Abstract
The great ape families are the species most closely related to our own, comprising chimpanzees, bonobos, gorillas, and orangutans. They live exclusively in tropical rainforests in Central Africa and the islands of Southeast Asia. Due to their close evolutionary relationship with humans, great apes share many cognitive, physiological, and morphological similarities with humans. The members of the great ape family make obvious models to facilitate the further understanding about humans' biology and history. This review will discuss how the recent addition of genome-wide data from great apes has furthered humans' understanding of these species and humanity, especially in the realm of evolutionary genetics.
Keywords: conservation genetics, demography, evolution, great apes, natural selection, population size
Introduction
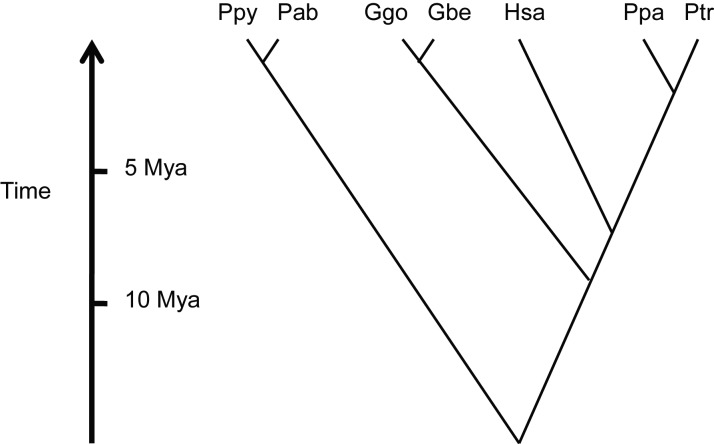
It has long been hypothesized that humans and the great apes share a recent common ancestry (Huxley 1863; Darwin 1871). This result has been confirmed by many molecular studies over the past several decades (Sibley and Ahlquist 1987; Goodman 1996; Satta et al. 2000; Chen and Li 2001). It is now known that common chimpanzees (Pan troglodytes) and bonobos (Pan paniscus) are our closest living relatives, followed by gorillas and orangutans (Figure 1). The sequence divergence at orthologous sites between humans and chimpanzees/bonobos is just 1.2% (Chimpanzee Sequencing and Analysis Consortium 2005; Prufer et al. 2012), which is much less than the divergence between two random fruit flies (Drosophila simulans) of the same species (Begun et al. 2007). Great apes are studied by geneticists for a wide variety of reasons, including intrinsic biological interest, interest in the horizontal transfer of pathogens such as HIV-1 (Keele et al. 2006), implications for conservation biology, and interest in recently evolved human-specific traits such as bipedality and large, complex brains. With assembled genome sequences now available from four different great ape species (Chimpanzee Sequencing and Analysis Consortium 2005; Locke et al. 2011; Prufer et al. 2012; Scally et al. 2012), as well as 100 additional genomes available for population genomic studies (Locke et al. 2011; Auton et al. 2012; Prado-Martinez et al. 2013), there is now an unprecedented ability to study the evolution of humanity's closest relatives. This review highlights how recent comparative studies have enhanced the knowledge of human genetics and human evolution. A comprehensive review is beyond the scope of this paper; rather, it highlights specific topics that represent major areas of current research.
Figure 1.
Schematic showing the evolutionary relationships and approximate divergence times among the great apes and humans. Ppy, Pongo pygmaeus; Pab, P. abelii; Ggo, Gorilla gorilla; Gbe, G. beringei; Hsa, Homo sapiens; Ppa, Pan paniscus; Ptr, P. troglodytes; Mya, Million years ago.
How Are Humans Unique?
By aligning the human reference sequence to the genomes of several closely related outgroup species, we can identify mutations that have occurred on the recent human branch and distinguish between ancestral and derived alleles at human single nucleotide polymorphisms (SNPs). Over the whole genome, these mutations occur at a roughly constant rate, consistent with the idea that the vast majority of substitutions are selectively neutral (i.e., nonfunctional) (Kimura 1983). Orangutans with a 2.4-fold higher sequence divergence (compared with chimpanzees) are expected to have a 2.4-fold longer divergence time from humans, in relation to the human-chimpanzee divergence time (Figure 1). Part of the challenge of identifying the genetic basis of human-specific morphological change is the difficulty in distinguishing between rare functional mutations and the more common selectively neutral mutations that separate humans from the great apes. For obvious ethical reasons, function can generally not be inferred in vivo. Rather, indirect computational techniques, as well as the utilization of a more distantly related animal model (e.g., mice and fruit flies) have been used to identify human-specific adaptations.
Coding Changes
One approach for identifying human-specific adaptions is to scan the exomes of humans (and various outgroups) to identify genes with patterns of genetic variation that are consistent with the action of recent natural selection. For example, Enard and colleagues (Enard et al. 2002) sequenced the FOXP2 gene, a forkhead class transcription factor, in humans, great apes, and mice. The gene is highly conserved among mammals, with just one non-synonymous (i.e., amino-acid changing) mutation separating the chimpanzee and mouse coding sequences. This conservation is not due to a low mutation rate, as there are 147 synonymous (i.e., non-amino acid changing) mutations between chimpanzees and mice. However, on the human-specific branch, there are two non-synonymous mutations and no synonymous mutations, which is significantly different from expectations (p<10−3, Fisher's exact test). Enard and colleagues interpreted this (and other evidence) as support for recent positive selection in the human lineage (Enard et al. 2002). Since mutations in FOXP2 can cause deficiencies in spoken language capacity, Enard and cohort speculated that the non-synonymous mutations in FOXP2 were related to humans' ability to develop complex forms of communication. Subsequent work has shown that both Neanderthals and Denisovans also contain these two recent non-synonymous mutations (Krause et al. 2007; Reich et al. 2010), though this observation can be interpreted in different ways (Coop et al. 2008; Ptak et al. 2009). Recent work has suggested that a mutation at a transcription factor binding site in intron 8 of the FOXP2 gene affects FOXP2 expression levels. These expression levels might have been recently selected for because they differ between modern humans and Neanderthals (Maricic et al. 2012).
