Abstract
Nonhuman primate (NHP) biomedical models are critical to our understanding of human health and disease, yet we are still in the early stages of developing sufficient tools to support primate genomic research that allow us to better understand the basis of phenotypic traits in NHP models of disease. A mere 7 years ago, the limited NHP transcriptome profiling that was being performed was done using complementary DNA arrays based on human genome sequences, and the lack of NHP genomic information and immunologic reagents precluded the use of NHPs in functional genomic studies. Since then, significant strides have been made in developing genomics capabilities for NHP research, from the rhesus macaque genome sequencing project to the construction of the first macaque-specific high-density oligonucleotide microarray, paving the way for further resource development and additional primate sequencing projects. Complete published draft genome sequences are now available for the chimpanzee ( Chimpanzee Sequencing Analysis Consortium 2005), bonobo ( Prufer et al. 2012), gorilla ( Scally et al. 2012), and baboon ( Ensembl.org 2013), along with the recently completed draft genomes for the cynomolgus macaque and Chinese rhesus macaque. Against this backdrop of both expanding sequence data and the early application of sequence-derived DNA microarrays tools, we will contextualize the development of these community resources and their application to infectious disease research through a literature review of NHP models of acquired immune deficiency syndrome and models of respiratory virus infection. In particular, we will review the use of -omics approaches in studies of simian immunodeficiency virus and respiratory virus pathogenesis and vaccine development, emphasizing the acute and innate responses and the relationship of these to the course of disease and to the evolution of adaptive immunity.
Keywords: genomics, human immunodeficiency virus [HIV], influenza, microarray, nonhuman primate, simian immunodeficiency virus [SIV], transcriptome
Development of Resources for Nonhuman Primate Biomedical Models
Hosting of the First Nonhuman Primate Genomics Workshop
The development of genomic resources for nonhuman primate (NHP) biomedical models was impelled after the first Primate Genomics Workshop in Seattle, which was organized by investigators from the Washington National Primate Research Center at the University of Washington and included attendees from all eight National Primate Research Centers (NPRCs) (Figure 1). The participants agreed on the clear need for the development of genomic resources for NHPs to advance research on the genetic basis for phenotypic traits, with a priority focus on the Indian-origin rhesus macaque. In the ensuing efforts to create these resources, a white paper proposal requesting the genome sequencing of the Indian-origin rhesus macaque and the generation of expressed sequence tag (EST) databases for macaque species in use at the NPRCs was submitted to the National Human Genome Research Institute. Since its commencement, the Primate Genomics Workshop has grown in stature to become the International Conference on Primate Genomics, with the Washington National Primate Research Center being instrumental in the organization of the first four conferences. The fourth conference, held in Seattle in 2010, highlighted systems biology approaches to NHP models of human health and disease. An outgrowth of this conference was another endeavor in generating a new resource: a compilation of transcriptome data for a panel of NHP species important in biomedical and evolutionary research—the NHP reference transcriptome resource (http://nhprtr.org) (Pipes et al. 2013). Moreover, since 2007, a Genetics and Genomics Working Group has been working on developing a framework for the analysis and comparison of genetic and genomic data across the eight NPRCs (Kanthaswamy et al. 2009).
Figure 1:
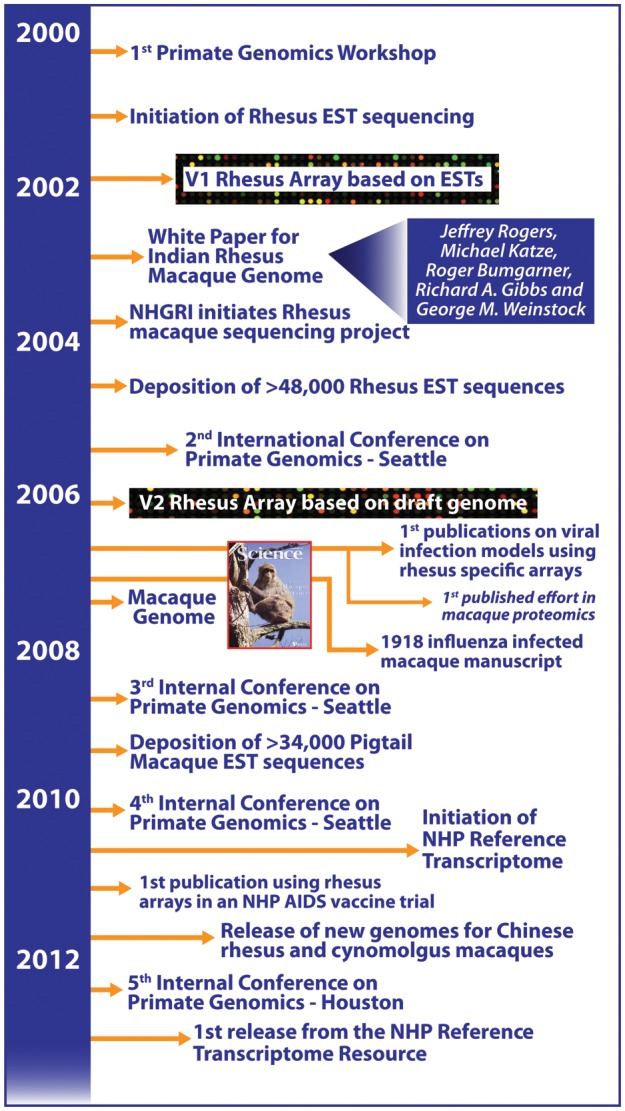
Advancements in the development of genomic resources that enable systems biology investigations in nonhuman primate (NHP) models of human disease. Efforts in NHP genomics have continued to grow from the simple origins in the 1st Primate Genomics Workshop. There was considerable acceleration in the application of genomics tools after the release of the genome sequence for the Indian-origin rhesus macaque. With the advent of next-generation sequencing technology, a rapid expansion occurred in the specific tools available for various macaque species that serve in models of infectious disease and other studies related to human health and disease. EST, expressed sequence tag.
Development of the First Rhesus Macaque Oligonucleotide Microarray
DNA microarray technology is an indispensable tool in molecular biology research; however, genome sequence and transcript sequence information is critical in the development of such research tools. Among the earliest efforts in NHP genomic resource development were EST sequencing projects for the rhesus macaque (Macaca mulatta) and the pig-tail macaque (M. nemestrina) to overcome the paucity of available sequence information for macaque species (Magness et al. 2005). Initial EST sequencing and comparative analysis of the rhesus macaque transcriptome included sequences from 11 tissues, nine animals, and three species (M. mulatta, M. fascicularis, and M. nemestrina), resulting in the generation of 48,642 rhesus EST sequence reads and, later, more than 34,000 ESTs for the pig-tail macaque. The EST data were used to assist genome assembly and annotation efforts and toward the production of the first macaque-specific, spotted complementary DNA (cDNA) microarray (V1 Rhesus Array) (Figure 1).
The first draft of the human genome was released in 2001, followed 18 months later by the draft genomic assembly for the mouse (Lander et al. 2001; Waterston et al. 2002). After the submission of a white paper proposal to the National Human Genome Research Institute, the sequencing effort for the rhesus genome was initiated in 2003, conducted by the Rhesus Macaque Genome Sequencing and Analysis Consortium under the leadership of the Human Genome Sequencing Center at the Baylor College of Medicine. After draft sequence data for the rhesus genome was made publically available in 2005, scientists quickly used it to design macaque oligonucleotide microarrays. For example, the Katze laboratory collaborated with Agilent Laboratories in the design of a rhesus-specific high-density oligonucleotide microarray, with annotation using the detailed sequence and exon structure of the human genome (Wallace et al. 2007), resulting in a commercially available Rhesus Macaque Gene Expression Microarray marketed by Agilent Technologies. This array format included the corresponding human gene symbols for the detected rhesus transcripts, allowing for functional analysis with the much richer body of annotation available for human genes. A separate macaque array was designed in association with Affymetrix, Inc. (Spindel et al. 2005). Because of the improved homology between the array probes and the target macaque transcripts, both these platforms provided superior performance versus arrays based on human transcript sequences (George et al. 2006; Wallace et al. 2007). With the release of the first draft of the rhesus genome sequence and the production of the more fully featured oligonucleotide rhesus microarray, the field was equipped to begin application of functional genomics in NHP models of human health and disease. As indicated in Figure 1, these applications actually preceded the formal publication of the rhesus genome in 2007 (Gibbs et al. 2007).
Genomic Analyses of NHP Models of Human Immunodeficiency Virus (HIV)/Acquired Immunodeficiency Syndrome (AIDS) and Respiratory Virus Infection
Given their genetic and physiologic similarities to humans, NHPs closely model the human response to HIV/AIDS and respiratory virus infections, among other clinically significant infectious diseases. For NHP models of viral respiratory disease, functional genomics measurements were undertaken to elucidate the molecular programs that underlie the severe pathogenesis (and morbidity) associated with particular strains of influenza, contrasting these with the host response initiated against milder seasonal variants.
Initial Genomic Characterization of Influenza in an NHP Model
The first use of genomics to profile NHP transcriptional responses to influenza virus infection was examined using human cDNA microarrays that contained approximately 13,000 unique cDNA clones (97% of which were annotated) and were integrated with clinical and pathology data (Baskin et al. 2004). Pig-tail macaques were infected with a low-pathogenicity human H1N1 influenza A/Texas/36/91 virus (Tx/91), and gene expression patterns were examined in the lung and tracheobronchial lymph nodes at days 4 and 7 after inoculation. Innate immune genes related to interferon (IFN) signaling and genes relevant to monocyte and macrophage function in the lung showed, in general, consistency with ancillary pathology, cytology, and cytotoxicity findings from immunohistochemistry of these tissues. Shortly thereafter, NHP lung and peripheral blood mononuclear cell (PBMC) gene expression changes were profiled using the macaque-specific oligonucleotide array that included an earlier time point at day 2 after inoculation (Baas et al. 2006). (This was one of the first applications of a species-specific microarray in an NHP model of human viral disease.) We hypothesized that host responses occurring at early time points after inoculation critically determined influenza-associated disease outcome. Indeed, at day 2 after inoculation, animals showed marked upregulation of IFN-stimulated genes, such as IRF7, IFIT2, and IFI44, in the lung, many of which were also induced in blood and showed sustained transcriptional activation in the periphery through day 7 after inoculation. Global protein abundance changes in lung samples after infection included IFN-stimulated antiviral proteins, such as STAT1, MX1, and GBP1, which complemented the transcriptomic data while creating an additional layer of complexity and further demonstrating the value of NHPs in functional genomics in NHP studies of influenza infection (Baas et al. 2006).
