Abstract
DNAzymes are known to bind metal ions specifically to carry out catalytic functions. Despite many studies since DNAzymes were discovered nearly two decades ago, the metal-binding sites in DNAzymes are not fully understood. Herein, we adopt uranyl photocleavage to probe specific uranyl-binding sites in the 39E DNAzyme with catalytically relevant concentrations of uranyl. The results indicate that uranyl binds between T23 and C25 in the bulge loop, G11 and T12 in the stem loop of the enzyme strand, as well as between T2.4 and G3 close to the cleavage site in the substrate strand. Control experiments using two 39E DNAzyme mutants revealed a different cleavage pattern of the mutated region. Another DNAzyme, the 8–17 DNAzyme, which has a similar secondary structure but shows no activity in the presence of uranyl, indicated a different uranyl-dependent photocleavage as well. In addition, a close correlation between the concentration-dependent photocleavage and enzymatic activities is also demonstrated. Together, these experiments suggest that uranyl photocleavage has been successfully used to probe catalytically relevant uranyl-binding sites in the 39E DNAzyme. As uranyl is the cofactor of the 39E DNAzyme as well as the probe, specific uranyl binding has now been identified without disruption of the structure.
INTRODUCTION
For many years, DNA was considered to be solely a carrier of genetic information. The discovery of short DNA molecules that possess catalytic activities, called deoxyribozymes or DNAzymes, changed the concept of DNA as a biomolecule (1). These DNAzymes are obtained by a technique known as in vitro selection, a combinatorial method for producing oligonucleotides that specifically catalyze certain reactions, ranging from phosphodiester transfer to porphyrin metallation (1–9). Not surprisingly, as polyanionic biopolymers, most of the DNAzymes selected to date require metal ions to function. An interesting property is the remarkable specificity shown by these DNAzymes for certain metal ions over other metal ions. This high specificity has been used by researchers in the field to convert these DNAzymes into highly selective biosensors for metal ions (10–13).
Even though high specificity for metal ions by DNAzymes has been established, and their application as metal sensors has been demonstrated, structural features responsible for such specificity remain to be understood, especially in comparison with those reported for metal interactions with nucleic acids (14–25) and ribozymes (26–32). Such an understanding of the role of metal ions and their specific interactions with the DNAzymes is important not only for advancing our fundamental knowledge of biocoordination chemistry, because DNAzymes are a new class of metalloenzymes next to metalloproteins and metallo-ribozymes, but also for making better sensors.
The DNAzyme that has been most studied so far is the 8–17 DNAzyme, which catalyzes the cleavage of a substrate strand containing a single ribonucleotide at the cleavage site in the presence of divalent metal ions, such as Mg2+, Ca2+ and Zn2+, showing the highest activity with Pb2+ (33,34). Extensive studies of the 8–17 DNAzyme have been carried out using various biochemical and biophysical methods (34–43), and a lock-and-key mechanism commonly observed in protein enzymes has been found to be responsible for the high selectivity for Pb2+. Another DNAzyme, called the 39E DNAzyme (Figure 1a), is even more selective, exhibiting over million-fold selectivity for uranyl over other metal ions (44,45), as compared with the ∼100-fold selectivity of Pb2+ over the next-best competing metal ion (Zn2+) for the 8–17 DNAzyme. Despite the demonstrated high selectivity and sensing applications (44,46–52), the source of the selectivity of the 39E DNAzyme for uranyl over other metal ions is unknown. To gain deeper insights into this selectivity, we have carried out a biochemical study to obtain conserved sequences responsible for the uranyl binding and enzymatic activity (45), and a fluorescence resonance energy transfer study to elucidate uranyl-dependent global folding of the 39E DNAzyme (53). Although these results contribute to our understanding of metal specificity, they provide information only on the global scale and do not provide detailed information about the metal-binding site. To overcome this limitation, we herein take advantage of uranyl-mediated photocleavage, commonly used as a general technique to probe the structure of nucleic acids and nucleic acid–protein complexes, to elucidate important uranyl-binding sites relevant to catalysis.
Figure 1.
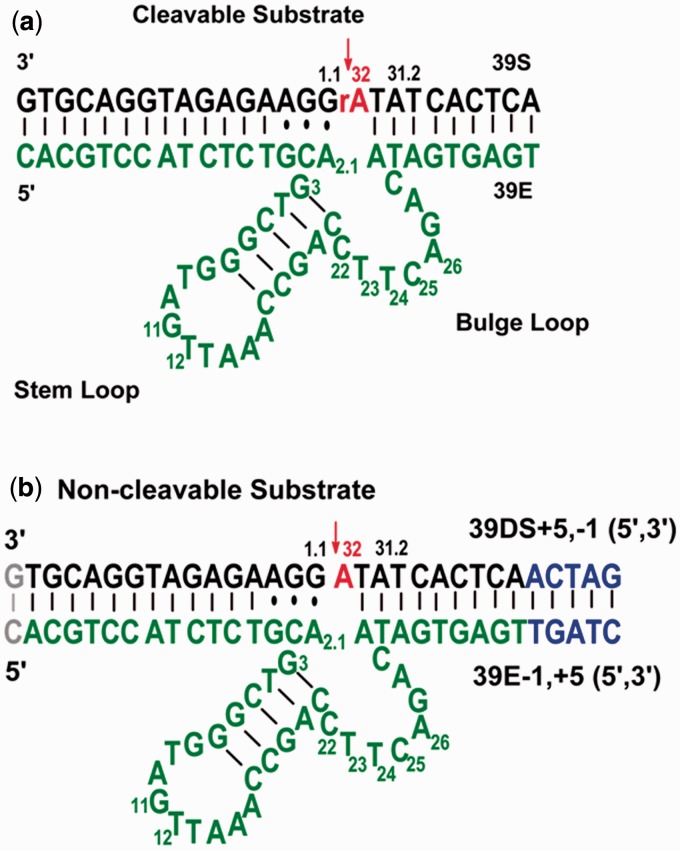
Predicted secondary structures of the trans-cleaving 39E DNAzyme used in this study. (a) The 39E DNAzyme consisting of a DNAzyme strand (in green) and a substrate strand (in black). The substrate strand contains a single riboadenosine at the cleavage site (in red). (b) A 39E DNAzyme variant used for the uranyl-mediated photocleavage experiments. The scissile riboadenosine was changed to deoxyriboadenosine to prevent DNAzyme-based cleavage. In addition, the arms of the DNAzyme-substrate at both ends are either extended (in blue) or deleted (in gray), making both arms more symmetrical to aid data collection and analysis. These changes do not affect the enzymatic activity.
