Abstract
Nonhuman primates (NHP) provide crucial biomedical model systems intermediate between rodents and humans. The vervet monkey (also called the African green monkey) is a widely used NHP model that has unique value for genetic and genomic investigations of traits relevant to human diseases. This article describes the phylogeny and population history of the vervet monkey and summarizes the use of both captive and wild vervet monkeys in biomedical research. It also discusses the effort of an international collaboration to develop the vervet monkey as the most comprehensively phenotypically and genomically characterized NHP, a process that will enable the scientific community to employ this model for systems biology investigations.
Keywords: African green monkey, genetics, genomics, phenomics, simian immunodeficiency virus [SIV], systems biology, transcriptomics, vervet
Introduction
The vervet monkey is a critical nonhuman primate (NHP) model system for biomedical research, and is particularly widely employed for investigations of brain and behavior, metabolism, and immunity. Its interest to immunologists and virologists derives mainly from its status as the most abundant natural host of simian immunodeficiency virus (SIV). Confusingly, primatologists typically employ the term “vervet” to refer to the entire genus Chlorocebus, a usage that we employ here; immunologists and virologists typically restrict the usage of this term to members of the subspecies (or species) C. pygerythrus, using the term “African green monkey” (AGM) to refer to the genus as a whole. In this review, we describe the steps that we and other members of an international collaborative effort have taken to develop this species into a comprehensive NHP model for systems biology. These steps include comprehensive phenotypic, genetic, and genomic characterization of large study samples of pedigreed vervets assessed in research colonies as well as independent vervets assayed in wild populations. In this article, we first review the motivation for creating an NHP systems biology model, then discuss the specific features of the vervet that are most relevant to the design and development of such a model, and finally, describe the progress so far in using the vervet to elucidate the biological underpinnings of phenotypes relevant to human diseases.
Why We Need NHP Systems Biology Models, and Why the Vervet Is Ideal for This Purpose
Trait Conservation between Humans and Model Animals
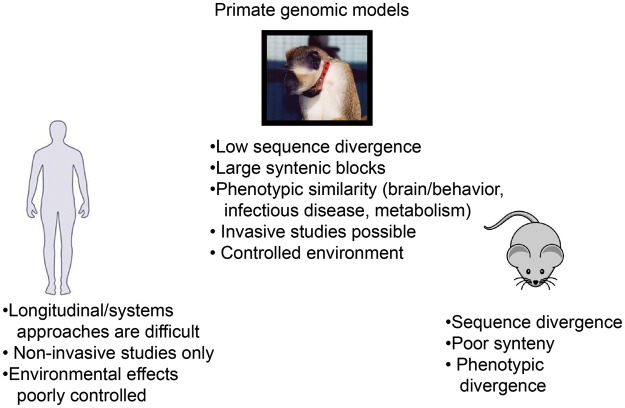
The importance of NHP models for biomedicine is becoming increasingly apparent, as are their advantages over rodent models (Figure 1). Humans and monkeys share extensive conservation ranging across various levels of biological organization—from genome sequence and structure through molecular pathways to complex physiological and behavioral traits. NHPs provide models of human biology, behavior, and disease that cannot be approximated by other model systems, which, for some traits, are simply too different from that of humans. For example, inflammatory processes that are central to a wide range of diseases use molecular pathways that are particularly divergent between humans and rodents. Such differences may explain the failure in human trials of recently developed agents for treating sepsis (Coe 1993; Maestripieri and Hoffman 2011; Seok et al. 2013; Williams et al. 2007). Other variables for which the wide divergence between rodents and humans may make NHPs better disease models include diet, circadian rhythms, social structure, and susceptibility to major pathogens. Additionally, Old World monkeys (OWMs), including vervets, share with humans endocrine characteristics such as a menstrual cycle and the experience of menopause, making them well suited to model hormonal effects on disease vulnerability and sex differences in disease expression, especially coronary artery atherosclerosis and type 2 diabetes. Most OWMs develop atherosclerosis if fed a typical American diet and develop obesity and impaired glucose tolerance that progresses in some individuals to type 2 diabetes. The inadequacy of rodent models of such phenotypes dictates the use of NHPs and mitigates their limitations.
Figure 1.
Nonhuman primates (NHPs) as intermediate models between human and rodents. NHPs are indispensable models for biomedical research because of their close phylogenetic relationship to humans, as reflected in their high conservation with humans in terms of genomic sequence and structure, physiology, behavior, susceptibility to diseases, and other phenotypes. At the same time, NHPs may be employed for longitudinal and invasive investigations that are impractical or unethical in humans.
Different Applications of NHP and Rodent Models
Mouse research mostly models human diseases by recapitulating human mutations and probing for their phenotypic impact. Despite the many discoveries obtained through such models, the biological differences between rodents and humans have often precluded the identification in mutant mice of phenotypic features that are central to human diseases. NHP disease models are intrinsically different: we do not expect to find exact vervet homologues of variants putatively associated with disorders such as autism, schizophrenia, or type 2 diabetes. Instead, research in vervet study populations, as with most NHP investigations, focuses on understanding the basis of quantitative phenotypes hypothesized to be important components of complex human disorders. These investigations include both epidemiologic and genetic studies, as well as preclinical evaluations and both drug and vaccine testing.
The Opportunity in NHPs to Conduct Studies That Are Infeasible in Humans
Although investigations of quantitative phenotypes in humans have been highly informative (e.g., in discovering genetic associations), they are limited by the infeasibility of controlling environmental exposures and of conducting invasive procedures, such as large-scale collection of tissue samples from internal organs. NHP models, by contrast, offer the opportunity to elucidate biological pathways (e.g., tissue-specific gene networks) underlying quantitative phenotypic variation. Such pathways can then become a focus for human translational research. In contrast with human studies, investigations of captive NHPs can control diet and other environmental factors, manipulate genetic relationships, sample all types of tissues, and employ physiologic and pharmacologic manipulations.
