Abstract
Mus81 resolvase and Sgs1 helicase have well-established roles in mitotic DNA repair. Moreover, Mus81 is part of a minor crossover (CO) pathway in the meiosis of budding yeast, plants and vertebrates. The major pathway depends on meiosis-specific synaptonemal complex (SC) formation, ZMM proteins and the MutLγ complex for CO-directed resolution of joint molecule (JM)-recombination intermediates. Sgs1 has also been implicated in this pathway, although it may mainly promote the non-CO outcome of meiotic repair. We show in Tetrahymena, that homologous chromosomes fail to separate and JMs accumulate in the absence of Mus81 or Sgs1, whereas deletion of the MutLγ-component Mlh1 does not affect meiotic divisions. Thus, our results are consistent with Mus81 being part of an essential, if not the predominant, CO pathway in Tetrahymena. Sgs1 may exert functions similar to those in other eukaryotes. However, we propose an additional role in supporting homologous CO formation by promoting homologous over intersister interactions. Tetrahymena shares the predominance of the Mus81 CO pathway with the fission yeast. We propose that in these two organisms, which independently lost the SC during evolution, the basal set of mitotic repair proteins is sufficient for executing meiotic recombination.
INTRODUCTION
Meiosis is the division by which germ progenitor cells reduce the somatic diploid chromosome set to the gametic haploid set. The chromosomes of the haploid set are mosaics assembled from corresponding parts of homologous parental chromosomes. The exchange of chromosome parts occurs by crossing over (CO). It contributes to the recombination of parental genes in the gametes and the genetic diversity of the offspring. At the same time, CO is instrumental in connecting homologous chromosomes by chiasmata, which are required for the correct bipolar orientation of bivalents during the first meiotic division. If CO is compromised, chromosomally unbalanced gametes may be formed.
CO is induced by programmed DNA double-strand breaks (DSBs). At a CO site, one of the four chromatids of a chromosome pair experiences a DSB made by a dedicated endonuclease, Spo11 (1). The DSB is widened to a gap, and DNA flanking the DSB is resected in the 5′–3′ direction, exposing single-stranded 3′ overhangs. These single-stranded DNA ends associate with strand exchange proteins Rad51 and Dmc1, and one end invades a DNA double strand, which results in a three-way DNA structure, the so-called displacement loop (D-loop). If strands within the D-loop are complementary, they form a heteroduplex, and the invading strand extends by DNA synthesis (2). Most heteroduplexes seem to be short-lived and become unwound by helicases. Other D-loops capture the second DSB end and expand into a stable joint molecule (JM). The standard model of CO, elaborated in budding yeast, invokes a JM consisting of two Holliday junctions (HJs) (3). To disengage, JMs must be resolved by endonucleases. Depending on the cleavage orientation of the two HJs, the ligation of nicked strands may result in a reciprocal exchange of two DNA molecules, corresponding to a CO, or alternatively, in a nonreciprocal exchange, a noncrossover (NCO) (4,5).
Based on their function in meiotic CO formation and their ability to cleave JMs in vitro, several potential eukaryotic HJ resolvases were identified. These were the Mlh1–Mlh3 (also known as MutLγ) complex, the Rad1–Rad10 complex, the Mus81–Mms4/Eme1 complex, the Slx1–Slx4 complex and Yen1/GEN1 (6–8). Of these, the MutLγ complex (with Mlh3 containing the nuclease motif) produces (in collaboration with Exo1) the majority of COs in budding yeast (9). The MutLγ complex seems to work on JMs that are generated in the context of a synaptonemal complex (SC) and the so-called ZMM proteins (Zip1,2,3, Msh4,5, Mer3, Spo16 and Spo22) (10,11). Paradoxically, Sgs1 (the BLM helicase ortholog), which has long been known for its mitotic anti-CO activity, functions as a CO promoting factor as well (9,12–14). There also exists a minor pathway in the budding yeast that is independent of ZMM proteins and MutLγ, and involves the Mus81–Mms4 nuclease complex (and, to a lesser degree, Yen1).
The fission yeast is believed to feature a nondouble HJ form of JM, either a single HJ (15) or a nonreciprocal (i.e. nicked HJ) CO precursor (16–18). JM resolution largely depends on the Mus81–Eme1 (the Mms4 ortholog) complex (16,19–21). Arabidopsis and mammals feature both a ZMM–MutLγ-dependent pathway and a Mus81-dependent pathway, with the former being predominant (22–24). Caenorhabditis elegans and Drosophila rely on different resolvase complexes (25,26). Because of the considerable diversity even within such a small selection of organisms, it is of interest to know whether other organisms use similar sets of resolvases or have come up with different solutions. Revealing the variability of CO pathways among different eukaryotes may help to understand the evolutionary flexibility of the meiotic process, and ultimately, the nature of primordial meiosis. To address these questions, we studied meiotic DSB processing in an evolutionarily distant model system, the ciliated protist Tetrahymena thermophila.
Tetrahymena is a unicellular organism with two functionally distinct nuclei. One is the polyploid somatic macronucleus, which is transcriptionally active and is propagated only during the vegetative life cycle. The other is the transcriptionally silent micronucleus, which functions as the germ line. Only the micronucleus undergoes meiosis and is passed on to the offspring during sexual reproduction [(27) and Supplementary Figure S1].
Pairs of mating Tetrahymena cells undergo synchronous meioses (28), and the progression of meiosis can be easily followed and staged (Figure 1). Early steps in meiotic recombination follow the canonical pathway with Spo11 inducing DSBs, and strand exchange requiring Rad51 and Dmc1 (29,30). A remarkable feature of Tetrahymena meiosis is the extreme elongation of nuclei during prophase. Nuclear elongation is triggered by DSB formation (31), and it begins ∼2 h after meiosis induction. Within an elongated nucleus, chromosomes are arranged in a stretched bouquet-like manner, with centromeres and telomeres attached to opposite ends. This ultimate bouquet arrangement is believed to promote the juxtapositioning of homologous regions and, thereby, homologous pairing and CO (32,33). Following this unusual pairing stage, nuclei shorten and DSBs become repaired (33). Condensed bivalents become discernible at the diplotene/diakinesis stage, which is followed by closed first and second meiotic divisions (Figure 1). Like the fission yeast (34), Tetrahymena also lacks an SC [(31) and literature citation therein], and here we propose that they adapted to this condition by using similar modes of CO generation.
Figure 1.
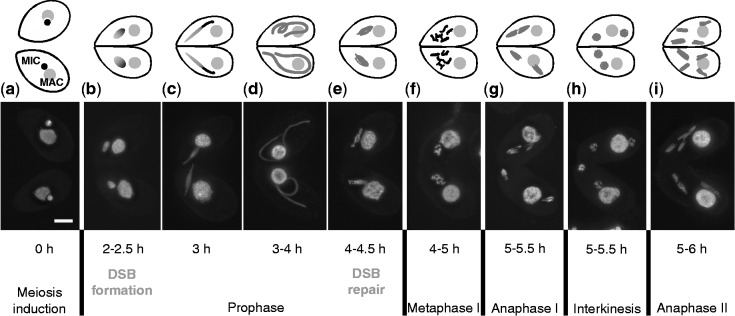
Meiotic stages in the WT (DAPI staining and schematic representation with nuclei not to scale). Each panel displays two mating cells in which meioses proceed synchronously. Each cell contains a macronucleus (MAC) and a micronucleus (MIC). Only the latter undergoes meiosis, and it becomes extremely elongated during meiotic prophase. Lower case letters a–i indicate the stages referred to in other figures. The approximate times after meiosis induction are given, at which the different stages are most abundant in our laboratory, according to (31) and (29). Stages of DSB formation and repair are indicated according to (31) and (33). Bar: 10 µm.
MATERIALS AND METHODS
Cell growth and induction of meiosis
Tetrahymena thermophila wild-type (WT) strains B2086 and Cu428, being of complementing mating types, served as controls and as the source material for the construction of knockdown and knockout strains. Strains were obtained from the Tetrahymena Stock Center at Cornell University; http://tetrahymena.vet.cornell.edu/) The spo11Δ and dmc1Δ knockout lines were described previously (30,31).