Assessment of NHP Immune Responses to 1918 Pandemic Influenza Virus
The reconstructed 1918 pandemic influenza virus is repeatedly cited as the archetype of highly pathogenic influenza, and NHP resources were instrumental in the study that characterized the aberrant immune response that attends this highly lethal infection in cynomolgus macaques (M. fascicularis) (Kobasa et al. 2007). The gene expression profiles for contemporary human H1N1 influenza A/Kawasaki/173/2001 virus (K173) show a dynamic course, with an early upregulation of the immune and inflammatory response that declines in later days after infection. Closer examination within the category of the immune response reveals that infection with 1918 virus exhibits distinct kinetics for the type I IFN response in macaque bronchial tissue, showing both a delay in onset and a reduction in magnitude of gene expression changes. For the less-pathogenic K173 virus, this latter stage is accompanied by increased expression of metabolic genes that may be associated with homeostasis or repair of the respiratory tissues. In contrast, the transcriptional profiles for the 1918 virus are sustained throughout the time course, and the inferred cellular processes for tissue remediation are not upregulated. The results of this study, published in Nature, indicate that atypical expression of innate immune response genes may be a critical determinant of the severity and outcome of infection by the 1918 virus, and continued investigations seek to determine specific 1918 virus genes eliciting these aberrant host responses (Cilloniz et al. 2009; Kobasa et al. 2007). Profiling lung gene expression changes to genetic reassortant viruses between the 1918 and the Tx/91 strains have revealed a role for 1918 virus envelope glycoproteins HA and NA toward 1918 pathogenesis. When expressed in the genetic background of the Tx/91 virus, the presence of 1918 HA and NA results in enhanced viral replication in macaque lung, more severe clinical disease and pathology, and stronger IFN, inflammatory, and innate immune transcriptional induction as early as 1 day after inoculation as compared with Tx/91 virus (Baskin et al. 2009).
Compelling genomic evidence from Baskin and colleagues showed highly pathogenic avian influenza (HPAI) H5N1 virus ignites an even more intense and incessant immune response than 1918 reassortant viruses in macaque lung (Baskin et al. 2009). Direct comparison of 1918- and H5N1-infected macaque bronchi early in infection revealed notable differences in apoptotic and inflammatory responses, including differences in the timing and extent of genes encoding NLRP3 and interleukin 1β, key components of the inflammasome, which were increased in expression in response to 1918 virus and decreased in expression in response to H5N1 virus (Cilloniz et al. 2009). These NHP genomic findings supported the notion that a rapid and intense host inflammatory response is the cause of severe lung damage caused by H5N1 virus in humans (de Jong et al. 2006; Peiris et al. 2004). Biphasic fever and lymphopenia patterns in pediatric patients infected with H5N1 virus are closely replicated in an NHP model of nonlethal viral pneumonia (Shinya et al. 2012). Shinya and colleagues reported a comprehensive analysis of pneumonia-induced pathogenesis in rhesus macaques infected with HPAI H5N1 influenza A/Anhui/2/2005 virus, integrating lung transcriptomic data collected at six time points, spanning the initial infection phase through recovery, with clinical, virologic, and histopathologic data. Transcriptomic profiling indicated different molecular mechanisms driving each phase of the biphasic fever, with proinflammatory mediators tumor necrosis factor and interleukin 1β responsible for the first wave and IFN-γ predominantly mediating the second wave.
Age-Related Transcriptomic Analysis of NHP Host Responses to 2009 Pandemic H1N1
The novel swine-origin influenza A virus (pH1N1) that emerged in Mexico in early 2009 resulted in a wide-spread pandemic, and the scientific community quickly mobilized to assess its pathogenicity in NHPs. One of the first US isolates, pH1N1 influenza A/California/04/2009 virus (CA04), was found to be more pathogenic than a contemporary human H1N1 virus in cynomolgus macaques (Itoh et al. 2009). Rapid assessment of macaque lung responses to two clinical pH1N1 isolates from Mexico during the early stage of the pandemic demonstrated more-pronounced transcriptional changes of genes associated with proinflammatory nuclear factor kappa B (NFκB) signaling and antigen presentation pathways in response to pH1N1 virus compared with K173 virus (Safronetz et al. 2011). In an investigation of age-related disease severity caused by pH1N1 virus, aged rhesus macaques, which have greater inflammatory cytokine production and T cell proliferation in the bronchial alveolar lavage (BAL) compared with young adults after CA04 virus infection, showed upregulated expression of genes encoding T cell markers (e.g., CD3D and CD3G) and B cell markers (e.g., CD20 and CD19) in BAL cells compared with young adult animals (Josset et al. 2012).
A longitudinal gene expression analysis of BAL cells from CA04-infected young adult and aged animals associated gene expression changes with immune and virologic measurements, such as intracellular cytokine staining, inhibition HA titer, and viral antigen stimulation (HA-NA and PA-NP) (Figure 2). The frequency of CA04-specific T cells in BAL cells correlated with genes directly involved in T lymphocyte function (iCOS–iCOSL signaling in T cells), whereas neutralizing antibody production was associated with genes directly involved in B lymphocyte accumulation and nuclear receptor signaling. Proliferation of memory B cells and viral titers were associated with genes directly involved in anti-influenza host responses. This study represents the first age-related genomic analysis after pH1N1 infection in a rhesus macaque model. Severe clinical disease caused by influenza virus is more frequent in elderly and immune-compromised individuals, and studies examining age-related effects have been made possible through current NHP genomic resources. Future applications will likely examine influenza vaccine efficacy in young adult and aged animals challenged with pH1N1 virus.
Figure 2:
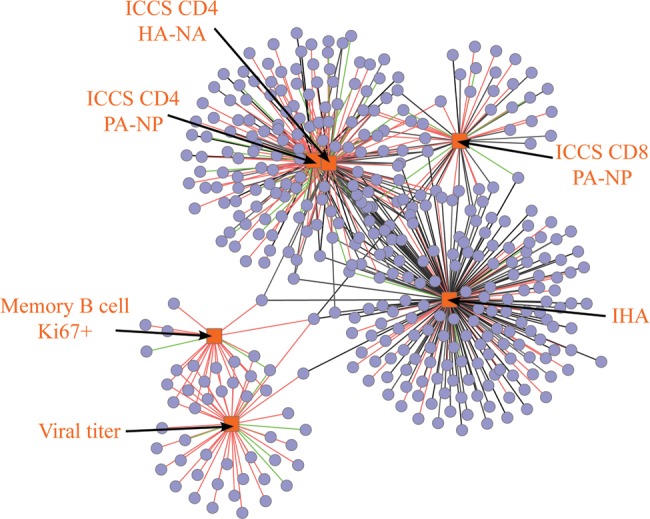
Cytoscape (Cline et al. 2007) rendering of Maximal Information Coefficient (MIC) (Reshef et al. 2011) results between gene expression data and the indicated immunologic and virologic measurements. All associations had MIC ≥ 0.8. Red/green edges indicate positive/negative correlations, respectively. Grey edges indicate nonlinear relationship. Measurements were performed on bronchial alveolar lavage samples from rhesus macaques infected with 2009 pandemic H1N1 influenza strain CA04 (exception of serum anitbody titers). HA-NA, viral hemagglutinin + neuraminidase; ICCS, intracellular cytokine staining; IHA, inhibition HA titer; PA-NP, viral polymerase component + nuclear protein.
Cross-species transcriptomic analyses have further enhanced our understanding of host responses to pH1N1 virus in NHPs by extending transcriptomic comparisons across different animal models of influenza infection. These include the well-established mouse model and the underused swine model, which is particularly relevant when considering the generation of novel influenza strains that often emerge in this intermediate host (Go et al. 2012). Notably, lung transcriptional changes in response to a single 2009 pandemic H1N1 influenza virus across macaque, swine, and mouse models showed significant differences in the expression of inflammatory and lipid metabolism genes, indicating that the nature of the host response, characterized by the timing of the acute phase and the kinetics of the gene expression changes, underlying overt clinical disease is distinct across different animal models. To fully appreciate the NHP host response in the context of respiratory infection, it will be important to profile transcriptional changes across different primate species and tissue compartments such as nasopharynx, trachea, bronchi, and lung.
Application of NHP Proteomics and Influenza Infection
Another ambition in the development of NHP resources for functional genomics studies was to demonstrate the feasibility and utility of employing proteomics techniques on NHP biomedical models. In the initial efforts, data from the rhesus genome were unavailable; nonetheless, using human protein sequences, differential protein abundance changes were identified in the lung tissues of macaques infected with a seasonal strain of H1N1 (Baas et al. 2006). A recent study with HPAI and 1918 reassortant viruses produced a reference database of macaque proteins based on the currently annotated transcripts of the rhesus genome that provided an increased number of identified peptides per protein and thus improved confidence in the protein identifications (Brown et al. 2010). Quantitative analysis identified 400 proteins that increased in abundance during viral infection, with differing viral strains exhibiting distinct temporal profiles for proteins involved in the innate response to viruses and inflammatory response and with patterns distinct from those observed by microarray measurements. This study is the most comprehensive NHP proteomics characterization to date.
Global Transcriptional Profiling in the Study of Simian Immunodeficiency Virus (SIV) Pathogenesis
As noted earlier, the importance of Indian-origin rhesus macaque for biomedical models of HIV pathogenesis and immunity was a prominent criterion for the genome sequencing of this species of NHP. However, the demands of the AIDS research community outpaced the availability of the species-specific genomic resources for characterizing the transcriptional changes that attend SIV infection in macaque species, and initial studies used expression arrays based on human sequence data, either in the form of spotted cDNA arrays or the Affymetrix human GeneChips. In hindsight, many of these studies appear quite technically limited, either from the smaller number of transcripts that were interrogated on the arrays or the small numbers of array measurements that constituted the experiment. Even in the early part of the last decade, human microarray technology was still on a steep slope of development despite the release of the high-quality human genome sequence in 2004 (International Human Genome Sequencing Consortium 2004). Small experimental designs were a consequence of the high cost of array measurements, the limited capacity of newly emerging microarray laboratories, and the high cost of investigations using NHPs.