Vervet as a Model NHP Species
Vervets are extraordinarily adaptable, inhabiting most of sub–Saharan Africa (except deserts and tropical forests) and are perhaps the most populous African NHP. For these reasons, vervets are good model for investigations of adaptations to local environments (e.g., physical conditions, diet, pathogens) and for large-scale population studies. Additionally, they are widely and increasingly employed in biomedical research internationally, with a PubMed citation record (over the past 10 years) close to that of rhesus macaque and greater than that of any other NHP. The growth in use of the vervet model derives in part from the fact that it provides an alternative to the Indian-origin rhesus macaque, of which there is now a critical shortage for biomedical research. In contrast, vervets (particularly those descended from feral Caribbean populations) are abundant, disease-free, and easy to handle. The vervet is similar to rhesus macaques in behavior and physiology (Coe 1993; Disotell 2000; Raleigh et al. 1992; Ziegler and Bercovitch 1990), is more accessible (from Caribbean populations), is less expensive, and is accompanied by fewer health and safety risks (Baulu et al. 2002; Gordon et al. 2005). Most important, several features of the vervet genome and population history make it uniquely valuable for genetic and evolutionary investigations, as discussed below.
Phylogeny of Vervets
Vervet Taxonomy
The Cercopithecidae or Old World Monkey (OWM) family includes the tribe Cercopithecini (which includes vervets) and the tribe Papioni (which includes macaques and baboons) and diverged from the family Hominidae (which includes humans and apes) approximately 23 million years ago (Goodman et al. 1998). The two OWM tribes separated from one another about 11 to 12 million years ago (Tosi et al. 2005).
The time of differentiation of the vervet from other members of its tribe is estimated to be about 1.5 to 3 million years ago, based on molecular phylogeny analyses from 54 nuclear genes in 191 primate taxa (Perelman et al. 2011) and similar analyses from mitochondrial genome data (Wertheim and Worobey 2007). Although there is growing consensus that Chlorocebus constitutes a distinct genus, this remains a matter of debate. Morphologic data have not resolved whether its members form a single species (aethiops) with five major subspecies (aethiops, AKA grivet; cynosuros, AKA malbrouck; pygerythrus, AKA vervet; sabaeus, AKA callithrix; tantalus, AKA tantalus) and one subspecies limited to a small mountainous zone in Ethiopia (djamjamensis, AKA the Bale monkey) or comprise distinct species. Documented inbreeding between subspecies supports the former taxonomy, which we use here; this has mainly occurred in narrow contact zones in areas of recent deforestation (Detwiler et al. 2005; Mekonnen et al. 2012). The new availability of genomic sequence for several members of each of the major vervet subspecies may soon enable clarification of these taxonomic controversies through population genetic analyses.
Vervet Populations of the Caribbean
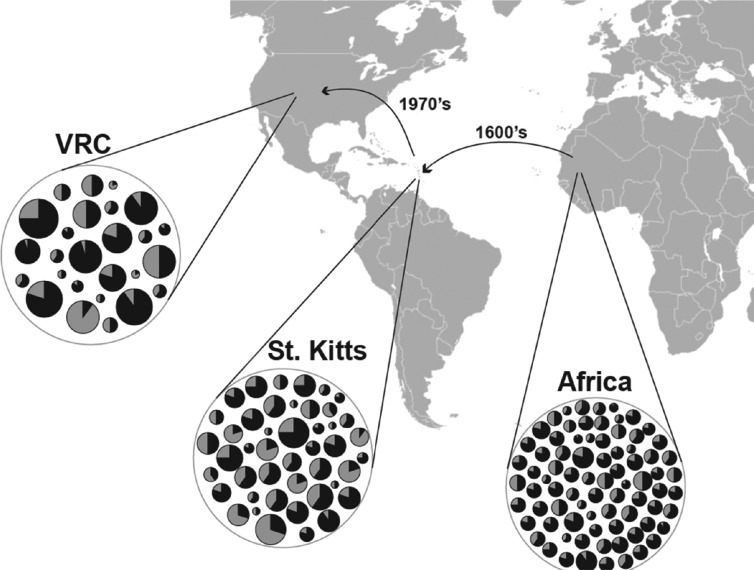
Much biomedical research on vervets focuses on descendants of a small number of monkeys brought as pets from Africa to the Caribbean islands of St. Kitts, Nevis, and Barbados in the 17th and 18th centuries (Allen 1911; Denham 1987; Horrocks 1986; Poirier 1972; van der Kuyl et al. 1995). Vervets on these islands established feral populations, which, in the absence of predators and major African pathogens, have expanded to levels estimated at more than 50,000 (Jasinska et al. 2012). These feral populations may represent one of the most dramatic examples in any primate species of expansion after a population bottleneck. This pattern of growth is similar to, but more extreme than, that observed in human population isolates that have been exceptionally valuable in genetic mapping of disease-related genes (Peltonen 2000). As in these human populations, in Caribbean vervets the degree of interindividual genetic variation that has survived the bottleneck is more than sufficient for the purposes of genetic mapping (Freimer et al. 2007; Jasinska et al. 2012). A second bottleneck occurred in the founding at the University of California–Los Angeles (UCLA) of the largest vervet research colony, the Vervet Research Colony (VRC), beginning in the 1970s, from 57 founders brought from St. Kitts and Nevis, as discussed in more detail below (Figure 2).
Figure 2.
Effect of genetic bottlenecks on genetic architecture in the Caribbean vervet. The Caribbean vervets employed in biomedical research include those living on the islands of St. Kitts, Nevis, and Barbados, as well as those brought form St. Kitts and Nevis to the University of California–Los Angeles to found the Vervet Research Colony (VRC). The history of the VRC includes two genetic bottlenecks, the first beginning in the 1600s when vervets were brought from West Africa to St. Kitts and Nevis, and the second beginning in the 1970s when wild caught animals from both islands were used as founders to establish the VRC. The figure provides a schematic representation of the effect of genetic bottlenecks on the genetic architecture of the Caribbean vervet. The pie charts within each circle depict polymorphic loci in each setting. The number of polymorphisms is presumed to be reduced through the progressive bottlenecks, as shown by the decreasing number of pie charts. The proportion of genetic variance within the population represented by a single locus is presumed to increase as the number of polymorphic loci decrease, as illustrated by the size of each pie chart. Genetic drift may dramatically alter the allele frequencies of the polymorphic alleles remaining after the bottleneck, as shown by the gray/black distributions in the pie charts.