Cells were cultured at 30°C according to standard methods [reviewed by (35)] and they were made competent for sexual reproduction by starvation in 10 mM Tris–Cl (pH 7.4) for at least 16 h. Meiosis was induced by mixing starved cultures of cells of complementing mating types at equal densities (∼2 × 105 cells/ml).
Gene knockdown by RNA interference
To create the mus81, sgs1 and mms4 RNA interference (RNAi) constructs, an ∼500 bp fragment of the corresponding open reading frame (ORF) was amplified from genomic DNA using compound polymerase chain reaction (PCR) primers (for primer sequences, see SupplementaryTable S1) to add appropriate restriction sites for cloning into the RNAi hairpin (hp) vector pREC8hpCYH (36) [For a map of the vector pREC8hpCYH, see Supplementary Figure S6 accompanying (36)]. This vector contains the Cd2+ inducible MTT1 metallothionein promoter and the homologous sequence to the native RPL29 gene, which provides cycloheximide resistance. The PCR products were used to replace the REC8 fragments in the RNAi hp vector. The finished hp construct was introduced into starved B2086 and Cu428 cells by biolistic transformation (37,38). Transformants were selected initially in media containing 10 µg/ml cycloheximide, and then were transferred to increasingly higher concentrations, up to 30 µg/ml. Expression of hp double-stranded RNA was induced by the addition of CdCl2 to transformed cells (36). To deplete proteins efficiently in meiosis, RNAi was induced in exponentially growing cells before meiosis. To this end, cultures were grown to high density, diluted to half, grown in medium with 0.25 μg/ml CdCl2 for 3 h (∼1 mitotic cycle) and starved in the presence of 0.075 μg/ml CdCl2. Before mixing, CdCl2 was washed out of the starvation medium. RNAi efficiency was confirmed by reverse transcription-PCR (RT-PCR) (Supplementary Figure S2).
To create mus81i spo11Δ, mus81i dmc1Δ and sgs1i dmc1Δ strains, the spo11 and dmc1 knockout strains were transformed with either MUS81 or SGS1 RNAi constructs as described above. spo11 RNAi was reported previously (36).
Macronuclear gene knockout
For the construction of mlh1 and TTHERM_00670390 knockout strains, ∼500 bp fragments of genomic Tetrahymena DNA upstream and downstream of the respective ORF were amplified using the primers listed in Supplementary Table S1. These fragments were then joined to each end of the neo4 selection cassette using overlapping PCR (39). The knockout construct was introduced into B2086 and CU428 cells by biolistic transformation as described previously (37). The NEO4 resistance gene is expressed under the Cd2+-inducible MTT1 metallothionein promoter (40). Transformants were selected in media containing 0.1–1 µg/ml CdCl2 and increasingly higher concentrations of paromomycin (from 120 µg to 10 mg/ml) until the WT chromosomes were completely replaced by the knockout chromosomes in the somatic nucleus (Supplementary Figure S2). Complete knockout was tested by PCR using primers flanking the ORFs (Supplementary Table S1).
Protein tagging
Sgs1-HA and Mlh1-GFP strains were created by tagging the respective genes at the C-terminus by the macronuclear knock-in method. Two short (∼500 bp) DNA fragments were amplified by PCR, one homologous to the 3′-end of the ORF (without the stop codon) and one to the flanking sequence downstream of the gene (primers listed in Supplementary Table S1). These fragments were joined by overlapping PCR with the tagging cassettes of the pHA-Neo4 and pEGFP-Neo4 vectors (gifts of Kazufumi Mochizuki). The constructs were introduced into the macronuclei of B2086 and CU428 cells by biolistic transformation. Transformants were selected using the same strategy as for the knockouts (see above). Somatic transformation introduces transgenes to the vegetative nucleus, which is ∼45-ploid. The more WT copies are replaced by tagged copies, the stronger the protein labeling is. The level of replacement was tested using primers listed in Supplementary Table S1.
Standard molecular techniques
cDNA for RT-PCR analysis was prepared as described in (29), and gene expression profiles were obtained by RT-PCR using the primers listed in Supplementary Table S1.
Cytological methods
Depending on further use, cells were prepared by one of the following methods [for details see (31,32)]: (A) 250 µl 10% Triton X-100 and 500 µl 37% formaldehyde were added to 5 ml of cell suspension. After 30 min, the fixed cell suspension was centrifuged and the pellet was resuspended in 500 µl of 4% paraformaldehyde + 3.4% sucrose solution. A drop of this mixture was spread onto a slide and air-dried. (B) 5 ml of cell suspension were fixed with 20 µl of partial Schaudinn’s fixative (saturated HgCl2, ethanol 2:1), centrifuged and resuspended in methanol. This suspension was dropped onto slides and air-dried. (C) Cells were pelleted, resuspended in Carnoy's fixative (methanol, chloroform, acetic acid 6:3:2), again pelleted and resuspended in 70% ethanol. This suspension was dropped onto slides and air-dried.
A polyclonal antibody against peptide FNIFSNDDPNDLINQ from the C-terminus of Mms4 was raised in rabbit and affinity purified by a commercial provider. Its specificity was confirmed by the lack of staining of mms4i meiotic nuclei (data not shown). For immunostaining, slides prepared after procedure (A) were washed 2 × 5 min with 1× phosphate buffered saline and 5 min with 1× phosphate buffered saline + 0.05% Triton X-100. The following primary antibodies were used: anti-Mms4 (1:100), anti-GFP (1:100) and anti-HA (1:100). After washing with buffer as before, appropriate fluorescence-labeled secondary antibodies were applied. Slides prepared after (B) were used for immunostaining with anti Cna1 antibody (1:200; a generous gift from Harmit Malik). For fluorescence in situ hybridization (FISH) on slides prepared according to (C), a compound FISH probe was produced by amplifying a 22.1 kb intercalary chromosomal locus by PCR (41). The purified PCR products were labeled with Cy3 by nick translation. The probe and chromosomal DNA were denatured by hot formamide and hybridized for 36 h at 37°C.
All preparations were mounted in Vectashield anti-fading agent (Vector Laboratories) supplemented with 0.05 µg/ml 4′,6′-diamidino-2-phenylindole (DAPI). A fluorescence microscope equipped with the appropriate filters was used for observation and documentation. Z stacks of pictures were recorded using MetaView software (Universal Imaging, Downingtown, PA, USA), deconvolved using AutoDeblur (AutoQuant Imaging, Watervliet, NY, USA) and projected with ImageJ (Wayne Rasband, N.I.H.; http://rsb.info.nih.gov/ij/) software.
Prepararation, digestion and pulsed-field gel electrophoresis of Mb-sized DNA
Chromosome-sized DNA was prepared in agarose plugs using disposable plug molds (Bio-Rad) following (29). Aliquots of the cultures used for DNA preparation were checked cytologically for efficient induction of meiosis and distribution of meiotic stages. The agarose plugs were washed in 1 M Tris–Cl (pH 7.5), treated with RNaseA and washed and stored in pulsed-field gel electrophoresis (PFGE) TE (0.05 M Na2EDTA, 0.01 M Tris–Cl) at 4°C (29).
For enzymatic treatment, plugs were incubated for 1 h in 1 mM PMSF in PFGE TE, washed twice with PFGE TE at 4°C for 1 h and three times in TE (0.001 M Na2EDTA, 0.01 M Tris–Cl, pH 7.5) for 30 min at room temperature. For the SmaI digest, plugs were equilibrated in 1× restriction enzyme buffer and treated with 30 U of SmaI per plug in restriction digest mixture (300 μl of 1× restriction buffer, 4 mM spermidine, 2 mM DTT, 0.2 mg/ml bovine serum albumin) at 30°C overnight, followed by addition of fresh 30 U of the enzyme and incubation for another 4 h.
