Abstract
Obtaining quantities of highly pure duplex DNA is a bottleneck in the biophysical analysis of protein–DNA complexes. In traditional DNA purification methods, the individual cognate DNA strands are purified separately before annealing to form DNA duplexes. This approach works well for palindromic sequences, in which top and bottom strands are identical and duplex formation is typically complete. However, in cases where the DNA is non-palindromic, excess of single-stranded DNA must be removed through additional purification steps to prevent it from interfering in further experiments. Here we describe and apply a novel reversed-phase ion-pair liquid chromatography purification method for double-stranded DNA ranging in lengths from 17 to 51 bp. Both palindromic and non-palindromic DNA can be readily purified. This method has the unique ability to separate blunt double-stranded DNA from pre-attenuated (n-1, n-2, etc) synthesis products, and from DNA duplexes with single base pair overhangs. Additionally, palindromic DNA sequences with only minor differences in the central spacer sequence of the DNA can be separated, and the purified DNA is suitable for co-crystallization of protein–DNA complexes. Thus, double-stranded ion-pair liquid chromatography is a useful approach for duplex DNA purification for many applications.
INTRODUCTION
The production of synthetic DNA molecules via phosphoramidite chemistry was first described >30 years ago (1), and although the method has been improved over time, it remains the standard for DNA oligonucleotide synthesis today (2). The specificity, high yield and relative ease of phosphoramidite DNA synthesis make this method attractive for many users, but it does have the disadvantage of producing chemical contaminants as well as pre-attenuated (n-1, n-2, etc) DNA or ‘failure products’ of shorter base pair lengths. Purifying oligonucleotides from the synthesis by-products and isolating full-length polynucleotides from n–products is a necessary procedure for many widely used experimental procedures such as X-ray crystallography (3), nuclear magnetic resonance and isothermal titration calorimetry (ITC).
Purification of highly pure double-stranded DNA (dsDNA) in milligram quantities has been achieved using several approaches, but these methods have both strengths and weaknesses in their implementation. Trityl-on reversed-phase high-pressure liquid chromatography has the advantage of specifically isolating single-stranded DNA (ssDNA) that have been tritylated at the 5′-oxygen of the terminal ribose. However, if the trityl-on protection reaction is not efficient, there is a probability of co-eluting protected impurities with trityl groups (4). Trityl-on purification has the associated risk of producing depurinated DNA owing to the use of acetic acid in the purification process (4). Trityl-on chromatography also has the disadvantage of producing lower yields of oligonucleotides (4). Furthermore, the trityl-on purification often requires a second trityl-off purification step to produce a pure oligonucleotide product (5). Agarose gel electrophoresis can be used to purify high quantities of DNA quickly, but this technique does not allow for separation of n–byproducts (6–10), and is only suitable when the ssDNA or dsDNA fragments are of different lengths. Denaturing size-exclusion chromatography can be effective for short ssDNA oligonucleotides (11). Polyacrylamide gel electrophoresis (PAGE) purification can produce highly pure ssDNA; however, only sub-milligram quantities can be readily purified, and a significant amount of DNA is often lost when recovering the DNA from the gel (12). Finally, anion-exchange chromatography can be used to purify moderate quantities of DNA, yet this method cannot separate n–products from full-length DNA (13). It is important to note that these methods are generally used to purify ssDNA oligos, which are later annealed. However, after annealing the ssDNA strands, ssDNA often remains and can be problematic in performing subsequent experiments.
Reversed-phase ion-pair liquid chromatography has been previously used to purify short (>50 bp) (14) and moderate length (50–2000 bp) (15–19) DNA fragments. Here, we present a facile method we term as the ‘double-stranded ion-pair (DSIP) method’ that expands on previous reversed-phase ion-pair methods for the purification of highly pure dsDNA. This approach is derived from the RNA purification method described by McCarthy et al. (20). This new method is a powerful technique because it is possible to obtain milligram quantities of purified dsDNA in just a few hours. In addition, the dsDNA is generally free from DNA synthesis contaminants, ssDNA and ds, n– and pre-attenuated DNA products. Furthermore, dsDNA molecules differing by a single base pair can be readily separated. The DNA purified using the DSIP method is capable of producing diffraction quality crystals of protein–DNA complexes (21,22).
MATERIALS AND METHODS
Materials
Forward and reverse ssDNA oligomers were purchased from Eurofins MWG Operon and Integrated DNA Technologies. Triethylammonium acetate (TEAA) buffer was purchased from Sigma-Fluka. Sep-Pak Classic C18 cartridges were purchased from Waters. High-performance liquid chromatography (HPLC) grade water and gold label acetonitrile were purchased from Fisher Scientific.
Single-stranded C18 cartridge DNA purification
The single-stranded oligonucleotides obtained from ∼1 µmol syntheses were resuspended in TE buffer [10 mM Tris-HCl, 1 mM ethylenediamine tetraacetic acid (EDTA), pH 8.0] with 150 mM NaCl and purified using Waters C18 Sep-Pak cartridges. The cartridges were prepared by flushing with 10 ml of acetonitrile and subsequently rinsing the cartridge two times with 10 ml of water, before flowing the resuspended DNA slowly through the cartridge. The flow-through was collected and passed through the cartridge two more times. The column was then washed with 10 ml of water, and a small amount of air was pushed through the cartridge to remove any excess water. The single-stranded oligomers were then eluted with 2 ml of 60% methanol and dried in a Savant Speed Vac.
Annealing single-stranded oligomers to form dsDNA
Single-stranded oligonucleotides were resuspended in TE buffer with 150 mM NaCl and the concentration was determined spectrophotometrically. Equimolar amounts of forward and reverse ssDNA oligomers were combined and annealed by heating at 95°C for 5 min followed by slow cooling to room temperature over the course of 5 h.
Ethanol precipitation of dsDNA to remove residual salts
The annealed dsDNA was ethanol-precipitated by addition of sodium acetate, pH 5.2, to 300 mM and addition of 3 volumes of 100% ethanol. The sample was gently mixed, and left at −80°C for 30 min to precipitate the DNA. Afterward, the precipitated DNA was pelleted in an Eppendorf 5415 R centrifuge by centrifugation at 16 100g at 4°C for 20 min. The ethanol was decanted and the pellet was washed with 70% ethanol and dried in a Savant Speed Vac or under nitrogen gas.
HPLC purification of ssDNA
ssDNA was purified using Sep-Pak C18 cartridges and then ethanol precipitated as described above. Each ssDNA sample of 0.25 mg was diluted in 0.1 M TEAA, pH 7.0, and 5% acetonitrile, and boiled using a heating block at 100°C for 2 min before immediately loading onto a Waters Xbridge MS C18 where each oligomer was purified individually on a Waters 1525 HPLC at 60°C. It is important to pre-equilibrate the column at this temperature and to load the sample onto the column hot to prevent hairpin formation that occurs due to the palindromic character of the DNA strands. An acetonitrile gradient of 5–12% in 0.1 M TEAA, pH 7.0, buffer over 40 min was used to separate the single strands from synthesis, n–DNA pre-attenuated products and possible hairpins that formed during the initial purification processes. The flow rate was 2 ml/min, and 0.5 ml fractions were collected. After purification, the DNA that eluted from the column was dried in a Savant Speed Vac and subsequently resuspended in TE buffer with 150 mM NaCl.