Subsequently, researchers have searched more systematically through many different genes to look for a similar signal of increased human-specific, non-synonymous substitutions relative to synonymous substitutions; first using unrooted trees (Clark et al. 2003; Nielsen et al. 2005) and later with one or more outgroups (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007; Lindblad-Toh et al. 2011). These studies have identified functional categories (i.e., the genes involved in spermatogenesis) that are enriched in genes showing evidence for positive selection, but the biological insight gained from these broad surveys is somewhat limited. Deeper insight generally requires additional information from physiological, behavioral, or targeted genetic studies. For example, great ape species differ in the size and structure of their social groups—chimpanzees live in multi-male, multi-female groups; gorillas live in single male, multi-female groups; and orangutans are mostly solitary. These life-history traits suggest that different great ape species undergo different sexual selection pressures. In particular, since receptive chimpanzee females mate with multiple males (unlike gorilla and orangutan females), it is expected that sperm competition will be more important for chimpanzee males than for males of other great ape species. Both physiological (Harcourt et al. 1981) and genetic (Dorus et al. 2004) studies provide support for this hypothesis, and may provide a partial explanation for the prevalence of genes that are involved in spermatogenesis that appear to be affected by recent positive selection.
Another approach for detecting the action of recent natural selection on the human lineage involves comparing synonymous (i.e., non-amino-acid changing) versus non-synonymous (i.e., amino-acid changing) polymorphisms and substitutions. If a gene has been subject to recent positive (i.e., adaptive) selection, this can lead to the fixation of several non-synonymous mutations, and an excess of non-synonymous substitutions relative to non-synonymous polymorphisms (McDonald and Kreitman 1991). Conversely, under continued purifying selection most non-synonymous mutations will be deleterious, and very few will rise in frequency to become fixed differences. This will tend to produce a deficit of non-synonymous substitutions relative to non-synonymous polymorphisms. Bustamante and colleagues examined the patterns of polymorphism and divergence in thirty-nine humans (20 European-Americans and 19 African-Americans) and one chimpanzee at more than 11,000 different protein-coding genes (Bustamante et al. 2005). As with other studies, they found certain functional categories were overrepresented (or underrepresented) among genes that they had inferred to be recently selected for, but functional insight for specific genes was minimal.
Regulatory Changes
One potential limitation of the studies described above is that they are confined to protein-coding regions, which comprise <2% of the human genome. It is possible that most of the genetic changes that “make us human” involve regulatory sequences rather than coding sequences (King and Wilson 1975). To identify the noncoding regions that are important to recent human evolution, Pollard, Siepel, and colleagues developed a likelihood-ratio test to estimate the statistical significance of regions that show an apparent increase in the substitution rate on the human evolutionary branch (Siepel, et al. 2006). Siepel and cohort then applied this method to whole genome sequence data from humans, and several other vertebrates, to identify 202 regions that showed significant rates of human-specific evolution (Pollard, Salama, King et al. 2006), which they called human accelerated regions (HARs). The most significant of these regions, HAR1, has 18 substitutions in 118 base pairs (relative to chimpanzees) and contains part of a novel RNA gene that is expressed in the human neocortex from seven to nineteen gestational weeks (Pollard, Salama, Lambert et al. 2006). Though the specific function of HAR1 is not known, it is tempting to speculate that it is related to the extreme growth in size and complexity of human brains in relation to other apes.
Pollard, Salama, King et al. (2006) found the substitutions at HARs show a weak-to-strong bias (i.e., tend to be from A or T to G or C), and that HARs tend to be in regions of high recombination. Both of these observations are consistent with the possibility that GC-biased gene conversion may play an important role in the creation of HARs (Galtier and Duret 2007; Katzman et al. 2010).
A complementary approach to identifying important regulatory changes involves analyses of gene expression levels using microarrays, or large-scale sequencing of RNA (RNA-seq) using next-generation sequencing (Romero et al. 2012). One intriguing hypothesis is that humans' unique cognitive abilities are due to changes in gene expression patterns in the brain during development (Caceres et al. 2003; Nowick et al. 2009; Xu et al. 2010), and there is some evidence in favor of this idea (Somel et al. 2009). More generally, measurements of gene expression within individuals of the same species, and between individuals of different species, are informative about the current or past action of natural selection for specific expression levels. For example, if between-species levels of gene expression variation are no larger than within-species levels, stabilizing selection on gene regulation is a likely explanation (Gilad et al. 2006). In contrast, if a higher expression level is observed specifically in one species, then directional selection for differential expression in that species is most likely operating. Recent studies have shown that the regulation of a majority of the genes in the genome evolve under evolutionary constraint, consistent with widespread stabilizing selection (Lemos et al. 2005; Rifkin et al. 2005). In addition, a substantial fraction of human and great ape genes show species-specific changes in gene expression levels that are best explained by recent positive selection (Gilad et al. 2006; Blekhman et al. 2008). Finally, it is important to note that gene expression levels are only an intermediate phenotype, and evidence of differential expression across species provides only a starting point for targeted studies to determine the genetic mechanism causing the difference and the function consequences (if any).
Gain or Loss of Genes
A related approach focuses on identifying and characterizing the gain or loss of genes in the human lineage subsequent to the divergence of humans and chimpanzees. Olson proposed that gene loss could be an important mechanism in the evolution of human-specific traits (Olson 1999). However, further studies have found that the rate of gene loss in the human-specific lineage is the same as the comparable rate in other great ape lineages (Kim et al. 2010; Prado-Martinez et al. 2013), suggesting that humans are not unusual in their rate of genic turnover. While this does not disprove Olson's hypothesis, it suggests that whatever role gene loss plays in species-specific traits is likely to be a general (rather than human-specific) phenomenon. In a few cases, more detailed studies have highlighted potential selective explanations for complete (Stedman et al. 2004) or partial (Xue et al. 2006) loss of genes in humans. Conversely, while several human-specific genes have been identified (Knowles and McLysaght 2009; Wu et al. 2011), most of these genes are of unknown function (but see [Buhl et al. 2006; Hu et al. 2012]), making their relevance to recent human evolution difficult to quantify.
Structural Changes
Humans have a diploid chromosome number of 2n = 46, while the great apes have 2n = 48 chromosomes (Yunis and Prakash 1982). Evolutionary genetic studies of the human chromosome 2 have highlighted additional evidence that this chromosome was formed by a fusion of two autosomes within the past several million years (Dreszer et al. 2007; Auton et al. 2012; Ventura et al. 2012). Additional human-specific inversions (Yunis and Prakash 1982) and copy number variations (Dumas et al. 2007) were identified by conventional banding techniques, while more recent array-based comparative genomic hybridization experiments (Linardopoulou et al. 2005; Goidts et al. 2006; Wilson et al. 2006) and next-generation sequencing studies (Alkan et al. 2009) have identified many more large rearrangements and segmental duplications that are unique to the human lineage. When all forms of variability are taken into account, the estimated divergence between humans and chimpanzees increases from 1.2% (single nucleotide polymorphisms at aligned orthologous sites) to ∼5% (Britten 2002). It is unclear what, if any, functional relevance many of these structural changes have in humans, though it may be relevant that regions near cytogenetically visible changes in the human genome are enriched for recent gene duplications and new genes (Fortna et al. 2004; Dumas et al. 2007).
