from yeast belonging to the Saccharomycetaceae phylum. Functional analyses indicate that the ability of Edc3 to interact with Rps28 is not required for its general function and for its role in the regulation of the YRA1 pre-mRNA decay. In contrast, this interaction appears to be exclusively required for the auto-regulatory mechanism controlling the RPS28B mRNA decay. These observations suggest a plausible model for the evolutionary appearance of a Rps28 binding motif in Edc3.
INTRODUCTION
Regulation of messenger RNA (mRNA) decay rates is an important mechanism to determine the abundance of cellular transcripts. Generally, eukaryotic mRNA degradation starts with the shortening of their poly(A)-tail. This is subsequently followed by decapping and 5′–3′ exonucleolytic degradation. Alternatively, 3′–5′ degradation by the exosome may take place after deadenylation (1–3). mRNAs as well as proteins involved in translation repression and mRNA decay assemble in cytoplasmic granules, often referred to as processing bodies (P-bodies), in which some transcript degradation was shown to occur (4–7). However, formation of these assemblies is not a prerequisite for initiation of mRNA decay, and some mRNA decay was shown to occur co-translationally (8). In yeast, decapping is mediated by Dcp2, but the latter needs to be stimulated by additional factors such as Lsm1-7 complex, Dhh1, Edc1-3, Scd6 and Pat1 (9–15). Dhh1 and Pat1 were reported to promote decapping mainly through translational repression (16), whereas Edc1-3 proteins were shown to activate Dcp1/Dcp2 enzyme directly (15,17,18). It has become evident that the control of mRNA stability and the control of mRNA translation are interconnected, although little is known about the physical and functional relationships between factors of mRNA degradation and the translation machinery. Interestingly, Edc3 protein was shown to interact with ribosomal protein S28 (Rps28) in yeast. This association is involved in an auto-regulatory mechanism controlling the production of Rps28 (19) but could be involved in other processes, in particular if Edc3 was able to interact with ribosome bound Rps28. Furthermore, molecular details of the mode of interaction of these two factors remain unclear.
Edc3 protein is a conserved eukaryotic factor that interacts genetically and/or physically with many proteins involved in mRNA decay (14,17,20,21). Edc3 is composed of three conserved domains, each being responsible for interaction with factors involved in mRNA decay: an N-terminal Sm-like domain was reported to bind Dcp1 and/or Dcp2 (17,21,22), an internal FDF motif was shown to bind Dhh1 (20), whereas its C-terminal YjeF-N domain was shown to promote homodimerization (23,24). The multi-domain organization of Edc3 was shown to contribute to its function as scaffold for decapping proteins during P-body assembly (23). In the yeast Saccharomyces cerevisiae, the EDC3 gene is not essential, and its inactivation does not result in detectable growth defect. Its deletion, however, enhances the growth phenotypes, and mRNA decay defects resulting from point mutation in the decapping enzyme Dcp2 or its close co-factor Dcp1 (25). Additional genetic screens also demonstrated that Edc3 acts synergistically with Scd6 and Pbp1 to control mRNA decapping (14). Transcriptome analyses revealed that EDC3 deletion causes accumulation of two specific RNAs: the RPS28B mRNA and the YRA1 pre-mRNA (19,26). Yra1 is an mRNA export factor encoded by a split gene. Excess Yra1 promotes cytoplasmic export of the Yra1 pre-mRNA, which, once in cytoplasm, is degraded by 5′–3′ decay mechanism dependent on Edc3 and on specific sequences in the YRA1 intron (19,26,27). The Rps28 protein is a highly conserved archaea- and eukaryote-specific component of the small ribosomal subunit (28–31). Nuclear magnetic resonance studies of archaeal Rps28 proteins revealed that the protein contains four β-strands, forming compact globular oligonucleotide/oligosaccharide binding (OB) fold followed by a non-structured C-terminal tail (28,31). X-ray structure of the yeast ribosome indicates that eukaryotic Rps28 proteins adopt the same overall fold, even though the region equivalent to the unstructured archaeal C-terminal tail is stabilized through interaction with other components of the small ribosomal subunit (29). Rps28 binds near the exit site of the mRNA and has been shown to crosslink with the mRNA (32). In yeast, Rps28 protein is encoded by two genes: RPS28A and RPS28B, which produce polypeptides differing by a single residue that can substitute for one another in the ribosome. When present in excess, the Rps28 protein (the pool of Rps28a and Rps28b) binds to a conserved hairpin structure present in the 3′ untranslated region (UTR) of the RPS28B mRNA and, in a manner dependent on Edc3, recruits the decapping machinery (19). The molecular details of this mechanism remain however unclear, as the mode of interaction of Rps28 with the RPS28B mRNA and Edc3 are currently unknown. It is also unclear whether this regulation is widely conserved in eukaryotes.
Here, we identify a region in yeast Edc3 that is necessary and sufficient for binding Rps28. Deletion of this region has no effect on yeast growth rate or accumulation of Edc3 in P-bodies, indicating that it is dispensable for the general RNA decay function of this factor. In contrast, this sequence is required for efficient degradation of the RPS28B mRNA. Degradation of YRA1 pre-mRNA, the second Edc3-regulated transcript, is not affected by deletion of this region, indicating that the mechanism of Edc3 action on YRA1 pre-mRNA is independent of its interaction with Rps28.