HPLC purification of duplex DNA
The dried dsDNA pellet was resuspended in a 0.1 M TEAA, pH 7.0, and 5% acetonitrile buffer, and further purified on a Waters Xbridge OST C18 column with a 2.5-µm particle size and a 10 × 50 mm2 column size (catalogue number 186003954) using a Waters 1525 HPLC with an acetonitrile gradient of 5–12% in 0.1 M TEAA, pH 7.0, buffer over the course of 40 min. It is important to note that this method was performed at room temperature to prevent denaturation of the dsDNA duplex; this method also reduces the amount of hairpin formation from palindromic oligonucleotides, which have the potential to co-elute with dsDNA palindromic sequences. The flow rate was 2 ml/min, and 0.5 ml fractions were collected. Elution of DNA was monitored by ultraviolet absorbance at 260 and 285 nm. Appropriate fractions were collected and ethanol precipitated as described above to remove any salts or organic solvent and dried in a Savant Speed Vac or under nitrogen gas at room temperature before resuspending in water or TE buffer for further use.
Polyacrylamide gel electrophoresis
The purity of DNA fractions was assessed using 8% non-denaturing polyacrylamide (40% 37.5:1 acrylamide:N,N′-methylene-bis-acrylamide, BioRad) gels and 16% polyacrylamide (40% 19:1 acrylamide:N,N′-methylene-bis-acrylamide, BioRad) 7 M urea denaturing gels. In all, 12.5 ng of DNA from appropriate fractions was loaded into each well. For 8% native gels, DNA was suspended in 10 µl of 1 × TBE (90 mM Tris, 90 mM boric acid, 2 mM EDTA) buffer with 6% glycerol. Native gels were electrophoresed at 125 V for 30 min in 0.33× TBE buffer at 4°C before DNA samples were loaded. After loading, the gel was electrophoresed for 45 min at 75 V at 4°C. For 16% acrylamide 7 M urea denaturing gels, DNA samples were suspended in 3 µl of 90% formamide, 10 mM EDTA and 0.1 M NaOH, before heating at 95°C for 5 min and gel loading. Denaturing gels were pre-electrophoresed for 1 h at 200 V in 1× TBE at 40°C. After pre-electrophoresis, the DNA samples were loaded, and electrophoresis proceeded for 1 h at 200 V at 40°C. Both types of gels were stained with SYBR Green I (Invitrogen) and imaged on a Storm 860 Scanner (Molecular Biosystems) with the following settings: excitation wavelength of 450 nm, emission cutoff filter of 520 nm and the photomultiplier tube set to 800 V.
Mass spectrometry
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) experiments were conducted to assess DNA purity and confirm the predicted molecular mass of the DNA samples. DNA samples were loaded onto pre-equilibrated G50 resin in a SpinX column and spun at 11 000g to remove excess TEAA buffer and trace amounts of small molecules such as salts. The DNA was then dried in a Savant Vac and resuspended in water at concentrations ranging from 2 to 9 mg/ml. The matrix consisted of 35 mg/ml 3-hydroxypicolinic acid (Sigma) and 2 mg/ml D-(–)-fructose in 50% acetonitrile (Fisher). The addition of D-(–)-fructose to the matrix improves mass resolution at the high laser fluence required to ionize the DNA (23). Matrix was first applied to a sample stage and air dried to form copious fine needles. A 1-µl aliquot of the DNA sample was then applied and allowed to dry. Negative ion mass spectrometry was performed using a QSTAR XL (Applied Biosystems/MDS SCIEX) mass spectrometer with an oMALDI ion source and a solid-state laser with light output at 355 nm.
TFAM purification
Recombinant TFAM protein lacking the mitochondrial localization sequence (21) was expressed and purified as previously described (24).
Crystallization
Crystallization of protein–DNA complexes was initiated by mixing pure protein with pure DNA fractions. The complex was concentrated to the appropriate concentration, and initial crystallization trials were performed using the commercially available JCSG Core Suite I (Qiagen) and Hampton Crystal Screen HT (Hampton Research).
RESULTS
Purification of double-stranded non-palindromic DNA
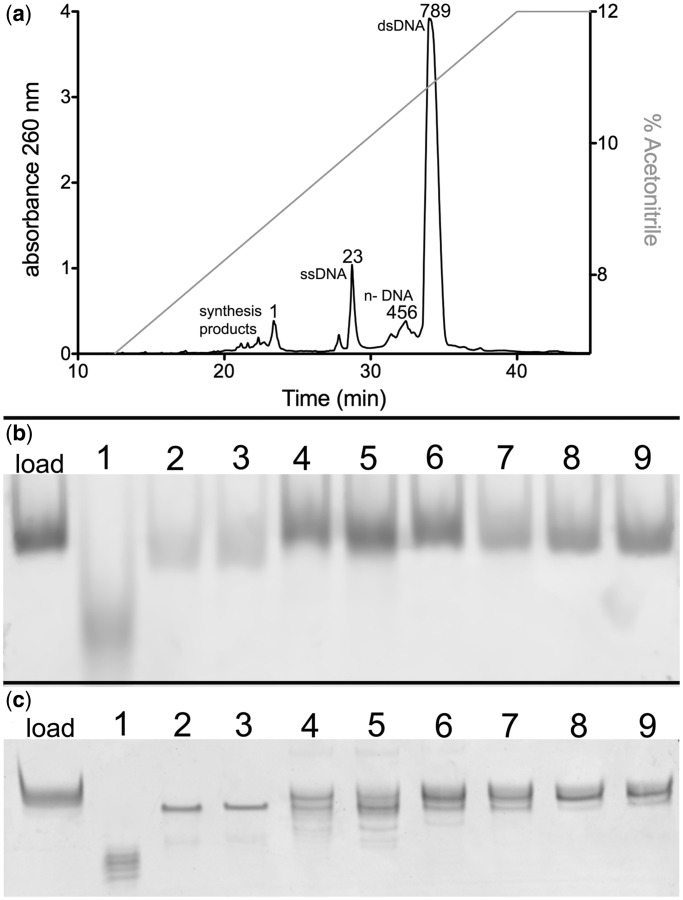
We adapted and applied the DSIP approach that had originally been used for RNA purification to the purification of duplex DNA oligonucleotides (20). We first describe results obtained with a 28-base long (28 mer) dsDNA binding site for the mitochondrial transcription factor A (LSP28mer; Table 1), which recognizes a non-palindromic DNA fragment that is long enough to be somewhat problematic for other purification methods, such as denaturing size exclusion and gel electrophoresis methods (7–11). After the Sep-Pak-purified single-stranded oligos were annealed, 0.5 mg of ds 28mer DNA was purified by reversed-phase ion-pair HPLC on a Waters C18 OST XBridge column with a 5–12% acetonitrile gradient over the course of 40 min (Figure 1a). As the acetonitrile gradient was applied, byproducts from the DNA synthesis were first eluted (1; Figure 1a), followed by ssDNA (2,3; Figure 1a), then n–products (4–6; Figure 1a) and finally the pure dsDNA (7–9; Figure 1a). Our assignments of the DNA species were initially based on previous findings for RNA purification (20).