In vitro experiments using rhesus PBMCs was one route to diminish the animal requirements, and an early report from our group took such an approach in assessing the changes attendant to synchronous infection with SIVmac239 (Thomas et al. 2006). As is typical with such infections in primary cells, the percentage of infected cells was quite low; nonetheless, the sensitivity of transcriptional profiling returned hundreds of differentially regulated genes at 3 and 6 days after infection. The heterogeneity that attends the use of outbred animals was also evident because PBMCs from one of the donor animals were relatively refractory to infection and consequently showed fewer changes in gene expression. Patterns from the other two donors showed early upregulation of immune response pathways, such as class I antigen presentation, apoptosis signaling, and cellular adhesion, as well as broad functional categories, such as protein metabolism and transcriptional regulation. It is interesting to contrast these results with a similar experiment we later reported examining transcriptional responses in PBMCs from pig-tail macaques after infection with either SIVmac239 or HIV-lLAI (Li Y. et al. 2007). Although the course of the SIV infection was comparable with our observation with rhesus PBMCs, the HIV-1 infection was quite constrained based on the percentage of infected cells and the extent of viral replication. Despite the lower percentage infection and replication with HIV-1 versus SIV, the early time point 4 days after infection with HIV-1 showed a much greater number of upregulated genes involved in antigen presentation, T cell signaling, and natural killer cell killing. This constraint on HIV replication is hardly surprising in light of our current understanding of lentiviral species restriction by host factors such as Trim5α (Nakayama and Shioda 2012). However, the results suggest the propagation of an immune stimulus attendant to the viral sensing/restriction by Trim5α or some other factor.
Other earlier studies that employed human gene expression arrays used longitudinal blood samples from SIV- or simian human immunodeficiency virus (SHIV)-infected macaques as a means to characterize the host response to pathogenic lentivirus infection (Bosinger et al. 2004; Chung et al. 2008; Vahey et al. 2003). In general, results from such in vivo models revealed the differential regulation of many more genes than observed with the simple ex vivo models. The expression data also revealed clear increases of type I IFN-stimulated genes (ISGs) and inflammatory response genes, commencing early after infection (1–3 weeks). And although these responses may decline as viral loads decline to set point levels, the expression hallmarks of activation are always present. These studies also showed that abatement of these activation signatures was often seen in spontaneous controllers, whereas they were exacerbated in animals that underwent rapid progression to elevated viremia and AIDS-like symptoms.
Early adopters of genomics studies for NHP models of AIDS also made focused efforts on specific organ systems. The Fox group used human GeneChip arrays to characterize expression changes in the brains of rhesus macaques, examining animals at 2 weeks after challenge, as well as during chronic stages, including subsets of animals with SIV encephalitis (i.e., neuroAIDS) (Roberts et al. 2003; Roberts et al. 2004). Perhaps the most surprising observation is the clear upregulation in the brain of ISGs and interleukin 6 as early as 2 weeks after challenge, at a stage far earlier than typically associated with neurological consequences of SIV/HIV infection. Guidance from immunohistochemistry then demonstrated that these antiviral responses largely originate from the microglia and monocytes, with the persistent inflammation progressively impacting astrocytes, endothelial cells, and the neuronal parenchyma.
The program by the Dandekar laboratory emphasized the pathogenic consequences in gut tissues that attend SIV infection. These studies paralleled the then-emerging appreciation that HIV and SIV severely deplete the CD4+ T cells from the gut-associated lymphoid tissue (Li Q. et al. 2005; Mehandru et al. 2004; Veazey et al. 1998). The expression analyses made clear that the immune activation in the gut onsets very early after infection (George et al. 2003; George et al. 2005). Moreover, there are changes in cell-cycle mediators and downregulation of genes associated with gut homeostasis and nutrient absorption. Although very early antiretroviral therapy can ameliorate the immune activation in the gut mucosa and lead to restoration of both the CD4+ T cell population and the gut homeostasis, these latter capacities are not restored when the therapy is administered later during the chronic stage of infection. This arc of work, coupled with related studies with human clinical samples, demonstrated the exactitude of the NHP model in recapitulating the course of the disease in the human gut, even to the extent of comparable gene expression characteristics of jejunal samples from either human or rhesus long-term nonprogressors (George et al. 2006; Guadalupe et al. 2003; Sankaran et al. 2005).
SIV Infections in Natural Hosts
In striking contrast with HIV infection in humans and SIV infection in Asian macaques, species of African NHPs that are naturally infected with SIV do not progress to AIDS (Chahroudi et al. 2012). SIV infection in these natural hosts produces high viral loads but is nonpathogenic, and animals maintain healthy CD4+ T cell counts (Apetrei et al. 2011; Silvestri et al. 2003). This is in contrast with progressive HIV/SIV infection, where high viral load leads to loss of CD4+ T cells, immune dysfunction, and progression to an AIDS-like state. Infection in natural hosts is also quite different from HIV/SIV infection in rare elite controllers, where the latter do not progress to AIDS by maintaining durable control of viral replication at very low levels (Deeks and Walker 2007). Contrasting the mechanisms contributing to protection from AIDS in natural hosts with mechanisms driving progression to AIDS in pathogenic SIV models could lead to new insights for HIV therapy or prevention. Natural hosts currently under study include sooty mangabeys and African green monkeys (AGMs) (Chahroudi et al. 2012).
Our own investigations into the host response in natural SIV infections compared the AGMs (Chlorocebus sabeus) versus the Asian macaque species pig-tail macaques after intravenous infection with the same inoculum of SIVagm.sab92018 (Favre et al. 2009; Lederer et al. 2009). Experimental infections with this strain conducted in AGMs derived from these Caribbean populations have been well characterized, with no evident pathogenesis despite high viral loads at both the acute peak and chronic stages of the infection (Diop et al. 2000; Pandrea et al. 2006). Infections with this same strain in pig-tail macaques result in viral replication kinetics and viral load levels similar to the natural hosts; however, this species experiences a rapid decline of CD4+ T cells in all compartments, and many animals experience severe immune compromise (Goldstein et al. 2005). For a detailed immunologic and functional genomics study, we performed a longitudinal analysis with four animals of each species, with blood, lymph node, and colon samples obtained at 14 days before inoculation and 10 and 45 days after inoculation. The array experiments employed the previously described rhesus-specific oligonucleotide DNA array that we developed in collaboration with Agilent Technologies. Note that despite the advances in NHP genomics that lead to the rhesus-specific arrays, no specific tools were available for the species in this study. At the time, in considering the existing knowledge of sequence similarities, the use of the macaque platform appeared to be less of a compromise than to make the expression measurements with human expression arrays (Magness et al. 2005; Wallace et al. 2007). Ratiometric array measurements were obtained for the postinfection samples, where the expression levels in an animal's tissue were compared with the individually matched baseline sample from 14 days before inoculation. Both species exhibited robust gene expression signatures after infection; this was especially pronounced in the lymph nodes where approximately 2500 genes showed a twofold or higher change relative to baseline regardless of species or time after challenge. Despite this nominal equivalence in the number of regulated transcripts, statistical comparisons at each time point showed the greatest number of differences between species for the day 10 lymph nodes (610 genes). The most prominent functional categories identified by gene ontology analysis were immune responses and cell death, and pig-tail macaques clearly exhibit greater upregulation of genes involved in caspase activation, DNA damage, and oxidative stress. Changes in the immune response genes for the pig-tail macaques implicate a Th1 response, cytotoxic T cell activity, and Ifn-γ signaling. In contrast, AGMs showed upregulation of the anti-inflammatory cytokine interleukin 10 and the inflammatory regulator NLRP3, with overall expression changes implicating a more active control of the inflammatory response and a shift to homeostasis of the lymphoid compartment. Perhaps the starkest contract between the pathogenic versus natural infection is the evident difference in the temporal expression of type I interferon α genes, with substantially increased expression of these transcripts in the lymph nodes of AGMs on day 10 after challenge versus significantly lower levels in this same compartment on day 45 after inoculation (Palermo and Fuller 2013). The lymph nodes from the infected pig-tail macaques do not show this consistent upregulation at the earlier time point, although as noted earlier, the pig-tail macaques exhibit great expression of type II INF-γ than do the AGMs at 10 days after inoculation. Moreover, this pattern does not recur in the type I IFN expression patterns for blood or colon of either species, highlighting the unique kinetics and localization in the AGMs. However, as we recently described, when looking at downstream ISGs, for AGMs we do observe similar kinetics, with generally elevated expression levels on day 10 and a decline by 45 days after inoculation; the pattern appears in lymph nodes and blood and is particularly conspicuous in the colon. On day 10, the expression ratios of these genes in the natural hosts are comparable with (for blood) or greater than (for colon and lymph node) the observations for the pathogenic context. The pig-tail macaques do show increased expression in some of these ISGs relative to their prechallenge state, but this elevated expression level persists even after the viral load has declined to set point at day 45 after inoculation. It also bears noting that the early upregulation of the ISGs in the pig-tail macaques happens in concert with the upregulation of many acute phase and inflammatory response genes. Network analysis of these genes finds many of them associated with NFκB signaling, and the presence of TLR2 and CD14 suggests a role for myeloid cells in this response.