Intraspecific Molecular Phylogeny of Vervet
The sequencing of the mitochondrial cytochrome b gene in more than 100 monkeys from Africa and the Caribbean has provided initial insight into the genetic diversity of Chlorocebus (Haus et al. 2013). They identified nine major clades, which reflect geographic distributions rather than taxa, and also confirmed hybridization between subspecies in contact zones. Consistent with historical evidence that St. Kitts monkeys originated from West Africa (van der Kuyl et al. 1995), the mitochondrial data revealed that Caribbean monkeys are most closely genetically related to sabaeus from Senegal and Mauritania and more distantly related to sabaeus from Ghana and Burkina Faso (Haus et al. 2013). The mitochondrial data provide an incomplete picture of vervet phylogeny, given that they reflect only maternal inheritance; a more complete understanding of this phylogeny will emerge from whole-genome sequencing studies currently underway in several vervet populations and the VRC (Table 1). The relationships between African vervet subspecies is of great importance to biomedical research; these relationships are important to our understanding of the host-dependent evolution of SIVagm, perhaps the most diverse lentivirus infecting NHPs (Kuhmann et al. 2001; Wertheim and Worobey 2007).
Table 1.
Ongoing whole genome sequencing (WGS) projects in wild and captive vervet populations
| Geographic group | 1× WGS* | 4× WGS* | 10× WGS* | 30× WGS* | Total No. of vervets |
|---|---|---|---|---|---|
| Caribbean | |||||
| sabaeus from St. Kitts | 0 | 32 | 0 | 0 | 32 |
| sabaeus from Nevis | 0 | 12 | 0 | 0 | 12 |
| sabaeus from Barbados | 0 | 4 | 1 | 0 | 5 |
| Africa | |||||
| sabaeus Gambia | 0 | 20 | 1 | 1 | 22 |
| sabaeus Ghana | 0 | 2 | 1 | 0 | 3 |
| tantalus CAR | 0 | 10 | 1 | 0 | 11 |
| aethiops Ethiopia | 0 | 15 | 1 | 0 | 16 |
| cynosuros Zambia | 0 | 15 | 1 | 0 | 16 |
| pygerythrus Tanzania | 0 | 1 | 1 | 0 | 2 |
| pygerythrus Botswana | 0 | 2 | 0 | 0 | 2 |
| pygerythrus Kenya | 0 | 4 | 0 | 0 | 4 |
| pygerythrus South Africa | 0 | 48 | 0 | 1 | 49 |
| Caribbean origin | |||||
| VRC pedigree | 301 | 409 | 0 | 17 | 727 |
The numbers in the column headers indicates the targeted depth of genomic coverage in WGS. VRC, Vervet Research Colony.
SIVagm Diversity in Vervets
Primate SIVs are very old (Worobey et al. 2010). Initial phylogenetic classifications of SIV described six distinct lineages (Courgnaud et al. 2001; Hirsch et al. 1995). More recent investigations, including molecular characterization of SIV strains from a much larger number of NHP species, suggest that these distinctions may be somewhat arbitrary. Classifications that include such recent data continue to show that SIVagm strains form a single clade in relation to strains from other NHPs (Bibollet-Ruche et al. 2004); however, the mosaic genome structure in viruses from West African sabaeus indicates that, in the past, recombination occurred with divergent lentiviruses from other NHP hosts (Jin et al. 1994). Each vervet subspecies is typically associated with a different strain of SIVagm: SIVagmVer in pygerythus, SIVagmSab in sabaeus, SIVagmTan in tantalus, and SIVagmGri in aethiops. Cynosorus is the only major vervet subspecies in which SIVagm has not yet been characterized. Little is known about virus diversity within each vervet subspecies except pygerythrus. Ma et al. (2013) recently conducted the first large-scale characterization of the SIVagmVer sequence in vervet populations in South Africa. This study revealed that two geographically distinct vervet populations, which are separated physically by the Drakensberg Mountains, carry different viral variants. Given the very old age of these mountains, this deep phylogenetic split between the two viral strains suggests that the virus was present in the ancestral vervet populations when they arrived in this region and since then have been effectively separated from each other by this physical barrier. Molecular clock analyses of the SIVagm env and pol gene sequences in South African pygerythrus, place the most recent common ancestor of South African SIVagm strains several hundred thousand to several million years ago (Ma et al. 2013). These results evidencing a very old origin of SIVagm suggest a long coexistence of the host and pathogen, which might have led to major adaptations to diminish virus pathogenecity. In this respect, the old and new hybridization zones represent interesting examples of not only host gene flow but also exposure to SIV strains from other subspecies.
The Vervet as a Biomedical Model System
The wide range of biomedical investigations of the vervet have focused on populations from the major subspecies in Africa, the vervets inhabiting the Caribbean islands, and the Caribbean-derived vervets brought to the United States to form the VRC. We describe here examples illustrating the use of the vervets for research in each of these three settings.
The Vervet in HIV/AIDS Research
Because the vervet is the most abundant natural host of SIV, a close relative of human immunodeficiency virus (HIV), it is of growing importance in HIV/AIDS research, forming the focus of several types of investigation, in large part conducted in African vervet populations.
Resistance to Immunodeficiency in SIV-Infected Vervets
Like other African natural hosts of SIV, when infected with their species-specific SIV, vervets typically do not progress to immunodeficiency (Hahn et al. 2000; Pandrea and Apetrei 2010; Pandrea et al. 2008; Pandrea et al. 2009). In stark contrast, humans, as recent HIV hosts, and rhesus macaques, as nonnatural hosts of SIV, almost always develop AIDS upon infection. In these “progressing” species, chronic immune activation drives CD4+ T cell loss and immunodeficiency, whereas natural hosts, despite high levels of viremia, sustain healthy levels of CD4+ T cells and low immune activation. The differences between pathogenic (in nonnatural or recent hosts) and benign (natural host) courses of lentiviral infection could be important for the development of novel therapeutic strategies for curing AIDS. It is believed that natural hosts exhibit a benign course of infection because they developed protective adaptations through at least several hundred thousand years of host–pathogen coevolution, and therefore they provide valuable model systems for investigating host factors restricting lentiviral infections (Worobey et al. 2010).