MATERIALS AND METHODS
Strains and plasmids
Yeast strains used in this study are listed in Table 1. All strains except BSY2475 are in the BMA64 background (33). The BSY2475 strain was derived from the MAV203 strain (Invitrogen) by PCR-based disruption of EDC3. Oligonucleotides OBS4144 and OBS4149 were used to amplify nourseothricine (NAT) resistance gene with flanking regions of yeast EDC3 gene. BSY2474 was constructed from BSY1664 using the same PCR product. BSY2831 strain was obtained by crossing strains BSY2474 and BSY2067. The construction of plasmids used in this study is described in the Supplementary Materials and Methods. These plasmids are listed in Supplementary Table S1. Oligonucleotides used for these constructions or for probing northerns are listed in Supplementary Table S2.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Origin |
|---|---|---|
| BMA64 | MATα ura3-1, delta trp1, ade2-1, leu2-3,112, his3-11,15 | (33) |
| BSY2474 | MATa ura3-1, Δtrp1, ade 2-1, leu2-3,112, his3-11,15, can1-100, Δedc3::NATR | This work |
| BSY2475 | MATα leu2-3, 112, trp1-901, his3-Δ200, ade2-101, gal4Δ, gal80Δ, SPAL10::URA3, GAL1::lacZ, HIS3UAS GAL1::HIS3@LYS2, can1R, cyh2R, Δedc3::NATR | This work |
| BSY2587 | MATα ura3-1, Δtrp1, ade2-1, leu2-3,112, his3-11,15, Δscd6::KanR | Gift of C. Gaudon |
| BSY2596 | MATα ura3-1, Δtrp1, ade2-1, leu2-3,112, his3-11,15, Δedc3::NATR, Δscd6::KanR | Gift of C. Gaudon |
| BSY2067 | MATα ura3-1, Δtrp1, ade 2-1, leu2-3,112, his3-11,15, can1-100, Δrps28B::HIS | Gift of T. van den Elzen |
| BSY2831 | MATα ura3-1, Δtrp1, ade 2-1, leu2-3,112, his3-11,15, can1-100, Δedc3::NATR, Δrps28B::HIS | This work |
Cell growth conditions
For growth analysis, cells were grown in exponential phase in synthetic complete (SC) medium lacking tryptophan complemented with 2% glucose. Cultures were diluted to the optical density at 600 nm (OD600) 0.1 with sterile water. Five microlitres of each culture as well as 5 µl of two 10-fold serial dilutions were plated on SC medium lacking tryptophan. Cell growth was monitored after 48 h at 25, 30 or 37°C. For microscopy, cells were grown at 30°C in SC medium lacking tryptophan and uracil complemented with 2% glucose, until an OD600 0.3 was reached. Cells were subsequently harvested by centrifugation, washed, resuspended in SC medium lacking tryptophan and uracil supplemented, or not, with a carbon source. Cells were incubated in a shaker at 30°C for 10 min before analysis. For total RNA isolation, yeasts were grown at 30°C in SC medium lacking tryptophan complemented with 2% galactose until an OD600 of 0.8–1 was reached. For western blot analysis, 1 ml of corresponding cell cultures was further grown till OD600 ≥ 2. For RPS28B mRNA half-life analysis, cells were grown in SC medium lacking tryptophan and uracil, supplemented with glucose. Cells were grown at 30°C until OD600 reached 0.8–1. Cultures were concentrated 10 times, doxycycline was added to the growth media to give a 40 μg/ml final concentration and 1 ml aliquots of cells were taken at the time points indicated for total RNA isolation.
Protein purification, protein–protein interaction assay and protein detection
Recombinant proteins were (co-)expressed in BL21 Codon+ cells grown overnight in 100 ml of auto-induction media at 37°C. Cells were harvested by centrifugation, resuspended in 5 ml of lysis buffer containing 50 mM Tris–HCl, 300 mM NaCl and 10 mM imidazole (pH 8.0) and sonicated. Two millilitres of cleared cell lysate was incubated with 100 µl of Ni-NTA agarose beads at 4°C with rotation for ∼1 h. Beads were washed with 500 µl of 50 mM Tris–HCl, 300 mM NaCl and 20 mM imidazole (pH 8.0) three times. Specifically bound proteins were eluted with 250 µl of 50 mM Tris–HCl, 300 mM NaCl and 200 mM imidazole (pH 8.0). Eluted proteins were separated in 12% Tris-Tricine gel and visualized with Coomassie-blue staining. Details of the analyses of protein interaction by size-exclusion chromatography with multi-angle laser light scattering measurements are described in the Supplementary Materials and Methods.
For western blot analysis of proteinA-tagged Edc3 proteins, total yeast extract was prepared as described previously (34). For detection, peroxidase-anti-peroxidase (3:10 000, Sigma) was used. For a loading control, the endogenous Stm1 proteins were detected using polyclonal anti-Stm1 antibody (1:1000, kind gift of Francoise Wyers).
Two-hybrid analysis
To avoid interference from the endogenous copy of Edc3, the BSY2475 strain was used. Beta-galactosidase activity was measured using Beta-Glo Assay system (Promega).
Microscopy
Microscopy analyses were performed as described previously (35). All images were acquired using Leica Microsystems Heidelberg GmbH microscope, using an objective HCX PL APO CS 63.0 × 1.40 OIL UV. ImageJ software was used to adjust all images to equal contrast ranges.