Two other systems-level, functional genomics investigations of natural infection models have yielded very similar outcomes. The study of Jacquelin and colleagues (2009) contrasted the responses of AGMs intravenously infected with SIVagm.sab92018 with the course for rhesus macaques challenged with SIVmac251. These investigators were able to obtain expression results on CD4+ T cells isolated from peripheral blood and lymph node samples taken as early as 1 day after challenge, when the infected AGMs already showed strong upregulation of a large number of ISGs typically associated with type I IFN response. However, in the rhesus macaques, increased expression of this category of genes was delayed until the ensuing time point at 6 days after inocultion. Using a highly sensitive functional assay for type I IFN, the authors were able to show AGMs exhibited an initial small peak in plasma levels at 2 days after inoculation, followed by a second much higher peak at day 9 after inoculation, before returning to baseline within days. Rapid control of the innate response in the natural infection of sooty mangabeys inoculated with SIVsmm was also the conclusion reached using a model contrasting the natural infection versus pathogenic challenges of rhesus macaques with either the same inoculum of SIVsmm or with highly virulent SIVmac239 (Bosinger et al. 2009), examining expression profiles in whole blood or lymph nodes using Affymetrix rhesus-specific expression arrays. Nonetheless, in these experiments, as in the aforementioned studies with pig-tail macaques, infected rhesus macaques continued to show persistent upregulation of ISGs and acute inflammatory response genes likely driven by NFκB transcriptional control. An interesting extension to the results reported by Bosinger and colleagues (2009) is the comparison of these gene expression features with those observed with samples from human HIV-1 infections that represent discordant phenotypes (i.e., rapid progressors vs. viremic nonprogressors). Whereas rapid progressors were characterized by a transcriptome profile of CD4+ and CD8+ T cells similar to that observed in pathogenic SIV-infected rhesus macaques, results for viremic nonprogressors exhibited gene expression features similar to those observed with nonpathogenic SIV-infected sooty mangabeys (Rotger et al. 2011).
Studies in NHP models for AIDS Vaccine Strategies
Our first application of microarrays to characterize an NHP trial of an AIDS vaccine strategy examined animals that had been immunized with replicating adenovirus constructs that expressed gag, nef, and env immunogens derived from SHIV89.6P , including one group of immunized animals receiving protein boosts with HIV89.6P gp140 to enhance the env-specific antibody response (Palermo et al. 2011; Patterson et al. 2008). It bears commenting that such NHP vaccine trails are a considerable expenditure of both resources and time, and it is not uncommon for the immunization phase of such studies to run 6 to 12 months before the protective efficacy is assessed by viral challenge. After such an investment in immunizing the animals, sampling procedures are generally limited to noninvasive, survival protocols. In such a context, we anticipated that peripheral blood was the only sample type that could be easily incorporated for the longitudinal evaluation of many animals. We also employed RNA from whole blood that had been drawn by methods that immediately stabilize the RNA, thereby avoiding any additional variability due to cellular stress that attends storage or ex vivo manipulation of the blood. In comparing the expression profiles of controls (n = 6 animals receiving only empty vectors) versus the vaccine groups (n = 6 per group; ± the gp140 boosts), the study demonstrated our hypothesis that genomic profiling of whole blood was sufficiently sensitive to distinguish the groups even 8 weeks after the last immunization and before viral challenge. Moreover, unsupervised 2D hierarchical clustering highlighted features unique to the best-protected vaccinees (i.e., those that received the protein boosts) that were consistent with a stronger induction of T cell and B cell memory.
Counter to expectation, at the peak of viremia on day 14 after intravenous SHIV89.6P challenge, the immunized animals did not show statistically significant differences in expression of genes associated with cell-mediated immunity, despite the presence of vaccine-induced antigen-specific CD4+ and CD8+ T cells. Transcripts for such genes were increased in all the groups, including the protein-boosted group, which had showed a significant reduction in peak viral load. Instead, the immunized animals exhibited significantly upregulated transcripts for many other acute-phase response genes relative to the levels observed in the controls. Finally, at week 12 after inoculation (early viral set point), the statistical comparison showed the greatest number of differences between the two immunized groups, despite both groups having comparable reductions in viral load relative to the control. This implied that these two vaccine groups were employing different mechanisms of immunological control at this stage in the study, despite their general phenotypic similarity. Moreover, the features at week 12 for the protein-boosted animals indicated mechanisms that would have been strongly linked to the higher env-binding antibodies in this group, such as antibody-dependent cellular cytotoxicity and cellular viral inhibition (ADCC, ADCVI) (Xiao et al. 2010).
An alternative approach to assess the immune response induced by particular vaccine regimens is to examine the transcriptional changes that attend specific antigenic stimulation of ex vivo cells from vaccinated animals. This approach was applied in the characterization of an SIV DNA vaccine, where animals were immunized with DNA plasmids that expressed SIV gag, pol, and env proteins, and additionally one group of animals also received a plasmid that expressed RANTES to serve as an adjuvant. Control animals receive empty DNA plasmids (Belisle et al. 2011). In vitro stimulation was performed with pol-peptides or mock-treatment using PBMCs isolated at 8 months after the last vaccination treatment and with PBMCs from 2 weeks after challenge with SIVmac251 (peak viremia). Using rhesus-specific gene expression arrays, the gene expression changes of each animal in response to SIV pol stimulation was calculated as the ratio to its own mock-stimulated gene expression at the concurrent time point. At both pre- and postchallenge time points, antigen stimulation of cells from vaccinated animals resulted in upregulation of genes with functions in cellular and innate immunity and an enhanced Th1 response. Cell lysis pathways were represented in those genes with increased expression in the prechallenge sample; however, at peak viremia such distinctions were lost because the expression of these genes was upregulated in all groups. Because the group that received the DNA + RANTES exhibited the greatest suppression of SIV viral load during the chronic phase of infection, we also examined genes that uniquely distinguished this group from the other vaccinated animals and the controls. Both pre- and postchallenge, approximately 160 genes showed unique expression patterns in this group, with functional associations to cell growth, cell death, inflammation, and cellular motility. Of particular interest was the distinct upregulation of CD69, a surface glycoprotein that can promote lymphocyte retention in lymphoid organs and may be related to the greater proportion of activated central memory CD8+ T cells observed in this group.
Given the sensitivity and discrimination of expression profiling, as well as the examples of how this sensitivity can lead to novel observations regarding protective immunity against HIV/SIV, it is surprising that these genomic methods are not more broadly applied in NHP trials of AIDS vaccines. This is especially true in light of the emphasis on such NHP evaluations after the unexpected outcomes from the STEP and Thai trails (Morgan et al. 2008). This begs the question of whether the inhibition arises from investigators who lack familiarity with the techniques or from financial limitations to what are already costly NHP protocols.
Future Genomic Resources and NHP Systems Biology
New Genome Resources
Next-generation sequencing (NGS) represents another paradigm-shift with transformative impacts throughout biomedical research (Mardis 2008). This was quite evident at the 4th International Conference on Primate Genomics, where preliminary results for three new macaque genomes were communicated before their public release: the Chinese-origin rhesus macaque, the Indonesian cynomolgus macaque, and the Mauritian cynomolgus macaque (Ebeling et al. 2011; Yan et al. 2011). Given the speed and lower cost of NGS, doubtless other NHP genomes will follow in short order. For example, the sooty mangabeys genome is an ongoing project at the Human Genome Sequencing Center–BCM (BCM Human Genome Sequencing Center 2013). Other efforts from that center include improvements in draft genomes for other species such as the baboon and marmoset. The current publically available genomes for these species were based on shotgun Sanger sequencing and represent typical draft sequencing depths of 5- to 6-fold genome coverage (Ensembl.org 2013; Ensembl.org 2011). The current Indian rhesus macaque draft genome is at this same coverage depth, and it has been noted that at this level there are assembly errors that have considerable impacts in the annotation of rhesus genes (Zhang et al. 2012). Sequencing genomes by short-read technologies will change the availability of genomes for many NHP species, including rarer endangered species such as the aye-aye (Perry, Melsted et al. 2012); however, assemblies based solely on short-read data rarely attain the contiguity and chromosomal anchoring achieved in earlier genome projects (Yandell and Ence 2012). Using reference genomes from other species can help circumvent such issues, as was done with the new macaque genomes cited above which employed the current rhesus genome as a reference. There is, of course, the danger of using a misassembled draft genome as a reference. Even in regions where the reference assembly is correct, this runs the risk of imposing false constraints that do not reflect the true underlying structure of the new genome, potentially failing to properly determine breakpoints or copy number variations between two species. Even if new technologies offer much longer read lengths, ultimately yielding longer genomic scaffolds, there is still the hurdle of chromosome anchoring for species that lack genetic markers or other hallmarks to assign genes to chromosomes. Nonetheless, with the advent of new genomes and new transcriptional databases, there is the near-term prospect of more species-specific tools, including expression arrays, reverse-transcription polymerase chain reaction assays, and proteins for generating antibody reagents. We can also expect the genotyping of research animals to become a major component of important NHP research models (Fang et al. 2011).
RNA-seq
NGS technology also supports sequence read acquisition for comparative messenger RNA (mRNA) profiling, global transcriptome profiling, and microRNA abundance measurements (Wang et al. 2009). These transcriptional profiling applications (RNA-seq) will revolutionize the measurement of functional genomic data to be obtained from NHP models. As described in the introduction, our group, in collaboration with Illumina, Inc., has created the Nonhuman Primate Reference Transcriptome Resource (NHPRTR) to generate deep compilations of RNA-seq data for 15 species of NHPs with importance in biomedical models and evolutionary research (Pipes et al. 2013). Another criterion for inclusion was either the existence of a current draft genome or the potential production of a genome in the near future. To generate transcriptomic results that would be very comprehensive for a species, the data were generated from pooled RNA representing approximately 20 tissues per species. The data and the transcript models produced by the NHPRTR will facilitate the genome assembly as well as serve as functional evidence when annotating the genomes for the species.
One advantage of NGS technology over microarrays is the greater dynamic range that can be achieved with RNA-seq, thereby allowing the accurate quantitation of low-abundance transcripts. However, unlike a microarray, data collection is not premised on prior knowledge of the transcripts to be measured. The result is a much-expanded concept of the host transcriptome; even from well-annotated genomes such as the human or mouse genome, RNA-seq data have revealed many novel transcripts, representing either novel splice forms or new genetic loci (Trapnell et al. 2010). Therefore such functional genomic measurements in NHPs also harbor the capacity for transcript discovery; within the datasets is the information for detecting mRNA transcripts for unannotated or misannotated genes, alternate splice forms, and untranslated regions of mRNAs. In addition, RNA-seq data have revealed that mammalian genomes express vast numbers and varieties of non-protein-coding RNAs (Cabili et al. 2011; Djebali et al. 2012). We have shown that HIV and respiratory virus infections result in the differential expression of a wide variety of short and long noncoding RNAs (Chang et al. 2011; Peng et al. 2010; Peng et al. 2011). A detailed knowledge of noncoding RNA regulation and function will therefore likely be at the forefront of NHP genomic resource development for a full understanding of gene regulation and viral pathogenesis in NHPs.