The sensitivity of putative branched DNA to Escherichia coli JM resolvase RuvC (Abcam) was assayed by digesting the DNA in plugs according to (15) and (42). After SmaI digestion, plugs were washed with TE, twice with RuvC buffer [50 mM Tris–acetate, pH 7.5, 10 mM Mg(OAc)2, 1 mM DTT, 50 µg of bovine serum albumin per ml] for 30 min at room temperature, and once for 30 min with RuvC buffer on ice. The plugs were incubated in 150 µl of buffer with 300 ng RuvC for 4 h at 4°C and then for 1 h at 55°C. The temperature increase promotes migration of JMs to a preferred DNA sequence context for RuvC activity (43). Reactions were stopped by adding 7.5 μl of 0.5 M EDTA, pH 8.0. Plugs with digested DNA were directly used for PFGE or stored in PFGE TE at 4°C.
PFGE was performed using a contour-clamped homogeneous electric field (CHEF) apparatus (Bio Rad Chef-DR III). DNA fragments were separated in a 0.8% agarose gel (pulsed-field certified agarose, Bio-Rad) in 1× TAE at 14°C for 24 h at 3.5 V/cm with a 300 s switch time followed by 48 h at 2.2 V/cm with a 1000 s switch time. After gel staining with ethidium bromide, DNA was transferred onto nylon membrane by Southern blotting. The majority of DNA in Tetrahymena cells is located in the somatic nuclei that do not undergo meiosis, and it obscures bands originating from meiotic DNA. Therefore, meiotic DNA was probed with a radioactively labeled Tlr DNA sequence, which is limited to germ line nuclei (44).
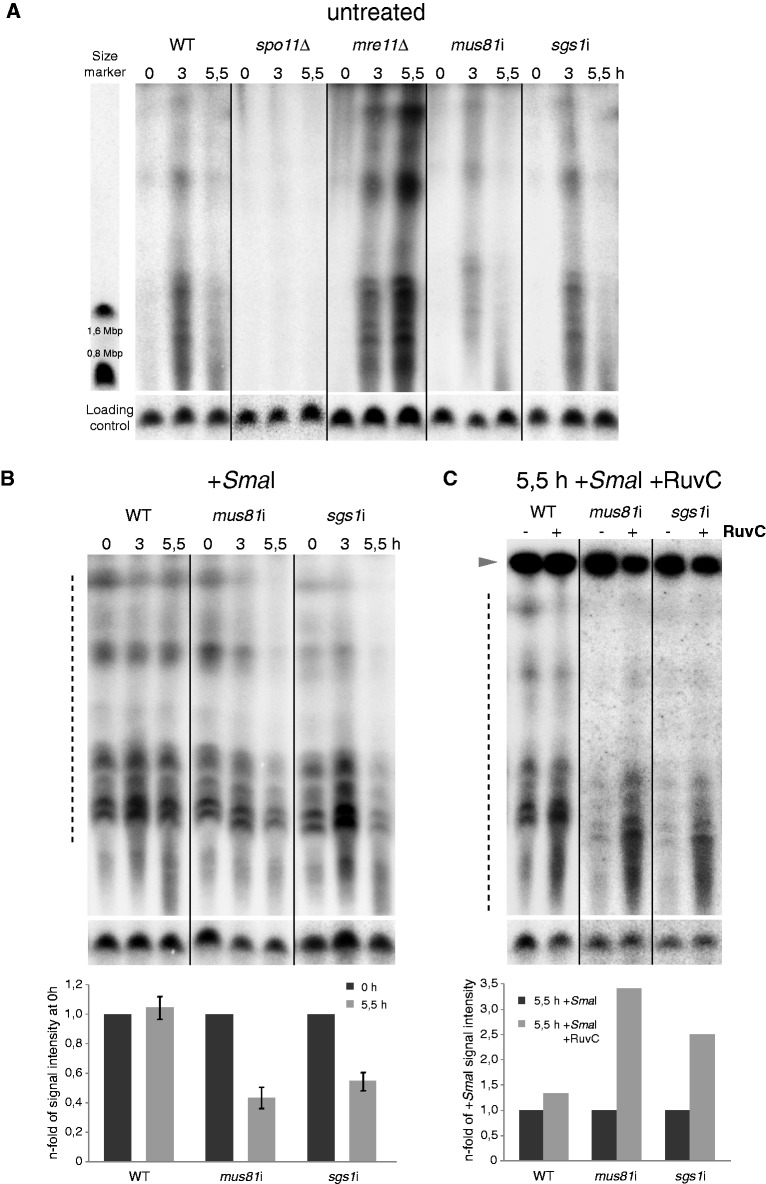
After hybridization and detection, membranes were stripped of the Tlr probe and reprobed with a sequence from a 306 kb macronuclear minichromosome, as a control for equal loading and Southern transfer. Band density of loading controls was measured, and Tlr signal intensities in the indicated areas of the gels in Figure 4B and C were integrated and measured using the ‘Analyze’ tool of ImageJ (Wayne Rasband, N.I.H.; http://rsb.info.nih.gov/ij/) software. Signal intensities were corrected according to the respective loading control intensities. For representation in Figure 4B and C, corrected signal intensities of timepoint t = 0 and –RuvC were arbitrarily assigned a value of 1, and corrected intensities at t = 5, 5 h and +RuvC were expressed as the n-fold.
Figure 4.
Detection of DSBs and JMs by PFGE. (A) A discontinuous smear in the range of ∼400 kb to 3 Mb represents DSB-generated micronuclear chromosome fragments. In the WT, no DSBs are present at the time when meiosis is induced (0 h). DSBs are maximally induced 3 h after meiosis induction, and have disappeared 5.5 h after meiosis induction. spo11Δ, where DSBs are not formed, and mre11Δ, where DSBs are not repaired (29), are shown as controls. In mus81i and sgs1i, DSB-generated fragments appear transiently, similar to the WT (see also Supplementary Figure S4). WT DNA was prepared from cultures grown in the presence of Cd to guarantee equal conditions to Cd-induced mus81i and sgs1i. Yeast chromosomes IV (1,6 Mb) and X (0,8 Mb) served as size markers. Discontinuity of the smear is caused by the incomplete separation of fragments of similar size. (B) Upper panel: After SmaI digestion, micronuclear chromosomes, which otherwise would be too large to enter the gel, run as a discontinuous smear of fragments at the time of meiosis induction (0 h). In the WT, the same SmaI fragments appear 5.5 h after meiosis induction, i.e. at a time when DSBs have been repaired (see A and Supplementary Figure S5). In mus81i and sgs1i, the amount of DNA entering the gel is reduced at 5.5 h after meiosis induction as compared with 0 h, suggesting that unlike in the WT, recombination is not completed. Lower panel: Quantification of DNA released into the gel at 0 h and 5.5 h. Error bars indicate SD of the mean from three biological repeats. (C) Upper panel: RuvC treatment releases DNA from the loading well in mus81i and sgs1i but not in the WT. This is seen by the appearance of a smear in the gel and by the reduced amount of DNA in the loading well (arrowhead). Lower panel: Quantification of DNA released into the gel after –RuvC and +RuvC treatment. In B and C, signal intensities were integrated from the gel regions indicated by dotted lines. Smaller fragments were excluded because they are formed at late timepoints owing to the degradation of postmeiotic nuclei. A signal produced by Southern hybridization to a 306 kb macronuclear chromosome, which served as a loading control, is shown underneath each lane. For the measurement of signal intensities and correction for the amount of DNA loaded, and the calculation of relative band intensities, see the ‘Materials and Methods’ section.
RESULTS
Candidates for JM resolving factors
Tetrahymena possesses homologs to a number of proteins that have been implicated in JM processing in other organisms. Tetrahymena Mus81 and Sgs1, encoded by ORFs TTHERM_00624870 and TTHERM_01030000, respectively, have been reported previously (31). Two Tetrahymena proteins, encoded by TTHERM_00127000 and TTHERM_01109940, are close homologs to budding yeast MutL family members, of which Mlh1 and Mlh3 are components of the MutLγ resolvase complex. TTHERM_01109940p is the first candidate in a BLAST search with Mlh3 as bait, but it is the best hit in a reciprocal BLAST search with Pms1. It is also the best hit with Mlh2 as bait. TTHERM_00127000p is the best hit in a reciprocal BLAST search with Mlh1. Altogether, it is likely that TTHERM_00127000p is the ortholog of Mlh1, whereas TTHERM_01109940p could be any of Pms1, Mlh2 or Mlh3, but an unambiguous assignment is not possible (31). (TTHERM_00127000 was given the official name TMLH1 in the Tetrahymena Genome Database, but it will be called MLH1 in this article.) Other members of the MutL family do not seem to exist in Tetrahymena [see also (45)].