Comparative Demography
The amount of divergence between two DNA sequences is, on average, directly proportional to the time since they last shared a common ancestor. By comparing patterns of genetic variation across multiple individuals from different populations, it is possible to make inferences about a species' demographic history including population migration, changes in population size, geographic structure, and admixture between diverged populations. By inferring demographic parameters in both humans and great apes, it can be determined if particular observations that are found in one species are common or unusual. This, in turn, allows for a more realistic assessment of the ways in which the human species might be unique.
Population Size
The current census size for the human population tops out at more than 7 billion individuals and continues to grow very rapidly. However, it is known that even as recently as several centuries ago, the human population size was orders of magnitude smaller. In fact, genetic studies have consistently found that two randomly chosen human chromosomes tend to differ from each other at roughly 1 in a 1000 base pairs, which is consistent with a long-term effective population size (Ne) of 10,000–15,000 (Li and Sadler 1991; Harding et al. 1997; Frisse et al. 2001). Here, Ne refers to the size of a randomly mating population that is expected to have the same levels of genetic diversity as what is observed. Although there are many reasons why Ne might be less than the census population size (Caballero 1994), the difference in magnitude is still immense, and requires explanation. The current belief is that for much of human history, the population sizes were much smaller, and it is only very recently (i.e., within the past 20-30 thousand years) that the population sizes have increased rapidly (Pluzhnikov et al.2002; Voight et al. 2005; Gutenkunst et al. 2009). Recent studies have identified an excess of very rare single nucleotide variants (minor allele frequency<0.1 %), consistent with explosive population growth over the past several thousand years, starting subsequent to the development and spread of agriculture (Coventry et al. 2010; Nelson et al. 2012).
In contrast, extant great apes are found exclusively in Old World tropical rain forests, and their ranges (and estimated census population sizes) have decreased dramatically over the past 200 years (Junker et al. 2012). While initial estimates of great ape genetic diversity from microsatellites were low (Wise et al. 1997; Cooper et al. 1998), subsequent studies of nuclear sequence variation have found that all great ape species, except for bonobos, have levels of genetic variation substantially higher than the levels of human genetic variation (Kaessmann et al. 2001; Fischer et al. 2006; Locke et al. 2011; Prado-Martinez et al. 2013). Since the underlying mutation rates are unlikely to be different between humans and great apes, the differences can be attributed to chimpanzees, gorillas, and orangutans having larger long-term Ne's than humans have. This finding highlights the value of a comparative demographic approach, as it emphasizes that the current human dominance (both in range and population size) is very recent—even though humans' ancestors had populated Eurasia and Africa for almost 2 million years, while the great apes have been confined to a much smaller area during that time. Most likely, there were more chimpanzees, gorillas, and orangutans around at any time prior to 50,000 years ago than there were humans.
Studies that estimate ancestral (effective) population sizes (Na) provide additional evidence that humans' current Ne is unusually low. Takahata (1993) examined genetic diversity in the human major histocompatibility complex (MHC), and concluded that the diversity was consistent with a Na of ∼ 100,000. Other approaches, such as using the variance in human-chimpanzee divergence across the genome, the discordance of gene trees across the genome, or maximum-likelihood techniques, have estimated Na≥50,000 for the population ancestral to humans and chimpanzees (Takahata and Satta 1997; Chen and Li 2001; O'hUigin et al. 2002; Wall 2003; Burgess and Yang 2008). While estimates of Na can be inflated due to past population structure, the basic picture appears to be that almost all extant and ancestral ape populations have (or had) substantially more genetic variation (and larger effective population sizes) than extant modern humans.
Population Structure
An additional way of viewing the low Ne in humans is the idea that all extant people are genetically similar when compared with the great apes. Another facet of this idea is that all great apes consist of groups of diverged populations that are taxonomically considered to be separate species or sub-species (Groves 2006). Currently, there are the chimpanzee and bonobo (P. troglodytes and P. paniscus respectively), two gorilla species (Gorilla gorilla and Gorilla beringei), and two orangutan species (Pongo pygmaeus and Pongo abelii), and additional subspecies from each group, compared with just a single extant species and subspecies of Homo sapiens sapiens. Sister great ape species within the same genus can interbreed and produce fertile offspring in zoos, but not in the wild where their ranges generally do not overlap. The taxonomic differences between great ape populations are not purely semantic though, in that the genetic distance between great ape species is substantially larger than the genetic distance between isolated human groups (Fischer et al. 2006; Wall et al. 2008). Broadly speaking, the existence of multiple diverged populations that can admix and interbreed is the general pattern in catarrhine primates, including gibbons, macaques, baboons, vervets, and guenons (Jolly 2001; Arnold and Meyer 2006), with humans and a few isolated species (e.g., the siamang) as outliers. It is unclear why this is so, but perhaps some aspect of life-history behavior, mate choice, or migration facilitates the isolation (and genetic divergence) of nearby populations. Interestingly, just 50,000 years ago, the human species coexisted with at least three other archaic human groups, including Neanderthals, Denisovans, and Homo floresiensis (Wall and Slatkin 2012), and potentially several other diverged populations (Harvati et al. 2011; Curnoe et al. 2012). H. floresiensis coexisted with modern humans in island Southeast Asia as recently as 12,000 years ago (Morwood et al. 2005). Perhaps not coincidentally, these archaic human groups all disappeared around the same time as modern human populations started to increase rapidly in size.
X Versus Autosome Comparisons
Comparative studies of humans and great apes can also shed light on sex-specific differences, and life-history traits in each species. In a randomly mating population with equal numbers of males and females, there are three X chromosomes for every four autosomes, leading to a 3:4 ratio in expected levels of diversity in X versus autosome comparisons. If a species is generally polygynous, there will be more breeding females than breeding males, which tends to increase the ratio of X-to-autosome diversity (Caballero 1994). Gorillas, who are strongly polygynous, are expected to have increased relative levels of X-linked diversity when compared with humans (weakly polygynous) and gibbons (monogamous). Concurrently recent positive selection, which decreases levels of genetic variation, is expected to affect the X chromosome more than the autosomes due to hemizygosity in males and the recessivity of some advantageous alleles. Analyses of human polymorphism data show the effects of both of these evolutionary processes (Hammer et al. 2008; Hammer et al. 2010; Keinan et al. 2009). Overall, the ratio of X-to-autosome diversity is much less than 0.75, which is consistent with the stronger effect of selection on the X chromosome. However, if the analysis is confined to regions far away from genes, where natural selection plays less of a role, the situation is reversed, with X-to-autosome ratios significantly higher than 0.75. A full analysis of whole genome sequencing data from the Great Ape Genomes Project (Prado-Martinez et al. 2013) is currently in process, and the results will be informative about the relative roles of natural selection, mating system, and sex-specific migration in shaping genome-wide patterns of genetic variation.