Table 1.
DNA sequences purified by the DSIP method and used in crystallization trials
| DNA | Sequence | Crystals/diffraction |
|---|---|---|
| LSP 22mer | 5′-TGTTAGTTGGGGGGTGACTCTT | 20.0 Å |
| 3′-ACAATCAACCCCCCACTGAGAA | ||
| LSP 23mer | 5′-GTGTTAGTTGGGGGGTGACTCTT | Yes |
| 3′-CACAATCAACCCCCCACTGAGAA | No diffraction | |
| LSP 24mer | 5′-GTGTTAGTTGGGGGGTGACTCTTA | Yes |
| 3′-CACAATCAACCCCCCACTGAGAAT | 20.0 Å | |
| LSP 25mer | 5′-GTGTTAGTTGGGGGGTGACTGTTAA | Yes |
| 3′-CACAATCAACCCCCCACTGACAATT | 7.1 Å | |
| LSP 25merGC | 5′-GTGTTAGTTGGGGGGTGACTGTTAC | Yes |
| 3′-CACAATCAACCCCCCACTGACAATG | 3.0 Å | |
| LSP 26mer | 5′-GTTAGTTGGGGGGTGACTGTTAAAAG | No |
| 3′-CAATCAACCCCCCACTGACAATTTTC | - | |
| LSP 27mer | 5′-TGTTAGTTGGGGGGTGACTGTTAAAAG | No |
| 3′-ACAATCAACCCCCCACTGACAATTTTC | - | |
| LSP 28mer | 5′-TGTTAGTTGGGGGGTGACTGTTAAAAGT | Yes |
| 3′-ACAATCAACCCCCCACTGACAATTTTCA | 8.2 Å | |
| LSP 29mer | 5′-TGTTAGTTGGGGGGTGACTGTTAAAAGTG | No |
| 3′-ACAATCAACCCCCCACTGACAATTTTCAC | – | |
| LSP 30mer | 5′-GTGTTAGTTGGGGGGTGACTGTTAAAAGTG | Yes |
| 3′-CACAATCAACCCCCCACTGACAATTTTCAC | None | |
| LSP 44mer | 5′-GTTAGTTGGGGGGTGACTGTTAAAAGTGCATACCGCCAAAAGAT | – |
| 3′-CAATCAACCCCCCACTGACAATTTTCACGTATGGCGGTTTTCTA | ||
| LSP 48mer | 5′-GTTAGTTGGGGGGTGACTGTTAAAAGTGCATACCGCCAAAAGATAAAA | – |
| 3′-CAATCAACCCCCCACTGACAATTTTCACGTATCGCGGTTTTCTATTTT | ||
| LSP 51mer | 5′-GTTAGTTGGGGGGTGACTGTTAAAAGTGCATACCGCCAAAAGATAAAATTT | – |
| 3′-CAATCAACCCCCCACTGACAATTTTCACGTATGGCGGTTTTCTATTTTAAA | ||
| PR 17mer | 5′-CAGAACAGTTTGTTCTG | – |
| 3′-GTCTTGTCAAACAAGAC | ||
| GR 17mer | 5′-CAGAACATGATGTTCTG | – |
| 3′-GTCTTGTACTACAAGAC | ||
| Pal AG 5′ | 5′-ACAACCTGCCCGATCGGGCAGGTT | Yes |
| 3′-TGTTGGACGGGCTAGCCCGTCCAA | 3.4 Å | |
| Pal AG 3′ | 5′-CAACCTGCCCGATCGGGCAGGTTGA | – |
| 3′-GTTGGACGGGCTAGCCCGTCCAACT |
Figure 1.
Purification of non-palindromic dsDNA. (a) DNA elution profile of 0.5 mg of 28-bp DNA corresponding to the human mitochondrial LSP sequence (LSP 28mer) from a Waters X-Bridge OST C18 column. DNA absorbance was monitored at 260 nm (black trace, left Y-axis), as DNA was eluted from the column using an acetonitrile gradient (gray trace, right Y-axis). Numbers correspond to fractions collected for purity analysis. Numbers correspond as follows: 1, phosphoramodite DNA synthesis material; 2–3, ssDNA 28 mer LSP DNA; 4–6, ds n–LSP DNA products; and 7–9, full length 28 mer LSP DNA. (b) Eight percent polyacrylamide native gel analysis and (c) 16% polyacrylamide 7 M urea denaturing gel analysis of fractions collected in (a) as indicated by numbers.
PAGE was used to distinguish the products of the 28mer DNA purification. We assessed the purity of the indicated fractions in Figure 1a using native PAGE (Figure 1b). The DNA species in lane 1 migrated faster than the other species in either gel (Figure 1b), indicating that this fraction includes impurities from the phosphoramidite synthesis. The DNA in lanes 2 and 3 migrated slightly faster than the DNA fractions in lanes 4–9, suggesting that these fractions are ssDNA (Figure 1b). The lower intensity of the bands in lanes 1, 2 and 3 is also indicative of single-stranded character, as SYBR Green I fluoresces with lower intensity when bound to ssDNA (25). Lanes 4–9 in Figure 1b fluoresced with similar intensity to the load lane, suggesting these fractions contain some form of dsDNA; however, no differences in gel migration were observed between lanes 4 and 9, suggesting these fractions are all of similar size. The eluted fractions from Figure 1a were analyzed using denaturing PAGE to identify differences between species in fractions 4–9 (Figure 1c). We were able to distinguish between the lighter top strand (8405.2 Da) and the heavier bottom strand (8769.7 Da), and as expected, the top strand migrated faster in the gel than the bottom strand (Supplementary Figure S1). In Figure 1c, the species in lane 1 migrated faster than any other lane, further confirming that this fraction corresponds to phosphoramidite DNA synthesis material. A single band corresponding to unannealed top strand DNA was observed in lanes 2 and 3. Lanes 4–6 contain top strand and bottom strand DNA as well as some shadowing below the main bands most likely due to n– failure products in the bottom strand (Figure 1c and Supplementary Figure S1). The presence of the n– products in the bottom oligonucleotide is likely caused by the presence of the 6-guanine base run in the middle of the oligonucleotide. Runs of ≥4 guanine bases in the phosphoramidite synthesis can lead to the formation of cruciform guanine complexes that can cause premature truncation of the DNA and form higher order structures (26). Lanes 7–9 in Figure 1c appear to show more of the bottom strand than the top. The higher intensity bands in lanes 7–9 could potentially be reannealed dsDNA, as dsDNA can form in denaturing gels (Supplementary Figure S1).