Uranyl ions ( ) are known to induce single-strand nicks in DNA on light irradiation (300–420 nm) (54). The photochemically excited state of the uranyl ion is a strong oxidant, capable of oxidizing a variety of substrates such as alcohols and phosphates (55). Although the molecular mechanism of uranyl-mediated photocleavage is not fully understood, it has been proposed that uranyl ions can bind strongly to the phosphate backbone of DNA because of electrostatic interactions, and then act as an efficient cleavage reagent on irradiation by means of oxidation of the proximal deoxyriboses via an electron transfer mechanism (56). In previous studies, uranyl has been used as an analog of other divalent metals, such as Mg2+, as an efficient photochemical probe for identifying protein–DNA interactions, exploring potential metal-binding sites in folded nucleic acids, and studying DNA and RNA conformations (54,56–63). As the 39E DNAzyme is highly selective for uranyl, uranyl-mediated photocleavage becomes a particularly powerful technique to directly probe uranyl binding. Our results using relatively low and catalytically relevant concentrations of uranyl in the photocleavage experiments suggest uranyl binding close to the bulge loop, the 5′ substrate-binding region close to the cleavage site and also to the stem loop. The specificity of these interactions is supported through citrate and magnesium competition experiments, as well as through the use of DNAzyme variants with mutations at selective locations.
) are known to induce single-strand nicks in DNA on light irradiation (300–420 nm) (54). The photochemically excited state of the uranyl ion is a strong oxidant, capable of oxidizing a variety of substrates such as alcohols and phosphates (55). Although the molecular mechanism of uranyl-mediated photocleavage is not fully understood, it has been proposed that uranyl ions can bind strongly to the phosphate backbone of DNA because of electrostatic interactions, and then act as an efficient cleavage reagent on irradiation by means of oxidation of the proximal deoxyriboses via an electron transfer mechanism (56). In previous studies, uranyl has been used as an analog of other divalent metals, such as Mg2+, as an efficient photochemical probe for identifying protein–DNA interactions, exploring potential metal-binding sites in folded nucleic acids, and studying DNA and RNA conformations (54,56–63). As the 39E DNAzyme is highly selective for uranyl, uranyl-mediated photocleavage becomes a particularly powerful technique to directly probe uranyl binding. Our results using relatively low and catalytically relevant concentrations of uranyl in the photocleavage experiments suggest uranyl binding close to the bulge loop, the 5′ substrate-binding region close to the cleavage site and also to the stem loop. The specificity of these interactions is supported through citrate and magnesium competition experiments, as well as through the use of DNAzyme variants with mutations at selective locations.
MATERIALS AND METHODS
All oligonucleotides were obtained in desalted form from Integrated DNA Technologies, Inc. (Coralville, IA) and were purified by polyacrylamide gel electrophoresis (PAGE) for kinetic assays and uranyl photocleavage. Uranyl Nitrate hexahydrate was purchased from Fischer Scientific. All other metal salts used were obtained from Alfa-Aesar (Ward Hill, MA) and were of Puratronic® grade (99.999% pure). [γ-32P]-adenosine 5′-triphosphate (ATP) was purchased from Perkin Elmer (Waltham, MA), and T4 kinase was purchased from New England Biolabs, Inc. (Ipswich, MA). All other chemicals were of at least ACS reagent grade and were purchased from either Sigma-Aldrich (St. Louis, MO) or Thermo Fisher Scientific, Inc. (Waltham, MA).
Uranyl photocleavage
DNA samples for uranyl photocleavage studies were purified by running them through a 20% polyacrylamide denaturing gel [20% w/v (29:1, acrylamide:bisacrylamide), 8 M urea, 1× TBE (Tris, Boric acid and EDTA) buffer]. The DNAzyme and substrate bands were cut out of the gel, crushed and soaked by shaking for 2–3 h in a soaking buffer [10 mM Tris buffer (pH 7.5), 1 mM EDTA, 100 mM NaCl]. The DNA samples were then recovered from the supernatant after centrifugation. Finally, the DNA was purified through ethanol precipitation. The substrate strand used for these studies contained a deoxyadenosine at the cleavage site instead of riboadenosine to render the substrate to be non-cleavable, a common practice in the field to focus on metal-binding studies. Conditions used were modified from those of Møllegaard, and Nielsen (62). After PAGE purification, the non-cleavable substrate (39DS) or enzyme oligonucleotide [e.g. 39E (−1,+5)] was labeled at the 5′-end with [γ-32P]-ATP using T4 kinase and purified by PAGE. The radiolabeled DNA strand was extracted from the gel as described previously, followed by ethanol precipitation, and subsequent storage at −20°C until use. The sequences were designed such that cytosine was not present at the 5' end, as it is known to lead to lower labeling efficiency (64). In addition, the original sequence of the 39E DNAzyme (44) was lengthened by five bases at the 3′ end as compared with the construct used for activity assays. This mutation was made to aid in gel analysis by giving a higher resolution of the catalytic core on the gel. The new construct, including modifications at both ends of the binding arms, was named 39E (−1,+5) to denote the deletion of one base and extension by five bases at the 5′ and 3′ ends, respectively. Previous studies of DNAzymes have shown that such modifications to the enzyme-substrate hybridization region away from the cleavage site do not have any effect on catalytic activity, as long as the base pairing is maintained (33).
The photocleavage reactions were performed by preparing a 90 µl of solution containing 5 nM 5′-radiolabeled strand and 50 nM complementary strand (experiments were performed with either enzyme or substrate labeled in turn) in 50 mM MES (pH 5.5) and 250 mM NaNO3, which is the same condition under which in vitro selection, biochemical studies and sensing applications of the 39E were carried out (33,34). (All concentrations were calculated based on a final reaction volume of 100 µl). The sample solution containing the enzyme and substrate in buffer was denatured at 95°C in a water bath and then annealed by gradual cooling to room temperature over ∼45 min. Uranyl nitrate was added to a final concentration ranging between 2.5 nM and 1 mM to result in a solution with a final volume of 100 µl. For competition experiments, either citrate (1 µM–1 mM) or Mg2+ (1 mM–100 mM) was added. Addition of citrate results in a competition for uranyl between the DNA and citrate, whereas addition of Mg2+ results in competition between the cations for binding sites on DNA. When Mg2+ was added, it was allowed to incubate with the annealed sample for a minimum of 10 min. When citrate was added, it was mixed with uranyl for a minimum of 10 min before being added to the reaction mixture. Samples were placed in a 96-well Matrix microplate (Thermo Fisher Scientific) and an Oriel long-wave Hg pen lamp was stationed ∼1 inch above the surface of the microplate. A maximum of 12 samples were run simultaneously to ensure even exposure to the pen lamp; exposure time was held at 20 min for all samples. Following exposure, samples were quenched by 20 µl of precipitation buffer containing 0.5 M sodium acetate (pH 4.5), followed by ethanol precipitation and a single wash with 70% ethanol. Afterwards, the samples were evaporated to dryness on a Speedvac (Concentrator Plus from Eppendorf, Hamburg, Germany) and stored at −20°C until analysis. A guanine-adenine (GA) ladder was prepared according to the method reported by Hampshire et al. (65). In preparation for gel analysis, samples were dissolved in 5 µl of 7:3 loading buffer:ultrapure water (loading buffer: 7 M urea, 40% v/v formamide, 12% w/v Ficoll, 30 mM EDTA, 100 mM Bis-tris and 0.05% w/v each bromophenol blue and xylene cyanol) (66), heated to >90°C for 2 min, plunged immediately into ice and separated on a 20% (or lower percentage, depending on sequence length) 29:1 acrylamide:bis-acrylamide gel. After this procedure, the gels were dried, exposed to a cassette, imaged on a Molecular Dynamics Storm 430 phosphorimager and quantified using Semi-Automated Footprinting Analysis (SAFA) (67). Each experiment was repeated at least twice. All data found are reproducible. Different control samples were probed to confirm that any cleavage observed was generated only by photocleavage. Neither self-cleavage, thermal damage caused by uranyl (1 mM uranyl, no light), nor light source damage (no uranyl) were found.