Unlike African vervet populations that are heavily infected with SIV, Caribbean vervets are free of SIV, and most likely have been removed from exposure to the virus for at least approximately 70 to 90 generations (since the time of their introduction to the Caribbean). Although there is no obvious difference in their response to SIV, compared with that of African vervets (Pandrea et al. 2006), it is hypothesized that without the selective pressure represented by SIV, Carribean vervets may display a different frequency of genetic alleles that protect against immunodeficiency after infection. Vervets, unlike other natural hosts, are not endangered and are simple to import into the United States. They thus provide an excellent model for experimental study of the immune response after initial infection.
Resistance to SIV Infection in Natural Vervet Populations
Ma et al. (2013) recently identified an additional protective process against SIV infection in wild African vervet populations—resistance to acquiring SIV infection. Human studies of HIV infections have shown that some at-risk individuals remain uninfected despite a high exposure to HIV. Investigation of such exposed seronegative (ESN) individuals could identify mechanisms of natural resistance to infection, which in turn could lead to novel strategies for prevention and treatment for HIV/AIDS (Fowke et al. 1996; Imagawa et al. 1989). ESN studies in human cohorts are limited by the lack of homogenous human cohorts large enough to address correlates of natural immunity (including genetic, mucosal, immune, and microbiome parameters), ethical constraints on invasive sampling and experimental interventions, and lack of feasibility of establishing prospective ESN cohorts. These limitations of human studies could be addressed by development of an NHP model for ESN studies, a need recognized by the US National Institutes of Health (Young et al. 2011).
The vervet is the only primate known to display a common ESN phenotype. Epidemiologic data on SIV prevalence in populations of South African pygerythrus have shown that heterosexual contacts are the main route of SIV transmission in natural vervet populations and adult reproductively active females form a high exposure group. Despite their massive SIV exposure through sexual activity, however, 20% of adult females remain uninfected, suggesting the presence of natural resistance to SIV infection in vervet populations.
The discovery that ESN is much more frequently observed in vervets than in humans creates an opportunity to develop new research strategies for viral resistance studies (Table 2) using the natural meta-population of African vervets as a model for ESN studies. A natural vervet model for ESN could enable identification of underlying genetic factors and other correlates of resistance (immune, microbial, environmental) by large-scale longitudinal studies of prospective ESN cohorts.
Table 2.
The vervet exposed seronegative (ESN) model: features that differentiate the investigation of ESN in vervets compared with humans.
| Issue | Human ESN studies | Vervet ESN model |
|---|---|---|
| Infection prevalence | Up to 30% of young adults are HIV infected in the most heavily infected countries (sub-Saharan Africa) | >70% adults are SIV infected in the African vervet metapopulation |
| ESN | Up to 5% among heavily exposed individuals | Approximately 20% among heavily exposed individuals |
| Power | The heterogeneity of human cohorts (different level of exposure, route of transmission, ethnic diversity) prevents combining cohorts for meta-analysis | The high prevalence of the SIV resistance phenotype permits establishment of sizable ESN cohorts |
| Systems biology approach | Ethical limitations on invasive sampling Unknown environmental variables Incompletely genetically characterized cohorts |
Flexibility in sampling of wild and captive animals Availability of captive colonies derived from local wild vervet populations Uniformity of conditions in vervet colonies reduces environmental variability and increases the accuracy of biological measures |
| Early response study | Early immune response is an important factor in determining the clinical course of disease but is difficult to measure in humans | Experimental infections in SIV-naive captive vervets will permit tight control of timing, dose and route, number of exposures, and multi-system mechanistic investigations |
| Prospective ESN cohorts | Not feasible | Already initiated through sampling of large cohorts of SIV-naive juvenile vervets, which were microchipped and returned to the wild to enable follow up in the future (Jasinska et al. 2012) |
HIV, human immunodeficiency virus; SIV, simian immunodeficiency virus.
Vervet Model for Neurodegenerative Processes
The Caribbean vervet on St. Kitts is a widely used model for neurodegeneration, in particular for Alzheimer's and Parkinson's disease. Caribbean vervets, as they age, naturally develop cerebral amyloid beta plaques; these are associated with gliosis and neuritic dystrophy, making the vervet a model for Alzheimer's disease research and treatment (Lemere et al. 2004). Vervet Parkinson's models are based on administration of the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), which destroys dopamine neurons in humans and monkeys and closely reproduces Parkinsonian pathology (Emborg 2007). This model has recently been extended to translational studies using neural stem cell approaches (Bjugstad et al. 2005; Redmond et al. 2007).
Vervet Populations for Genetic Mapping of Complex Traits
The genetic mapping studies in the vervets have focused on the VRC. Geneticists have long been interested in identifying human study samples characterized by relative genetic homogeneity; both linkage and association approaches depend on the probability that affected individuals share disease susceptibility genetic variants and marker loci identical by descent from a common ancestor. In genetic isolates, such as the Caribbean vervet populations, one or a few founders introduce disease-susceptibility variants into the population, which may drift to high frequency (Sabatti et al. 2009; Service et al. 2006), therefore potentially increasing the power of genetic mapping studies. Examples of the use of Caribbean ancestry vervets in the VRC to map quantitative traits are discussed below. The bottlenecked origin of Caribbean-origin monkey populations makes them valuable not only for genetic mapping studies but also for interventional investigations (e.g., vaccine studies) that take advantage of their homogenous genetic background.
Vervet Genomics
The recent progress in characterizing the vervet genome—in particular the generation of a high-quality reference assembly and whole-genome sequencing of vervets from all of the major subspecies—will transform the opportunities for vervet investigation. It is now feasible to undertake studies to elucidate the genetic and genomic underpinnings of traits relevant to human health and disease and to conduct comparative genomics projects that will inform our understanding of primate evolution. In this section, we review the progress of vervet genetics and genomics investigations to date and the resources that they have made available to the scientific community, many of which can be used through the Integrated Vervet/AGM Research & Resources website (www.genomequebec.mcgill.ca/compgen/vervet_research/genomics_genetics/).