Comparative Population Genetics
As hinted at above, between-species comparisons can also be informative about other population genetic processes such as natural selection, mutation, and recombination. Two examples are discussed in greater detail below.
Recombination rates vary dramatically between individuals and between different regions of the genome. In particular, it is now well established that most recombination events occur in narrow 1–2 kilobase (kb) wide “hot spots”, with almost two-thirds of all crossovers occurring in 10% of the human genome (Myers et al. 2005). Roughly 40% of these recombination hot spots contain a (partially degenerate) thirteen base pair sequence motif, which matches the predicted binding site of the PRDM9 gene, and is thought to be involved in double strand break formation (Baudat et al. 2010; Myers et al. 2010). PRDM9 evolves rapidly in metazoans (Oliver et al. 2009), and specific single nucleotide variants have been shown to strongly affect male recombination rates (Berg et al. 2010).
Recent studies of recombination in chimpanzees have shown that humans and chimpanzees generally do not share recombination hot spots (Wall et al. 2003; Ptak et al. 2004; Winckler et al. 2005; Auton et al. 2012), but most hot spots are shared across different human populations. Further, there is not a simple sequence motif that appears to be associated with the location of chimpanzee hot spots (Auton et al. 2012). This situation is not surprising, since there is a single dominant PRDM9 allele at high frequency in humans (corresponding to the thirteen base pair [bp] sequence motif found near human recombination hot spots) but many low frequency PRDM9 alleles in chimpanzees. Even if PRDM9 plays a primary role in hot spot localization, it appears that the allelic diversity at PRDM9 in chimpanzees promotes a wide array of different hot spot locations in different chimpanzee individuals. Since large-scale detection of recombination hot spots is based on patterns of linkage disequilibrium, which reflect long-term average rates of recombination, it is unlikely that a particular sequence motif will be associated with hot spots unless there is a dominant (i.e., high-frequency) PRDM9 allele. It remains to be seen how common hot spot motifs are, but ongoing studies of recombination rates in all great ape species will provide a partial answer to this question (L Stevison and J Wall, unpublished data).
Whole genome sequence polymorphism data in great apes also help facilitate the search for polymorphisms that are shared by descent between multiple species. Theory suggests that such ancient shared polymorphisms are unlikely to arise by chance (Leffler et al. 2013), and are more likely due to long-lived balancing or frequency-dependent selection (e.g., selection for rare alleles, or selection for heterozygotes). One well-known example of ancient balancing selection is from the MHC, which has a primary role in the active immune system (Klein et al. 1993). Mutations in the MHC have been shown to be present before the split of humans from the great apes, and to persist as polymorphisms in multiple species due to selection favoring heterozygous genotypes for over 12 million years (Ayala 1995). Beyond this one well-known example, the evidence for ancient balancing selection is rare, in part because of the difficulty in distinguishing mutations that are identical by descent (i.e., arose before the split of humans and chimpanzees) from those that are identical by state (i.e., parallel mutations that happened at the same nucleotide site in multiple species). Whole genome sequence data makes it possible to distinguish between the two by looking for multi-SNP haplotypes shared across species (Leffler et al. 2013). Leffler and colleagues utilized complete genome sequences from fifty-nine humans and ten chimpanzees to identify 125 regions with haplotypes shared between humans and chimpanzees. These regions are significantly enriched for membrane glycoproteins, suggesting that genes involved in host-pathogen interactions are more likely to have been subject to ancient balancing selection. This idea is in keeping with previous studies of ancient balancing selection in humans (Klein et al. 1993; Segurel et al. 2012) as well as more recent balancing selection in humans and other species (Stahl et al. 1999; Hedrick 2011).
Great Apes as Reservoirs of Disease
One area where great ape genomics is of direct relevance to human health involves infectious diseases that are shared across species (i.e., zoonosis). The most well known example concerns HIV-1, a strain of human immunodeficiency virus that is pandemic in humans (Sharp and Hahn 2011). Initial genetic studies of HIV found that the disease was closely related to other lentiviruses found in African apes and monkeys. It has been shown that HIV-1 is similar to SIVcpz, a simian immunodeficiency virus found in the central chimpanzee Pan troglodytes troglodytes (Huet et al. 1990). Subsequent studies of natural chimpanzee populations confirmed P. t. troglodytes as a natural reservoir of HIV-1, and indicated that there had been multiple transmission events of HIV from chimpanzees to humans (Keele et al. 2006). Chimpanzees in turn are thought to have acquired SIVcpz relatively recently from eating infected monkeys (mangabeys, guenons, or both). There is some evidence that chimpanzees infected with SIVcpz can develop AIDS (Sharp and Hahn 2011), but there is a possibility that studies of how chimpanzees (and gorillas who are infected by SIVgor) physiologically respond to SIV may provide some insight into how humans can treat current HIV-1 infections.
Conservation Biology
One of the primary motivations for studying great ape demography is to help inform advocacy and policy decisions for promoting the conservation, health, and genetic diversity of our evolutionary cousins. All great ape species are considered to be Endangered or Critically Endangered by the International Union for Conservation of Nature (IUCN 2012) due to the recent drastic reduction of their rainforest habitat (Junker et al. 2012). From a conservation perspective, the priority is obviously to increase (or maintain) suitable habitats for great apes, and to eliminate (or reduce) human-mediated mortality, but there is an important role for genomics in helping conservation biologists focus their efforts. DNA-based studies can help identify populations that are genetically distinct from all others, which can elevate the importance of preserving them as a previously unknown reservoir of genetic diversity. For example, a study of orangutan mtDNA helped the recognition of the Sumatran orangutan as a separate species (Xu and Arnason 1996), and a study of autosomal SNP variation argued for the creation of a fourth chimpanzee subspecies, Pan troglodytes ellioti (Bowden et al. 2012). Genetic studies have also estimated the extent of recent population decline in orangutans (Goossens et al. 2006) and chimpanzees (Campbell et al. 2008), and identified important evidence for long-range migration in the Sumatran orangutan (Nater et al. 2013). Finally, additional analyses are possible with widespread whole-genome sequence data in great apes. To some extent, the provenance of captive animals can be inferred from large-scale genetic data, and recent work suggests that most captive individuals are genetically admixed from two, or more, distinct wild-born populations (Prado-Martinez et al. 2013). Prado-Martinez and colleagues also utilized runs of homozygosity (McQuillan et al. 2008) to quantify the extent of inbreeding in different great ape species. Bonobos had large homozygous tracts similar to isolated human groups that are known to have small population sizes (e.g., the Karitiana from Brazil), suggesting that inbreeding is an ongoing concern in extant bonobo populations.