RESULTS
Uranyl photocleavage of the 39E DNAzyme and 39S substrate complex
Uranyl-mediated photocleavage when the 39E DNAzyme strand is labeled
To locate the site of uranyl binding to the enzyme strand, the uranyl photocleavage reactions were first conducted with the 39E DNAzyme strand labeled by 32P at the 5′ end in the enzyme/substrate complex (see Figure 1b). The addition of the unlabeled substrate strand allows formation of the enzyme/substrate complex, even though the substrate strand is not detectable in gels because it is not radioactively labeled.
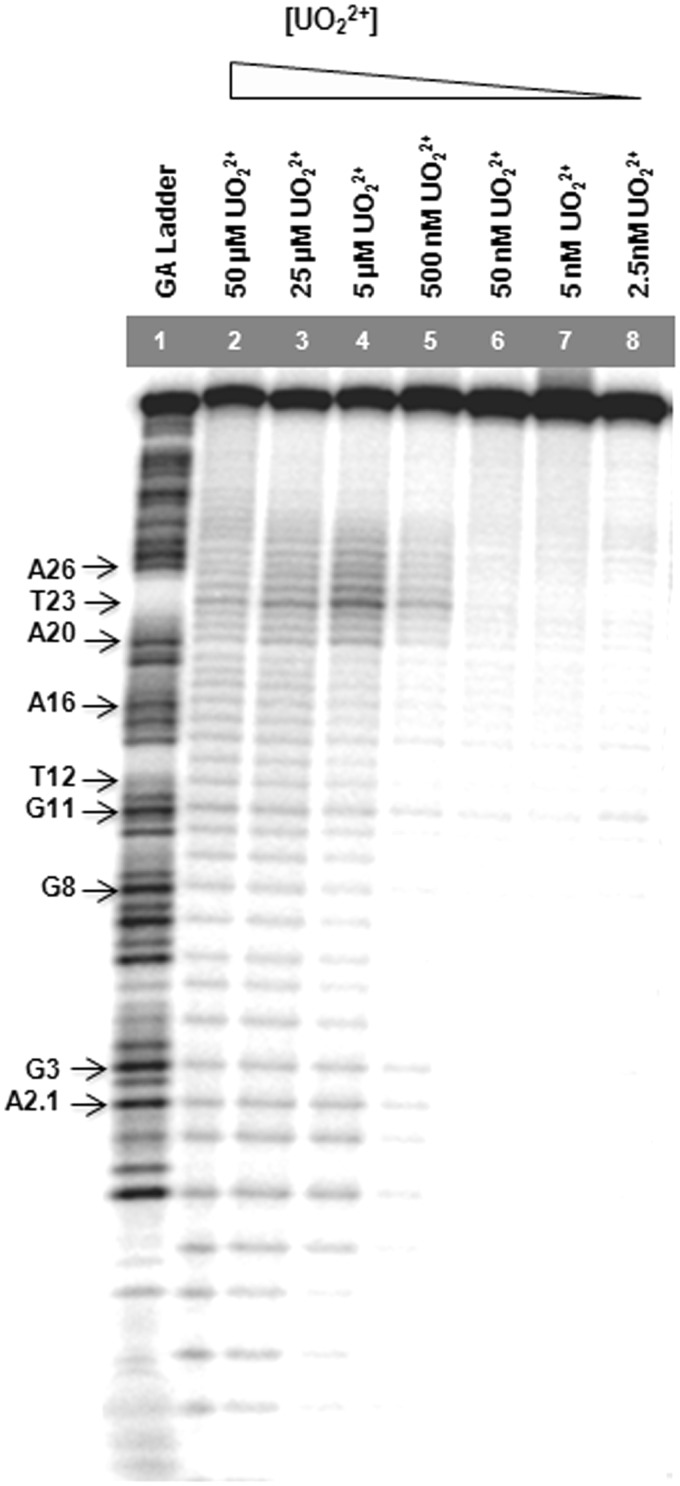
Previous uranyl photocleavage studies used uranyl concentrations from 50 µM to 1 mM to see a protection effect indicative of protein binding (54,56–57,60,62). These concentrations are significantly higher than what is required for enzymatic activity of the 39E DNAzyme, as it is known to have a high affinity for uranyl with an apparent dissociation constant of 496 nM (45). Therefore, to find the optimal concentrations of uranyl for this study that are relevant to the enzymatic activities, a range of uranyl concentrations from 2.5 nM to 1 mM was tested. At high concentrations of uranyl (500 µM and 1 mM), an even cleavage pattern was obtained across all positions, indicating non-specific uranyl-mediated photocleavage (see Supplementary Figure S1). This result is not surprising, as previous enzymatic assays also showed a largely inactive 39E DNAzyme in the presence of 1 mM uranyl (45). The photocleavage reactions were therefore carried out at lower concentrations (between 2.5 nM and 50 µM), and the data obtained using PAGE are shown in Figure 2. In contrast to the uniform cleavage pattern at high uranyl concentration (e.g. 50 µM), only certain nucleotides are selectively cleaved at low uranyl concentrations. The nucleotides denoted in bold were identified through the use of a GA ladder.
Figure 2.
Gel images of uranyl photocleavage reactions of 39E (−1, +5) DNAzyme with the enzyme strand labeled and in the presence of varying concentrations of uranyl.
To investigate the effects of either a chelator for uranyl (citrate) or a competing metal ion (Mg2+) on the cleavage pattern, the same photocleavage reactions were repeated in the presence of increasing concentrations of citrate or Mg2+, and the data are shown in Supplementary Figure S2a and b, respectively. The presence of either competitor is able to reduce the overall cleavage, demonstrating the high affinity of uranyl by the 39E DNAzyme. The gel image shows a distinctive cleavage pattern. An intense cleavage was observed in the bulge loop region, and in the 5′ substrate-binding region close to the cleavage site. A less intense degree of cleavage was observed at the stem loop region.
The photocleavage pattern was analyzed, and the band intensities were quantified using the SAFA program (67). The photocleavage intensities obtained after quantification with SAFA were first normalized to a specific band located at the 5′ arm, where no specific cleavage pattern was observed. This normalization makes every line comparable with each other, regardless of possible artifacts such as differences in intensity, owing to disparity in the amount of radiolabeled DNA strand loaded in the gel after the entire experimental procedure is complete for all concentrations of uranyl under study (2.5 nM–1 mM). Next, the mean values of the normalized intensities were calculated from the three individual uranyl-mediated photocleavage experiments for the 39E (−1,+5) DNAzyme, which showed high reproducibility in terms of specific cleavage pattern. Data reported are mean values of the results.