Vervet Karyotype and Chromosomal Organization
The karyotype of higher primates, generally, is very conservative. A small number of lineages, however, show particular tendencies toward karyotype divergence at the level of chromosome number and possibly also intrachromosomal organization, both within the lineage and in comparison with other closely related groups. The Cercopithecini tribe that includes the vervet provides one of the most striking examples of such a lineage-specific tendency toward increasing karyotype diversity. Although the Papionini show a stable diploid chromosome number (2N = 42), Cercopithecini vary widely between species in the diploid number of chromosomes (2N = 48–72). In vervets, the diploid chromosome number is 2N = 60. The mechanism for this increased rate of chromosome rearrangements in the Cercopithecini remains unclear, although they mostly occur through breakpoints outside of centromeres (Finelli et al. 1999). Additionally, it is not yet known whether a similarly increased rate of rearrangement occurs at a finer-scale level in Cercopithecini genomes. However, the newly available vervet genomic tools will permit investigations across the different subspecies that may clarify this point. Identifying the mechanisms through which increased diploid number occurs during species divergence and the consequences of such rearrangements for epigenetic processes, gene expression, and the function of regulatory genomic elements may shed light on aspects of cancer pathogenesis involving epigenetic deregulation and genomic instability.
Higher resolution studies of the vervet genome, at the level of subchromosomal level rearrangements, first became possible with the development of a vervet genetic linkage map comprising 360 microsatellite markers distributed genome-wide (Jasinska et al. 2007); the use of this map for genetic mapping of quantitative trait loci (QTL) is discussed below. Comparison of genetic markers order between vervet and other primate chromosomes revealed 12 rearrangements compared with human, 16 compared with chimpanzee, and 12 compared with rhesus macaque. Nine human chromosomes showed full colinearity with vervet, 10 contained one or two simple marker rearrangements, and one (human chromosome 3) showed a more complex rearrangement (Jasinska et al. 2007). These extensive regions of synteny between vervet and human enable a lift-over of the human gene content in these regions to obtain a provisional annotation of important regions. All of these rearrangements except that observed for human chromosome 4 were also seen in comparisons between vervet and chimpanzee; these comparisons revealed an additional four rearrangements resulting from pericentric inversions that occurred in a chimpanzee lineage. Rearrangements observed between vervet and rhesus macaque comprise a relatively balanced number of inversions and single-marker rearrangements, whereas rearrangements between vervet and both human and chimpanzee are mostly inversions, resulting from the tendency to inversion that characterizes hominid but not OWM evolution (Dutrillaux 1979).
The Generation of the Vervet Reference Genome Sequence
As already noted, the vervet genome is distinguished from most other primate genomes by a higher chromosome number (n = 30) and by a large number of intrachromosomal inversions and rearrangements. Both types of events are evolutionarily recent, having occurred since the time of vervet divergence from the other members of the OWM lineage. Thus, a high-quality chromosome level genome assembly is important for delineating and understanding rearrangements, and it is especially crucial for the vervet as a biomedical model, where complex trait mapping is dependent upon extremely accurate knowledge of positions and orders of genetic mark.
The sequencing and assembly of large genomes has become a feasible option since the introduction of next-generation sequencing technologies, which combine microfluidics and massive parallelization to lower sequencing costs and increase sequencing capacity. These technologies continue to evolve and improve in terms of cost, output, and quality and have spurred the development of increasingly more sophisticated and accurate assembly software packages. Nonetheless, mammalian genome assemblies remain vulnerable to various problems, including regions recalcitrant to amplification and or sequencing and repetitive regions that cannot be disentangled from one another and definitively placed in their correct genomic location. Experimental strategies for the best possible assemblies currently blend a variety of different technologies and software to allow cross-validations and to compensate for weaknesses in one technology that can be balanced with another.
The vervet genome assembly is founded upon the principles of a truly de novo assembly that will recreate a faithful reconstruction of the vervet genome, such that the core data for mapping results and rearrangement studies will not have been distorted by changes in syntenic placements between vervets and any other species. As illustrated in Figure 3 and summarized in Tables 3 and 4, the vervet genome is not contiguous with that of the rhesus or human, and levels of structural variation differ across different chromosomes from both species. Thus, using a separate species as a guide to organizing the vervet genome poses risks to many of the key reasons why a genome assembly is needed.
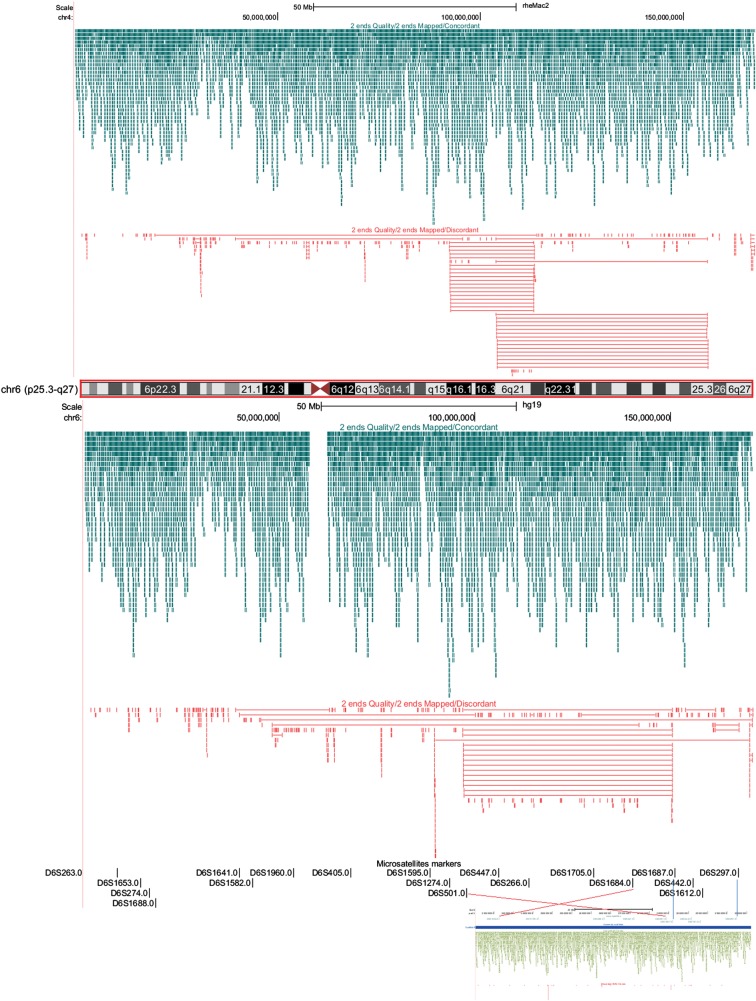
Figure 3.