Conclusions
Given the dire situation that all great ape populations currently face, there is added motivation for researchers to learn as much as possible from their genomes, both to provide additional justification for conservation efforts and because such studies may be impossible in the not-too-distant future. As highlighted above, great ape genomic studies have already been highly informative in regards to human biology, human history, and the effects of natural selection. The great apes are likely to remain of critical importance for future human genetic studies, even without the benefits of manipulative studies that are used in model organisms such as the macaque or the mouse.
Acknowledgments
This work was supported in part by NIH grant R01 HG005226.
References
- Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, Sahinalp SC, Gibbs RA, Eichler EE. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML, Meyer A. Natural hybridization in primates: One evolutionary mechanism. Zoology (Jena) 2006;109:261–276. doi: 10.1016/j.zool.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Auton A, Fledel-Alon A, Pfeifer S, Venn O, Segurel L, Street T, Leffler EM, Bowden R, Aneas I, Broxholme J, Humburg P, Iqbal Z, Lunter G, Maller J, Hernandez RD, Melton C, Venkat A, Nobrega MA, Bontrop R, Myers S, Donnelly P, Przeworski M, McVean G. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala FJ. The myth of Eve: Molecular biology and human origins. Science. 1995;270:1930–1936. doi: 10.1126/science.270.5244.1930. [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh YP, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, Pachter L, Myers E, Langley CH. Population genomics: Whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050310. e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000271. e1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden R, MacFie TS, Myers S, Hellenthal G, Nerrienet E, Bontrop RE, Freeman C, Donnelly P, Mundy NI. Genomic tools for evolution and conservation in the chimpanzee: Pan troglodytes ellioti is a genetically distinct population. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002504. e1002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ. Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels. Proc Natl Acad Sci U S A. 2002;99:13633–13635. doi: 10.1073/pnas.172510699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhl AM, Jurlander J, Jorgensen FS, Ottesen AM, Cowland JB, Gjerdrum LM, Hansen BV, Leffers H. Identification of a gene on chromosome 12q22 uniquely overexpressed in chronic lymphocytic leukemia. Blood. 2006;107:2904–2911. doi: 10.1182/blood-2005-07-2615. [DOI] [PubMed] [Google Scholar]
- Burgess R, Yang Z. Estimation of hominoid ancestral population sizes under bayesian coalescent models incorporating mutation rate variation and sequencing errors. Mol Biol Evol. 2008;25:1979–1994. doi: 10.1093/molbev/msn148. [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, Civello D, Adams MD, Cargill M, Clark AG. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Caballero A. Developments in the prediction of effective population size. Heredity (Edinb) 73 (Pt 6) 1994:657–679. doi: 10.1038/hdy.1994.174. [DOI] [PubMed] [Google Scholar]
- Caceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci U S A. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G, Kuehl H, N'Goran Kouame P, Boesch C. Alarming decline of West African chimpanzees in Cote d'Ivoire. Curr Biol. 2008;18:R903–904. doi: 10.1016/j.cub.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, Ferriera S, Wang G, Zheng X, White TJ, Sninsky JJ, Adams MD, Cargill M. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Coop G, Bullaughey K, Luca F, Przeworski M. The timing of selection at the human FOXP2 gene. Mol Biol Evol. 2008;25:1257–1259. doi: 10.1093/molbev/msn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G, Rubinsztein DC, Amos W. Ascertainment bias cannot entirely account for human microsatellites being longer than their chimpanzee homologues. Hum Mol Genet. 1998;7:1425–1429. doi: 10.1093/hmg/7.9.1425. [DOI] [PubMed] [Google Scholar]
- Coventry A, Bull-Otterson LM, Liu X, Clark AG, Maxwell TJ, Crosby J, Hixson JE, Rea TJ, Muzny DM, Lewis LR, Wheeler DA, Sabo A, Lusk C, Weiss KG, Akbar H, Cree A, Hawes AC, Newsham I, Varghese RT, Villasana D, Gross S, Joshi V, Santibanez J, Morgan M, Chang K, Iv WH, Templeton AR, Boerwinkle E, Gibbs R, Sing CF. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat Commun. 2010;1:131. doi: 10.1038/ncomms1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curnoe D, Xueping J, Herries AI, Kanning B, Tacon PS, Zhende B, Fink D, Yunsheng Z, Hellstrom J, Yun L, Cassis G, Bing S, Wroe S, Shi H, Parr WC, Shengmin H, Rogers N. Human remains from the Pleistocene-Holocene transition of southwest China suggest a complex evolutionary history for East Asians. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031918. e31918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. London, England: John Murray; 1871. The Descent of Man, and Selection in Relation to Sex. [Google Scholar]
- Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT. Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat Genet. 2004;36:1326–1329. doi: 10.1038/ng1471. [DOI] [PubMed] [Google Scholar]
- Dreszer TR, Wall GD, Haussler D, Pollard KS. Biased clustered substitutions in the human genome: The footprints of male-driven biased gene conversion. Genome Res. 2007;17:1420–1430. doi: 10.1101/gr.6395807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L, Kim YH, Karimpour-Fard A, Cox M, Hopkins J, Pollack JR, Sikela JM. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17:1266–1277. doi: 10.1101/gr.6557307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- Fischer A, Pollack J, Thalmann O, Nickel B, Paabo S. Demographic history and genetic differentiation in apes. Curr Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, Karimpour-Fard A, Glueck D, McGavran L, Berry R, Pollack J, Sikela JM. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet. 2001;69:831–843. doi: 10.1086/323612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23:273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006;440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- Goidts V, Armengol L, Schempp W, Conroy J, Nowak N, Muller S, Cooper DN, Estivill X, Enard W, Szamalek JM, Hameister H, Kehrer-Sawatzki H. Identification of large-scale human-specific copy number differences by inter-species array comparative genomic hybridization. Hum Genet. 2006;119:185–198. doi: 10.1007/s00439-005-0130-9. [DOI] [PubMed] [Google Scholar]