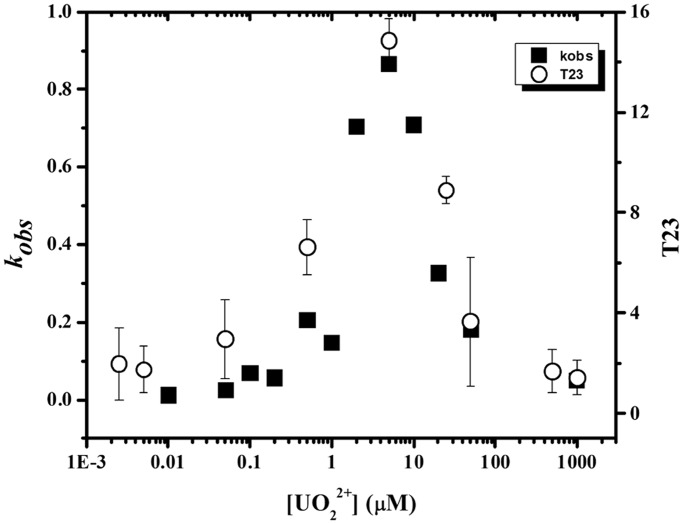
After quantification with SAFA (67), we proceeded to evaluate the correlation between the photocleavage intensity and the catalytic enzymatic activity of 39E. Previously, kinetics studies under single-turnover conditions demonstrated that 39E shows a bell-shaped curve depending on the concentration of uranyl, with the highest activity found at ∼5 µM uranyl. Remarkably, the photocleavage intensities showed the same trend. Figure 3 shows these results for the uranyl-mediated photocleavage at T23, located at the bulge loop of the 39E DNAzyme, which demonstrates the highest photocleavage intensity. This bell-shape curve matches well with the bell-shaped curve for the enzymatic activity of the same enzyme reported previously, as both data sets follow the same pattern, with a maximum uranyl photocleavage and enzymatic activity peak at 5 µM uranyl (45). These results strongly suggest that the uranyl-mediated photocleavage study is an excellent method to probe the uranyl binding to the DNAzyme that is highly relevant to the enzymatic activities.
Figure 3.
Comparison between the normalized uranyl-dependent photocleavage intensity at T23 and the enzymatic activity of 39E DNAzyme. The intensity values correspond to the mean value calculated from three individual gels.
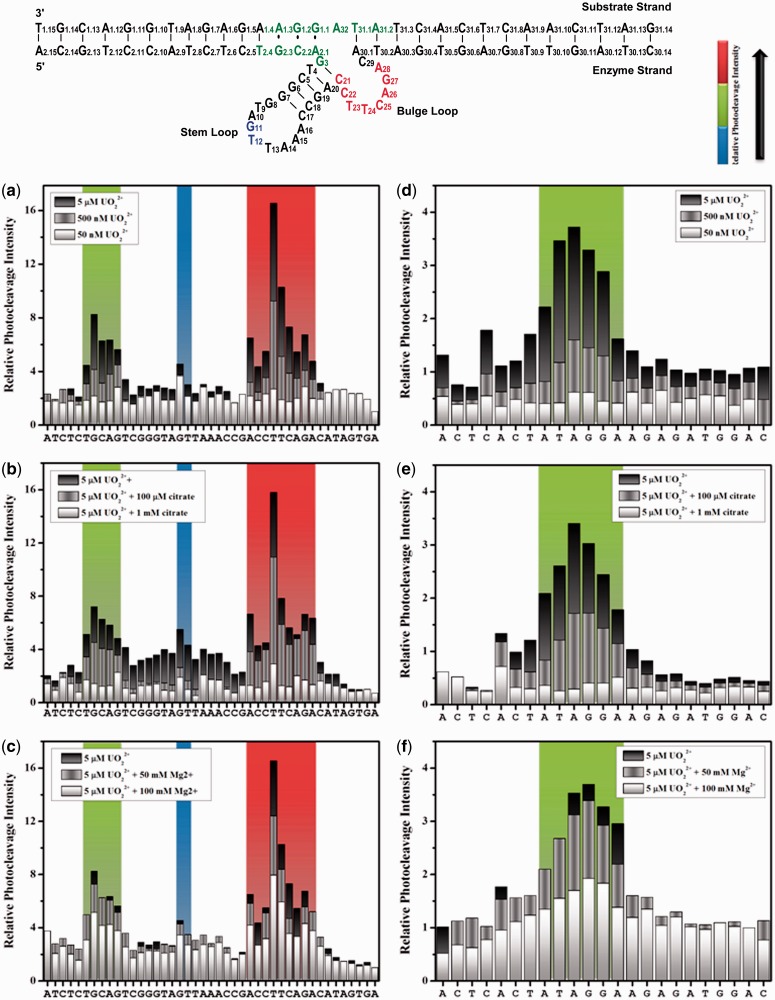
Encouraged by the strong correlation between the uranyl-dependent photocleavage and enzymatic activity, we used the SAFA program to analyze the pattern of the cleavage to locate sites of uranyl binding. Figure 4a shows relative photocleavage intensities for each nucleotide in the enzyme strand at 50 nM, 500 nM and 5 µM uranyl, as this is the concentration range relevant to the enzymatic activity (see Figure 3). A high degree of cleavage was observed in the bulge loop region (C21, C22, T23, T24, C25, A26 and G27) and in the 5′ substrate-binding region close to the cleavage site (T2.4, G2.3, C2.2, A2.1 and G3), suggesting specific uranyl binding at these locations. A much less intense degree of cleavage was observed at the stem loop region (G11 and T12).
Figure 4.
Uranyl photocleavage of 39E (−1, +5) DNAzyme with either the enzyme strand (a–c) or the substrate labeled (d–f). Plots display strand sequences from 5′ end to 3′ end. Plots (a–c) show the 39E (−1,+5) enzyme strand sequence from A2.9 to A30.7. Plots (d–f) show the 39DS (+5,−1) substrate strand sequence from A31.9 to C1.13. The gel images were quantified by SAFA, with the horizontal axis indicating the bands from 5′ to 3′ end according to Figure 1b. Colors represent the intensity of photocleavage, red (high), green (intermediate) and blue (low) according to regions in the secondary structure of the 39E/S complex. (a) Uranyl-mediated photocleavage of 39E DNAzyme with the enzyme strand labeled and in the presence of 5 µM (Black), 500 nM (gray) and 50 nM (white) of uranyl. (b) Uranyl-mediated photocleavage of 39E DNAzyme with the enzyme strand labeled and in the presence of 5 µM uranyl in addition to increasing concentrations of sodium citrate. 5 µM uranyl (black), 5 µM uranyl in addition to 100 µM sodium citrate (gray) and 5 µM uranyl in addition to 1 mM sodium citrate (white). (c) Uranyl-mediated photocleavage of 39E DNAzyme with the enzyme strand labeled and in the presence of 5 µM uranyl (black) in addition to magnesium chloride, 1 mM Mg2+ (gray) and 10 mM (white). (d) Uranyl-mediated photocleavage of 39E DNAzyme with 39DS substrate strand labeled and in the presence of 5 µM (Black), 500 nM (gray) and 50 nM (white) uranyl. (e) Uranyl-mediated photocleavage of 39E DNAzyme with the 39DS substrate strand labeled and in the presence of 5 µM uranyl in addition to an increasing concentration of sodium citrate, 100 µM citrate (gray) and 1.0 mM citrate (white). (f) Uranyl-mediated photocleavage of the 39E DNAzyme with 39DS substrate strand labeled and in the presence of 5 µM uranyl in addition to magnesium chloride, 1 mM Mg2+ (gray) and 10 mM (black).