TTHERM_00194130p was found as the best Mms4/Eme1-homolog candidate in a reciprocal BLAST search of the Tetrahymena, budding yeast and fission yeast proteomes. BLAST searches for protein homologs to budding yeast Yen1 and its human ortholog GEN1 both produced TTHERM_00670390p as the first hit with limited local similarities. Moreover, the comparison of protein domains using the Feature Architecture Comparison Tool [FACT; http://fact.cibiv.univie.ac.at/index.php; (46)] revealed that Tetrahymena TTHERM_00670390p shares with Yen1 and GEN1 a similar arrangement of XPGN and XPGI regions. This makes TTHERM_00670390p a possible Yen1/GEN1 candidate. We failed to identify good homologs of Slx1 and Slx4 proteins.
To assess potential roles of the above-mentioned proteins in meiosis, gene expression was studied by reverse transcription-PCR (RT-PCR). As can be seen from Figure 2A, expression of SGS1 is strongest at ∼3 h after induction of meiosis, which coincides with the time when DSBs are generated and Dmc1 foci form on chromatin (30). These temporally coordinated activities are reflected by similar timing of the transcription of SPO11 and DMC1 genes. MUS81, MMS4 and MLH1 display a basal level of expression in vegetatively growing cells but are upregulated throughout meiosis. The patterns of MUS81 and MMS4 expression, with maximums at 4–5 h, are similar, which is consistent with the possibility that, like their yeast homologs, they act as a complex. Expression of the YEN1 homolog, TTHERM_00670390 peaks at 6 h, which is too late in meiosis to act as a JM resolvase. Thus, the transcription profiles of MUS81, MMS4, SGS1 and MLH1 candidates, which are in good agreement with a microarray analysis of gene expression (47), suggest their potential involvement in meiotic recombination, and we went on to characterize them further.
Figure 2.
Expression of HJ resolvase candidate genes, Sgs1 and Mms4 localization during meiosis, and aberrant metaphase I in mus81i and sgs1i. (A) RT-PCR. Expression patterns of meiosis-specific genes SPO11 and DMC1 and the ubiquitously expressed control gene RPL21 are shown for comparison. V = vegetative cells, 0 = starving cells at the time of meiosis induction, 1–10 = h after meiosis induction. (B) Sgs1-HA (red) localizes to meiotic nuclei throughout prophase. By metaphase I it disappears. (C) Immunostained Mms4 is visible at diakinesis. (D) mus81i and sgs1i cells do not show condensed metaphase bivalents, yet association of FISH signal pairs (red) demonstrates that homologous chromosomes are paired in most nuclei. FISH signal distances were measured in 100 nuclei of the WT, mus81i and sgs1i, each. Nuclei were plotted along the X-axis according to increasing distances. Single FISH signals, indicating close pairing, were assigned a zero distance on the Y-axis. Bars in B and D: 10 µm.
Mms4, Sgs1 and Mlh1 localize to meiotic nuclei
To study the presence and localization of candidate JM-processing proteins during the course of meiosis, we constructed strains expressing tagged proteins and/or had antibodies raised. We failed to detect Mus81 by immunostaining or protein tagging both in situ and in western blot experiments. However, we obtained an antibody against its putative partner Mms4, and we were able to visualize Sgs1 and Mlh1 by tagging with HA and GFP epitopes, respectively.
Little Sgs1-HA was detected in the micronucleus of vegetatively growing cells, but it increased in meiotic nuclei from an early stage onward and was most abundant in elongated prophase nuclei (Figure 2B). By meiotic metaphase, a signal was no longer visible. Mms4 was hardly detectable cytologically in elongated prophase nuclei and was most abundant in nuclei exiting from the elongated state, i.e. after the onset of Sgs1 expression (Figure 2C). This appearance of Mms4 and Sgs1 at different stages reflects the 1–2 h temporal difference in maximal mRNA transcription of the respective genes (Figure 2A). Mlh1–GFP was present in vegetative micronuclei, produced weak signals in meiotic prophase nuclei and appeared to be slightly more abundant only in nuclei that had completed the first and second meiotic divisions (Supplementary Figure S3).
Knockdown of MUS81 or SGS1 causes abnormal bivalent formation
Despite their upregulation in meiotic cells, a basal expression level of MUS81 and SGS1 during vegetative growth (Figure 2A) suggests their requirement for mitotic propagation. Indeed, MUS81 appears to be essential because we were not able to generate a full MUS81 knockout by random gene assortment. To study the function of Mus81 and of Sgs1 in meiosis, we resorted to the meiosis-specific depletion of the proteins by inducible RNAi. We constructed strains that display a robust reduction of mRNA on induction of double-stranded RNA transcription (Supplementary Figure S2). In the following, we will denote RNAi–knocked-down genes and RNAi mutant cells by the suffix ‘i’.
As a first indication of whether mus81i or sgs1i might cause a meiotic defect, we tested the fertility of these mutants. We found that mus81i and sgs1i failed to produce any viable sexual progeny (Supplementary Table S2).
Next, we cytologically monitored the progression of mus81i and sgs1i strains through meiosis. In both mutants, prophase looked normal, and nuclei elongated as in the WT. However, at metaphase, chromosomes did not condense to the same extent as in the WT, and they formed a single mass of chromatin (Figure 2D). To determine whether these chromatin masses consisted of unpaired chromosomes or bivalents, we applied FISH to an intercalary chromosomal locus (41) and scored tightly connected (single signal) versus separate (2 signals) homologous regions (Figure 2D). In both mutants, nuclei with single or closely associated FISH dots prevailed, which confirms that, despite the lack of distinct condensed structures, homologous chromosomes had formed bivalents. We next assessed the closeness of homolog associations by scoring the frequency of nuclei with a single FISH signal and measuring the distances between pairs of FISH signals. In 100 nuclei of the WT, we found 25% with a single FISH signal and the rest with two signals side-by-side. This relaxed association is owing to the fact that diplotene–metaphase I bivalents are not intimately paired along their entire lengths, and only loci in the vicinity of a chiasma are closely aligned. Interestingly, in the mus81i mutant, 54 of 100 nuclei displayed fused FISH signals. Also, the average distance (center to center) of FISH foci was 0.47 µm in the WT and 0.25 µm in the mus81i mutant. This suggests that mus81i bivalents are linked at more sites than WT bivalents. No such enforced connection was observed in the sgs1i mutant with an average distance of foci of 0.51 µm (n = 100) (Figure 2D).
Homologs do not separate in the absence of Sgs1 and Mus81
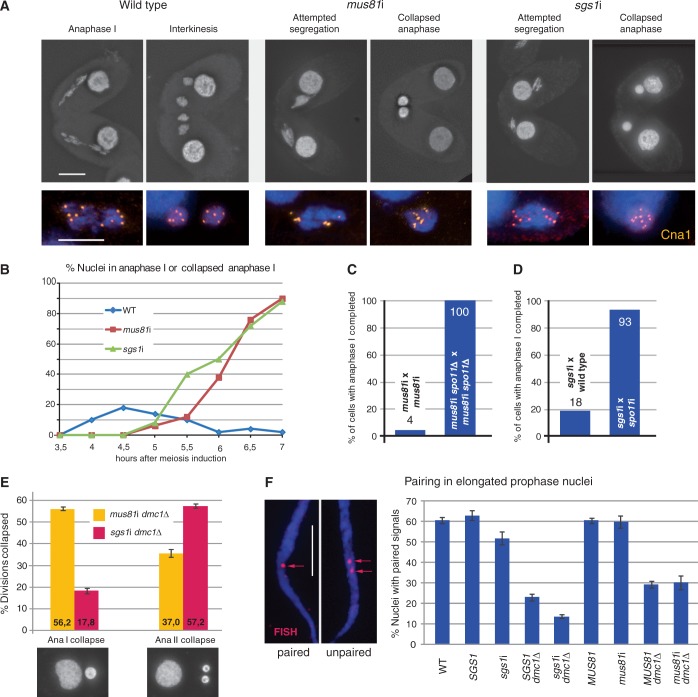
We next studied the segregation behaviour of the abnormal mus81i and sgs1i bivalents and found that they completely failed to undergo anaphase separation. Nuclei were seen to elongate as if they were trying to separate chromosomes, but ultimately they collapsed back into a single nucleus and remained arrested in that stage (Figure 3A and B). Immunostaining with an antibody against the centromeric histone Cna1 (48) revealed that the centromeres were correctly bioriented during attempted anaphase (Figure 3A). However, while the WT formed two haploid late anaphase/telophase ensembles with 5 centromeres, mutants retained nuclei with a diploid set of 10 centromeres. This indicates that the spindle was pulling, but was unable to separate homologous chromosomes.