- Goodman M. Epilogue: A personal account of the origins of a new paradigm. Mol Phylogenet Evol. 1996;5:269–285. doi: 10.1006/mpev.1996.0021. [DOI] [PubMed] [Google Scholar]
- Goossens B, Chikhi L, Ancrenaz M, Lackman-Ancrenaz I, Andau P, Bruford MW. Genetic signature of anthropogenic population collapse in orang-utans. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040025. e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves CP. Order Primates. In: DE Wilson, DM Reeder., editors. Mammal Species of the World. Baltimore: Johns Hopkins University Press. p. 111–184; 2006. [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000695. e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Mendez FL, Cox MP, Woerner AE, Wall JD. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000202. e1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Woerner AE, Mendez FL, Watkins JC, Cox MP, Wall JD. The ratio of human X chromosome to autosome diversity is positively correlated with genetic distance from genes. Nat Genet. 2010;42:830–831. doi: 10.1038/ng.651. [DOI] [PubMed] [Google Scholar]
- Harcourt AH, Harvey PH, Larson SG, Short RV. Testis weight, body weight and breeding system in primates. Nature. 1981;293:55–57. doi: 10.1038/293055a0. [DOI] [PubMed] [Google Scholar]
- Harding RM, Fullerton SM, Griffiths RC, Clegg JB. A gene tree for beta-globin sequences from Melanesia. J Mol Evol 44 Suppl. 1997;1 doi: 10.1007/pl00000063. pS133-138. [DOI] [PubMed] [Google Scholar]
- Harvati K, Stringer C, Grun R, Aubert M, Allsworth-Jones P, Folorunso CA. PLoS One. Vol. 6. Nigeria: Morphology and chronology; 2011. The Later Stone Age calvaria from Iwo Eleru. e24024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. Population genetics of malaria resistance in humans. Heredity (Edinb) 2011;107:283–304. doi: 10.1038/hdy.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HY, He L, Fominykh K, Yan Z, Guo S, Zhang X, Taylor MS, Tang L, Li J, Liu J, Wang W, Yu H, Khaitovich P. Evolution of the human-specific microRNA miR-941. Nat Commun. 2012;3:1145. doi: 10.1038/ncomms2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- Huxley TH. Evidence as to Man's Place in Nature. London England: Williams and Norgate; 1863. [Google Scholar]
- IUCN. International Union for Conservation of Nature Red List. 2012 [Google Scholar]
- Jolly CJ. A proper study for mankind: Analogies from the Papionin monkeys and their implications for human evolution. Am J Phys Anthropol Suppl. 2001;33:177–204. doi: 10.1002/ajpa.10021. [DOI] [PubMed] [Google Scholar]
- Junker J, Blake S, Boesch C, Campbell G, du Toit L, Duvall C, Ekobo A, Etoga G, Galat-Luong A, Gamys J, Ganas-Swaray J, Gatti S, Ghiurghi A, Granier N, Hart J, Head J, Herbinger I, Hicks TC, Huijbregts B, Imong IS, Kuempel N, Lahm S, Lindsell J, Maisels F, McLennan M, Martinez L, Morgan B, Morgan D, Mulindahabi F, Mundry R, N'Goran KP, Normand E, Ntongho A, Okon DT, Petre CA, Plumptre A, Rainey H, Regnaut S, Sanz C, Stokes E, Tondossama A, Tranquilli S, Sunderland-Groves J, Walsh P, Warren Y, Williamson EA, Kuehl HS. Recent decline in suitable environmental conditions for African great apes. Diversity and Distributions. 2012;18:1077–1091. [Google Scholar]
- Kaessmann H, Wiebe V, Weiss G, Paabo S. Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nat Genet. 2001;27:155–156. doi: 10.1038/84773. [DOI] [PubMed] [Google Scholar]
- Katzman S, Kern AD, Pollard KS, Salama SR, Haussler D. GC-biased evolution near human accelerated regions. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000960. e1000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Mullikin JC, Patterson N, Reich D. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nat Genet. 2009;41:66–70. doi: 10.1038/ng.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HL, Igawa T, Kawashima A, Satta Y, Takahata N. Divergence, demography and gene loss along the human lineage. Philos Trans R Soc Lond B Biol Sci. 2010;365:2451–2457. doi: 10.1098/rstb.2010.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Cambridge. United Kingdom: Cambridge University Press; 1983. The Neutral Theory of Molecular Evolution. [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Klein J, Satta Y, Takahata N, O'hUigin C. Trans-specific Mhc polymorphism and the origin of species in primates. J Med Primatol. 1993;22:57–64. [PubMed] [Google Scholar]
- Knowles DG, McLysaght A. Recent de novo origin of human protein-coding genes. Genome Res. 2009;19:1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J, Lalueza-Fox C, Orlando L, Enard W, Green RE, Burbano HA, Hublin JJ, Hanni C, Fortea J, de la Rasilla M, Bertranpetit J, Rosas A, Paabo S. The derived FOXP2 variant of modern humans was shared with Neandertals. Curr Biol. 2007;17:1908–1912. doi: 10.1016/j.cub.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Leffler EM, Gao Z, Pfeifer S, Segurel L, Auton A, Venn O, Bowden R, Bontrop R, Wall JD, Sella G, Donnelly P, McVean G, Przeworski M. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 2013;339:1578–1582. doi: 10.1126/science.1234070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Meiklejohn CD, Caceres M, Hartl DL. Rates of divergence in gene expression profiles of primates, mice, and flies: Stabilizing selection and variability among functional categories. Evolution. 2005;59:126–137. [PubMed] [Google Scholar]