To support the aforementioned findings of a uranyl binding site, the same photocleavage reactions were repeated in the presence of increasing concentrations of citrate (Supplementary Figure S2a). A few other studies have used several citrate concentrations (59,61). We carried out titrations with a wide range of concentrations (1 µM–1 mM) of citrate as competitor of the DNAzyme in the presence of 5 µM uranyl. As the citrate concentration increases relative to the uranyl concentration, only the high affinity sites in the DNAzyme are capable of competing to bind any remaining uranyl. At 5 µM uranyl, the addition of a low concentration of citrate (1–5 µM) results in minimal perturbations of the relative photocleavage intensity (Supplementary Figure S2a, lanes 2–6). Even though concentrations of citrate higher than 10 µM lead to an overall decrease in the relative cleavage intensity, the specific pattern remains observable and is analogous to the cleavage pattern seen with lower uranyl concentrations (Supplementary Figure S2a, lanes 7–10). Furthermore, the relative cleavage intensities between the specific uranyl binding sites and those positions that uranyl does not bind specifically increase with increasing concentrations of citrate. For example, Supplementary Figure S3 shows the changes in the ratio of the relative intensities of T23 located at the bulge loop and C18 located in the stem on addition of citrate. The ratio increases with increasing concentrations of citrate until ∼50 µM, indicating that the citrate preferentially reduced uranyl binding and thus cleavage at lower affinity binding sites such as C18. Once the concentration of citrate is higher than 50 µM, the ratio between these intensities starts decreasing, attributable to citrate being able to out-compete the DNAzyme for uranyl binding at higher concentrations. Ten equivalents of citrate and above minimize the overall photocleavage by shifting the equilibrium in favor of citrate binding to uranyl, leaving much less free uranyl available to bind to the DNAzyme. The DNAzyme may still have access to uranyl even at higher citrate concentrations, although the amount of uranyl bound may be insufficient to show cleavage due to the low yield of the uranyl-mediated photocleavage at low concentrations of uranyl (56). Figure 4b shows relative photocleavage intensities for each nucleotide in the enzyme strand at 5 µM uranyl in the presence of 100 µM and 1 mM citrate.
A similar competition experiment was performed in the presence of Mg2+. After the enzyme and substrate strands were annealed, Mg2+ was added and allowed to incubate with the DNAzyme for 10 min to ensure full association of Mg2+ with the DNAzyme. Uranyl was then added, followed by irradiation. In the presence of 5 µM uranyl, addition of 1–10 mM Mg2+ did not perturb the specific binding of uranyl to the DNAzyme (Supplementary Figure S2b, lanes 1–5). When the Mg2+ concentration was increased from 50 to 100 mM, the high susceptibility of these sites towards photocleavage was reduced, suggesting direct competition between the two cations for binding to the DNAzyme (Figure 4c). Addition of either citrate or Mg2+ results in a trend similar to that of low concentrations of uranyl alone, confirming specific interactions with uranyl at the bulge loop, the 5′ binding region adjacent to the cleavage site and the stem loop. These results demonstrate the high affinity of uranyl for the 39E DNAzyme because uranyl is able to bind and cleave the enzyme in the presence of 200 times more citrate or 20 000 fold excess of Mg2+.
Uranyl-mediated photocleavage when the 39DS substrate is labeled
We further investigated the uranyl binding sites of the substrate strand. Similar to experiments performed with the enzyme strand labeled with 32P, the photocleavage experiments were performed with the substrate strand labeled at the 5′ end, in the presence of uranyl alone (Supplementary Figure S4), and in the presence of citrate or Mg2+ (Supplementary Figure S5). A strong interaction between uranyl and 39DS was observed between A31.2 and G1.2, with the highest intensity at A32, which is the cleavage site. As the adenosine ribonucleotide in the middle of the substrate strand was replaced with an adenosine deoxyribonucleotide (called 39DS) to render it non-cleavable in the presence of uranyl and in the absence of light, any cleavage observed in the photocleavage experiments here must have been due to binding of uranyl and subsequent photocleavage rather than catalytic activity. Analogous quantification of the cleavage intensities was performed using SAFA (67). Figure 4d–f shows the normalized photocleavage intensities in the presence of uranyl alone and in the presence of citrate or Mg2+. The results from addition of either citrate (Figure 4e) or Mg2+ (Figure 4f) show a similar trend.
Uranyl-mediated photocleavage of 39E mutants
A previous mutational study carried out by Brown et al. (45) revealed that the nucleotides in the bulge region of the 39E DNAzyme (between C22 and G27) are highly conserved. In this study, a 39E variant called Mutant 14, in which the CTTC sequence was mutated to a TCCT sequence, showed no activity after the mutation. Motivated by this information, we carried out the uranyl-mediated photocleavage of two 39E variants with mutations at the bulge region for the purpose of analyzing the sequence requirement for uranyl binding in this single-stranded region. Based on the aforementioned report (45), we chose Mutant 14 and designed a second variant we called Mutant 14+T, where A26 was mutated into thymine (Figure 1). We performed parallel uranyl-mediated photocleavage of the 39E DNAzyme along with Mutant 14 and Mutant 14+T in the presence of 5 µM uranyl only and 5 µM uranyl with an increasing concentration of citrate (5 µM, 100 µM and 1 mM) under the same condition as in Figure 4. The gel images of the uranyl photocleavage for the original 39E DNAzyme and the variants under study, shown in Supplementary Figure S6a, demonstrate that the two mutants display a different cleavage pattern in comparison with the 39E DNAzyme and do so at a lower binding affinity. To obtain a detailed comparison, we quantified the gels using the SAFA program. As shown in Supplementary Figure S6b, the results obtained from the quantification confirm the observation that the distinctive cleavage pattern shown by the 39E DNAzyme is changed when mutations are introduced in the region of highest affinity for uranyl. Similar to what was observed for 39E (−1,+5) in Figure 4 and Supplementary Figure S2, when an excess of citrate was added to the mutants, the overall relative photocleavage intensity was decreased.
To improve base resolution around the mutated bulge region, we repeated the experiments by splitting the samples in half and then loading them in different lanes on the same gel at different time intervals. As shown in Supplementary Figure S7, the well-resolved bands in the bulge region clearly indicate the differences in cleavage patterns between the 39E DNAzyme and its mutants in the bulge region. Not only are different patterns present but the relative photocleavage intensity in the bulge loop is also lower for the mutants. Together, these results strongly support the conclusion that the specific pattern observed for the photocleavage of the 39E DNAzyme can be directly associated with specific uranyl binding to this DNAzyme.