Northern blot analysis
Total RNA was extracted by a hot phenol method (36). Usually, 15 µg of total RNA were loaded per lane on a 3% formaldehyde agarose gel. Nucleic acids were transferred to the Hybond N membrane by passive transfer overnight. Random-prime labeling with NEBlot kit (NEB) was used to generate probes specific to RPS28 and YRA1 mRNA. The Yra1-specific PCR product was obtained with OBS4425 and OBS4426, whereas the Rps28-specific PCR product was obtained with OBS4427 and OBS4262. As a loading control, the signal recognition particle 7S RNA (SCR1), a stable RNA polymerase III transcript, was detected by a labeled probe generated by random priming. The amount of YRA1 mRNA and pre-mRNA, as well as RPS28A and RPS28B mRNA was quantified using the Typhoon Phosphoimager. Mean values of the ratios of these species and standard deviation were calculated from three independent experiments. For RPS28B mRNA half-life analysis, 6 μg of total RNA were loaded on gel and detected as indicated earlier in the text. The quantity of RPS28B mRNA normalized for the loading control was plotted as a function of time and fitted for exponential decay. Mean values of half-lives and standard deviations were calculated from two independent experiments.
RESULTS
A new conserved motif in yeast Edc3 proteins
Amino acid sequence of Edc3 from different species (S.cerevisiae, Human, Mouse and Drosophila) were reported to share significant conservation (25). This early analysis was improved with the identification of three conserved motifs and domains: Lsm, FDF and YjeF-N. The availability of additional putative Edc3 sequences deduced from genome-sequencing projects lead us to perform new Edc3 sequence alignments. Although confirming the conservation of the Lsm domain, FDF motif and YjeF-N domain, those revealed the presence of a conserved region located between the FDF motif and the YjeF-N domain in Edc3 proteins from various Saccharomycetaceae species, but not from the other subdivisions of the Saccharomycetales phylum or more divergent species (Figure 1). This correlated with the previous report of the presence of a conserved hairpin in the 3′ UTR of the RPS28B mRNA in the former group of organisms (19). This additional conserved region of Edc3, which corresponds to amino acids 209–222 of the S.cerevisiae protein, is hereafter referred to as the RB motif (for Rps28 binding motif). The presence of this motif in a specific subset of Edc3 proteins suggested that it could be involved in the auto-regulatory feedback loop that controls the stability of the RPS28B mRNA, for example by mediating interaction with Rps28 or facilitating interaction of the latter with the hairpin structure present in the RPS28B mRNA 3′ UTR.
Figure 1.
Localization of a new conserved motif in yeast Edc3 protein. Upper part: Domain architecture of Saccharomycetaceae Edc3 protein composed of an Lsm domain, a FDF motif and an YjeF-N domain. Numbers above the schematic representation of the protein indicate the amino acid positions of domain boundaries for the S. cerevisiae protein. Lower part: Amino acid alignment of Edc3 proteins from several Saccharomycetaceae species encompassing the RB motif. The RB motif is often preceded by an acidic region that is not absolutely conserved.
Recombinant Edc3 protein binds directly Rps28, and this interaction requires region 201–231 of Edc3
Experimental evidences supporting an interaction between Edc3 and Rps28 were derived from two-hydrid and co-immunoprecipitation analyses (19). These data did not indicate whether Rps28 and Edc3 interact directly, possibly in a manner stabilized by protein and/or RNA partners, or whether their interaction was exclusively bridged by other factor(s). To study the role of the newly defined Edc3 motif, we, thus, first sought to develop a more direct protein interaction assays. For this purpose, we constructed plasmids to overexpress Edc3, either alone or together with Rps28a in Escherichia coli (Figure 2A). Although plasmids encoding full-length Edc3 produced insoluble protein (data not shown), a construct carrying an operon encoding a 6His-tagged Rps28 protein and Edc3 lacking its C-terminal YjeF-N domain [Edc3(1–277)] expressed both proteins in a soluble form (Figure 2B). Moreover, affinity chromatography on Ni-NTA beads demonstrated a co-purification of recombinant Rps28 and Edc3 (Figure 2B). A control construct lacking the 6His-Rps28 demonstrated that Edc3(1–277) did not bind non-specifically to Ni-NTA beads. These results indicate that Rps28 and Edc3 interact directly forming a stable complex without requirement for additional RNA or protein partners, and further that the YjeF-N domain of Edc3 is not required for this interaction.
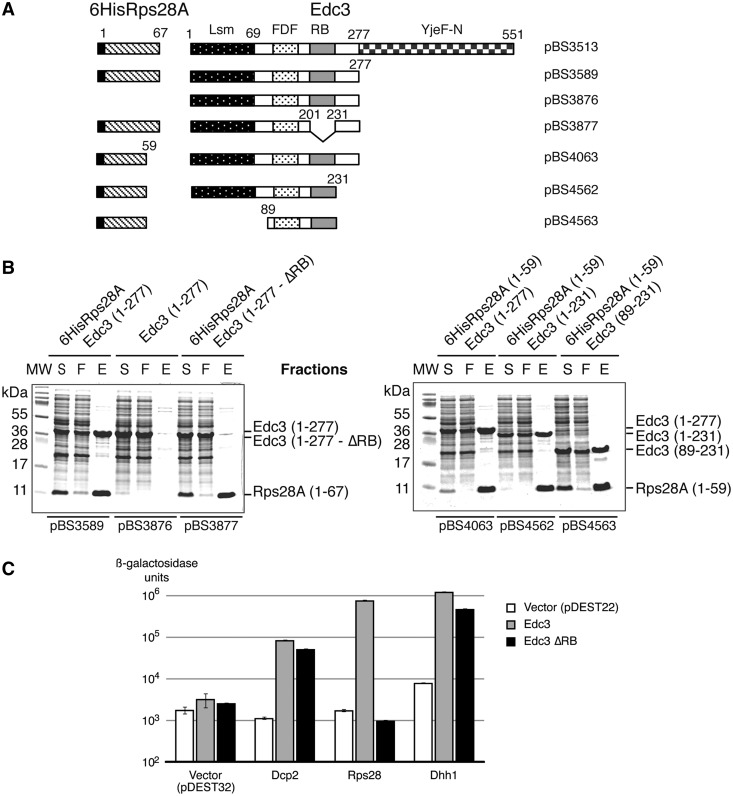
Figure 2.