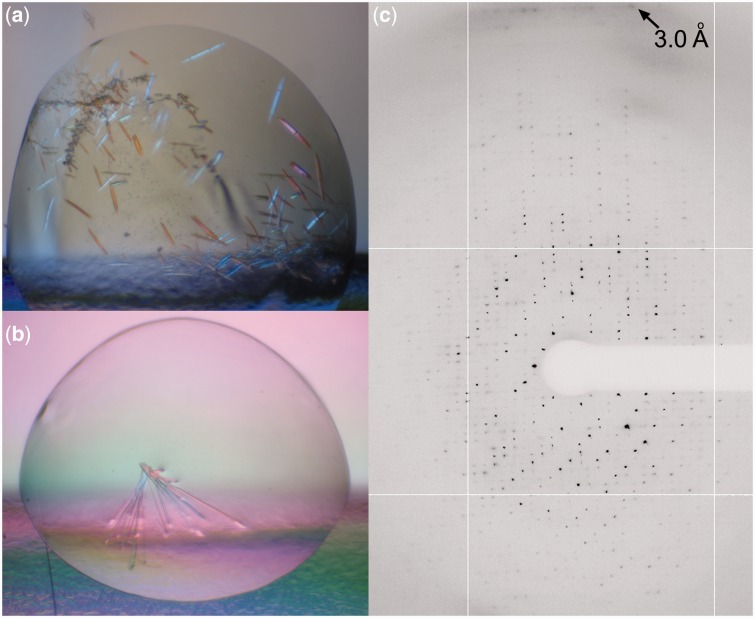
To determine whether DNA purified by this method was suitable for protein–DNA complex crystallization, we combined TFAM and LSP 25mer GC (Table 1) to form a 15.6-mg/ml complex that was then used for crystallography experiments. Using the commercially available JCSG Core Suite I (Qiagen) screen, we obtained flat and rod-shaped crystals of the TFAM-LSP 25mer GC complex (Figure 2a and b). In addition to obtaining crystals with the LSP 28mer sequence used here and by Ngo et al. to obtain crystals of a TFAM-LSP 28mer complex (22), we have obtained diffraction quality crystals of TFAM-LSP complexes with various LSP DNA sequences (Table 1). Using this method, we were able to obtain TFAM-LSP crystals with every LSP sequence we tried; however, whether or not the crystals diffracted X-rays was largely dependent on the DNA sequence (Table 1), as the crystal packing of DNA molecules can have dramatic effects on the resolution of the structure (27). The crystals shown in Figure 2b were sent to the Advanced Photon Source (beamline NE-CAT: 24-ID-C) at Argonne National Laboratory (Argonne, IL), and data were collected. A representative image from a dataset is shown in Figure 2c with X-rays diffracting to 3.0 Å. The slight shadowing below the 3.0 Å label in Figure 2c is indicative of the presence of DNA. Indeed, when we solved the structure of this complex, DNA was present in the structure (data not shown). These findings demonstrate the feasibility of this DNA purification method for producing X-ray diffraction quality protein–DNA crystals.
Figure 2.
Protein–DNA crystals and X-ray diffraction pattern. Crystals of 15.6 mg/ml TFAM bound to 25-bp LSP DNA in (a) 0.2 M calcium acetate, 0.1 M imidazole, pH 8.0, and 20% polyethylene glycol 1000, and (b) 0.1 M Tris, pH 7.0, 20% polyethylene glycol 2000 obtained by sitting drop vapor diffusion. The well solution for both images was 1 M NaCl. (c) X-ray diffraction image of a rod-like TFAM-25mer GC crystal shown in (b) to 3.0 Å.
Purification of pseudopalindromic 25mer DNA duplexes
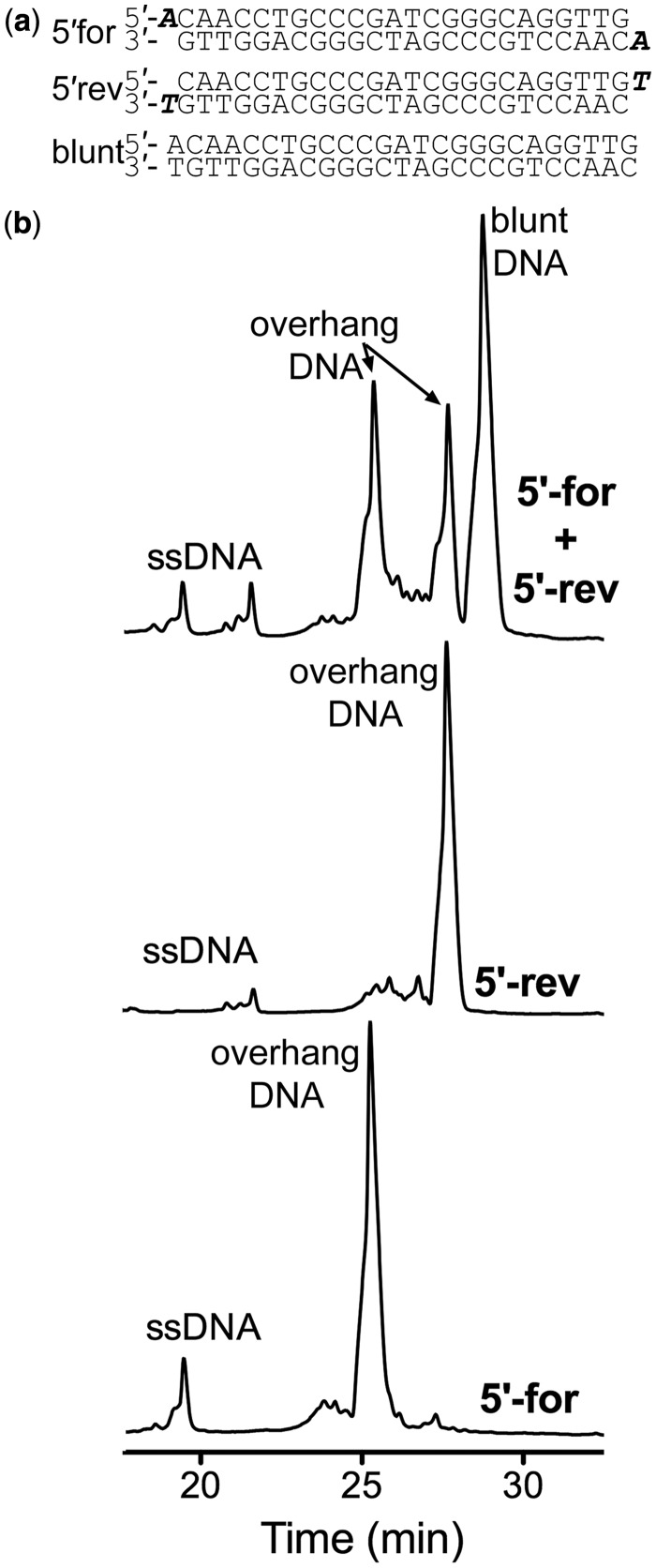
We further investigated the potential of this dsDNA purification method to discriminate between duplex DNA species that differed by only a single base pair. The pseudopalindromic DNA sequence has an additional adenine and thymine base pair at the 5′end of the sequence (Figure 3a), so we have named this DNA sample ATPal. Thus, the palindromic 25mer has the potential to form three distinct duplex DNA products as depicted in Figure 3a: two duplex DNA species will have 5′-A or 3′-T overhangs (5′for and 3′rev), as denoted by the top and bottom strands illustrated in Figure 3a) and one will be a blunt-ended dsDNA (blunt). After loading 0.25 mg of the annealed DNA onto the Waters C18 XBridge column and applying a gradient of 5–12% acetonitrile over 40 min at room temperature, we obtained five peaks on the chromatogram (Figure 3b, top). ssDNA eluted first, followed by the ds overhang DNA corresponding to the 5′for annealed strand and then the 5′rev annealed strand (Figure 3b, top). The blunt-ended DNA was the last species to elute off the column (Figure 3b, top). To confirm the identities of the DNA within these peaks, we loaded 0.25 mg of the 5′for top strand and the 5′rev bottom strand individually and repurified the dsDNA. These DNA oligonucleotides, when annealed individually, generated dsDNA species with overhangs (Figure 3a). When these were purified, the ssDNA peak, the dsDNA peak of the 5′rev dsDNA (Figure 3b, middle) and the 5′for dsDNA (Figure 3b, bottom), respectively, were the major peaks. These peaks obtained from purifying the 5′for top strand and 5′rev bottom strand DNA sequences alone have identical retention times to the overhang DNA peaks presented in Figure 3b (top), confirming the identities of all DNA species in this purification. These experiments demonstrate that this method is able to separate DNA fragments that are highly similar in length and sequence, even DNA with slightly different overhangs, on an otherwise palindromic DNA sequence.