Figure 3.
Depletion of Mus81 and Sgs1 causes DSB-dependent meiotic segregation defects. (A) In the WT, homologous chromosomes separate in anaphase I and form two distinct chromatin masses in telophase I–interkinesis. In mus81i and sgs1i meiosis, chromosomes collapse back into a single diploid nucleus after an attempted anaphase I. Staining of the centromere marker Cna1 (orange) shows the orientation of five centromeres to each of the opposite poles and five centromeres in each daughter nucleus after complete separation of the homologs in the WT. In the mutants, collapsed nuclei contain 10 centromeres. (B) Quantification of the segregation defect. WT meiotic nuclei pass through anaphase I, whereas mutants arrest with collapsed anaphase I nuclei. Hundred cells were evaluated for each genotype and timepoint. (C) Spo11 depletion rescues the mus81i segregation defect. mus81i strains were depleted of Spo11 by SPO11 knockout. All mus81i spo11Δ cells completed anaphase I 5.5 h after induction of meiosis (n = 50 cells for each phenotype). (D) Spo11 depletion rescues the sgs1i segregation defect. sgs1i cells were depleted of Spo11 by spo11 RNAi of their mating partner. spo11 RNAi is transferable, i.e. if one of the mating partners expresses interfering RNA, both cells display the depletion phenotype. spo11 RNAi efficiency was monitored by the failure of micronuclei to elongate during meiotic prophase (see main text). The sgs1i phenotype is limited to the cell expressing siRNA and is attenuated by mating to a non-sgs1i partner. Therefore, the frequency of successful anaphases was scored in the partner affected by spo11 sgs1 double RNAi and compared with sgs1i × WT pairs. n = 100 cells for each genotype. (E) dmc1Δ mus81i and dmc1Δ sgs1i double mutants are partially rescued from the anaphase I defect and undergo defective anaphase II. Shown for each genotype are the mean values ± S.D. from three experiments with 100 nuclei counted in each. An unaccounted-for percentage of cells contained three or four nuclei, where one or both products of a first division may have undergone a second division or where nonseparating bivalents or chromosomes had formed extra nuclei. (F) Pairing in the elongated prophase nucleus. Examples of paired (FISH signals fused) and unpaired (FISH signals separate) homologous loci (red arrows) are shown. SGS1 and MUS81 genotypes carry the respective RNAi hp constructs, but RNAi was not induced; hence, they resemble the WT control. In the absence of Sgs1 but not of Mus81, homologous pairing is reduced both in the WT and in the dmc1Δ background, in which recombination takes place primarily between sisters (30). Values are means of three repeats with 100 nuclei evaluated, each. Error bars indicate standard deviation. Bars in A and F: 10 µm.
Depletion of Mms4 caused an anaphase arrest identical to the one of mus81i (Supplementary Figure S4). This makes it likely that Mus81 and Mms4 cooperate in Tetrahymena as they do in the yeasts, and given the similar behavior of the two mutants, we continued with the further characterization of Mus81 only.
DSB-dependent physical links prevent the separation of homologs in mus81i and sgs1i
Because metaphase bivalents of mus81i and sgs1i fail to condense properly, it is conceivable that their separation was inhibited by the twisting and entangling of abnormally long chromosomes. Alternatively, segregation could be prevented by inappropriately processed recombination intermediates. To test if the mus81i and sgs1i anaphase arrest was due to persistent interchromosomal connections formed during recombination, we created strains depleted of Spo11. In the absence of Spo11, no DSBs and hence no COs are formed (1). In Tetrahymena spo11Δ, micronuclei do not fully elongate, and univalents are formed and segregate randomly in the first meiotic anaphase (31).
spo11Δ strains were transformed with the MUS81 hp construct to create spo11Δ mus81i double mutants (see ‘Materials and Methods’ section), and we found that the mus81i segregation defect was fully rescued by the spo11Δ mutation (Figure 3C). Similarly, we simultaneously inactivated SGS1 and SPO11 by RNAi. WT strains were transformed with either SGS1 or SPO11 hp constructs, selected with cycloheximide, and RNAi was induced by adding cadmium (see ‘Materials and Methods’ section). spo11 RNAi is mostly transferable and penetrant, i.e. when a spo11i strain is mated to a WT strain, the Spo11 depletion phenotype is usually elicited in both partners (30). (Here, micronuclear elongation was abolished in 82 of 100 mating pairs, indicating that spo11 RNAi was efficient.) sgs1 RNAi is not systemic, and if an sgs1i cell is mated to WT, the Sgs1 depletion phenotype is limited to the sgs1i partner (data not shown). Therefore, in spo11i × sgs1i pairs, one partner is Spo11−, the other mostly Spo11−/Sgs1−. Of 100 spo11i × sgs1i pairs, where one partner (spo11i) had completed anaphase I, there were 93, where the other partner had also completed anaphase I (Figure 3D). Thus, in sgs1 spo11 double RNAi cells, rescue of anaphase segregation was robust. These results indicate that both in mus81i and sgs1i, persistent Spo11-dependent recombination intermediates are responsible for the anaphase collapse.
The separation of sister chromatids is inhibited in mus81i and in sgs1i meiosis
While the anaphase I arrest revealed the persistence of recombination-dependent interhomolog links, we wanted to know if intersister links are also formed and remain unresolved. To this end, we created a situation where cells overcame the anaphase I arrest. In Tetrahymena, like in most other eukaryotes, the meiosis-specific recombination protein Dmc1 is required to direct recombinational repair toward the homolog [(30) and literature citation therein]. Hence, in a dmc1Δ mutant, there is little interhomolog CO, and mostly univalents are formed at metaphase I. The random segregation of univalents at anaphase I is followed by the separation of sister chromatids at anaphase II (30). However, anaphase II segregation of sisters will be impaired if unresolved intersister links are present.
We scored meiotic configurations in a mus81i dmc1Δ × mus81i dmc1Δ crossing 5.5 h after induction of meiosis, i.e. at a time when, in the WT, ∼90% of cells have completed the second meiotic division. We found that about half of the cells (56%, n = 150) had abolished anaphase I (Figure 3E). Thus, while a residual CO is formed in only 9% of dmc1Δ cells (30), unresolved NCO intermediates may contribute to the substantial anaphase I arrest in double mutants. Nevertheless, we found that 37% of cells had completed anaphase I but became arrested at anaphase II.
Next, we quantified anaphase I and II segregation in a sgs1i dmc1Δ × sgs1i dmc1Δ double mutant crossing and found that only 18% of cells abolished anaphase I, whereas 57% arrested at anaphase II (Figure 3E). The anaphase II defect in both double mutants suggests that links are formed between sisters and that Mus81 and Sgs1 are required for resolving them.
Homologous pairing is slightly reduced in the absence of Sgs1
The above experiment showed that sgs1i bypasses the anphase I segregation defect more efficiently than mus81i in the dmc1Δ background. We wondered whether this might be owing to a lower abundance of interhomolog recombination intermediates in the sgs1i dmc1Δ mutant. Previous experiments have shown that interhomolog strand exchange promotes pairing in elongated prophase nuclei (30). Therefore, we determined the pairing frequency of a chromosomal locus by FISH (Figure 3F). We found that mus81i in the dmc1Δ mutant background has not reduced pairing as compared with the uninduced RNAi control. In contrast, sgs1i in the dmc1Δ background reduced pairing by ∼50%. This suggests that reduced interhomolog connections may be the reason for the relatively successful anaphase I segregation in the sgs1i dmc1Δ double mutant. Next, we evaluated pairing frequency in mus81i and sgs1i single mutants. Again, Mus81 depletion did not reduce pairing, whereas in sgs1i it was reduced by 20% (Figure 3F). This is consistent with the possibility that Sgs1 promotes or stabilizes interhomolog interactions.