Comparative Primate Genomics
For the understanding and improvement of human health, the most paramount cross-species comparison is between Homo sapiens and NHPs used in biomedical models of human disease. Our own research in infectious disease has resulted in our focus on numerous genes that are annotated as orthologs to human genes in innate and adaptive immunity. Although this orthologous relationship is a reasonable premise for the assumption that the genes fulfill the same function in the NHP species as they do in humans, this is by no means assured. And it is the rare instance where detailed experiments establish the complete equivalence in both species. One example of such imperfect orthology is the helper T cell marker CD4. This human gene has unambiguous orthologs in the genomes of numerous Old World primates (chimpanzee, rhesus macaque, orangutan, gibbon) and even prosimians such as the mouse lemur (Ensembl.org 2010a; Ensembl.org 2010b; Ensembl.org 2012a; Ensembl.org 2012b; Ensembl.org 2012c), and cross-reactive antibodies are used to identify CD4+ lymphocytes for humans, macaques, and AGMs (Beaumier et al. 2009). However, a protracted deficit of CD4+ T cells has no impact on the health of AGMs, whereas such a condition would result in fatal immunodeficiency in humans or macaques (Murayama et al. 1999), a difference of evident significance in the course of natural infection of AGMs by SIV (Beaumier et al. 2009). A different challenge attends understanding orthologous genes where there has been an expansion of the gene family, for example as noted in the rhesus macaque genome for the MHC class I B genes and killer immunoglobulin-like receptor genes (Gibbs et al. 2007), presenting the possibility that members of the expanded gene family may be serving specialized roles instead of a strict orthology of function. However, genomic analysis of NHPs provides the information to cope with these issues. In one instance, the sequence of the NHP gene enables the production of the corresponding protein that can be used for the generation of antibodies, and the latter can then be used to characterize the function of the protein or, with immunophenotyping assays, the cells that express it. For expanded gene loci, the gene model provides the framework for understanding the more-complex association of differing haplotypes with disease outcome (Aarnink et al. 2011; Sambrook et al. 2005; Sauermann et al. 2008). As annotation improves, particularly for less-characterized species, we will likely be able to probe and characterize host responses to viral infection more deeply and at greater resolution.
Genetic diversity in NHPs parallels that in humans, which has prompted researchers to identify and characterize functionally significant intra- and intergenetic variation among individuals within a primate colony and across different species. Genetic variation being realized with RNA-seq includes single nucleotide polymorphisms (SNPs), segmental duplications, and mutation rates in a lineage, which benefits studies of adaptive evolutionary histories (i.e., speciation events), risk factors associated with disease outcomes, as well as conservation efforts of endangered species. Perry and colleagues used RNA-seq combined with de novo gene assembly to characterize liver transcriptomes and assess natural genetic variation at the gene regulatory level in a wide range of mammalian species, including primate suborders: haplorhines (humans, chimpanzees, Old and New World monkeys) and strepsirrhines (lemurs and lorises) (Perry, Reeves et al. 2012). They reported the regulatory role of peroxisome genes in liver metabolism between primate and other mammalian species that possibly evolved under directional selection in the ancestral primate lineage. In addition to RNA-seq, genetic polymorphism data in NHPs have also been generated using an SNP array that has identified more than 3 million SNPs, and these data are being analyzed for greater genome-wide knowledge of genetic variation segregating within research colonies of rhesus macaques (Fawcett et al. 2011). This will be particularly valuable in gaining information of potentially significant effects on gene function and physiologic processes relevant to human disease.
Future Directions—NHP Systems Virology
Systems biology is a process of integration, both of high-dimensional datasets such as global expression profiles and of other data types that represent phenotypic data from the biological system (e.g., viral load, plasma cytokine levels, antigen-specific cellular immune responses) (Tisoncik and Katze 2010). The grail of such endeavors is not merely to define correlations between a gene expression value and a phenotypic observation but to mathematically model the molecular networks and establish relationships that are predictive of how the system will behave when perturbed. Such perturbations could represent this change in the level of a specific transcript or protein, an altered level of a metabolite, or pharmacologic intervention to alter the biochemical activity of a system component. One requirement for the most effective modeling is voluminous, high-quality data under many different conditions (Cassman 2005).
NHP models have exceptional merit in replicating many features of human immunology and the host response to viral infection, but despite declining costs for expression profiling, aspects of the NHP model still pose challenges to systems biology approaches. The veterinary expenses for such models are a constraint on the size and flexibility of such models. And because species such as rhesus macaques are genetically diverse, outbred individuals, sufficient statistical power, and data volume require the use of more animals (“biological replicates”). The irony is that accessing a large set of human clinical samples at times appears more approachable than accessing a set of NHP samples, although this fails to consider the greater variability of demographic and clinical aspects that would require a human study to be much larger than the NHP experimental model. Likewise, certain scientific questions require protocols that cannot be replicated with human subjects.
The good news is that high-throughput molecular profiling, especially transcriptional profiling by arrays or RNA-seq (vide infra) continues to become more robust, faster, and cheaper. We can expect the application of these methods in NHP research, especially in priority areas such as AIDS vaccines, emerging pathogens, and biodefense. And although individual studies may lack the scope of “really big data” (barring the rare NHP study based on many animals), large integrative modeling approaches may depend on combining multiple studies from different investigators. In this respect, the sharing of genomic data and model systems between the NPRCs could be an important enabling infrastructure. Another alternative is the use of NHP models as one component in a systems biology approach that spans multiple model systems, where some models (cell lines, inbred mice) can be iterated more cheaply and rapidly (Figure 3). A recent publication from our systems virology effort illustrates such an application, where inferred regulatory influences from numerous influenza cell line infections were then used in predicting the temporal expression patterns in mouse and macaque (McDermott et al. 2011). Either of these tactics for NHP systems biology may be applicable in major campaigns, such as vaccine development, particularly in the AIDS vaccine field (Sekaly and Pulendran 2012).
Figure 3.
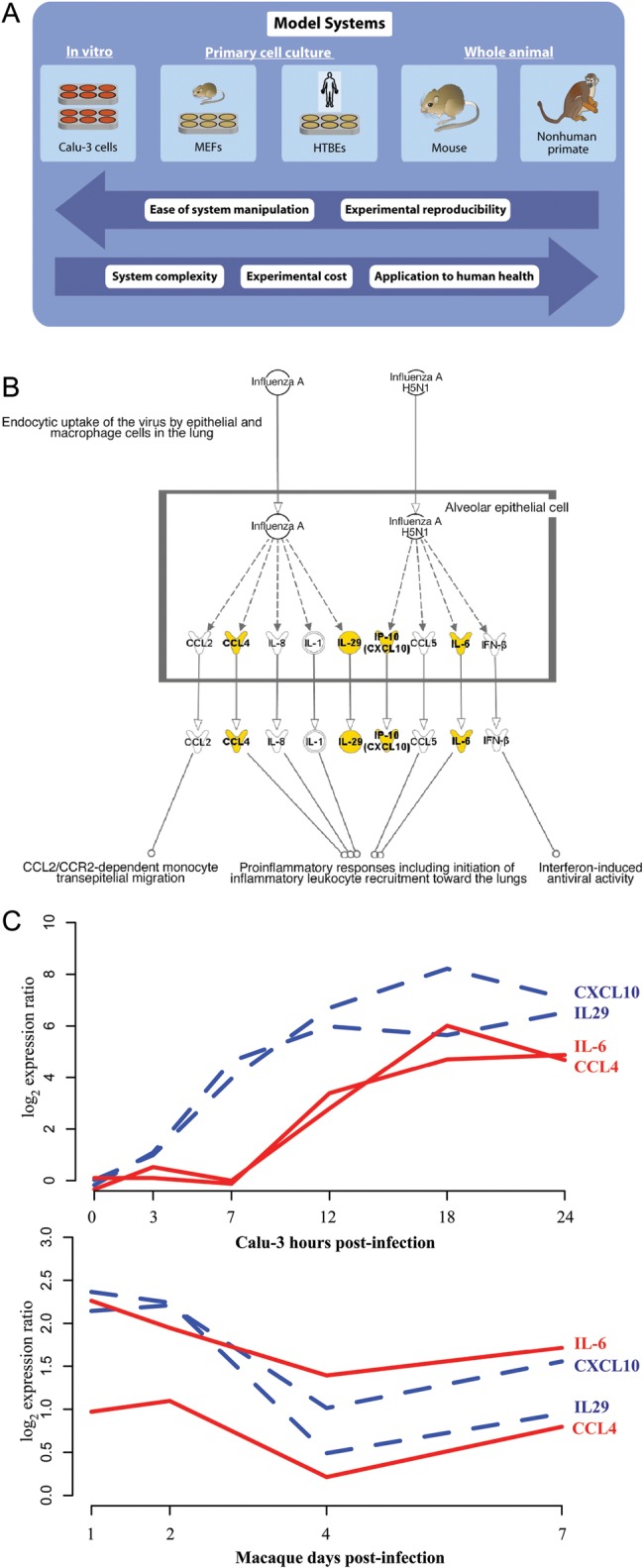
Absent large datasets from nonhuman primate (NHP) studies, smaller instances of such models are still important in systems biology endeavors. (A) NHP models can be considered on a continuum of model systems, where in vitro experiments and rodent models can be iterated more rapidly and at lower cost, resulting in computational predictions that can be evaluated in the more translationally relevant NHP setting. An exemplar of this strategy is shown in panels (B) and (C) (McDermott et al. 2011). (B) Computational analyses on longitudinal transcription profiles of Calu-3 cells infected with influenza strains were used to infer regulatory influences on the expression of the highlighted genes in the indicated signaling pathway. (C) Expression pattern of highly predicted genes in Calu-3 cells and macaque lungs after H5N1 infection. The inferred regulatory network predicts the temporal course in both these settings, despite their different timescales and different shapes. IFN, interferon; IL, interleukin.