Figure 3.
Separation of 5′ single base pair overhang palindromic DNA. (a) Top: 5′-forward palindromic strand annealed to itself with the adenine overhang base highlighted in bold italics. Middle: 5′-reverse palindromic strand annealed to itself with the thymine overhang base highlighted in bold italics. Bottom: Blunt DNA sequence of the 5′-forward and reverse palindromic strands. (b) Top: DNA elution profile of 0.25 mg of 5′-forward and 5′-reverse DNA after annealing. Blunt dsDNA can be separated from overhang dsDNA as well as ssDNA. The left overhang peak corresponds to 5′-forward self-annealed palidromic DNA, whereas the right overhang peak corresponds to 5′-reverse self-annealed palindromic DNA. Middle: DNA elution profile of 0.25 mg of 5′-reverse self-annealed palindromic DNA alone. Bottom: the DNA elution profile of 0.25 mg 5′-forward self-annealed palindromic DNA alone. The acetonitrile gradient for all spectra was 5–12% over the course of 40 min.
In a classic DNA purification used for biophysical techniques, ssDNA oligomers are individually purified using similar HPLC methods at high temperatures (28), and then subsequently annealed to form dsDNA. However, this leaves open the possibility of having unannealed ssDNA in the sample. To show that unannealed ssDNA is in the final sample after purifying the individual oligomers and later annealing them, we used this traditional high temperature (60°C) approach to purify each ssDNA oligo, annealed the purified oligos to form the desired palindromic blunt-ended DNA and analyzed the resulting dsDNA using this new reversed-phase ion-pair method. Our results show that the desired blunt-ended DNA product contains ssDNA and overhang DNA, and that the DNA did not anneal to form a single homogeneous blunt-ended DNA product. Supplementary Figures S2A and S2C show the ssDNA HPLC chromatograms using the high temperature method. Supplementary Figures S2B and S2D show the purity of the DNA using a denaturing PAGE method. Thus, our technique can be applied at high temperatures to purify ssDNA. Each lane is mapped to the fractions collected in the chromatograms. Lane 1 of both Supplementary Figures S2B and S2D show DNA species that have migrated further in the gel that are therefore smaller in size in comparison with the 25mer DNA. These are n– pre-attenuated failure products that have been separated from the DNA of interest. Lanes 3 and 4 of Supplementary Figures S2B and S2D are indistinguishable by denaturing PAGE, which contain the desired 25-bp palindromic ssDNA. The two purified single-stranded oligonucleotides were then annealed and analyzed by the dsDNA ion-pair procedure (Supplementary Figure S3A). Again, the characteristic five peaks appeared for this annealed dsDNA, indicating that certain palindromic ssDNA cannot be individually purified and subsequently annealed to form a single blunt-ended DNA product owing to the single strands annealing to themselves to form overhang DNA. The DSIP method separates all of these annealed products to obtain the desired blunt-ended product. Native (Supplementary Figure S3B) and denaturing (Supplementary Figure S3C) PAGE analysis of the DNA species in each peak of the chromatogram from Supplementary Figure S3A shows that lanes 1 and 2 correspond to ssDNA, but the species in lanes 3–8 of Supplementary Figure S3B are all indistinguishable from one another. However, they are clearly different forms of the dsDNA. Thus, using both the ssDNA and the dsDNA purification methods, we obtained similar results of obtaining five characteristic DNA peaks in the chromatogram. The advantage of the DSIP method is the ability to separate these uniquely annealed DNA fragments in a single purification step.
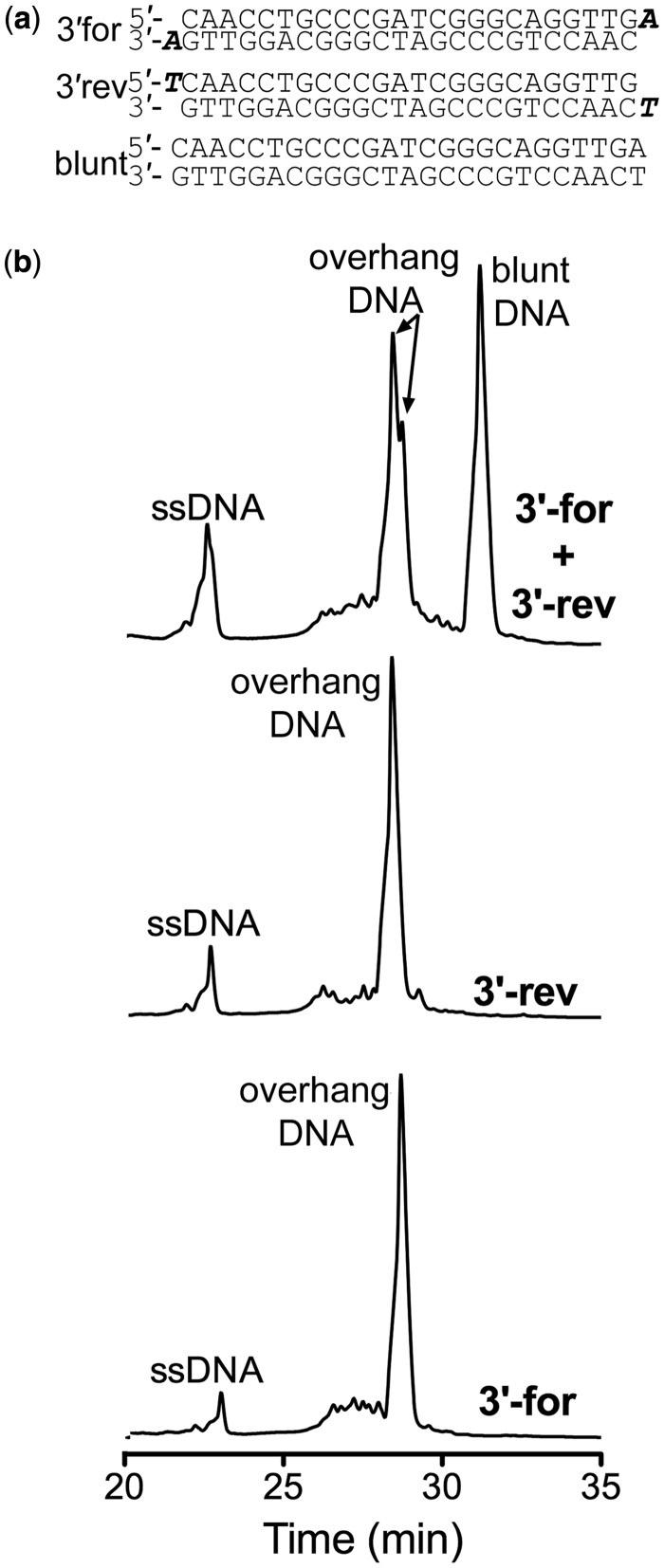
To determine whether the DSIP purification method could discriminate single base overhangs on the 3′ end of the same DNA, we tested the sequence named PalAT, where we added an A/T base pair at the 3′end of the palindromic DNA sequence used in Figure 3. We hypothesized that three distinctive DNA peaks would appear in the chromatogram corresponding to two overhang duplex DNA species and one duplex DNA peak. However, we saw differences in the overhang DNA versus the blunt-ended DNA, but did not resolve the two overhang DNA species (Figure 4b) that were seen in Figure 3b. We confirmed these results by annealing the 3′end forward top strand to itself (3′for) and annealing the 3′end reverse bottom strand to itself (3′rev) and purifying these species using the same method. In both the purifications for 3′rev (Figure 4b, middle) and 3′for (Figure 4b, bottom), a single-stranded un-annealed DNA peak and an overhang self-annealed duplex DNA peak that corresponded to the retention times shown in Figure 4b (top) were observed, thus demonstrating that 3′ modified palindromic overhang DNA can be separated from the blunt-ended DNA.