- Li WH, Sadler LA. Low nucleotide diversity in man. Genetics. 1991;129:513–523. doi: 10.1093/genetics/129.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou EV, Williams EM, Fan Y, Friedman C, Young JM, Trask BJ. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, Ward LD, Lowe CB, Holloway AK, Clamp M, Gnerre S, Alfoldi J, Beal K, Chang J, Clawson H, Cuff J, Di Palma F, Fitzgerald S, Flicek P, Guttman M, Hubisz MJ, Jaffe DB, Jungreis I, Kent WJ, Kostka D, Lara M, Martins AL, Massingham T, Moltke I, Raney BJ, Rasmussen MD, Robinson J, Stark A, Vilella AJ, Wen J, Xie X, Zody MC, Broad Institute Sequencing Platform and Whole Genome Assembly Team, Baldwin J, Bloom T, Chin CW, Heiman D, Nicol R, Nusbaum C, Young S, Wilkinson J, Worley KC, Kovar CL, Muzny DM, Gibbs RA, Baylor College of Medicine Human Genome Sequencing Center Sequencing Team, Cree A, Dihn HH, Fowler G, Jangiani S, Joshi V, Lee S, Lewis LR, Nazareth LV, Okwuonu G, Santibanez J, Warren WC, Mardis ER, Weinstock GM, Wilson RK, Genome Institute at Washington University, Delehaunty K, Dooling D, Fronik C, Fulton L, Fulton B, Graves T, Minx P, Sodergren E, Birney E, Margulies EH, Herrero J, Green ED, Haussler D, Siepel A, Goldman N, Pollard KS, Pedersen JS, Lander ES, Keller M. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, Mitreva M, Cook L, Delehaunty KD, Fronick C, Schmidt H, Fulton LA, Fulton RS, Nelson JO, Magrini V, Pohl C, Graves TA, Markovic C, Cree A, Dinh HH, Hume J, Kovar CL, Fowler GR, Lunter G, Meader S, Heger A, Ponting CP, Marques-Bonet T, Alkan C, Chen L, Cheng Z, Kidd JM, Eichler EE, White S, Searle S, Vilella AJ, Chen Y, Flicek P, Ma J, Raney B, Suh B, Burhans R, Herrero J, Haussler D, Faria R, Fernando O, Darré F, Farré D, Gazave E, Oliva M, Navarro A, Roberto R, Capozzi O, Archidiacono N, Della Valle G, Purgato S, Rocchi M, Konkel MK, Walker JA, Ullmer B, Batzer MA, Smit AF, Hubley R, Casola C, Schrider DR, Hahn MW, Quesada V, Puente XS, Ordoñez GR, López-Otín C, Vinar T, Brejova B, Ratan A, Harris RS, Miller W, Kosiol C, Lawson HA, Taliwal V, Martins AL, Siepel A, Roychoudhury A, Ma X, Degenhardt J, Bustamante CD, Gutenkunst RN, Mailund T, Dutheil JY, Hobolth A, Schierup MH, Ryder OA, Yoshinaga Y, de Jong PJ, Weinstock GM, Rogers J, Mardis ER, Gibbs RA, Wilson RK. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic T, Gunther V, Georgiev O, Gehre S, Curlin M, Schreiweis C, Naumann R, Burbano HA, Meyer M, Lalueza-Fox C, de la Rasilla M, Rosas A, Gajovic S, Kelso J, Enard W, Schaffner W, Pääbo S. A Recent Evolutionary Change Affects a Regulatory Element in the Human FOXP2 Gene. Mol Biol Evol. 2013;4:844–852. doi: 10.1093/molbev/mss271. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, Smolej-Narancic N, Janicijevic B, Polasek O, Tenesa A, Macleod AK, Farrington SM, Rudan P, Hayward C, Vitart V, Rudan I, Wild SH, Dunlop MG, Wright AF, Campbell H, Wilson JF. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morwood MJ, Brown P, Jatmiko, Sutikna T, Saptomo EW, Westaway KE, Due RA, Roberts RG, Maeda T, Wasisto S, Djubiantono T. Further evidence for small-bodied hominins from the Late Pleistocene of Flores, Indonesia. Nature. 2005;437:1012–1017. doi: 10.1038/nature04022. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nater A, Arora N, Greminger MP, van Schaik CP, Singleton I, Wich SA, Fredriksson G, Perwitasari-Farajallah D, Pamungkas J, Krutzen M. Marked population structure and recent migration in the critically endangered Sumatran orangutan (Pongo abelii) J Hered. 2013;104:2–13. doi: 10.1093/jhered/ess065. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Shen J, Tang Z, Bacanu SA, Fraser D, Warren L, Aponte J, Zawistowski M, Liu X, Zhang H, Zhang Y, Li J, Li Y, Li L, Woollard P, Topp S, Hall MD, Nangle K, Wang J, Abecasis G, Cardon LR, Zöllner S, Whittaker JC, Chissoe SL, Novembre J, Mooser V. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz MJ, Fledel-Alon A, Tanenbaum DM, Civello D, White TJ, J Sninsky J, Adams MD, Cargill M. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030170. e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, Gernat T, Almaas E, Stubbs L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci U S A. 2009;106:22358–22363. doi: 10.1073/pnas.0911376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'hUigin C, Satta Y, Takahata N, Klein J. Contribution of homoplasy and of ancestral polymorphism to the evolution of genes in anthropoid primates. Mol Biol Evol. 2002;19:1501–1513. doi: 10.1093/oxfordjournals.molbev.a004213. [DOI] [PubMed] [Google Scholar]
- Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Ponting CP. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000753. e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MV. When less is more: Gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluzhnikov A, Di Rienzo A, Hudson RR. Inferences about human demography based on multilocus analyses of noncoding sequences. Genetics. 2002;161:1209–1218. doi: 10.1093/genetics/161.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, Rosenbloom KR, Kent J, Haussler D. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020168. e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M, Jr, Vanderhaeghen P, Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O'Connor TD, Santpere G, Cagan A, Theunert C, Casals F, Laayouni H, Munch K, Hobolth A, Halager AE, Malig M, Hernandez-Rodriguez J, Hernando-Herraez I, Prüfer K, Pybus M, Johnstone L, Lachmann M, Alkan C, Twigg D, Petit N, Baker C, Hormozdiari F, Fernandez-Callejo M, Dabad M, Wilson ML, Stevison L, Camprubí C, Carvalho T, Ruiz-Herrera A, Vives L, Mele M, Abello T, Kondova I, Bontrop RE, Pusey A, Lankester F, Kiyang JA, Bergl RA, Lonsdorf E, Myers S, Ventura M, Gagneux P, Comas D, Siegismund H, Blanc J, Agueda-Calpena L, Gut M, Fulton L, Tishkoff SA, Mullikin JC, Wilson RK, Gut IG, Gonder MK, Ryder OA, Hahn BH, Navarro A, Akey JM, Bertranpetit J, Reich D, Mailund T, Schierup MH, Hvilsom C, Andrés AM, Wall JD, Bustamante CD, Hammer MF, Eichler EE, Marques-Bonet T. Great ape genetic diversity and population history. Nature. 2013;449:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, Munch K, Hellmann I, Akagi K, Miller JR, Walenz B, Koren S, Sutton G, Kodira C, Winer R, Knight JR, Mullikin JC, Meader SJ, Ponting CP, Lunter G, Higashino S, Hobolth A, Dutheil J, Karakoç E, Alkan C, Sajjadian S, Catacchio CR, Ventura M, Marques-Bonet T, Eichler EE, André C, Atencia R, Mugisha L, Junhold J, Patterson N, Siebauer M, Good JM, Fischer A, Ptak SE, Lachmann M, Symer DE, Mailund T, Schierup MH, Andrés AM, Kelso J, Pääbo S. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak SE, Enard W, Wiebe V, Hellmann I, Krause J, Lachmann M, Paabo S. Linkage disequilibrium extends across putative selected sites in FOXP2. Mol Biol Evol. 2009;26:2181–2184. doi: 10.1093/molbev/msp143. [DOI] [PubMed] [Google Scholar]