The RB motif of Edc3 is needed for interaction with Rps28a protein. (A) Schematic representation of the operon constructs used for expressing variants of S. cerevisiae Edc3 and Rps28a in E. coli. A 6His tag was fused to the N-terminus of Rps28a. Numbers above the schematic representation of the proteins correspond to the amino acid boundaries. Pattern codes for Edc3 domains are as in Figure 1. (B) Coomassie blue-stained Tris–Tricine–SDS–PAGE showing co-purification of recombinant Edc3 and Rps28a proteins. Supernatants of lysed E. coli cells expressing recombinant yeast 6His-Rps28a and Edc3 protein fragments were incubated with Ni-NTA beads and subsequently washed before elution of bound proteins with imidazole. S, supernatant of lysed cells; F, flow through; E, elution. (C) Beta-galactosidase activity measurements to monitor interactions in the two-hybrid assay. Full-length wild-type Edc3 and derivative Edc3ΔRB were fused to the GAL4 activation domain, whereas Dcp2, Dhh1 and Rps28 proteins were fused to the GAL4 DNA-binding domain. In each case, the matching vector was used as negative control.
Using this assay, we tested deletion derivatives of Rps28 and/or Edc3 to define sequences required or dispensable for heterodimer formation. Nuclear magnetic resonance and X-ray crystallography studies have shown that archaeal and eukaryotic Rps28 proteins contain four β-strands, forming compact globular part, followed by a less structured C-terminal tail (28,29,31). Deletion of the eight residues forming this tail in yeast Rps28 did not affect its association with Edc3 (Figure 2B). This indicates that Edc3 interacts with the globular OB fold of Rps28. An Edc3 construct covering residues 1–231 interacted well with Rps28, indicating that none of the residues located between the RB motif and the YjeF-N domain are required for this interaction. Similarly, deletion of the Lsm domain of Edc3 (construct 89-231) did not affect the formation of the Edc3-Rps28 heterodimer. In contrast, only background level of an Edc3(1–277) variant lacking residues 201–231 [Edc3(1–277 -ΔRB)] were recovered with Rps28 (Figure 2B), indicating that this region, encompassing the RB motif, is required for heterodimer formation.
We used two-hybrid assay as a second strategy to validate these results in vivo. As Edc3 is known to multimerize (23,24), we preventively deleted the edc3 gene in the host strain MAV203 to avoid any interference from wild-type Edc3 protein encoded by the endogenous chromosomal gene. We analysed the interaction of full-length wild-type Edc3, or derivative Edc3 lacking residues 201–231, fused to the GAL4 activation domain with either Dcp2, Dhh1 and Rps28 proteins fused to GAL4 DNA-binding domain. Interactions were monitored by ß-galactosidase production. Wild-type Edc3 interacted with Dcp2, Rps28 and Dhh1 as expected (23,37) (Figure 2C). Deletion of residues 201–231 of Edc3 resulted in only slightly lower ß-galactosidase production when Dcp2 or Dhh1 were present but reduced this activity to background level with Rps28 (Figure 2C). Altogether, these data demonstrated that Edc3 binds directly the Rps28 core, and that this interaction requires amino acids 201–231 of Edc3.
The Edc3(201–231) fragment is sufficient for interaction with Rps28
The 201–231 region of Edc3, encompassing the conserved RB motif, was too small by itself to test whether it contains the necessary information to bind Rps28, using the co-purification strategy described earlier in the text. To assay whether this region is sufficient for interaction with Rps28, we constructed operons encoding a 6His-tagged GST carrier protein fused to amino acids 201–231 of Edc3 and untagged Rps28 (Figure 3A). Chromatography on Ni-NTA revealed co-purification of Rps28 with the 6His-GST-Edc3(201–231) fusion (Figure 3B). Untagged Rps28 by itself was not retained on the Ni-NTA resin when co-expressed with 6His-GST, indicating that Edc3 fragment 201–231 is responsible for interaction with ribosomal protein. Consistent with the results reported earlier in the text, a C-terminally truncated Rps28 behaved like the full-length protein (Figure 3B). We conclude that residues 201–231 of Edc3 are necessary and sufficient to interact with the core of Rps28 protein.
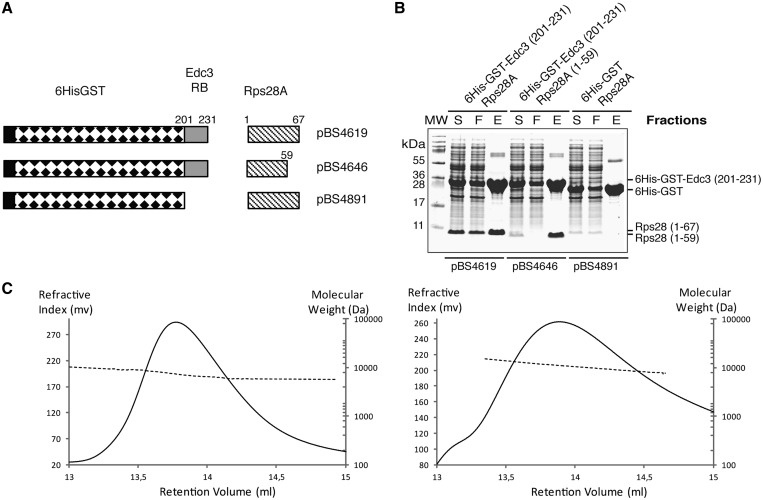
Figure 3.