Vervet genome rearrangements provide a new perspective on primate genome evolution. The panels show UCSC Genome Browser views of vervet bacterial artificial chromosome (BAC) paired ends aligned to rhesus chr6 (rheMac.2, top) human chr6 (hg19, middle), and the vervet draft assembly (bottom). Green lines delineate vervet BAC paired ends concordant for interval distance and sequence orientations. Red lines delineate vervet BAC end pairs, discordant for either interval distance, sequence orientations, or both. Clusters of concordant BACs indicate cross-species syntenic regions, whereas clusters of discordant BACs indicate rearrangements. In the example, the vervet orthologous region is organized differently from both rhesus and human. Vervet BAC end pairs aligned to a 70-Mb vervet scaffold (bottom) demonstrate that the rearrangements have been accurately reconstructed in the vervet genome assembly.
Table 3.
Vervet BAC discordancy rates to human (hg19)
| Human Chromosome | Length | Concordant BACs | Discordant BACs | Discordant BACs/Mb |
|---|---|---|---|---|
| 1 | 249,250,621 | 10,220 | 454 | 1.8 |
| 2 | 243,199,373 | 12,394 | 352 | 1.4 |
| 3 | 198,022,430 | 10,439 | 324 | 1.6 |
| 4 | 191,154,276 | 10,344 | 522 | 2.7 |
| 5 | 180,915,260 | 9117 | 337 | 1.9 |
| 6 | 171,115,067 | 8793 | 312 | 1.8 |
| 7 | 159,138,663 | 7109 | 370 | 2.3 |
| 8 | 146,364,022 | 7688 | 244 | 1.7 |
| 9 | 141,213,431 | 5183 | 210 | 1.5 |
| 10 | 135,534,747 | 6197 | 718 | 5.3 |
| 11 | 135,006,516 | 5752 | 258 | 1.9 |
| 12 | 133,851,895 | 6384 | 199 | 1.5 |
| 13 | 115,169,878 | 5094 | 252 | 2.2 |
| 14 | 107,349,540 | 4479 | 130 | 1.2 |
| 15 | 102,531,392 | 3525 | 135 | 1.3 |
| 16 | 90,354,753 | 2923 | 153 | 1.7 |
| 17 | 81,195,210 | 2601 | 149 | 1.8 |
| 18 | 78,077,248 | 3907 | 175 | 2.2 |
| 19 | 59,128,983 | 1037 | 189 | 3.2 |
| 20 | 63,025,520 | 2524 | 206 | 3.3 |
| 21 | 48,129,895 | 1559 | 69 | 1.4 |
| 22 | 51,304,566 | 912 | 118 | 2.3 |
| X | 155,270,560 | 3232 | 301 | 1.9 |
| Y | 59,373,566 | 37 | 42 | 0.7 |
| Assembly | 3,095,677,412 | 131,450 | 6219 | 2.0 |
The vervet reference assembly: Chromosomes with discordancy rates higher than the genome average are highlighted in bold italics; chromosomes with discordance rates lower than the genome average are highlighted in boldface. BAC, bacterial artificial chromosome.
Table 4.
Vervet BAC discordancy rates to rhesus chromosomes (rheMac2)
| Rhesus Chromosome | Length | Concordant BACs | Discordant BACs | Discordant BACs/Mb |
|---|---|---|---|---|
| 1 | 228,252,215 | 11,051 | 333 | 1.5 |
| 2 | 189,746,636 | 10,793 | 271 | 1.4 |
| 3 | 196,418,989 | 9421 | 416 | 2.1 |
| 4 | 167,655,696 | 9358 | 277 | 1.7 |
| 5 | 182,086,969 | 10,950 | 247 | 1.4 |
| 6 | 178,205,221 | 9697 | 411 | 2.3 |
| 7 | 169,801,366 | 8560 | 340 | 2.0 |
| 8 | 147,794,981 | 8085 | 358 | 2.4 |
| 9 | 133,323,859 | 6598 | 476 | 3.6 |
| 10 | 94,855,758 | 3610 | 339 | 3.6 |
| 11 | 134,511,895 | 6886 | 153 | 1.1 |
| 12 | 106,505,843 | 6266 | 108 | 1.0 |
| 13 | 138,028,943 | 6869 | 193 | 1.4 |
| 14 | 133,002,572 | 6630 | 210 | 1.6 |
| 15 | 110,119,387 | 5563 | 214 | 1.9 |
| 16 | 78,773,432 | 2829 | 165 | 2.1 |
| 17 | 94,452,569 | 5380 | 206 | 2.2 |
| 18 | 73,567,989 | 4092 | 143 | 1.9 |
| 19 | 64,391,591 | 1124 | 199 | 3.1 |
| 20 | 88,221,753 | 3134 | 120 | 1.4 |
| X | 153,947,521 | 3743 | 302 | 2.0 |
| Assembly | 2,863,665,185 | 140,639 | 5481 | 1.9 |
This table shows the same metrics as in Table 3, except that it compares vervet to rhesus macaque rather than human. Chromosomes with discordancy rates higher than the genome average are highlighted in bold italics; chromosomes with discordance rates lower than the genome average are highlighted in boldface.