Uranyl-mediated photocleavage of the 39DS Duplex and the 8–17 DNAzyme
To further confirm that the results are relevant to uranyl-specific activity of the 39E DNAzyme, we repeated the experiment using a duplex containing the same sequence of the enzyme substrate 39DS and its complementary strand (39DS duplex) and the 8–17 DNAzyme under the same condition. As the 39DS duplex is purely double stranded, it is used to check for potential bias in terms of sequence. As the 8–17 DNAzyme forms a tertiary structure with single-stranded regions, any preference of uranyl for single-stranded regions should become evident in this control. The cleavage patterns for these two constructs are shown in Supplementary Figure S8. Photocleavage of the 39DS Duplex showed negligible cleavage with 5 µM uranyl, in contrast to 39E (−1,+5) DNAzyme, which demonstrated maximum cleavage intensity at this concentration. When photocleavage was carried out with 500 µM and 1 mM uranyl, the intensity of the cleavage improved, although no evidence of specific binding was observed.
On the other hand, photocleavage of the 8–17 DNAzyme showed a different cleavage pattern with uranyl between 500 nM and 1 mM in comparison with the 39E DNAzyme. Although the secondary structures of both the 39E and 8–17 DNAzymes are similar, the cleavage patterns are markedly different (Supplementary Figure S9). The 8–17 DNAzyme exhibits strong cleavage at T2.1 located in the region close to the cleavage site (Supplementary Figure S8a) with a generally lower signal to background ratio. Contrary to what we observed for the 39E DNAzyme, the 8–17 DNAzyme did not show a strong cleavage at the bulge region between T12 and A15, despite the similarities between their secondary structures. These results demonstrate that uranyl cleavage does not occur unselectively in single-stranded regions of the secondary structure of the 39E DNAzyme. These results support the conclusion that the specific pattern observed for the photocleavage of the 39E DNAzyme can be directly associated with specific uranyl binding to this DNAzyme.
DISCUSSION
The uranyl-mediated photocleavage studies on the 39E DNAzyme (Figure 4a–c) indicate a significant photocleavage within the bulge loop of the 39E enzyme strand (in red), with the highest cleavage efficiency between C22 and A28, suggesting a strong binding of uranyl to this bulge loop of the 39E DNAzyme. Previous mutational studies have shown the bulge region of 39E to be highly conserved (45). When mutations were carried out on the pyrimidine-rich loop containing C22, T23, T24 and C25, the activity was highly perturbed. Mutations of the two central thymines into two cytosines resulted in a less active DNAzyme, whereas the simultaneous replacement of C22 and C25 by two thymines produced a completely inactive mutant. The same study also showed that deletion of any of these bases resulted in a complete loss of activity. Moreover, the photocleavage results observed for the 39E variants confirm the sequence-dependent binding of uranyl to the catalytic core of this DNAzyme. Therefore, the uranyl-mediated photocleavage studies presented here not only confirm the importance of the conserved loop in enzymatic activities based on the mutational studies but also pinpoint this loop as a site for uranyl binding.
The uranyl photocleavage experiments also reveal strong interaction with uranyl in the substrate-binding region close to the cleavage site on the 39E DNAzyme, specifically between the bases T2.4 and G3 (Figure 4a–c). This finding suggests that these bases may be involved in uranyl binding. Previous mutagenesis studies have established that A2.1 is a highly conserved base; mutations of this base to thymine and cytosine render the enzyme completely inactive (45). This A2.1 base forms a wobble base pair with the G1.1 nucleotide on the substrate strand. The importance of non-canonical base pairs in DNAzyme-substrate complexes has been noted previously by several groups (34,40,45). Interestingly, the relevance of the presence of wobble pairs adjacent to the cleavage site has also been shown for the 8–17 DNAzyme. This DNAzyme has a G·T wobble base pair, which is crucial for the enzyme function. These kinds of wobble pairs may allow flexibility to help achieve the conformation for correct binding of the metal ions to perform catalysis (68–70).
A less intense, though reproducible, interaction between uranyl and the stem loop of the 39E DNAzyme, specifically at G11 and T12, was observed. In the original selection of the 39E DNAzyme, 49 clones could be aligned against the secondary structure of the 39E DNAzyme (44). In all these sequences, the stem loop region was highly conserved. However, variations of sequence length in this region could be allowed without compromising the DNAzyme activity; a complete deletion of the stem loop resulted in little inhibition of the DNAzyme. We found that both G11 and T12 in this stem loop region may be involved in uranyl binding, likely as a result of being in proximity to the active site, as these bases are unlikely to bind uranyl independently.
Previous structural studies of this highly uranyl-specific 39E DNAzyme focused mostly on metal ion-dependent global folding or conformation changes (53). In this work, we use uranyl photocleavage to zoom in on the uranyl-binding site in this DNAzyme. We took advantage of a general footprinting method for mapping nucleic acids and used it to find specific uranyl-binding sites in this DNAzyme by using uranyl concentrations (2.5 nM–1 μM) that are much lower than those used in previous uranyl photocleavage studies (50 μM–1 mM) and more relevant to the DNAzyme activity. This method has produced results with higher resolution than previous folding studies. By adopting a uranyl-mediated photocleavage method, we identified three regions in the 39E DNAzyme involved in binding the uranyl ion: the bulge loop between T23, C25 and A28, the 5′ binding region close to the cleavage site between T2.4 and C3, and the stem loop at G11 and T12 (see Figure 5). It is also shown that uranyl binds specifically at the cleavage site of the substrate strand. The results presented here provide important information about the positions that are crucial for uranyl binding in the 39E DNAzyme.
Figure 5.
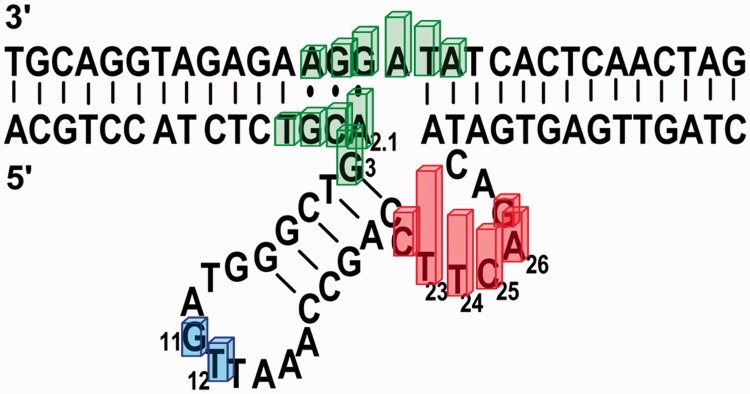
Schematic representation of the 39E DNAzyme (original sequence), summarizing the results from the uranyl photocleavage studies. Bars indicate bases that showed specific binding of uranyl in the photocleavage studies, color code is based on Figure 4.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The U.S. National Institutes of Health [ES016865] and the U.S. Department of Energy [DE-FG02-08ER64568]. Funding for open access charge: U.S. National Institutes of Health [ES016865]; U.S. Department of Energy [DE-FG02–08ER64568].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
M.C.-P. thanks CONICYT Chile for her postdoctoral scholarship.