The RB motif of Edc3 is sufficient for interaction with Rps28a protein. (A) Schematic representation of the operon constructs used to express the RB domain of Edc3 protein (fragment 201–231) fused downstream of a 6His-GST carrier protein and Rps28a protein in E. coli. Numbers above the schematic representation of the proteins correspond to the amino acid boundaries. (B) Coomassie blue-stained Tris–Tricine–SDS–PAGE showing co-purification of recombinant yeast proteins. Supernatants of lysed E. coli cells expressing recombinant 6His-GST-Edc3(201–231) and Rps28A, 6His-GST-Edc3(201–231) and Rps28A(1–59), or 6His-GST and Rps28A, were incubated with Ni-NTA beads and subsequently washed before elution of bound proteins with imidazole. S, supernatant of lysed cells; F, flow through; E, eluate. Rps28A expressed with 6His-GST accumulates to a lower level, probably as a result of instability, in the absence of its partner 6His-GST-Edc3(201–231). Yet, the protein can be detected [compare with lanes where Rps28A(1–59) is present]. Furthermore, Rps28A(1–59) also accumulates at a low level; yet, its interaction with Edc3(201–231) can readily be detected, indicating that the absence of RPS28A in the eluate fraction when it is expressed with 6His-GST results from a lack of interaction. Consistently, when expressed with 6His-GST, Rps28A remains in the flow-through fraction. (C) Size-exclusion chromatograms of Rps28a (left panel) and Rps28-Edc3(201–231) complex (right panel) are shown. For clarity, only the refractive index (RI, solid line, left y-axis) for the eluted samples and the molecular mass calculated from light scattering (right y-axis, dashed line, logarithmic scale) are shown.
We next checked that the observed complex formation was not due to formation of soluble aggregates between both partners. For this purpose, we purified the untagged Rps28a protein as well as the Rps28a-Edc3(201–231) complex lacking the His-GST tag to analyse their quaternary structure in solution using size exclusion chromatography coupled online to light scattering (Figure 3C). This yielded calculated molecular weights of 7142 Da for free Rps28 (theoretical molecular weight of 7592 Da) and of 10 836 Da for the Rps28a-Edc3(201–231) complex (theoretical molecular weight of 11 267 Da). Interestingly, the molecular weight difference (3694 Da) between the Rps28a-Edc3(201–231) complex and free Rps28 corresponds to the molecular weight of the isolated Edc3(201–231) peptide (3693 Da theoretical), confirming complex formation. This further indicates that Rps28a and the Edc3(201–231) region interact with a 1:1 stoichiometry to form a heterodimeric complex in solution, and that this complex is stable as it resists to a 3-steps purification protocol.
The Rps28 binding motif of Edc3 is not required for the general activity of Edc3 nor for its targeting to P-bodies
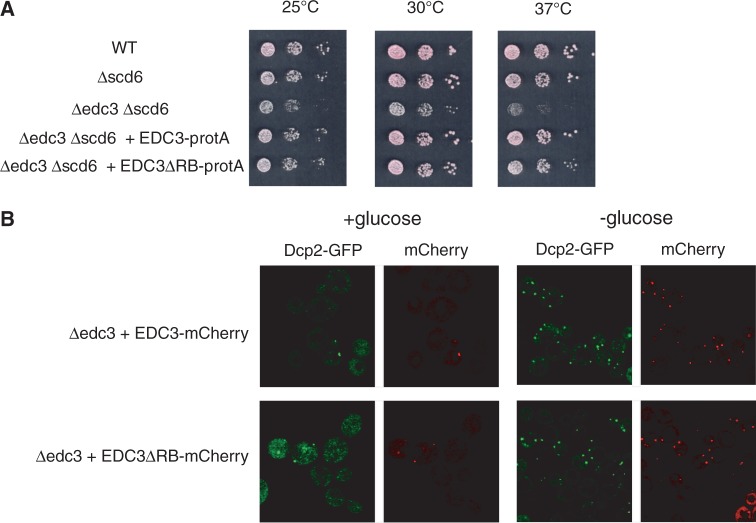
To analyse the functional role of interaction between Edc3 and Rps28, we constructed a centromeric plasmid carrying the EDC3 gene in which residues 201–231 were deleted. To facilitate protein-level monitoring, a Protein A tag (protA) was fused to the C-terminus of the protein (EDC3ΔRB-ProtA construct). A plasmid encoding protA tagged wild-type Edc3 was built to serve as a positive control, whereas the vector without insert served as a negative control. We tested the ability of Edc3ΔRB-protA protein to complement the slow growth phenotype resulting from the poor RNA decay in a Δedc3Δscd6 strain (14). Indeed, deletion of edc3 by itself has no impact on cell division or RNA decay (25). In the BMA yeast strain background, the reduced growth rate of the Δedc3Δscd6 strain is especially apparent at 37°C (Figure 4A). Introduction of the wild-type EDC3 gene in this strain restored a growth comparable with that of Δscd6 strain, which was itself equivalent to a wild-type strain (Figure 4A, expression of untagged Edc3 protein had the same effect, data not shown). Expression of Edc3ΔRB-protA fusion restored the growth rate of the Δedc3Δscd6 strain almost to the wild-type level, indicating that the mutant gene is functional and that interaction between Edc3 and Rps28 is not prerequisite for normal Edc3 function. Consistent with this efficient complementation, western blot analysis confirmed that Edc3ΔRB-protA and Edc3-protA accumulate to the same level in cells (data not shown).