Acknowledgments
We are grateful to Laurence Josset for supplying Figure 2. Research was supported by the Public Health Service (grants R2400011172, R2400011157, P30DA015625, P51RR00166, and U54AI081680) and by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (contract HHSN272200800060C). Funding was also provided by the NIAID Reagent Resource Support Program for AIDS Vaccine Development (Division of AIDS contract N01-A30018).
References
- Aarnink A, Dereuddre-Bosquet N, Vaslin B, Le Grand R, Winterton P, Apoil PA, Blancher A. Influence of the mhc genotype on the progression of experimental SIV infection in the mauritian cynomolgus macaque. Immunogenetics. 2011;63:267–274. doi: 10.1007/s00251-010-0504-6. [DOI] [PubMed] [Google Scholar]
- Apetrei C, Sumpter B, Souquiere S, Chahroudi A, Makuwa M, Reed P, Ribeiro RM, Pandrea I, Roques P, Silvestri G. Immunovirological analyses of chronically simian immunodeficiency virus SIVmnd-1- and SIVmnd-2-infected mandrills (Mandrillus sphinx) J Virol. 2011;85:13077–13087. doi: 10.1128/JVI.05693-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas T, Baskin CR, Diamond DL, Garcia-Sastre A, Bielefeldt-Ohmann H, Tumpey TM, Thomas MJ, Carter VS, Teal TH, Van Hoeven N, Proll S, Jacobs JM, Caldwell ZR, Gritsenko MA, Hukkanen RR, Camp DG, 2nd, Smith RD, Katze MG. Integrated molecular signature of disease: Analysis of influenza virus-infected macaques through functional genomics and proteomics. J Virol. 2006;80:10813–10828. doi: 10.1128/JVI.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin CR, Bielefeldt-Ohmann H, Tumpey TM, Sabourin PJ, Long JP, Garcia-Sastre A, Tolnay AE, Albrecht R, Pyles JA, Olson PH, Aicher LD, Rosenzweig ER, Murali-Krishna K, Clark EA, Kotur MS, Fornek JL, Proll S, Palermo RE, Sabourin CL, Katze MG. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc Natl Acad Sci U S A. 2009;106:3455–3460. doi: 10.1073/pnas.0813234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin CR, Garcia-Sastre A, Tumpey TM, Bielefeldt-Ohmann H, Carter VS, Nistal-Villan E, Katze MG. Integration of clinical data, pathology, and cdna microarrays in influenza virus-infected pigtailed macaques (Macaca nemestrina) J Virol. 2004;78:10420–10432. doi: 10.1128/JVI.78.19.10420-10432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BCM Human Genome Sequencing Center. Genome data. 2013 Available online (www.hgsc.bcm.tmc.edu/content/genome-data. ), accessed on March 21, 2013. [Google Scholar]
- Beaumier CM, Harris LD, Goldstein S, Klatt NR, Whitted S, McGinty J, Apetrei C, Pandrea I, Hirsch VM, Brenchley JM. CD4 downregulation by memory CD4+ t cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat Med. 2009;15:879–885. doi: 10.1038/nm.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle SE, Yin J, Shedlock DJ, Dai A, Yan J, Hirao L, Kutzler MA, Lewis MG, Andersen H, Lank SM, Karl JA, O'Connor DH, Khan A, Sardesai N, Chang J, Aicher L, Palermo RE, Weiner DB, Katze MG, Boyer J. Long-term programming of antigen-specific immunity from gene expression signatures in the PBMC of rhesus macaques immunized with an SIV DNA vaccine. PloS One. 2011;6 doi: 10.1371/journal.pone.0019681. e19681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosinger SE, Hosiawa KA, Cameron MJ, Persad D, Ran L, Xu L, Boulassel MR, Parenteau M, Fournier J, Rud EW, Kelvin DJ. Gene expression profiling of host response in models of acute HIV infection. J Immunol. 2004;173:6858–6863. doi: 10.4049/jimmunol.173.11.6858. [DOI] [PubMed] [Google Scholar]
- Bosinger SE, Li Q, Gordon SN, Klatt NR, Duan L, Xu L, Francella N, Sidahmed A, Smith AJ, Cramer EM, Zeng M, Masopust D, Carlis JV, Ran L, Vanderford TH, Paiardini M, Isett RB, Baldwin DA, Else JG, Staprans SI, Silvestri G, Haase AT, Kelvin DJ. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest. 2009;119:3556–3572. doi: 10.1172/JCI40115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JN, Palermo RE, Baskin CR, Gritsenko M, Sabourin PJ, Long JP, Sabourin CL, Bielefeldt-Ohmann H, Garcia-Sastre A, Albrecht R, Tumpey TM, Jacobs JM, Smith RD, Katze MG. Macaque proteome response to highly pathogenic avian influenza and 1918 reassortant influenza virus infections. J Virol. 2010;84:12058–12068. doi: 10.1128/JVI.01129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding rnas reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassman M. Barriers to progress in systems biology. Nature. 2005;438 doi: 10.1038/4381079a. 1079. [DOI] [PubMed] [Google Scholar]
- Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G. Natural SIV hosts: Showing aids the door. Science. 2012;335:1188–1193. doi: 10.1126/science.1217550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang ST, Sova P, Peng X, Weiss J, Law GL, Palermo RE, Katze MG. Next-generation sequencing reveals HIV-1–mediated suppression of T cell activation and RNA processing and regulation of noncoding RNA expression in a CD4+ T cell line. mBio 2:e00134–11. 2011 doi: 10.1128/mBio.00134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Chung HK, Pise-Masison CA, Radonovich MF, Brady J, Lee JK, Cheon SY, Markham P, Cristillo A, Pal R. Cellular gene expression profiles in rhesus macaques challenged mucosally with a pathogenic R5 tropic simian human immunodeficiency virus isolate. Viral Immunol. 2008;21:411–423. doi: 10.1089/vim.2008.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilloniz C, Shinya K, Peng X, Korth MJ, Proll SC, Aicher LD, Carter VS, Chang JH, Kobasa D, Feldmann F, Strong JE, Feldmann H, Kawaoka Y, Katze MG. Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000604. e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. Fatal outcome of human influenza a (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks SG, Walker BD. Human immunodeficiency virus controllers: Mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Diop OM, Gueye A, Dias-Tavares M, Kornfeld C, Faye A, Ave P, Huerre M, Corbet S, Barre-Sinoussi F, Muller-Trutwin MC. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in African green monkeys. J Virol. 2000;74:7538–7547. doi: 10.1128/jvi.74.16.7538-7547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebeling M, Kung E, See A, Broger C, Steiner G, Berrera M, Heckel T, Iniguez L, Albert T, Schmucki R, Biller H, Singer T, Certa U. Genome-based analysis of the nonhuman primate Macaca fascicularis as a model for drug safety assessment. Genome Res. 2011;21:1746–1756. doi: 10.1101/gr.123117.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensembl.org. 2010a Macaque assembly and gene annotation -mmul_1. Available online (http://uswest.ensembl.org/Macaca_mulatta/Info/Annotation#assembly. ), accessed June 11, 2013. [Google Scholar]
- Ensembl.org. 2010b Mouse lemur assembly and gene annotation -micmur1. Available online (http://uswest.ensembl.org/Microcebus_murinus/Info/Annotation#assembly. ), accessed June 11, 2013. [Google Scholar]
- Ensembl.org. 2011 Marmoset assembly and gene annotation. Available online (http://www.ensembl.org/Callithrix_jacchus/Info/Annotation/#assembly. ), accessed March 20, 2013. [Google Scholar]
- Ensembl.org. 2012a Chimpanzee assembly and gene annotation -chimp2.1.4. Available online (http://uswest.ensembl.org/Pan_troglodytes/Info/Annotation#assembly. ), accessed June 11, 2013. [Google Scholar]
- Ensembl.org. 2012b Gibbon assembly and gene annotation -nleu1.0. Available online (http://uswest.ensembl.org/Nomascus_leucogenys/Info/Annotation#assembly. ), Genebuild released Apr 2011. Accessed June 11, 2013. [Google Scholar]
- Ensembl.org. 2012c Orangutan assembly and gene annotation -ppyg2. Available online (http://uswest.ensembl.org/Pongo_abelii/Info/Annotation#assembly. ), accessed June 11, 2013. [Google Scholar]
- Ensembl.org. 2013 Hamadryas baboon (Papio hamadryas) description. Available online (http://pre.ensembl.org/Papio_hamadryas/Info/Index. ), accessed March 20, 2013. [Google Scholar]
- Fang X, Zhang Y, Zhang R, Yang L, Li M, Ye K, Guo X, Wang J, Su B. Genome sequence and global sequence variation map with 5.5 million snps in chinese rhesus macaque. Genome Biol. 2011;12 doi: 10.1186/gb-2011-12-7-r63. R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre D, Lederer S, Kanwar B, Ma ZM, Proll S, Kasakow Z, Mold J, Swainson L, Barbour JD, Baskin CR, Palermo R, Pandrea I, Miller CJ, Katze MG, McCune JM. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000295. e1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett GL, Raveendran M, Deiros DR, Chen D, Yu F, Harris RA, Ren Y, Muzny DM, Reid JG, Wheeler DA, Worley KC, Shelton SE, Kalin NH, Milosavljevic A, Gibbs R, Rogers J. Characterization of single-nucleotide variation in indian-origin rhesus macaques (Macaca mulatta) BMC Genomics. 2011;12:311. doi: 10.1186/1471-2164-12-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George MD, Reay E, Sankaran S, Dandekar S. Early antiretroviral therapy for simian immunodeficiency virus infection leads to mucosal CD4+ T-cell restoration and enhanced gene expression regulating mucosal repair and regeneration. J Virol. 2005;79:2709–2719. doi: 10.1128/JVI.79.5.2709-2719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George MD, Sankaran S, Reay E, Gelli AC, Dandekar S. High-throughput gene expression profiling indicates dysregulation of intestinal cell cycle mediators and growth factors during primary simian immunodeficiency virus infection. Virology. 2003;312:84–94. doi: 10.1016/s0042-6822(03)00207-1. [DOI] [PubMed] [Google Scholar]