Our approach to the vervet combined a series of independent sequence datasets and data types, all of which were derived from DNA obtained from an adult male from the VRC, representing the Caribbean population of sabaeus. In addition to a bacterial artificial chromosome (BAC) library constructed and maintained at BACPAC Resources, next-generation sequencing libraries were generated for Roche/454 and Illumina sequencing. Overall, the datasets included Roche/454 long-reads (approximately 450 nt) and 8 kb paired ends, and a hierarchy of Illumina paired end and mate-pair datasets for insert sizes ranging from 200 nt to 8 kb. Additionally, the CH252 vervet BAC library clones were end sequenced to provide a paired-end dataset of 150 to 170 kb insert sizes. As the datasets were developed, we also used a strategy to enhance the assembly strengths of different software, notably Newbler for Roche/454 data and ALLPATHS for Illumina data, which were then integrated with each other and with the BAC end data to produce a subchromosomal level scaffold assembly. As depicted in Figure 4, our results were then cross-referenced with vervet genetic map information to build chromosome-sized assemblies. Although the reference assembly is still awaiting annotation, the draft version is available to the scientific community through the Integrated Vervet/AGM Research & Resources website.
Figure 4.
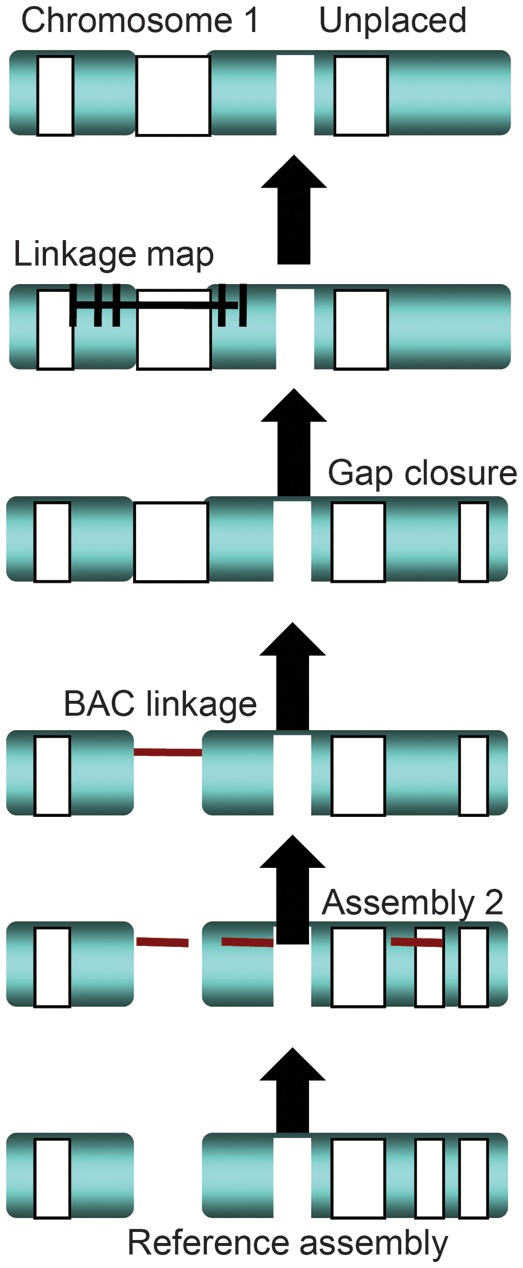
A phased assembly process for multiple data types. Lined white boxes represent gaps in scaffolds. Gray blocks represent nongapped consensus sequence. Blank gaps are areas where no linkage or sequence data are available.
Despite the known higher number of chromosomal structural variants in the vervet genome compared with that of other NHPs, the vervet reference assembly demonstrates a very high quality of sequence base accuracy, order, and orientation. We used cytogenetic, genome synteny, and long-range paired end concordance data to ensure such quality. Moreover, the careful manual validation of discordant sequence links allowed us to break misjoins where appropriate. Overall, the vervet genome displays a degree of contiguity that is second only to human among primate genomes; 50% of the assembled genome sequence is greater than 45 Mb in length with the largest scaffold being 127 Mb. The genome is organized in chromosome files with more than 149 assigned scaffolds of 1 Mb or higher. Another measure of assembly quality is gene representation. We BLAT (BLAST-like alignment tool) aligned assembled transcripts derived from a comprehensive set of diverse tissue types against this reference at a 90% identity threshold and found that more than 95% of transcripts were represented in the assembly at 95% of their length.
Measurement of Vervet Gene Expression
This section describes the importance of vervet transcriptomics resources for a wide range of investigations. We first describe the creation of resource datasets that are already widely available to the scientific community. We then illustrate the utility of transcriptomic studies for advancing the vervet model for HIV/AIDS research and for comparative genomics analyses.
NHP models provide an opportunity to assess gene expression in tissues (such as brain) from which high-quality RNA is often unavailable in humans. Invasive sampling feasible in captive NHPs gives an opportunity to obtain insight into gene expression in internal tissues requiring invasive (postmortem) sampling, such as brain, that typically are difficult to obtain at high quality or in large samples from human subjects. In NHPs, such tissues can be collected from individuals from a uniform environmental background (e.g., with similar diets) under standardized conditions, with a minimal postmortem interval and with immediate storage under conditions that preserve RNA profiles in these tissues.
Given the high degree of conservation between the vervet and human genomes and the ready availability of inexpensive human gene expression microarrays, such arrays have provided a cost effective means for initial genome-wide profiling of vervet gene expression. Data from two annotated array-based vervet gene expression resources, described below, are now freely available through the Integrated Vervet/AGM Research & Resources Website.
Illumina Gene Expression Resource
This resource was created using data from gene expression in eight brain regions (cerebellar vermis, pulvinar, head of caudate, hippocampus, frontal pole, dorsolateral prefrontal cortex, orbital frontal cortex, and occipital pole) in 12 VRC vervets and blood gene expression pedigree-wide, both using the Illumina HumanRef-8 v2 chip (Jasinska et al. 2009). Analysis of regional brain gene expression revealed that narrowly dissected cortical tissues showed extensive similarities with respect to gene expression profile; however, some genes were observed to be uniquely expressed in each region.