Figure 4.
Deletion of the RB motif does not affect the general function of Edc3 or its intracellular location. (A) Assaying Edc3 activity by complementation of the slow growth phenotype of the Δedc3Δscd6 strain at various temperatures. Ten-fold serial dilutions of Δedc3Δscd6 strain transformed with a control empty vector (vector), a plasmid expressing wild-type Edc3 fused to the protA tag (Edc3-protA), or with a plasmid expressing Edc3ΔRB-protA were spotted on -TRP selective media and incubated at 25, 30 or 37°C for 2 days. As positive controls, the isogenic wild-type and Δscd6 strains transformed with a control empty vector were spotted on the same plate. (B) Intracellular localization of Dcp2-GFP and Edc3-mCherry or Edc3ΔRB-mCherry proteins in Δedc3 strain during exponential growth in selective media lacking uracil and tryptophan with or without glucose.
To test another potential general role of the Edc3-Rps28 interaction, we analysed the effect of deleting Edc3 residues 201–231 upon P-bodies assembly. Indeed, a strong reduction of microscopically visible P-bodies on glucose deprivation was observed in a Δedc3 strain (23). We analysed the co-localization of Dcp2-GFP with Edc3-mCherry fusions, using either wild-type Edc3 or Edc3ΔRB. No difference in P-bodies number and/or pattern was observed upon expression of either wild-type or mutant Edc3 (Figure 4B). This indicates that interaction of Edc3 with Rps28 is not needed to address Edc3 to P-bodies.
The region 201–231 of Edc3 is needed for the regulation of the RPS28B mRNA decay
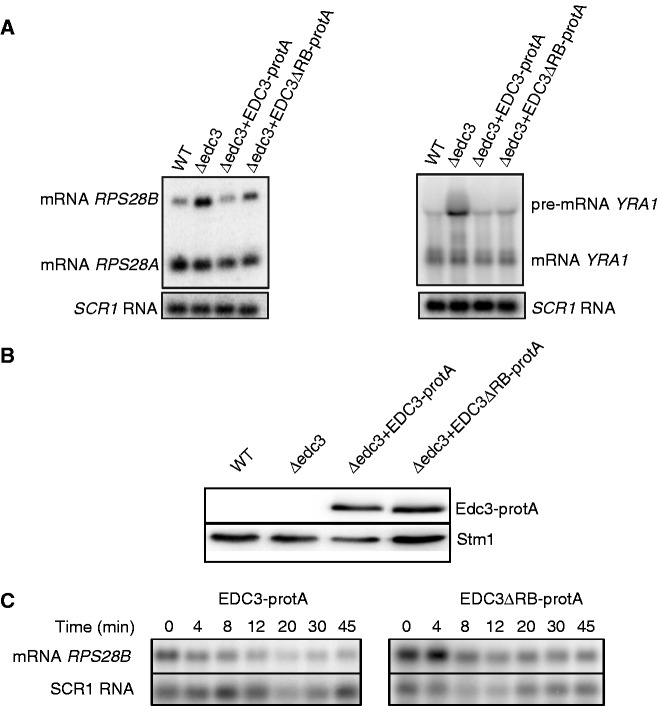
Deletion of the EDC3 gene blocks the degradation of two specific RNAs: the RPS28B mRNA and the YRA1 pre-mRNA (19,26). We analysed whether the deletion of the Edc3 RB motif influenced the stability of these RNAs. Total RNA was extracted from Δedc3 cells transformed either with an empty vector, with a vector coding for the wild-type Edc3-protA fusion, or with Edc3ΔRB-protA. As a control, total RNA from wild-type cells transformed with an empty vector was used. We observed that deletion of the RB motif of Edc3 that abrogates interaction of the latter with Rps28 significantly stabilized the RPS28B mRNA (Figure 5A, left panel). Quantification revealed that deletion of the edc3 gene led to more than 2-fold (2.8 ± 0.29) increase of the amount of RPS28B mRNA relative to RPS28A mRNA compared with the wild-type strain. Expression of the wild-type Edc3 restored a normal ratio of RPS28A versus RBS28B mRNA (1.3 ± 0.25). Deletion of residues 201–231 of Edc3 resulted in a significant accumulation of the RPS28B mRNA, although this effect was not as strong as in the complete absence of the protein (2.2- ± 0.36-fold increase compared with 2.8- ± 0.29-fold). Western blot analysis of total protein extract from yeast cells indicated that the Edc3ΔRB-protA protein was expressed at least as well as the wild-type factor excluding the possibility that lower levels of the former was responsible for the poor RPS28B mRNA degradation (Figure 5B).
Figure 5.