Branched DNA molecules accumulate in the absence of Mus81 or Sgs1
To test if the persistent links consist of DNA JMs, we analyzed the mobility of meiotic DNA species in pulsed-field gel electrophoresis. Chromosomes of generative nuclei are too big to enter a pulsed-field gel. They will do so only when they are fragmented by meiotic DSBs (29). Therefore, in the WT, a smear diagnostic of DSBs appeared transiently (3 h after meiosis induction), and disappeared later when DSBs were repaired (5.5 h after meiosis induction) (Figure 4A). (For the timing of DSB appearance and disappearance, see also Supplementary Figure S5.) By contrast, no such smear appeared in the spo11Δ control, which does not form DSBs. A smear formed at 3 h and became stronger at 5.5 h in the mre11Δ control, which we showed previously to accumulate unrepaired DSBs (29). In meiotic cultures of mus81i or sgs1i, the DSB smear appeared and disappeared similarly to the WT (Figure 4A). We speculated that in these cases the loss of the smear was not due to the restoration of intact chromosomes but due to the persistence of branched molecules that would not enter the gel. To test this hypothesis, we treated DNA with SmaI, which cleaves the Tetrahymena genome into fragments with an average size of 1.5 Mb (M. Novatchkova, personal communication). Equal amounts of SmaI-generated fragments entered the gel in WT samples from before and after meiotic DSB formation (0 and 5.5 h after meiosis induction—Figure 4B). In contrast, in the mus81i and sgs1i samples, the amount of DNA entering the gel at t = 5.5 h was reduced to about half that of t = 0 h (Figure 4B). It was shown previously that branched DNA molecules remain trapped in the loading well, whereas linear molecules of the same size enter the gel (49,50). Therefore, it is likely that mus81i and sgs1i meioses arrest with branched DNA.
Persistent branched DNA molecules are RuvC sensitive
To study in more detail the molecular nature of the branched molecules that fail to reenter the gel after the disappearance of DSB-generated fragments, we treated the samples with RuvC. RuvC has been shown to cleave HJs in vitro (15,42) and hence to produce linear pieces of DNA, which would migrate into the gel. Indeed, RuvC digestion restored entry of DNA fragments into the gel (Figure 4C). [To exclude the possibility that the handling of samples during RuvC treatment caused DNA fragmentation or JM separation, we did a control mock treatment in the absence of RuvC (Supplementary Figure S6).] This indicates that the retainment of post-DSB DNA in the loading well is caused by the presence of JMs, which are RuvC-sensitive. RuvC cleaves mature HJs as well as three-stranded DNA molecules that are structurally similar to D loops or nicked HJs (51). Therefore, the precise nature of JMs that accumulate in mus81i and sgs1i meioses remains unclear.
mlh1 and TTHERM_00670390 mutant cells do not undergo meiotic catastrophe
The inability of the mus81i mutant to segregate homologs is not a sufficient indication for the exclusive function of Mus81 in resolving JMs because a small number of unresolved JMs is sufficient to block nuclear division (52). Therefore, it is possible that another activity contributes to the completion of meiotic recombination in Tetrahymena. A potential candidate is the MutLγ (Mlh1–Mlh3) complex which, in the budding yeast, plays a prominent role in the same pathway as the ZMM proteins in JM to CO transformation (9,52,53). Another candidate is Yen1, which may resolve JMs that escape from the main pathway (54).
Despite a possible role of Mlh1 and the Yen1 homolog TTHERM_00670390p for cell viability or vegetative propagation, we were able to produce stable mlh1 and TTHERM_00670390 knockout lines (Supplementary Figure S2). When two mlh1Δ strains or two TTHERM_00670390Δ strains were mated, both failed to initiate postmeiotic mitoses of the gametic nuclei (for WT postmeiotic development of the gametes, see Supplementary Figure S1). Importantly, however, the segregation of chromatids at anaphase I and II was uninhibited (Figure 5A and B). This was expected in the case of TTHERM_00670390Δ, given that the peak expression of the protein occurs only at t = 6 h after meiosis induction (see above). In mlh1Δ, on the other hand, anaphase could be largely rescued by the activity of Mus81. However, mlh1Δ could have a more subtle meiotic phenotype. If Mus81 was not fully compensating for the lack of MutLγ, Sgs1 might step in and convert persistent JMs to NCOs, which would result in an increase in univalents. Therefore we used FISH to score for univalent formation at metaphase (Supplementary Figure S7). We found 100% bivalent formation both in the mutant and in the WT (N = 50 metaphases, each).
Figure 5.

Deletion of MLH1 and of ORF TTHERM_00670390, encoding a Yen1 homolog, does not affect meiotic divisions. mlh1Δ (A) and TTHERM_00670390Δ (B) undergo normal first and second anaphases (for comparison, see the WT in Figure 1). Bar: 10 µm.
Altogether, our results suggest that Mlh1 (and a potential MutLγ complex of which it may be part) and the Yen1-like TTHERM_00670390p may have roles in nuclear development after fertilization, but play a nonessential role, if any, in the resolution of HJs.
DISCUSSION
Mus81 is an essential resolvase of recombination intermediates
Here, we showed that in the absence of Mus81, DNA JMs (probably HJs) persist in meiosis and homologs do not separate. Moreover, at diplotene/metaphase I, chromosomes are less condensed and homologs are more tightly connected than in the WT. This confirms that an abnormally high number of interhomolog connections are present. In the absence of Mlh1, normal-looking intact bivalents are formed, and both meiotic divisions progress uninhibited. Together, our observations indicate that Mus81 is an essential, if not the only, HJ resolvase in Tetrahymena. This raises the question of whether Mus81-dependent COs in Tetrahymena are interfering.
COs within a certain distance tend to suppress each other. This is called CO interference, and it is likely related to the mechanism ensuring that each pair of chromosomes obtains at least one CO (CO assurance) [reviewed in (5,55,56)]. ZMM–MutLγ-dependent COs were found to be interfering, whereas residual COs in the absence of ZMM proteins in budding yeast, Arabidopsis and mouse, and COs in fission yeast are noninterfering. Thus, in the organisms where both pathways co-exist, Mus81-dependent COs have been generally assumed to be noninterfering (22–24,57). In the fission yeast, which has only the Mus81 pathway, interference does not exist (58), and in C.elegans, where COs are mainly ZMM-dependent, interference is strong (59,60).
In Tetrahymena, there are an estimated number of >200 DSBs (30). As in most eukaryotes, only a subset of DSBs may give rise to mature COs. Because of the link between CO interference and assurance, low numbers of COs/chiasmata are considered a sign of interference. Unfortunately, the number of interhomolog COs or chiasmata could not yet be reliably determined in Tetrahymena. In the future, we hope to be able to test the link between the mode of CO generation and CO interference.
A possible role of Sgs1 in NCO and a proposed additional function in discouraging intersister recombination
The RecQ family helicases of budding yeast and humans (Sgs1 and BLM) are known as anti-CO factors in mitosis. They act by reversing or destabilizing early strand invasion intermediates (D-loop structures) (61) and by dissolving double HJs in collaboration with DNA topoisomerase III (62) (5,14). In meiosis, destabilization of recombination intermediates by Sgs1 and other helicases is assumed to channel the majority of DSBs into a NCO pathway to avoid the adverse effects of excessive COs on homolog segregation (52,63–65). Another function of Sgs1 is to eliminate multichromatid or other aberrant recombination intermediates, to prevent potentially deleterious COs (66,67). Conversely, a pro-CO activity of Sgs1 (9,52) and its orthologs in fission yeast, worm and Drosophila (68–70) has become evident. Also in the budding yeast, Sgs1 was suggested to contribute directly to CO formation by being part of the major ZMM-dependent CO pathway (9). Alternatively, it is possible that its pro-CO role is owing to its recycling the above-mentioned toxic intermediates and feeding them into the CO pathway.