Figure 4.
Separation of 3′ single base pair overhang palindromic DNA. (a) Top: 3′-forward palindromic strand annealed to itself with the adenine overhang base highlighted in bold italics. Middle: 3′-reverse palindromic strand annealed to itself with the thymine overhang base highlighted in bold italics. Bottom: Sequences of the 3′-forward, reverse and blunt palindromic DNA. (b) Top: DNA elution profile of 0.25 mg of 3′-forward and 3′-reverse DNA after annealing. Blunt dsDNA can be separated from overhang dsDNA as well as ssDNA. The left overhang peak corresponds to 3′-forward self-annealed palindromic DNA, whereas the right overhang peak corresponds to 3′-reverse self-annealed palindromic DNA. (Middle) DNA elution profile of 3′-reverse self-annealed palindromic DNA alone. Bottom: the DNA elution profile of 3′-forward self-annealed palindromic DNA alone. The acetonitrile gradient for all spectra was 5–12% over the course of 40 min.
Purification of partially palindromic 17mer duplex DNA fragments
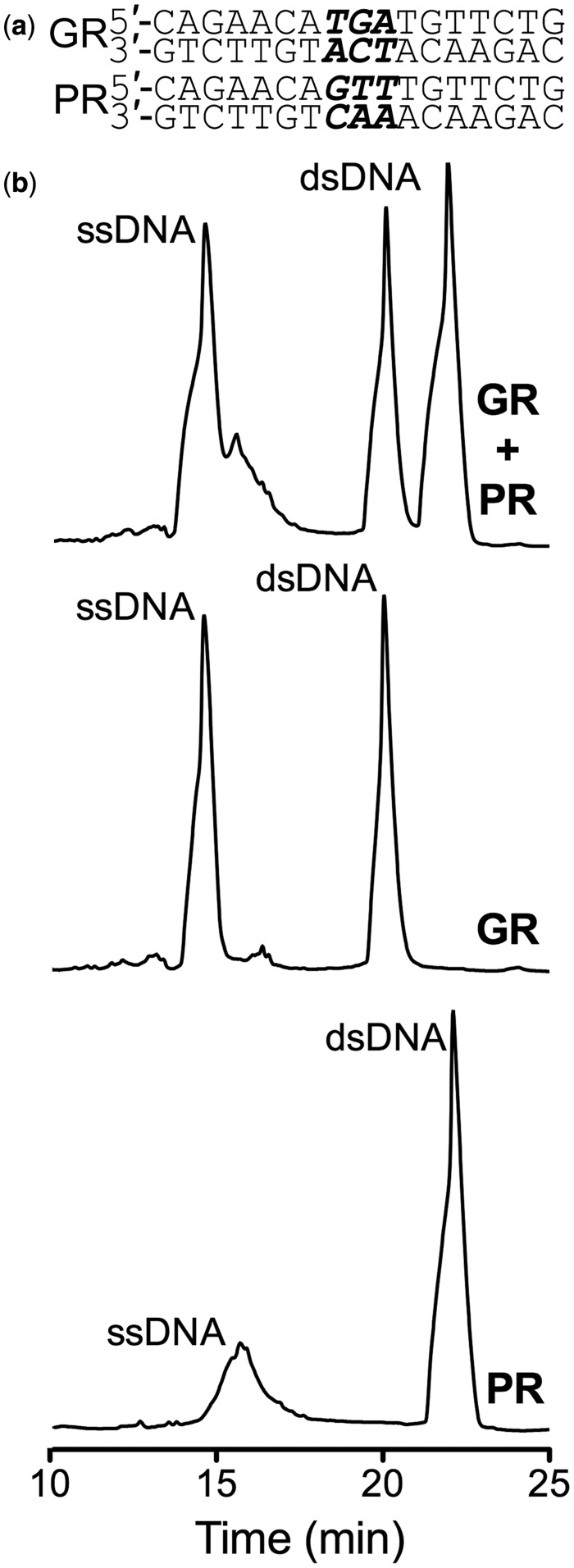
We have found that the DSIP technique is very powerful for purifying duplex DNA as well as pseudopalindromic DNA species with single base pair differences on the 5′ and 3′ ends. We further assessed this method to purify other partially palindromic sequences of DNA. These sequences, as depicted in Figure 5a, are recognized by the glucocorticoid receptors (GR) and progesterone receptors (PR), differing only by 3 bp in the center of the promoter DNA sequences. After annealing these DNA duplexes and analyzing them using the DSIP method, we found that the different DNA duplexes could be resolved as shown in Figure 5b. The top spectrum in Figure 5b demonstrates that the similar dsDNA GR and PR sequences can not only be separated from ssDNA, but from highly similar dsDNA as well. We performed MALDI-MS on the PR fractions and confirmed the presence of only PR DNA in each peak (Supplementary Figure S4). The middle and bottom elution profiles indicate the elution times for GR and PR DNA that were individually annealed, respectively. It is interesting to note that both DNA sequences have the same purine and pyrimidine content (Figure 5b), yet these dsDNA can still be separated. On the other hand, under the conditions used here, we were not able to separate any potential DNA with unpaired DNA in the spacer region (bubble DNA) should it have formed in this case. The potential inability to separate dsDNA with unpaired sequences might represent a limitation of the DSIP method. Again, we have shown that it is possible to obtain highly pure dsDNA that can be used for biophysical and analytical techniques, and that this technique has the potential to discriminate between different DNA species with very few differences in their base pair sequences.