REFERENCES
- 1.Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 2.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–498. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 3.Breaker RR. DNA enzymes. Nat. Biotechnol. 1997;15:427–431. doi: 10.1038/nbt0597-427. [DOI] [PubMed] [Google Scholar]
- 4.Sen D, Geyer CR. DNA enzymes. Curr. Opin. Chem. Biol. 1998;2:680–687. doi: 10.1016/s1367-5931(98)80103-8. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Breaker RR. Deoxyribozymes: new players in the ancient game of biocatalysis. Curr. Opin. Struct. Biol. 1999;9:315–323. doi: 10.1016/S0959-440X(99)80042-6. [DOI] [PubMed] [Google Scholar]
- 6.Emilsson GM, Breaker RR. Deoxyribozymes: new activities and new applications. Cell. Mol. Life Sci. 2002;59:596–607. doi: 10.1007/s00018-002-8452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Y. New transition-metal-dependent DNAzymes as efficient endonucleases and as selective metal biosensors. Chem. Eur. J. 2002;8:4588–4596. doi: 10.1002/1521-3765(20021018)8:20<4588::AID-CHEM4588>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 8.Peracchi A. DNA catalysis: potential, limitations, open questions. ChemBioChem. 2005;6:1316–1322. doi: 10.1002/cbic.200500098. [DOI] [PubMed] [Google Scholar]
- 9.Schlosser K, Li Y. Biologically inspired synthetic enzymes made from DNA. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Lu Y. A highly sensitive and selective catalytic DNA biosensor for lead ions. J. Am. Chem. Soc. 2000;122:10466–10467. [Google Scholar]
- 11.Navani NK, Li Y. Nucleic acid aptamers and enzymes as sensors. Curr. Opin. Chem. Biol. 2006;10:272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Cao Z, Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X-B, Kong R-M, Lu Y. Metal ion sensors based on DNAzymes and related DNA molecules. Annu. Rev. Anal. Chem. 2011;4:105–128. doi: 10.1146/annurev.anchem.111808.073617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barton J, Lippard S. In: Metal Ions in Biology Series. Spiro TG, editor. Vol. 1. New Jersey: John Wiley & Sons; 1980. pp. 31–113. [Google Scholar]
- 15.Sigel H. Interactions of metal ions with nucleotides and nucleic acids and their constituents. Chem. Soc. Rev. 1993;22:255–267. [Google Scholar]
- 16.Feig AL, Uhlenbeck OC. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Vol. 37. New York, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 287–319. [Google Scholar]
- 17.Guo Z, Sadler PJ. Metals in medicine. Angew. Chem., Int. Ed. 1999;38:1512–1531. doi: 10.1002/(SICI)1521-3773(19990601)38:11<1512::AID-ANIE1512>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 18.Boerner LJK, Zaleski JM. Metal complex–DNA interactions: from transcription inhibition to photoactivated cleavage. Curr. Opin. Chem. Biol. 2005;9:135–144. doi: 10.1016/j.cbpa.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Müller J, Lippert B. Imposing a three-way junction on DNA or recognizing one: a metal triple helicate meets double helix. Angew. Chem., Int. Ed. 2006;45:2503–2505. doi: 10.1002/anie.200600031. [DOI] [PubMed] [Google Scholar]
- 20.Lippert B. In: RSC Biomolecular Sciences. Hud NV, editor. Cambridge: Royal Soc Chemistry; 2009. pp. 39–74. [Google Scholar]
- 21.Sigel RKO, Sigel H. A stability concept for metal ion coordination to single-stranded nucleic acids and affinities of individual sites. Acc. Chem. Res. 2010;43:974–984. doi: 10.1021/ar900197y. [DOI] [PubMed] [Google Scholar]
- 22.Dong X, Wang X, Lin M, Sun H, Yang X, Guo Z. Promotive effect of the platinum moiety on the DNA cleavage activity of copper-based artificial nucleases. Inorg. Chem. 2010;49:2541–2549. doi: 10.1021/ic100001x. [DOI] [PubMed] [Google Scholar]
- 23.Dhar S, Lippard SJ. In: Bioinorganic Medicinal Chemistry. Alessio E, editor. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2011. pp. 73–95. [Google Scholar]
- 24.Sigel A, Sigel H, Sigel RKO. Structural and Catalytic Roles of Metal Ions in RNA. Cambridge: Royal Soc Chemistry; 2011. [PubMed] [Google Scholar]
- 25.Sontz PA, Muren NB, Barton JK. DNA charge transport for sensing and signaling. Acc. Chem. Res. 2012;45:1792–1800. doi: 10.1021/ar3001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeRose VJ. Metal ion binding to catalytic RNA molecules. Curr. Opin. Struct. Biol. 2003;13:317–324. doi: 10.1016/s0959-440x(03)00077-0. [DOI] [PubMed] [Google Scholar]
- 27.Sigel RKO, Pyle AM. Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem. Rev. 2007;107:97–113. doi: 10.1021/cr0502605. [DOI] [PubMed] [Google Scholar]
- 28.Freisinger E, Sigel RKO. From nucleotides to ribozymes—a comparison of their metal ion binding properties. Coord. Chem. Rev. 2007;251:1834–1851. [Google Scholar]
- 29.Shepotitiovskaya IV, Uhlenbeck OC. Catalytic diversity of extended hammerhead ribozymes. Biochemistry. 2008;47:7034–7042. doi: 10.1021/bi7025358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakano S, Kitagawa Y, Karimata HT, Sugimoto N. Nucleic Acids Symposium Series. Vol. 52. Oxford: Oxford University Press; 2008. pp. 519–520. [DOI] [PubMed] [Google Scholar]
- 31.Hunsicker-Wang L, Vogt M, DeRose VJ. In: Methods Enzymol. Herschlag D, editor. Vol. 468. Walthman, Germany: Academic Press; 2009. pp. 335–367. [DOI] [PubMed] [Google Scholar]
- 32.Nakano S, Karimata HT, Kitagawa Y, Sugimoto N. Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes. J. Am. Chem. Soc. 2009;131:16881–16888. doi: 10.1021/ja9066628. [DOI] [PubMed] [Google Scholar]
- 33.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown AK, Li J, Pavot CMB, Lu Y. A lead-dependent DNAzyme with a two-step mechanism. Biochemistry. 2003;42:7152–7161. doi: 10.1021/bi027332w. [DOI] [PubMed] [Google Scholar]
- 35.Bonaccio M, Credali A, Peracchi A. Kinetic and thermodynamic characterization of the RNA-cleaving 8–17 deoxyribozyme. Nucleic Acids Res. 2004;32:916–925. doi: 10.1093/nar/gkh250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruz RPG, Withers JB, Li Y. Dinucleotide junction cleavage versatility of 8–17 deoxyribozyme. Chem. Biol. 2004;11:57–67. doi: 10.1016/j.chembiol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Peracchi A, Bonaccio M, Clerici M. A mutational analysis of the 8–17 deoxyribozyme core. J. Mol. Biol. 2005;352:783–794. doi: 10.1016/j.jmb.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 38.Schlosser K, Gu J, Sule L, Li Y. Sequence-function relationships provide new insight into the cleavage site selectivity of the 8–17 RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2008;36:1472–1481. doi: 10.1093/nar/gkm1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HK, Li J, Nagraj N, Lu Y. Probing Metal Binding in the 8–17 DNAzyme by Tb(III) Luminescence Spectroscopy. Chem. Eur. J. 2008;232:8696–8703. doi: 10.1002/chem.200701789. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Sen D. A contact photo-cross-linking investigation of the active site of the 8–17 deoxyribozyme. J. Mol. Biol. 2008;381:845–859. doi: 10.1016/j.jmb.2008.06.036. [DOI] [PubMed] [Google Scholar]