It is conceivable that Sgs1 serves similar functions in Tetrahymena. Because, like in most other organisms, there seems to be an excess of recombination intermediates compared with interhomolog COs (see above), Sgs1 could play a role in limiting CO number by redirecting DSB repair to NCO outcomes, such as synthesis-dependent strand annealing (SDSA), or by HJ dissolution, in a subset of intermediates. In the absence of Sgs1, excess persisting JMs could overwhelm Mus81's capacity of JM resolution, hence causing the sgs1i phenotype observed here.
However, Sgs1 may have an additional earlier function. In its absence, we noticed reduced homologous pairing and, revealed by the dmc1Δ background, decreased interhomolog JM formation. Therefore, we propose that Sgs1 promotes interhomolog over intersister recombination. In the majority of eukaryotes, axial element components of the SC pose a barrier against intersister exchange [see (71) and literature citation therein]. This may even be true for the truncated axial element-like structures of fission yeast (72). In Tetrahymena, homologous chromosomes become aligned within the stretching prophase nucleus. In the absence of axial elements, DSB ends would tend to invade sister molecules, when homologs are not yet aligned. Sgs1 was present in prophase nuclei from the beginning to after the end of the elongation stage (Figure 2B). This corresponds to the entire ≥90-min interval from DSB formation to recombination-related DNA synthesis (Supplementary Figure S8). Its presence before recombination-related DNA synthesis makes a role in SDSA or CO during this period unlikely. Instead, it might destabilize D-loops, the majority of which would be intersister, and recycle DSBs for use in homologous recombination once homologs have become aligned. Consistent with this proposed function of Sgs1, increased intersister double HJ formation in the absence of Sgs1 was observed in budding yeast (66). This suggests that the role of Sgs1 in discouraging intersister recombination might be conserved.
Tetrahymena and fission yeast have independently evolved streamlined meioses
Studies in model systems revealed that there exist different pathways for the transformation of recombination intermediates into COs (8). In most model systems tested so far, the majority of COs is dependent on ZMM proteins, which are also required for SC formation. Furthermore, they need the MutLγ complex for JM resolution, and thus it seems that they are components of a single pathway, which may also include Exo1 and Sgs1 (9,53). Only a subset of COs requires the Mus81–Eme1(Mms4) complex (8), which seems to be part of an independent pathway. In contrast, in Schizosaccharomyces pombe, which lacks the ZMM proteins, and hence an SC, and MutLγ, CO formation depends almost entirely on Mus81–Eme1 (see ‘Introduction’ section). Besides SC-less Aspergillus (73), of which little is known, Tetrahymena is the only other organism so far where the SC and a full set of ZMM proteins are missing. Moreover, although the MutLγ-component Mlh1 is present, this complex may not play a prominent role in meiotic segregation. These parallels lead us to suggest that Tetrahymena and the fission yeast have similarly developed Mus81-dependent meiotic recombination as their primary (or only) CO pathway. However, they differ in the use of Sgs1, which is essential in Tetrahymena, but only has a moderate contribution to spore viability in the fission yeast (68,74). While Sgs1 has been considered by some to be part of the ZMM–MutLγ-dependent pathway (9), its importance for Tetrahymena CO formation may mainly come from its role in discouraging intersister recombination.
Tetrahymena and the fission yeast possess meiosis-specific factors, such as Spo11 and Dmc1, for allowing programmed DSB formation and homolog-directed recombinational repair. However, unlike the majority of eukaryotes, they depend exclusively on mitotic repair factors for the processing of recombination intermediates. Another parallel, which may also be related to the loss of an SC, is the homology searching and pairing by peculiar forms of bouquet arrangement (33). It will be interesting in future work to detect further parallels between these two evolutionarily distant organisms and to find out which adaptations led them to their parsimonious way of meiotic recombination.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online, including [75–78].
FUNDING
University of Vienna Initiativkolleg [I031-B] and Austrian Science Fund (FWF) [P21859 and P23802]. Funding for open access charge: Austrian Science Fund (FWF) [P21859 and P23802].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Maria Novatchkova and Tina Koestler for bioinformatic advice and to Kazufumi Mochizuki for continuous support and constructs, and permission to use his biolistic gun.
REFERENCES
- 1.Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Topics Dev. Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 2.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 3.Bell LR, Byers B. Homologous association of chromosomal DNA during yeast meiosis. Cold Spring Harbor. Symp. Quant. Biol. 1983;47:829–840. doi: 10.1101/sqb.1983.047.01.095. [DOI] [PubMed] [Google Scholar]
- 4.Ehmsen KT, Heyer WD. In: Recombination and Meiosis. Models, Means, and Evolution. Egel R, Lankenau DH, editors. Berlin, Heidelberg: Springer-Verlag; 2008. pp. 91–164. [Google Scholar]
- 5.Youds JL, Boulton SJ. The choice in meiosis—defining the factors that influence crossover or non-crossover formation. J. Cell Sci. 2011;124:501–513. doi: 10.1242/jcs.074427. [DOI] [PubMed] [Google Scholar]
- 6.Wang TF, Kleckner N, Hunter N. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl Acad. Sci. USA. 1999;96:13914–13919. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svendsen JM, Harper JW. GEN1/Yen1 and the SLX4 complex: solutions to the problem of Holliday junction resolution. Genes Dev. 2010;24:521–536. doi: 10.1101/gad.1903510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynn A, Soucek R, Börner V. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 11.Shinohara M, Oh SD, Hunter N, Shinohara A. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 2008;40:299–309. doi: 10.1038/ng.83. [DOI] [PubMed] [Google Scholar]
- 12.Rockmill B, Fung JC, Branda SS, Roeder GS. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 2003;13:1954–1962. doi: 10.1016/j.cub.2003.10.059. [DOI] [PubMed] [Google Scholar]
- 13.Jessop L, Rockmill B, Roeder GS, Lichten M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2006;2:e1000155. doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein HL, Symington LS. Sgs1—the maestro of recombination. Cell. 2012;149:257–259. doi: 10.1016/j.cell.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 15.Cromie G, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz EK, Wright WD, Ehmsen KT, Evans JE, Stahlberg H, Heyer WD. Mus81-Mms4 functions as a single heterodimer to cleave nicked intermediates in recombinational DNA repair. Mol. Cell. Biol. 2012;32:3065–3080. doi: 10.1128/MCB.00547-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boddy MN, Gaillard PHL, McDonald WH, Shanahan P, Yates JR, Russell P. Mus81-Eme1 are essential components of a Holliday junction. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 20.Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81·Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitby MC. Making crossovers during meiosis. Biochem. Soc. Transact. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- 22.Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 2007;3:e1000132. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 2008;4:e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins JD, Buckling EF, Franklin FC, Jones GH. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 2008;54:152–162. doi: 10.1111/j.1365-313X.2008.03403.x. [DOI] [PubMed] [Google Scholar]
- 25.Yildiz O, Majumder S, Kramer B, Sekelsky JJ. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell. 2002;10:1503–1509. doi: 10.1016/s1097-2765(02)00782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito TT, Youds JL, Boulton SJ, Colaiácovo MP. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 2009;5:e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karrer KM. Nuclear dualism. In: Collins K, editor. Tetrahymena thermophila. Vol. 109. San Diego: Academic Press; 2012. pp. 29–52. [Google Scholar]
- 28.Cole E, Sugai T. Developmental progression of Tetrahymena through the cell cycle and conjugation. In: Collins K, editor. Tetrahymena thermophila. Vol. 109. San Diego: Academic Press; 2012. pp. 177–236. [DOI] [PubMed] [Google Scholar]
- 29.Lukaszewicz A, Howard-Till RA, Novatchkova M, Mochizuki K, Loidl J. MRE11 and COM1/SAE2 are required for double-strand break repair and efficient chromosome pairing during meiosis of the protist Tetrahymena. Chromosoma. 2010;119:505–518. doi: 10.1007/s00412-010-0274-9. [DOI] [PubMed] [Google Scholar]