Figure 5.
Separation of palindromic DNA with altered central base pairs. (a) Top: a 17-bp region of the consensus GR DNA recognition sequence; and Bottom: a 17-bp region of the consensus progesterone receptor (PR) DNA recognition sequence. Differences between the two DNA sequences are highlighted in bold italics. (b) Top: DNA elution profile of 0.25 mg of GR and PR DNA. The left peak corresponds to ssDNA. The right dsDNA peak corresponds to PR DNA and the left dsDNA corresponds to GR DNA. Middle: DNA elution profile of 0.25 mg of GR DNA; and Bottom: 0.25 mg of PR DNA indicating the retention times for these dsDNA species. The acetonitrile gradient was 5–12% over the course of 40 min.
Purification of 44mer, 48mer and 51mer duplex DNA fragments
To further examine the versatility of this technique, we tested the purification of longer DNA fragments. Individual forward and reverse strands of DNA sequences of 44, 48 and 51 bp in length corresponding to the human mitochondrial light strand promoter (LSP) region (Table 1) (21,29) were purified using Sep-Pak cartridges. After annealing the ssDNA species and ethanol precipitating them, 0.5 mg of each duplex was loaded to the Waters Xbridge OST C18 column and a gradient of 5–20% acetonitrile was applied over the course of 40 min at room temperature. We achieved variable degrees of purification of the dsDNA, with most being separated from n– pre-attenuated synthesis products and other synthesis material (Supplementary Figure S5). The data in Supplementary Figure S5 show, as expected, that the retention times for the LSP DNA increase as DNA length increases. We have not attempted purification of dsDNA longer than 51 bp using this method, but one could theoretically do so assuming a larger acetonitrile concentration was used for elution of the DNA.
DISCUSSION
The DSIP method of DNA purification provides a versatile approach for obtaining highly pure dsDNA that is capable of producing X-ray diffraction quality crystals on the semi-preparative (>100 µg) scale from a single purification. Separation of dsDNA from ssDNA, n– products, and DNA synthesis material can be achieved in a few hours. In addition, the purification of non-palindromic, palindromic, short 17 bp sequences of dsDNA, dsDNA sequences that differ by only a few base pairs with the same purine:pyrimidine content, 44mer and 51mer dsDNA all can be purified using this novel method. The use of purified homogeneous DNA is also of great interest to those researchers conducting biophysical experiments such as circular dichroism, electrophoretic mobility shift assays (EMSAs), fluorescence, differential scanning calorimetry and ITC (21,24,30). For example, if a researcher were to examine the ability of a protein to bind a certain DNA sequence specifically using EMSA, using DNA that contains contaminants could lead a researcher to make incorrect conclusions about the specificity of the protein of interest binding to DNA. In an EMSA experiment, low-purity DNA can cause smearing on the gel, and this smearing could be interpreted as a non-specific interaction with a protein of interest. On the other hand, if DNA of high purity is used in the EMSA experiment, specific shifts in the protein–DNA complex or non-specific smears can readily be interpreted.
With the DSIP method, we were able to separate DNA sequences differing by a single base pair. In Figure 3b, we observed that palindromic DNA containing a 5′ adenine had a shorter retention time than a similar DNA containing 3′ thymine; furthermore, both these species eluted before the blunt palindromic DNA. In Figure 4b, the adenine was placed at the 3′ position and the thymine at the 5′ end of the DNA, and we did not achieve the same level of separation observed in Figure 3b. Interestingly, when the positions of the adenine and thymine overhangs are reversed (Figures 3b and 4b), the 5′ thymine DNA elutes before the 3′ adenine (Figure 4b), whereas the 5′ adenine has a shorter retention time than the 3′ thymine overhang DNA.
The position of a purine or pyrimidine at the 5′ or 3′ overhang position clearly alters the retention time (Figures 3b and 4b). However, there does not seem to be a discernable pattern for the position (3′ versus 5′) and identity (purine versus pyrimidine) of the overhang base in predicting which overhang will have a shorter retention time. Retention time is governed by the hydrophobicity of the molecule, which, in this case, is similar between the DNA fragments being compared. Therefore, the subtle variations in hydrophobicity possibly due to slight conformational differences at the DNA ends must contribute to the ability to separate otherwise similar DNA fragments. Whatever the molecular mechanism, single overhang base pair resolution surprisingly can be readily achieved using the DSIP method (Figures 3b and 4b). Finally, in Figure 5b, we were able to separate palindromic DNA with altered central base pairs. The PR DNA molecule eluted after the GR DNA. Perhaps, this is due to the AA purine run in the differing central regions of the DNA molecules that could increase the hydrophobic character PR molecule compared with GR.
Several DNA purification methods are at the disposal for researchers, including DEAE anion exchange chromatography (13), denaturing size exclusion chromatography (11), PAGE purification (12), agarose gel purification (6–10) and trityl-on/trityl-off purification (5). The DSIP method has many of the advantages of these methods without several of the disadvantages. For example, the DSIP method can be run quickly as with agarose gel and ion exchange chromatography; however, the DSIP method can separate desired DNA from n– products with a degree of separation that cannot be achieved by agarose gel purification or DEAE anion exchange chromatography. The DSIP method can be modified to purify single strands of DNA (Supplementary Figure S2) as can denaturing size exclusion or PAGE purification; however, the DSIP method can be used on the semi-preparative scale. Furthermore, if one desires pure dsDNA, the DSIP method can be used after the ssDNA has been annealed (Figure 3), and still achieve the same level of separation as when the single strands are purified separately (Supplementary Figure S3). Finally, the DSIP method is the only method that has been shown to separate DNA sequences differing by a single base pair at the semi-preparative scale (Figures 3–5).
This method has been applied to several other lengths of non-palindromic DNA, and we have been able to obtain diffraction quality protein–DNA complex crystals for several of these DNA fragments (Table 1). Furthermore, we have used this method to purify DNA to study the interaction of TFAM with promoter and non-promoter DNA (21) using the purified DNA in circular dichroism and ITC studies.