- George MD, Verhoeven D, McBride Z, Dandekar S. Gene expression profiling of gut mucosa and mesenteric lymph nodes in simian immunodeficiency virus–infected macaques with divergent disease course. J Med Primatol. 2006;35:261–269. doi: 10.1111/j.1600-0684.2006.00180.x. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y, Dinh HH, Dugan-Rocha S, Fulton LA, Gabisi RA, Garner TT, Godfrey J, Hawes AC, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Kirkness EF, Cree A, Fowler RG, Lee S, Lewis LR, Li Z, Liu YS, Moore SM, Muzny D, Nazareth LV, Ngo DN, Okwuonu GO, Pai G, Parker D, Paul HA, Pfannkoch C, Pohl CS, Rogers YH, Ruiz SJ, Sabo A, Santibanez J, Schneider BW, Smith SM, Sodergren E, Svatek AF, Utterback TR, Vattathil S, Warren W, White CS, Chinwalla AT, Feng Y, Halpern AL, Hillier LW, Huang X, Minx P, Nelson JO, Pepin KH, Qin X, Sutton GG, Venter E, Walenz BP, Wallis JW, Worley KC, Yang SP, Jones SM, Marra MA, Rocchi M, Schein JE, Baertsch R, Clarke L, Csuros M, Glasscock J, Harris RA, Havlak P, Jackson AR, Jiang H, Liu Y, Messina DN, Shen Y, Song HX, Wylie T, Zhang L, Birney E, Han K, Konkel MK, Lee J, Smit AF, Ullmer B, Wang H, Xing J, Burhans R, Cheng Z, Karro JE, Ma J, Raney B, She X, Cox MJ, Demuth JP, Dumas LJ, Han SG, Hopkins J, Karimpour-Fard A, Kim YH, Pollack JR, Vinar T, Addo-Quaye C, Degenhardt J, Denby A, Hubisz MJ, Indap A, Kosiol C, Lahn BT, Lawson HA, Marklein A, Nielsen R, Vallender EJ, Clark AG, Ferguson B, Hernandez RD, Hirani K, Kehrer-Sawatzki H, Kolb J, Patil S, Pu LL, Ren Y, Smith DG, Wheeler DA, Schenck I, Ball EV, Chen R, Cooper DN, Giardine B, Hsu F, Kent WJ, Lesk A, Nelson DL, O'Brien W E, Prufer K, Stenson PD, Wallace JC, Ke H, Liu XM, Wang P, Xiang AP, Yang F, Barber GP, Haussler D, Karolchik D, Kern AD, Kuhn RM, Smith KE, Zwieg AS. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Go JT, Belisle SE, Tchitchek N, Tumpey TM, Ma W, Richt JA, Safronetz D, Feldmann H, Katze MG. 2009 pandemic H1N1 influenza virus elicits similar clinical course but differential host transcriptional response in mouse, macaque, and swine infection models. BMC Genomics. 2012;13:627. doi: 10.1186/1471-2164-13-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein S, Ourmanov I, Brown CR, Plishka R, Buckler-White A, Byrum R, Hirsch VM. Plateau levels of viremia correlate with the degree of CD4+-T-cell loss in simian immunodeficiency virus SIVagm-infected pigtailed macaques: Variable pathogenicity of natural SIVagm isolates. J Virol. 2005;79:5153–5162. doi: 10.1128/JVI.79.8.5153-5162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, Dandekar S. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, Sakabe S, Imai M, Hatta Y, Watanabe S, Li C, Yamada S, Fujii K, Murakami S, Imai H, Kakugawa S, Ito M, Takano R, Iwatsuki-Horimoto K, Shimojima M, Horimoto T, Goto H, Takahashi K, Makino A, Ishigaki H, Nakayama M, Okamatsu M, Warshauer D, Shult PA, Saito R, Suzuki H, Furuta Y, Yamashita M, Mitamura K, Nakano K, Nakamura M, Brockman-Schneider R, Mitamura H, Yamazaki M, Sugaya N, Suresh M, Ozawa M, Neumann G, Gern J, Kida H, Ogasawara K, Kawaoka Y. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquelin B, Mayau V, Targat B, Liovat AS, Kunkel D, Petitjean G, Dillies MA, Roques P, Butor C, Silvestri G, Giavedoni LD, Lebon P, Barre-Sinoussi F, Benecke A, Muller-Trutwin MC. Nonpathogenic siv infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119:3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset L, Engelmann F, Haberthur K, Kelly S, Park B, Kawoaka Y, Garcia-Sastre A, Katze MG, Messaoudi I. Increased viral loads and exacerbated innate host responses in aged macaques infected with the 2009 pandemic H1N1 influenza a virus. J Virol. 2012;86:11115–11127. doi: 10.1128/JVI.01571-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanthaswamy S, Capitanio JP, Dubay CJ, Ferguson B, Folks T, Ha JC, Hotchkiss CE, Johnson ZP, Katze MG, Kean LS, Kubisch HM, Lank S, Lyons LA, Miller GM, Nylander J, O'Connor DH, Palermo RE, Smith DG, Vallender EJ, Wiseman RW, Rogers J. Resources for genetic management and genomics research on non-human primates at the national primate research centers (NPRCs) J Med Primatol. 2009;38:17–23. doi: 10.1111/j.1600-0684.2009.00371.x. [DOI] [PubMed] [Google Scholar]
- Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, Hatta Y, Kim JH, Halfmann P, Hatta M, Feldmann F, Alimonti JB, Fernando L, Li Y, Katze MG, Feldmann H, Kawaoka Y. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lederer S, Favre D, Walters KA, Proll S, Kanwar B, Kasakow Z, Baskin CR, Palermo R, McCune JM, Katze MG. Transcriptional profiling in pathogenic and non-pathogenic siv infections reveals significant distinctions in kinetics and tissue compartmentalization. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000296. e1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Duan L, Estes JD, Ma ZM, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, Haase AT. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- Li Y, Chan EY, Katze MG. Functional genomics analyses of differential macaque peripheral blood mononuclear cell infections by human immunodeficiency virus-1 and simian immunodeficiency virus. Virology. 2007;366:137–149. doi: 10.1016/j.virol.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magness CL, Fellin PC, Thomas MJ, Korth MJ, Agy MB, Proll SC, Fitzgibbon M, Scherer CA, Miner DG, Katze MG, Iadonato SP. Analysis of the Macaca mulatta transcriptome and the sequence divergence between macaca and human. Genome Biol. 2005;6 doi: 10.1186/gb-2005-6-7-r60. R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. Next-generation DNA sequencing methods. Ann Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- McDermott JE, Shankaran H, Eisfeld AJ, Belisle SE, Neumann G, Li C, McWeeney S, Sabourin C, Kawaoka Y, Katze MG, Waters KM. Conserved host response to highly pathogenic avian influenza virus infection in human cell culture, mouse and macaque model systems. BMC Syst Biol. 2011;5:190. doi: 10.1186/1752-0509-5-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, Boden D, Racz P, Markowitz M. Primary HIV-1 infection is associated with preferential depletion of CD4(+) t lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Marthas M, Miller C, Duerr A, Cheng-Mayer C, Desrosiers R, Flores J, Haigwood N, Hu SL, Johnson RP, Lifson J, Montefiori D, Moore J, Robert-Guroff M, Robinson H, Self S, Corey L. The use of nonhuman primate models in HIV vaccine development. PLoS Med. 2008;5 doi: 10.1371/journal.pmed.0050173. e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Mukai R, Inoue-Murayama M, Yoshikawa Y. An African green monkey lacking peripheral CD4 lymphocytes that retains helper T cell activity and coexists with SIVagm. Clin Exper Immunol. 1999;117:504–512. doi: 10.1046/j.1365-2249.1999.00999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama EE, Shioda T. Trim5alpha and species tropism of HIV/SIV. Frontiers Microbiol 3:13. 2012 doi: 10.3389/fmicb.2012.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo RE, Fuller DH. 'Omics investigations of HIV and SIV pathogenesis and innate immunity. Curr Topics Microbiol Immunol. 2013;363:87–116. doi: 10.1007/82_2012_255. [DOI] [PubMed] [Google Scholar]
- Palermo RE, Patterson LJ, Aicher LD, Korth MJ, Robert-Guroff M, Katze MG. Genomic analysis reveals pre- and postchallenge differences in a rhesus macaque aids vaccine trial: Insights into mechanisms of vaccine efficacy. J Virol. 2011;85:1099–1116. doi: 10.1128/JVI.01522-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandrea I, Apetrei C, Dufour J, Dillon N, Barbercheck J, Metzger M, Jacquelin B, Bohm R, Marx PA, Barre-Sinoussi F, Hirsch VM, Muller-Trutwin MC, Lackner AA, Veazey RS. Simian immunodeficiency virus SIVagm.Sab infection of Caribbean African green monkeys: A new model for the study of SIV pathogenesis in natural hosts. J Virol. 2006;80:4858–4867. doi: 10.1128/JVI.80.10.4858-4867.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson LJ, Beal J, Demberg T, Florese RH, Malkevich N, Venzon D, Aldrich K, Richardson E, Kalyanaraman VS, Kalisz I, Lee EM, Montefiori DC, Robey FA, Robert-Guroff M. Replicating adenovirus HIV/SIV recombinant priming alone or in combination with a gp140 protein boost results in significant control of viremia following a SHIV89.6p challenge in mamu-a*01 negative rhesus macaques. Virology. 2008;374:322–337. doi: 10.1016/j.virol.2007.12.037. [DOI] [PubMed] [Google Scholar]