- Ptak SE, Roeder AE, Stephens M, Gilad Y, Paabo S, Przeworski M. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020155. e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, Maricic T, Good JM, Marques-Bonet T, Alkan C, Fu Q, Mallick S, Li H, Meyer M, Eichler EE, Stoneking M, Richards M, Talamo S, Shunkov MV, Derevianko AP, Hublin JJ, Kelso J, Slatkin M, Pääbo S. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Houle D, Kim J, White KP. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438:220–223. doi: 10.1038/nature04114. [DOI] [PubMed] [Google Scholar]
- Romero IG, Ruvinsky I, Gilad Y. Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet. 2012;13:505–516. doi: 10.1038/nrg3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satta Y, Klein J, Takahata N. DNA archives and our nearest relative: The trichotomy problem revisited. Mol Phylogenet Evol. 2000;14:259–275. doi: 10.1006/mpev.2000.0704. [DOI] [PubMed] [Google Scholar]
- Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, McCarthy S, Montgomery SH, Schwalie PC, Tang YA, Ward MC, Xue Y, Yngvadottir B, Alkan C, Andersen LN, Ayub Q, Ball EV, Beal K, Bradley BJ, Chen Y, Clee CM, Fitzgerald S, Graves TA, Gu Y, Heath P, Heger A, Karakoc E, Kolb-Kokocinski A, Laird GK, Lunter G, Meader S, Mort M, Mullikin JC, Munch K, O'Connor TD, Phillips AD, Prado-Martinez J, Rogers AS, Sajjadian S, Schmidt D, Shaw K, Simpson JT, Stenson PD, Turner DJ, Vigilant L, Vilella AJ, Whitener W, Zhu B, Cooper DN, de Jong P, Dermitzakis ET, Eichler EE, Flicek P, Goldman N, Mundy NI, Ning Z, Odom DT, Ponting CP, Quail MA, Ryder OA, Searle SM, Warren WC, Wilson RK, Schierup MH, Rogers J, Tyler-Smith C, Durbin R. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurel L, Thompson EE, Flutre T, Lovstad J, Venkat A, Margulis SW, Moyse J, Ross S, Gamble K, Sella G, Ober C, Przeworski M. The ABO blood group is a trans-species polymorphism in primates. Proc Natl Acad Sci U S A. 2012;109:18493–18498. doi: 10.1073/pnas.1210603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1 doi: 10.1101/cshperspect.a006841. a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CG, Ahlquist JE. DNA hybridization evidence of hominoid phylogeny: Results from an expanded data set. J Mol Evol. 1987;26:99–121. doi: 10.1007/BF02111285. [DOI] [PubMed] [Google Scholar]
- Siepel A, Pollard KS, Haussler D. New methods for detecting lineage-specific selection. Research in Computational Molecular Biology, Proceedings. 2006;3909:190–205. [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, Webster MJ, Weickert CS, Lachmann M, Pääbo S, Khaitovich P. Transcriptional neoteny in the human brain. Proc Natl Acad Sci U S A. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- Stedman HH, Kozyak BW, Nelson A, Thesier DM, Su LT, Low DW, Bridges CR, Shrager JB, Minugh-Purvis N, Mitchell MA. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature. 2004;428:415–418. doi: 10.1038/nature02358. [DOI] [PubMed] [Google Scholar]
- Takahata N. Allelic genealogy and human evolution. Mol Biol Evol. 1993;10:2–22. doi: 10.1093/oxfordjournals.molbev.a039995. [DOI] [PubMed] [Google Scholar]
- Takahata N, Satta Y. Evolution of the primate lineage leading to modern humans: Phylogenetic and demographic inferences from DNA sequences. Proc Natl Acad Sci U S A. 1997;94:4811–4815. doi: 10.1073/pnas.94.9.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Catacchio CR, Sajjadian S, Vives L, Sudmant PH, Marques-Bonet T, Graves TA, Wilson RK, Eichler EE. The evolution of African great ape subtelomeric heterochromatin and the fusion of human chromosome 2. Genome Res. 2012;22:1036–1049. doi: 10.1101/gr.136556.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci U S A. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD. Estimating ancestral population sizes and divergence times. Genetics. 2003;163:395–404. doi: 10.1093/genetics/163.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Cox MP, Mendez FL, Woerner A, Severson T, Hammer MF. A novel DNA sequence database for analyzing human demographic history. Genome Res. 2008;18:1354–1361. doi: 10.1101/gr.075630.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Frisse LA, Hudson RR, Di Rienzo A. Comparative linkage-disequilibrium analysis of the beta-globin hotspot in primates. Am J Hum Genet. 2003;73:1330–1340. doi: 10.1086/380311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Slatkin M. Paleopopulation genetics. Annu Rev Genet. 2012;46:635–649. doi: 10.1146/annurev-genet-110711-155557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GM, Flibotte S, Missirlis PI, Marra MA, Jones S, Thornton K, Clark AG, Holt RA. Identification by full-coverage array CGH of human DNA copy number increases relative to chimpanzee and gorilla. Genome Res. 2006;16:173–181. doi: 10.1101/gr.4456006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler W, Myers SR, Richter DJ, Onofrio RC, McDonald GJ, Bontrop RE, McVean GA, Gabriel SB, Reich D, Donnelly P, Altshuler D. Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- Wise CA, Sraml M, Rubinsztein DC, Easteal S. Comparative nuclear and mitochondrial genome diversity in humans and chimpanzees. Mol Biol Evol. 1997;14:707–716. doi: 10.1093/oxfordjournals.molbev.a025810. [DOI] [PubMed] [Google Scholar]
- Wu DD, Irwin DD, Zhang YP. De novo origin of human protein-coding genes. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002379. e1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu AG, He L, Li Z, Xu Y, Li M, Fu X, Yan Z, Yuan Y, Menzel C, Li N, Somel M, Hu H, Chen W, Pääbo S, Khaitovich P. Intergenic and repeat transcription in human, chimpanzee and macaque brains measured by RNA-Seq. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000843. e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Arnason U. The mitochondrial DNA molecule of Sumatran orangutan and a molecular proposal for two (Bornean and Sumatran) species of orangutan. J Mol Evol. 1996;43:431–437. doi: 10.1007/BF02337514. [DOI] [PubMed] [Google Scholar]
- Xue Y, Daly A, Yngvadottir B, Liu M, Coop G, Kim Y, Sabeti P, Chen Y, Stalker J, Huckle E, Burton J, Leonard S, Rogers J, Tyler-Smith C. Spread of an inactive form of Caspase-12 in humans is due to recent positive selection. Am J Hum Genet. 2006;78:659–670. doi: 10.1086/503116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O. The origin of man: A chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]