The motif of Edc3 is required for regulation of the RPS28B mRNA degradation but not for the decay of the YRA1 pre-mRNA. (A) Northern blot analysis of total yeast RNA isolated from the Δedc3 strain transformed with empty vector (vector), with a plasmid expressing Edc3-protA, or with a plasmid expressing Edc3ΔRB-protA. The isogenic wild-type strain transformed with empty vector was included as a positive control. Left panel: hybridization with a Rps28 probe that reveals both the RPS28A and RPS28B mRNAs. Right panel: hybridization with a Yra1 probe to detect the YRA1 mRNA and pre-mRNA. To control for equal loading, the blots were hybridized with a probe recognizing the SCR1 RNA. (B) Western blot analysis of total protein extract from yeast cells used for total RNA isolation for northern blot hybridization. Presence of protA tagged Edc3 was analysed with peroxidase-anti-peroxidase complexes. Antibodies against Stm1 were used to control for equal loading. (C) Northern blot analysis of total yeast RNA isolated from the Δedc3Δrps28B strain transformed with a vector bearing RPS28B gene under a tetracycline-repressible transcription activator and a plasmid expressing Edc3-protA, or a plasmid expressing Edc3ΔRB-protA. Total RNA was isolated at indicated time points after addition of doxycycline. Hybridization was done with a Rps28 probe and a Scr1 probe (which serves as a loading control).
Analysis of the YRA1 pre-mRNA degradation gave opposite results. Deletion of the edc3 gene resulted in an almost 4-fold (3.7 ± 1.86) increase in the amount of YRA1 pre-mRNA relative to the mRNA compared with the wild-type strain (Figure 5A, right panel). Expression of wild-type Edc3-protA led to an almost complete restoration of the YRA1 pre-mRNA/YRA1 mRNA ratio (1.1 ± 0.66). Expression of Edc3ΔRB-protA had the same effect (1.2 ± 0.42), indicating that interaction between Edc3 and Rps28 is not needed for the degradation of the YRA1 pre-mRNA. This observation demonstrates further that the effect of the removal of residues 201–231 of Edc3 on RPS28B mRNA was highly specific.
To verify that the observed changes in RPS28B mRNA steady-state level are related to autoregulation of its stability, we analysed the half-life of RPS28B mRNA using a strain where the endogenous RPS28B and EDC3 genes were deleted. Cells were a transformed with a vector bearing RPS28B gene under a tetracycline-repressible transcription activator (19) and either a vector coding for wild-type EDC3-protA fusion or a vector coding for EDC3ΔRB-protA fusion. Total RNA was isolated at different time points after doxycycline-induced transcriptional repression of RPS28B and analysed by northern blotting. The results obtained (Figure 5C) clearly show the increase of the half-life time of RPS28B mRNA in the strain expressing EDC3ΔRB-protA fusion (22.5 ± 2.1 min compared with 8 ± 1 min in the strain expressing the wild-type EDC3-protA fusion). These changes are similar to those reported when the autoregulation loop of RPS28B mRNA level was identified (19). These data indicate that the interaction between Rps28 and Edc3 mediated by RB motif is implicated in this autoregulatory process.
DISCUSSION
We have identified a novel motif within yeast Edc3 protein that is required and sufficient for binding to the ribosomal protein Rps28. Moreover, we have shown that Rps28 and Edc3 interact directly, forming a stable heterodimeric complex in the absence of other specific RNA or protein partners. In vivo analyses demonstrated that this region of Edc3 is crucial for the auto-regulatory feedback loop that controls the RPS28B mRNA level, but that it is dispensable for other functions of Edc3, including its ability to regulate the YRA1 mRNA level by inducing degradation of YRA1 pre-mRNA. The latter result indicates that the two processes in which Edc3 participates to the auto-regulation of RNA levels by controlling their decay rates rest on different mechanisms involving different protein–protein interaction interfaces. The binding of Edc3 to Rps28 will ensure a rapid induction of RPS28B mRNA decay if free Rps28 protein starts to accumulate.
One can wonder whether the ability of Edc3 to interact with Rps28 extends to situation where the latter is incorporated in ribosomes. Our data indicate that Edc3 interacts with the Rps28 core rather than its C-terminal tail. Interaction of Edc3 with ribosome-bound Rps28 may be limited by the presence of Rps5 and ribosomal RNA that mask large share of the Rps28 core surface (29). If decapping was shown to occur in part on polysome-associated mRNAs (8) and several mRNA decay factors were reported to sediment in polysomes (38,39), the presence of Edc3 in polysomes was, to the best of our knowledge, never reported. We also failed to detect such an association by polysome analyses or pull-down experiments (data not shown). Thus, it is probable that Rps28 present in ribosome is not accessible to Edc3, even if we cannot exclude that such association may take place temporarily or under specific conditions. If it would occur, association of Edc3 with ribosome-bound Rps28 would be restricted to a small group of organisms, arguing against a general role for such an interaction in mRNA decay. Detailed structural characterization of the Edc3-Rps28 dimer would provide more insights into this issue.
The limited conservation of the region of Edc3 involved in Rps28 binding suggests that it was acquired recently during evolution. This would explain its exclusive presence in proteins encoded by Saccharomycetaceae species where it would have been co-opted to finely tune the Rps28 protein production. Given the small size of the RB motif, one can easily imagine how it may naturally have evolved by incremental selection of residues improving the binding affinity in an otherwise poorly conserved linker region. Other events of auto-regulation of yeast ribosomal protein production, either through modulation of translation, splicing and/or control of transcription termination are known (40–42). The frequent observations of such mechanisms suggest that they evolve rapidly, taking advantage of the ability of proteins and RNA to create new interaction interfaces from short linear stretches of amino acid and oligonucleotide sequences. Their recursive occurrence indicates that finely tuning the production of ribosome subunits provides a key evolutionary advantage to yeast cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [43–45].