- Peiris JS, Yu WC, Leung CW, Cheung CY, Ng WF, Nicholls JM, Ng TK, Chan KH, Lai ST, Lim WL, Yuen KY, Guan Y. Re-emergence of fatal human influenza a subtype H5N1 disease. Lancet. 2004;363:617–619. doi: 10.1016/S0140-6736(04)15595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, Bradel-Tretheway BG, Korth MJ, Castle JC, Biery MC, Bouzek HK, Haynor DR, Frieman MB, Heise M, Raymond CK, Baric RS, Katze MG. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio 1:e00206–10. 2010 doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Gralinski L, Ferris MT, Frieman MB, Thomas MJ, Proll S, Korth MJ, Tisoncik JR, Heise M, Luo S, Schroth GP, Tumpey TM, Li C, Kawaoka Y, Baric RS, Katze MG. Integrative deep sequencing of the mouse lung transcriptome reveals differential expression of diverse classes of small RNAs in response to respiratory virus infection. mBio 2:e00198–11. 2011 doi: 10.1128/mBio.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Melsted P, Marioni JC, Wang Y, Bainer R, Pickrell JK, Michelini K, Zehr S, Yoder AD, Stephens M, Pritchard JK, Gilad Y. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 2012;22:602–610. doi: 10.1101/gr.130468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Reeves D, Melsted P, Ratan A, Miller W, Michelini K, Louis EE, Jr, Pritchard JK, Mason CE, Gilad Y. A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar. Genome Biol Evol. 2012;4:126–135. doi: 10.1093/gbe/evr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipes L, Li S, Bozinoski M, Palermo R, Peng X, Blood P, Kelly S, Weiss JM, Thierry-Mieg J, Thierry-Mieg D, Zumbo P, Chen R, Schroth GP, Mason CE, Katze MG. The non-human primate reference transcriptome resource (NHPTR) for comparative functional genomics. Nucleic Acids Res. 2013;41:D906–D914. doi: 10.1093/nar/gks1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, Munch K, Hellmann I, Akagi K, Miller JR, Walenz B, Koren S, Sutton G, Kodira C, Winer R, Knight JR, Mullikin JC, Meader SJ, Ponting CP, Lunter G, Higashino S, Hobolth A, Dutheil J, Karakoc E, Alkan C, Sajjadian S, Catacchio CR, Ventura M, Marques-Bonet T, Eichler EE, Andre C, Atencia R, Mugisha L, Junhold J, Patterson N, Siebauer M, Good JM, Fischer A, Ptak SE, Lachmann M, Symer DE, Mailund T, Schierup MH, Andres AM, Kelso J, Paabo S. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M, Sabeti PC. Detecting novel associations in large data sets. Science. 2011;334:1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts ES, Burudi EM, Flynn C, Madden LJ, Roinick KL, Watry DD, Zandonatti MA, Taffe MA, Fox HS. Acute SIV infection of the brain leads to upregulation of IL6 and interferon-regulated genes: Expression patterns throughout disease progression and impact on neuroaids. J Neuroimmunol. 2004;157:81–92. doi: 10.1016/j.jneuroim.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Roberts ES, Zandonatti MA, Watry DD, Madden LJ, Henriksen SJ, Taffe MA, Fox HS. Induction of pathogenic sets of genes in macrophages and neurons in neuroaids. Am J Pathol. 2003;162:2041–2057. doi: 10.1016/S0002-9440(10)64336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotger M, Dalmau J, Rauch A, McLaren P, Bosinger SE, Martinez R, Sandler NG, Roque A, Liebner J, Battegay M, Bernasconi E, Descombes P, Erkizia I, Fellay J, Hirschel B, Miro JM, Palou E, Hoffmann M, Massanella M, Blanco J, Woods M, Gunthard HF, de Bakker P, Douek DC, Silvestri G, Martinez-Picado J, Telenti A. Comparative transcriptomics of extreme phenotypes of human HIV-1 infection and SIV infection in sooty mangabey and rhesus macaque. J Clin Invest. 2011;121:2391–2400. doi: 10.1172/JCI45235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safronetz D, Rockx B, Feldmann F, Belisle SE, Palermo RE, Brining D, Gardner D, Proll SC, Marzi A, Tsuda Y, Lacasse RA, Kercher L, York A, Korth MJ, Long D, Rosenke R, Shupert WL, Aranda CA, Mattoon JS, Kobasa D, Kobinger G, Li Y, Taubenberger JK, Richt JA, Parnell M, Ebihara H, Kawaoka Y, Katze MG, Feldmann H. Pandemic swine-origin H1N1 influenza A virus isolates show heterogeneous virulence in macaques. J Virol. 2011;85:1214–1223. doi: 10.1128/JVI.01848-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, Parham P, Carrington M, Beck S. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran S, Guadalupe M, Reay E, George MD, Flamm J, Prindiville T, Dandekar S. Gut mucosal T cell responses and gene expression correlate with protection against disease in long-term HIV-1-infected nonprogressors. Proc Natl Acad Sci U S A. 2005;102:9860–9865. doi: 10.1073/pnas.0503463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauermann U, Siddiqui R, Suh YS, Platzer M, Leuchte N, Meyer H, Matz-Rensing K, Stoiber H, Nurnberg P, Hunsmann G, Stahl-Hennig C, Krawczak M. MHC class I haplotypes associated with survival time in simian immunodeficiency virus (SIV)–infected rhesus macaques. Gene Immun. 2008;9:69–80. doi: 10.1038/sj.gene.6364448. [DOI] [PubMed] [Google Scholar]
- Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, McCarthy S, Montgomery SH, Schwalie PC, Tang YA, Ward MC, Xue Y, Yngvadottir B, Alkan C, Andersen LN, Ayub Q, Ball EV, Beal K, Bradley BJ, Chen Y, Clee CM, Fitzgerald S, Graves TA, Gu Y, Heath P, Heger A, Karakoc E, Kolb-Kokocinski A, Laird GK, Lunter G, Meader S, Mort M, Mullikin JC, Munch K, O'Connor TD, Phillips AD, Prado-Martinez J, Rogers AS, Sajjadian S, Schmidt D, Shaw K, Simpson JT, Stenson PD, Turner DJ, Vigilant L, Vilella AJ, Whitener W, Zhu B, Cooper DN, de Jong P, Dermitzakis ET, Eichler EE, Flicek P, Goldman N, Mundy NI, Ning Z, Odom DT, Ponting CP, Quail MA, Ryder OA, Searle SM, Warren WC, Wilson RK, Schierup MH, Rogers J, Tyler-Smith C, Durbin R. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekaly R, Pulendran B. Systems biology in understanding HIV pathogenesis and guiding vaccine development. Curr Opin HIV AIDS. 2012;7:1–3. doi: 10.1097/COH.0b013e32834e0667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya K, Gao Y, Cilloniz C, Suzuki Y, Fujie M, Deng G, Zhu Q, Fan S, Makino A, Muramoto Y, Fukuyama S, Tamura D, Noda T, Eisfeld AJ, Katze MG, Chen H, Kawaoka Y. Integrated clinical, pathologic, virologic, and transcriptomic analysis of H5N1 influenza virus-induced viral pneumonia in the rhesus macaque. J Virol. 2012;86:6055–6066. doi: 10.1128/JVI.00365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri G, Sodora DL, Koup RA, Paiardini M, O'Neil SP, McClure HM, Staprans SI, Feinberg MB. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity. 2003;18:441–452. doi: 10.1016/s1074-7613(03)00060-8. [DOI] [PubMed] [Google Scholar]
- Spindel ER, Pauley MA, Jia Y, Gravett C, Thompson SL, Boyle NF, Ojeda SR, Norgren RB., Jr. Leveraging human genomic information to identify nonhuman primate sequences for expression array development. BMC Genomics. 2005;6:160. doi: 10.1186/1471-2164-6-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MJ, Agy MB, Proll SC, Paeper BW, Li Y, Jensen KL, Korth MJ, Katze MG. Functional gene analysis of individual response to challenge of SIVmac239 in M. mulatta PBMC culture. Virology. 2006;348:242–252. doi: 10.1016/j.virol.2005.12.015. [DOI] [PubMed] [Google Scholar]
- Tisoncik JR, Katze MG. What is systems biology? Future Microbiol 5. 2010:139–141. doi: 10.2217/fmb.09.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahey MT, Nau ME, Taubman M, Yalley-Ogunro J, Silvera P, Lewis MG. Patterns of gene expression in peripheral blood mononuclear cells of rhesus macaques infected with SIVmac251 and exhibiting differential rates of disease progression. AIDS Res Hum Retroviruses. 2003;19:369–387. doi: 10.1089/088922203765551728. [DOI] [PubMed] [Google Scholar]
- Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- Wallace JC, Korth MJ, Paeper B, Proll SC, Thomas MJ, Magness CL, Iadonato SP, Nelson C, Katze MG. High-density rhesus macaque oligonucleotide microarray design using early-stage rhesus genome sequence information and human genome annotations. BMC Genomics. 2007;8:28. doi: 10.1186/1471-2164-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Xiao P, Zhao J, Patterson LJ, Brocca-Cofano E, Venzon D, Kozlowski PA, Hidajat R, Demberg T, Robert-Guroff M. Multiple vaccine-elicited nonneutralizing antienvelope antibody activities contribute to protective efficacy by reducing both acute and chronic viremia following simian/human immunodeficiency virus SHIV89.6p challenge in rhesus macaques. J Virol. 2010;84:7161–7173. doi: 10.1128/JVI.00410-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Zhang G, Fang X, Zhang Y, Li C, Ling F, Cooper DN, Li Q, Li Y, van Gool AJ, Du H, Chen J, Chen R, Zhang P, Huang Z, Thompson JR, Meng Y, Bai Y, Wang J, Zhuo M, Wang T, Huang Y, Wei L, Li J, Wang Z, Hu H, Yang P, Le L, Stenson PD, Li B, Liu X, Ball EV, An N, Huang Q, Zhang Y, Fan W, Zhang X, Li Y, Wang W, Katze MG, Su B, Nielsen R, Yang H, Wang J, Wang X, Wang J. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and chinese rhesus macaques. Nat Biotechnol. 2011;29:1019–1023. doi: 10.1038/nbt.1992. [DOI] [PubMed] [Google Scholar]
- Yandell M, Ence D. A beginner's guide to eukaryotic genome annotation. Nat Rev Genet. 2012;13:329–342. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]
- Zhang X, Goodsell J, Norgren RB., Jr. Limitations of the rhesus macaque draft genome assembly and annotation. BMC Genomics. 2012;13:206. doi: 10.1186/1471-2164-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]