In summary, this method can be used to purify DNA sequences ranging in length from 17 bp to 51 bp (Table 1). Although we have not used the DSIP method to purify DNA sequences shorter than 17 bp, this method could likely be optimized to purify dsDNA <17 bp in length. Specifically, the acetonitrile concentrations used to elute the shorter DNA fragments will be <12% upper limit used for mid-sized (17–30 base pairs) used in this study. On the other hand, purification of DNA sequences longer than 51 bp will require an acetonitrile concentration >20%, which was the upper limit of the gradient used in this study. The DSIP method described here provides a robust tool for the production of highly pure short DNA sequences with minimal time and effort. We expect that research requiring large quantities of pure DNA will greatly benefit from this DSIP method.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
[NIH R56AI081872, R01CA46938 and AHA-11GRNT7380064 to M.E.A.C.]; [AHA-13POST16330009 to C.S.M]; The University of Colorado Cancer Center Support Grant [P30CA046934] partially supported the usage of core facilities. Funding for open access charge: National Institutes of Health [1 R01 AI098417].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Beaucage SL, Caruthers MH. Deoxynucleoside phosphoramidites-a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 2.Hayakawa Y, Wakabayashi S, Kato H, Noyori R. The allylic protection method in solid-phase oligonucleotide synthesis. An efficient preparation of solid-anchored DNA oligomers. J. Am. Chem. Soc. 1990;112:1691–1696. [Google Scholar]
- 3.Mooers BH. Crystallographic studies of DNA and RNA. Methods. 2009;47:168–176. doi: 10.1016/j.ymeth.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Brown T, Brown DJ. Purification of synthetic DNA. Methods Enzymol. 1992;211:20–35. doi: 10.1016/0076-6879(92)11004-3. [DOI] [PubMed] [Google Scholar]
- 5.Gilar M, Bouvier ES. Purification of crude DNA oligonucleotides by solid-phase extraction and reversed-phase high-performance liquid chromatography. J. Chromatogr. A. 2000;890:167–177. doi: 10.1016/s0021-9673(00)00521-5. [DOI] [PubMed] [Google Scholar]
- 6.Dingman CW, Peacock AC. Analytical studies on nuclear ribonucleic acid using polyacrylamide gel electrophoresis. Biochemistry (Mosc) 1968;7:659–668. doi: 10.1021/bi00842a022. [DOI] [PubMed] [Google Scholar]
- 7.Tabak HF, Flavell RA. A method for the recovery of DNA from agarose gels. Nucleic Acids Res. 1978;5:2321–2332. doi: 10.1093/nar/5.7.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith HO. Recovery of DNA from gels. Methods Enzymol. 1980;65:371–380. doi: 10.1016/s0076-6879(80)65048-4. [DOI] [PubMed] [Google Scholar]
- 9.Chen CW, Thomas CA., Jr Recovery of DNA segments from agarose gels. Anal. Biochem. 1980;101:339–341. doi: 10.1016/0003-2697(80)90197-9. [DOI] [PubMed] [Google Scholar]
- 10.Vogelstein B, Gillespie D. Preparative and analytical purification of DNA from agarose. Proc. Natl Acad. Sci. USA. 1979;76:615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy FV, Sweet RM, Churchill ME. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 1999;18:6610–6618. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurien BT, Scofield RH. Extraction of nucleic acid fragments from gels. Anal. Biochem. 2002;302:1–9. doi: 10.1006/abio.2001.5526. [DOI] [PubMed] [Google Scholar]
- 13.Ohmiya Y, Kondo Y, Kondo T. Separation of DNA fragments by high-resolution ion-exchange chromatography on a nonporous QA column. Anal. Biochem. 1990;189:126–130. doi: 10.1016/0003-2697(90)90057-g. [DOI] [PubMed] [Google Scholar]
- 14.Andrus A, Kuimelis RG. Analysis and purification of synthetic nucleic acids using HPLC. Curr. Protoc. Nucleic Acid Chem. 2001 doi: 10.1002/0471142700.nc1005s01. Chapter 10, Unit 10 15. [DOI] [PubMed] [Google Scholar]
- 15.Wong LY, Belonogoff V, Boyd VL, Hunkapiller NM, Casey PM, Liew SN, Lazaruk KD, Baumhueter S. General method for HPLC purification and sequencing of selected dsDNA gene fragments from complex PCRs generated during gene expression profiling. Biotechniques. 2000;28:776–783. doi: 10.2144/00284rr07. [DOI] [PubMed] [Google Scholar]
- 16.Shaw-Bruha CM, Lamb KA. Ion pair-reversed phase HPLC approach facilitates subcloning of PCR products and screening of recombinant colonies. Biotechniques. 2000;28:794–797. doi: 10.2144/00284pf02. [DOI] [PubMed] [Google Scholar]
- 17.Huber CG, Oefner PJ, Bonn GK. Rapid and accurate sizing of DNA fragments by ion-pair chromatography on alkylated nonporous poly(styrene-divinylbenzene) particles. Anal. Chem. 1995;67:578–585. [Google Scholar]
- 18.Huber CG, Oefner PJ, Preuss E, Bonn GK. High-resolution liquid chromatography of DNA fragments on non-porous poly(styrene-divinylbenzene) particles. Nucleic Acids Res. 1993;21:1061–1066. doi: 10.1093/nar/21.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecker KH, Green SM, Kobayashi K. Analysis and purification of nucleic acids by ion-pair reversed-phase high-performance liquid chromatography. J. Biochem. Biophys. Methods. 2000;46:83–93. doi: 10.1016/s0165-022x(00)00133-0. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy SM, Gilar M, Gebler J. Reversed-phase ion-pair liquid chromatography analysis and purification of small interfering RNA. Anal. Biochem. 2009;390:181–188. doi: 10.1016/j.ab.2009.03.042. [DOI] [PubMed] [Google Scholar]
- 21.Malarkey CS, Bestwick M, Kuhlwilm JE, Shadel GS, Churchill ME. Transcriptional activation by mitochondrial transcription factor A involves preferential distortion of promoter DNA. Nucleic Acids Res. 2012;40:614–624. doi: 10.1093/nar/gkr787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ngo HB, Kaiser JT, Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 2011;18:1290–1296. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahgholi M, Garcia BA, Chiu NH, Heaney PJ, Tang K. Sugar additives for MALDI matrices improve signal allowing the smallest nucleotide change (A:T) in a DNA sequence to be resolved. Nucleic Acids Res. 2001;29:E91. doi: 10.1093/nar/29.19.e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gangelhoff TA, Mungalachetty PS, Nix JC, Churchill ME. Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A. Nucleic Acids Res. 2009;37:3153–3164. doi: 10.1093/nar/gkp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004;32:e103. doi: 10.1093/nar/gnh101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poon K, Macgregor RB., Jr Unusual behavior exhibited by multistranded guanine-rich DNA complexes. Biopolymers. 1998;45:427–434. doi: 10.1002/(SICI)1097-0282(199805)45:6<427::AID-BIP2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 27.Wahl MC, Sundaralingam M. Crystal structures of A-DNA duplexes. Biopolymers. 1997;44:45–63. doi: 10.1002/(SICI)1097-0282(1997)44:1<45::AID-BIP4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 28.Gilar M, Fountain KJ, Budman Y, Neue UD, Yardley KR, Rainville PD, Russell RJ, II, Gebler JC. Ion-pair reversed-phase high-performance liquid chromatography analysis of oligonucleotides: retention prediction. J. Chromatogr. A. 2002;958:167–182. doi: 10.1016/s0021-9673(02)00306-0. [DOI] [PubMed] [Google Scholar]
- 29.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dragan AI, Klass J, Read C, Churchill ME, Crane-Robinson C, Privalov PL. DNA binding of a non-sequence-specific HMG-D protein is entropy driven with a substantial non-electrostatic contribution. J. Mol. Biol. 2003;331:795–813. doi: 10.1016/s0022-2836(03)00785-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.