FUNDING
CERBM-IGBMC, the Ligue Contre le Cancer (Equipe Labellisée 2011) (to B.S.) as well as the CNRS and the Agence Nationale de la Recherche [ANR 11 BSV8 009 02 to M.G. and B.S.]. Funding for open access charge: Agence Nationale de la Recherche [ANR 11 BSV8 009 02].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Authors are grateful to Ysaline Billier, Laetitia Cormier, Claudine Gaudon, Nicolas Cougot, Françoise Wyers, Gwenael Badis, Alain Jacquier and Sylvie Friant for providing plasmids, strains and/or antibodies, and to all laboratory members for discussions. They thank the IGBMC mass-spectrometry, peptide synthesis and imaging platforms for their help and IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire) for assistance. They are indebted to Dr Noureddine Lazar for his help with the MALLS experiments.
REFERENCES
- 1.Balagopal V, Fluch L, Nissan T. Ways and means of eukaryotic mRNA decay. Biochim. Biophys. Acta. 2012;1819:593–603. doi: 10.1016/j.bbagrm.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene. 2012;500:10–21. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cougot N, Babajko S, Séraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erickson SL, Lykke-Andersen J. Cytoplasmic mRNP granules at a glance. J. Cell Sci. 2011;124:293–297. doi: 10.1242/jcs.072140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnerot C, Boeck R, Lapeyre B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tharun S, He W, Mayes AM, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 14.Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle JC, Fromont-Racine M, Jacquier A. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl Acad. Sci. USA. 2008;105:5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz D. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA. 2003;9:239–251. doi: 10.1261/rna.2171203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harigaya Y, Jones BN, Muhlrad D, Gross JD, Parker R. Identification and analysis of the interaction between Edc3 and Dcp2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2010;30:1446–1456. doi: 10.1128/MCB.01305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 20.Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol. Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O. A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol. Cell. Biol. 2007;27:8600–8611. doi: 10.1128/MCB.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fromm SA, Truffault V, Kamenz J, Braun JE, Hoffmann NA, Izaurralde E, Sprangers R. The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex. EMBO J. 2012;31:279–290. doi: 10.1038/emboj.2011.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling SH, Decker CJ, Walsh MA, She M, Parker R, Song H. Crystal structure of human Edc3 and its functional implications. Mol. Cell. Biol. 2008;28:5965–5976. doi: 10.1128/MCB.00761-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kshirsagar M, Parker R. Identification of Edc3p as an Enhancer of mRNA Decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong S, Li C, Zenklusen D, Singer RH, Jacobson A, He F. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell. 2007;25:559–573. doi: 10.1016/j.molcel.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong S, Jacobson A, He F. Degradation of YRA1 Pre-mRNA in the cytoplasm requires translational repression, multiple modular intronic elements, Edc3p, and Mex67p. PLoS Biol. 2010;8:e1000360. doi: 10.1371/journal.pbio.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aramini JM, Huang YJ, Cort JR, Goldsmith-Fischman S, Xiao R, Shih LY, Ho CK, Liu J, Rost B, Honig B, et al. Solution NMR structure of the 30S ribosomal protein S28E from Pyrococcus horikoshii. Protein Sci. 2003;12:2823–2830. doi: 10.1110/ps.03359003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 30.Marquez V, Frohlich T, Armache JP, Sohmen D, Donhofer A, Mikolajka A, Berninghausen O, Thomm M, Beckmann R, Arnold GJ, et al. Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins. J. Mol. Biol. 2011;405:1215–1232. doi: 10.1016/j.jmb.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 31.Wu B, Yee A, Pineda-Lucena A, Semesi A, Ramelot TA, Cort JR, Jung JW, Edwards A, Lee W, Kennedy M, et al. Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum. Protein Sci. 2003;12:2831–2837. doi: 10.1110/ps.03358203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pisarev A, Kolupaeva V, Yusupov MM, Hellen CUT, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baudin-Baillieu A, Guillemet E, Cullin C, Lacroute F. Construction of a yeast strain deleted for the TRP1 promoter and coding region that enhances the efficiency of the polymerase chain reaction-disruption method. Yeast. 1997;13:353–356. doi: 10.1002/(SICI)1097-0061(19970330)13:4<353::AID-YEA86>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 34.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 35.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daugeron MC, Mauxion F, Séraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fromont-Racine M, Mayes AE, Brunet-Simon A, Rain JC, Colley A, Dix I, Decourty L, Joly N, Ricard F, Beggs JD, et al. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast. 2000;17:95–110. doi: 10.1002/1097-0061(20000630)17:2<95::AID-YEA16>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangus DA, Jacobson A. Linking mRNA turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods. 1999;17:28–37. doi: 10.1006/meth.1998.0704. [DOI] [PubMed] [Google Scholar]
- 39.Sweet T, Kovalak C, Coller J. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012;10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dabeva MD, Warner JR. Ribosomal protein L32 of Saccharomyces cereuisiae regulates both splicing and translation of its own transcript. J. Biol. Chem. 1993;268:19669–19674. [PubMed] [Google Scholar]
- 41.Fewell SW, Woolford JL., Jr Ribosomal Protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B Pre-mRNA and to 18S rRNA. Mol. Cell. Biol. 1999;19:826–834. doi: 10.1128/mcb.19.1.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gudipati RK, Neil H, Feuerbach F, Malabat C, Jacquier A. The yeast RPL9B gene is regulated by modulation between two modes of transcription termination. EMBO J. 2012;31:2427–2437. doi: 10.1038/emboj.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ekstrom F, Stier G, Sauer UH. Crystallization of the actin-binding domain of human alpha-actinin: analysis of microcrystals of SeMet-labelled protein. Acta Crystallogr. D Biol. Crystallogr. 2003;59:724–726. doi: 10.1107/s0907444903002063. [DOI] [PubMed] [Google Scholar]
- 45.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.