- 41.Mazumdar D, Nagraj N, Kim H-K, Meng X, Brown AK, Sun Q, Li W, Lu Y. Activity, folding and Z-DNA formation of the 8–17 DNAzyme in the presence of monovalent Ions. J. Am. Chem. Soc. 2009;131:5506–5515. doi: 10.1021/ja8082939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Sen D. Local rather than global folding enables the lead-dependent activity of the 8–17 deoxyribozyme: evidence from contact photo-crosslinking. J. Mol. Biol. 2010;395:234–241. doi: 10.1016/j.jmb.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 43.Sekhon GS, Sen D. A stereochemical glimpse of the active site of the 8–17 deoxyribozyme from iodine-mediated cross-links formed with the substrate's scissile site. Biochemistry. 2010;49:9072–9077. doi: 10.1021/bi1013547. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl Acad. Sci. USA. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown AK, Liu J, He Y, Lu Y. Biochemical characterization of a uranyl ion-specific DNAzyme. ChemBioChem. 2009;10:486–492. doi: 10.1002/cbic.200800632. [DOI] [PubMed] [Google Scholar]
- 46.Wernette DP, Mead C, Bohn PW, Lu Y. Surface immobilization of catalytic beacons based on ratiometric fluorescent DNAzyme sensors: A systematic study. Langmuir. 2007;23:9513–9521. doi: 10.1021/la701303k. [DOI] [PubMed] [Google Scholar]
- 47.Lee JH, Wang Z, Liu J, Lu Y. Highly sensitive and selective colorimetric sensors for uranyl (UO22+): development and comparison of labeled and label-free DNAzyme-gold nanoparticle systems. J. Am. Chem. Soc. 2008;130:14217–14226. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moshe M, Elbaz J, Willner I. Sensing of UO22+ and design of logic gates by the application of supramolecular constructs of ion-dependent DNAzymes. Nano Lett. 2009;9:1196–1200. doi: 10.1021/nl803887y. [DOI] [PubMed] [Google Scholar]
- 49.Xiang Y, Wang Z, Xing H, Wong NY, Lu Y. Label-free fluorescent functional DNA sensors using unmodified DNA: a vacant site approach. Anal. Chem. 2010;82:4122–4129. doi: 10.1021/ac100244h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang Y, Lu Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziółkowski R, Górski Ł, Oszwałdowski S, Malinowska E. Electrochemical uranyl biosensor with DNA oligonucleotides as receptor layer. Anal. Bioanal. Chem. 2012;402:2259–2266. doi: 10.1007/s00216-011-5510-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo Y, Zhang Y, Xu L, Wang L, Wen G, Liang A, Jiang Z. Colorimetric sensing of trace UO22+ by using nanogold-seeded nucleation amplification and label-free DNAzyme cleavage reaction. Analyst. 2012;137: 1866–1871. doi: 10.1039/c2an00039c. [DOI] [PubMed] [Google Scholar]
- 53.He Y, Lu Y. Metal-ion-dependent folding of a uranyl-specific DNAzyme: insight into function from fluorescence resonance energy transfer studies. Chem. Eur. J. 2011;17:13732–13742. doi: 10.1002/chem.201100352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nielsen PE, Jeppesen C, Buchardt O. Uranyl salts as photochemical agents for cleavage of DNA and probing of protein-DNA contacts. FEBS Lett. 1988;235:122–124. doi: 10.1016/0014-5793(88)81245-6. [DOI] [PubMed] [Google Scholar]
- 55.Burrows HD, Kemp TJ. The photochemistry of the uranyl ion. Chem. Soc. Rev. 1974;3:139–165. [Google Scholar]
- 56.Nielsen PE, Hiort C, Sonnichsen SH, Buchardt O, Dahl O, Norden B. DNA binding and photocleavage by uranyl(VI)(UO22+) salts. J. Am. Chem. Soc. 1992;114:4967–4975. [Google Scholar]
- 57.Jeppesen C, Nielsen PE. Uranyl mediated photofootprinting reveals strong E. coli RNA polymerase-DNA backbone contacts in the + 10 region of the DeoP1 promoter open complex. Nucleic Acids Res. 1989;17:4947–4956. doi: 10.1093/nar/17.13.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bassi GS, Moellegaard N-E, Murchie AIH, von Kitzing E, Lilley DMJ. Ionic interactions and the global conformations of the hammerhead ribozyme. Nat. Struct. Biol. 1995;2:45–55. doi: 10.1038/nsb0195-45. [DOI] [PubMed] [Google Scholar]
- 59.Nielsen PE, Møllegaard NE. Sequence/structure selective thermal and photochemical cleavage of yeast-tRNAPhe by UO 22+ J. Mol. Recognit. 1996;9:228–232. doi: 10.1002/(SICI)1099-1352(199605)9:3%3C228::AID-JMR243%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 60.Møllegaard NE, Nielsen PE. Uranyl photoprobing of DNA structures and drug-DNA complexes. Methods Mol. Biol. 1997;90:43–49. doi: 10.1385/0-89603-447-X:43. [DOI] [PubMed] [Google Scholar]
- 61.Bassi GS, Møllegaard NE, Murchie AIH, Lilley DMJ. RNA folding and misfolding of the hammerhead ribozyme. Biochemistry. 1999;38:3345–3354. doi: 10.1021/bi982985r. [DOI] [PubMed] [Google Scholar]
- 62.Møllegaard NE, Nielsen PE. Applications of uranyl cleavage mapping of RNA structure. Methods Enzymol. 2000;318:43–47. doi: 10.1016/s0076-6879(00)18043-7. [DOI] [PubMed] [Google Scholar]
- 63.Møllegaard NE, Lindemose S, Nielsen PE. Uranyl photoprobing of nonbent A/T- and bent A-tracts. A difference of flexibility? Biochemistry. 2005;44:7855–7863. doi: 10.1021/bi0502083. [DOI] [PubMed] [Google Scholar]
- 64.van Houten V, Denkers F, van Dijk M, van den Brekel M, Brakenhoff R. Labeling efficiency of oligonucleotides by T4 polynucleotide kinase depends on 5′-nucleotide. Anal. Biochem. 1998;265:386–389. doi: 10.1006/abio.1998.2900. [DOI] [PubMed] [Google Scholar]
- 65.Hampshire AJ, Rusling DA, Broughton-Head VJ, Fox KR. Footprinting: a method for determining the sequence selectivity, affinity and kinetics of DNA-binding ligands. Methods. 2007;42:128–140. doi: 10.1016/j.ymeth.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 66. Updyke,T.V., Bogoev,R.A. and Ke,S.-H. (1999) Sample buffer and methods for high resolution gel electrophoresis of denatured nucleic acids patent no. 9937813.
- 67.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cate JH, Doudna JA. Metal-binding sites in the major groove of a large ribozyme domain. Structure. 1996;4:1221–1229. doi: 10.1016/s0969-2126(96)00129-3. [DOI] [PubMed] [Google Scholar]
- 69.Perrotta AT, Been MD. Core sequences and a cleavage site Wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 1996;24:1314–1321. doi: 10.1093/nar/24.7.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varani G, McClain WH. The G center dot U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.