- 30.Howard-Till RA, Lukaszewicz A, Loidl J. The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS Genet. 2011;7:e1001359. doi: 10.1371/journal.pgen.1001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mochizuki K, Novatchkova M, Loidl J. DNA double-breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena. J. Cell Sci. 2008;121:2148–2158. doi: 10.1242/jcs.031799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loidl J, Scherthan H. Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J. Cell Sci. 2004;117:5791–5801. doi: 10.1242/jcs.01504. [DOI] [PubMed] [Google Scholar]
- 33.Loidl J, Lukaszewicz A, Howard-Till RA, Koestler T. The Tetrahymena meiotic chromosome bouquet is organized by centromeres and promotes interhomolog recombination. J. Cell Sci. 2012;125:5873–5880. doi: 10.1242/jcs.112664. [DOI] [PubMed] [Google Scholar]
- 34.Loidl J. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma. 2006;115:260–271. doi: 10.1007/s00412-006-0047-7. [DOI] [PubMed] [Google Scholar]
- 35.Orias E, Hamilton EP, Orias JD. Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. In: Asai DJ, Forney JD, editors. Tetrahymena thermophila. Vol. 62. San Diego: Academic Press; 2000. pp. 189–211. [DOI] [PubMed] [Google Scholar]
- 36.Howard-Till RA, Lukaszewicz A, Novatchkova M, Loidl J. A single cohesin complex performs mitotic and meiotic functions in the protist Tetrahymena. PLoS Genet. 2013;9:e1003418. doi: 10.1371/journal.pgen.1003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruns P, Cassidy-Hanley D. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 2000;62:501–512. doi: 10.1016/s0091-679x(08)61553-8. [DOI] [PubMed] [Google Scholar]
- 38.Yao MC, Yao CH. Accurate processing and amplification of cloned germ line copies of ribosomal DNA injected into developing nuclei of Tetrahymena thermophila. Mol. Cell. Biol. 1989;9:1092–1099. doi: 10.1128/mcb.9.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mochizuki K. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene. 2008;425:79–83. doi: 10.1016/j.gene.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky MA. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl Acad. Sci. USA. 2002;99:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loidl J, Mochizuki K. Tetrahymena meiotic nuclear reorganization is induced by a checkpoint kinase-dependent response to DNA damage. Mol. Biol. Cell. 2009;20:2428–2437. doi: 10.1091/mbc.E08-10-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wehrkamp-Richter S, Hyppa RW, Prudden J, Smith GR, Boddy MN. Meiotic DNA joint molecule resolution depends on Nse5-Nse6 of the Smc5-Smc6 holocomplex. Nucleic Acids Res. 2012;40:9633–9646. doi: 10.1093/nar/gks713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West SC. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 44.Wuitschick JD, Gershan JA, Lochowicz AJ, Li S, Karrer KM. A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila. Nucleic Acids Res. 2002;30:2524–2537. doi: 10.1093/nar/30.11.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM. An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS One. 2008;3:e2879. doi: 10.1371/journal.pone.0002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koestler T, von Haeseler A, Ebersberger I. FACT: functional annotation transfer between proteins with similar feature architecture. BMC Bioinformatics. 2010;11:417. doi: 10.1186/1471-2105-11-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cervantes MD, Xi X, Vermaak D, Yao MC, Malik HS. The CNA1 histone of the ciliate Tetrahymena thermophila is essential for chromosome segregation in the germline micronucleus. Mol. Biol. Cell. 2006;17:485–497. doi: 10.1091/mbc.E05-07-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho CK, Mazón G, Lam AF, Symington LS. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell. 2010;40:988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun WL, Lorenz A, Osman F, Whitby MC. A failure of meiotic chromosome segregation in a fbh1Δ mutant correlates with persistent Rad51-DNA associations. Nucleic Acids Res. 2011;39:1718–1731. doi: 10.1093/nar/gkq977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benson FE, West SC. Substrate specificity of the Escherichia coli RuvC protein. Resolution of three- and four-stranded recombination intermediates. J. Biol. Chem. 1994;269:195–201. [PubMed] [Google Scholar]
- 52.De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell. 2012;46:43–53. doi: 10.1016/j.molcel.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zakharyevich K, Ma YM, Tang SM, Hwang PYH, Boiteux S, Hunter N. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol. Cell. 2010;40:1001–1015. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matos J, Blanco MG, Maslen S, Skehel JM, West SC. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berchowitz LE, Copenhaver GP. Genetic interference: don't stand so close to me. Curr. Genomics. 2010;11:91–102. doi: 10.2174/138920210790886835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Globus ST, Keeney S. The joy of six: how to control your crossovers. Cell. 2012;149:11–12. doi: 10.1016/j.cell.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 57.de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohli J, Bähler J. Homologous recombination in fission yeast—Absence of crossover interference and synaptonemal complex. Experientia. 1994;50:295–306. doi: 10.1007/BF01924013. [DOI] [PubMed] [Google Scholar]
- 59.Zalevsky J, Duffy JB, Kemphues KJ, Villeneuve AM. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153:1271–1283. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hillers KJ, Villeneuve AM. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 2003;13:1641–1647. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 61.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, Oneil NJ, Rose AM, West SC, Meyer BJ, Boulton SJ. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lorenz A, Osman F, Sun W, Nandi S, Steinacher R, Whitby MC. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science. 2012;336:1585–1588. doi: 10.1126/science.1220111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R. FANCM limits meiotic crossovers. Science. 2012;336:1588–1590. doi: 10.1126/science.1220381. [DOI] [PubMed] [Google Scholar]
- 66.Oh SD, Lao JP, Hwang PYH, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cromie GA, Hyppa RW, Smith GR. The fission yeast BLM homolog Rqh1 promotes meiotic recombination. Genetics. 2008;179:1157–1167. doi: 10.1534/genetics.108.088955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wicky C, Alpi A, Passannante M, Rose A, Gartner A, Muller F. Multiple genetic pathways involving the Caenorhabditis elegans Bloom's syndrome genes him-6, rad-51, and top-3 are needed to maintain genome stability in the germ line. Mol. Cell. Biol. 2004;24:5016–5027. doi: 10.1128/MCB.24.11.5016-5027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McVey M, Andersen SL, Broze Y, Sekelsky J. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics. 2007;176:1979–1992. doi: 10.1534/genetics.106.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sheridan S, Bishop DK. Red-Hed regulation: recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev. 2006;20:1685–1691. doi: 10.1101/gad.1447606. [DOI] [PubMed] [Google Scholar]
- 72.Latypov V, Rothenberg M, Lorenz A, Octobre G, Csutak O, Lehmann E, Loidl J, Kohli J. Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe. Mol. Cell. Biol. 2010;30:1570–1581. doi: 10.1128/MCB.00919-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Egel-Mitani M, Olson LW, Egel R. Meiosis in Aspergillus nidulans: another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas. 1982;97:179–187. doi: 10.1111/j.1601-5223.1982.tb00761.x. [DOI] [PubMed] [Google Scholar]
- 74.Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song XY, Gjoneska E, Ren QH, Taverna SD, Allis CD, Gorovsky MA. Phosphorylation of the SQ H2A.X motif is required for proper meiosis and mitosis in Tetrahymena thermophila. Mol. Cell. Biol. 2007;27:2648–2660. doi: 10.1128/MCB.01910-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cole ES, Cassidy-Hanley D, Hemish J, Tuan J, Bruns PJ. A mutational analysis of conjugation in Tetrahymena thermophila. 1. Phenotypes affecting early development: meiosis to nuclear selection. Dev. Biol. 1997;189:215–232. doi: 10.1006/dbio.1997.8648. [DOI] [PubMed] [Google Scholar]
- 77.Orias E, Cervantes MD, Hamilton E. Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res. Microbiol. 2011;162:578–586. doi: 10.1016/j.resmic.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bruns PJ, Brussard TEB. Nullisomic Tetrahymena: eliminating germinal chromosomes. Science. 1981;213:549–551. doi: 10.1126/science.213.4507.549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.