Using duplicate blood samples matched with brain specimens from the same individuals, 32 transcripts displayed stable expression profiles in peripheral blood that correlated strongly with their expression profiles in brain. This resource demonstrates the utility of NHP models for development of peripheral markers of gene expression in internal tissues. Such peripheral biomarkers can be assessed in large populations (e.g., whole pedigree or wild populations) without complicated or invasive sampling.
Affymetrix U133 Plus 2.0 Gene Expression Resource
A vervet tissue gene expression atlas was constructed by measuring expression from a subset of 24 brain regions from 3 male vervets and 3 female vervets from the VRC and by conducting basic comparisons of gene expression between different brain regions, which can be accessed through the Integrated Vervet/AGM Research and Resources website
The completion of the vervet reference assembly has enhanced the utility of the above expression array datasets. Vervet genome sequence is highly conserved with those of other primates (human and rhesus macaque) with well-developed reference genomes. However, measuring and interpreting vervet gene expression based on heterologous probes may still be significantly affected by vervet genome divergence at the level of both sequence (1–2%) and structure (the numerous chromosome rearrangements in the vervet lineage, a phenomenon often associated with changes in gene content between species). Probes from the human expression arrays that are most suitable for analysis of the vervet transcriptome have been identified by determining probe specificity, complementarity to vervet transcripts, and sensitivity unbiased by intrinsic sequence variants by aligning the probe sequences to the vervet draft assembly. By using whole-genome sequencing data from the VRC pedigree, it has been possible to conservatively select probes as high-quality tools for expression assays if they were uniquely mapping to the vervet genome, showed no known vervet SNPs, and showed no indels between human probe and vervet transcript. Probe characteristics can be found at the Integrated Vervet/AGM Research & Resources website (see above).
Vervet Transcriptomics for HIV/AIDS Research
One focus of investigations of global gene expression in vervets is on the course of SIV infection in vivo, in particular with respect to their similarities to other nonprogressing species and dissimilarities with species susceptible to AIDS. A series of studies yielded a concordant observation that SIV infection induces expression of a broad array of innate immune antiviral genes, including interferon-stimulated genes, both in progressing and nonprogressing species (Bosinger et al. 2012; Favre et al. 2009; Jacquelin et al. 2009; Lederer et al. 2009; Manches and Bhardwaj 2009). Although this initial induction of innate immunity during acute infection, including the interferon system, persists in pathogenic hosts (rhesus macaques), in vervet (and sooty mangabey) it is resolved to the baseline level in the chronic infection stage, occurring less than 30 days after infection (Manches and Bhardwaj 2009). The ability of natural hosts to downregulate the initial induction of interferon-stimulated genes may indicate a possible mechanism by which these species avoid progression to immunodeficiency.
Transcriptome characterization through cDNA sequencing (including RNA sequencing) has clear advantages over approaches based on microarrays. Although extensive tissue repositories from various vervet study samples will be ideal for large-scale RNA sequencing, such studies remain in a preliminary phase and we will not discuss them in detail here. Sequence-level comparative transcriptomics investigations in vervet have been so far conducted through expression sequence tags sequencing in peripheral blood, a tissue providing direct insight into immune response to SIV infection. Tchitchek and colleagues (2012) covered about 12% of the vervet transcriptome using cDNA libraries derived from untreated, stimulated blood cells from SIV-infected and -uninfected individuals to maximize the discovery of immune defense genes, which often remain inactive until induced by infection. In this reference set of vervet expression sequence tags, including more than 500 nearly complete transcript sequences, approximately 9000 assembled sequences were annotated based on known primate orthologues. Interestingly, more than 6500 expression sequence tags do not match any known primate genes from 10 primate species that were assayed; these possibly represent novel transcripts involved in immunity and activated upon infection, but they also may reflect the fact that annotation of primate genes remains rudimentary (Tchitchek et al. 2012).
Vervet Resources: Study Populations and Biorepositories
Vervet biomedically related research has mainly been conducted in several research colonies, which we describe briefly. The recent efforts of a multidisciplinary international collaboration have enabled, in different vervet subspecies, the first large-scale biomedical investigations of wild NHP populations. From both colony and wild-living vervets, a wide range of biological samples are now available to the scientific community through US National Institutes of Health-supported biorepositories.
Captive Vervet Populations
Wild Caribbean vervet populations of sabaeus from St. Kitts, Nevis, and Barbados are perhaps the largest source of vervets for biomedical research. Monkeys captured from these populations, as well as their colony-born descendants, are housed primarily at the following colonies, all of which make monkeys available to the scientific community more broadly: the VRC, now located at Wake Forest University in North Carolina, the main breeding colony in the United States; the New Iberia Research Center, in Louisiana; the St. Kitts Biomedical Research Foundation; the Behavioral Science Foundation, also on St. Kitts; and the Barbados Primate Research Center.
Pygerythrus is the second most commonly used vervet subspecies in biomedical research. The US-based vervet colony located in the National Institutes of Health Animal Center was established from monkeys of this subspecies captured from Tanzania. The Primate Unit and Delft Animal Centre in the Medical Research Council of South Africa (Cape Town) holds pedigreed vervets established from South African monkeys captured in the Eastern Cape region and conducts studies mostly in the area of reproductive biology, cardiometabolic diseases, and HIV/AIDS. This Medical Research Council pygerythrus colony is genetically related to the wild populations that were recently epidemiologically characterized with respect to SIV-related measures (Ma et al. 2013) and provides an opportunity for studies under well-controlled conditions of traits identified in wild monkeys, such as ESN, described previously.
Wild Vervet Populations
Vervet monkeys have a natural distribution that ranges across savannah woodland environments throughout sub-Saharan Africa and in the Caribbean (Struhsaker 1967). Within this range, they can be found in wild environments and on farmlands and other exurban human-impacted areas and can even thrive in major cities. Ready accessibility, high abundance, and “least concern” conservation status of the major vervet subspecies make wild vervet populations ideal for a wide range of biomedical investigations (IUCN 2013).
Vervets typically live in multimale, multifemale cohesive social groups ranging in size from 5 to more than 75 individuals (Isbell et al. 1991; Turner et al. 1997) and aggressively defend group territories that range in size from 0.6 km2 to 1.78 km2 (Harrison 1983; Kavanagh 1981). Females are philopatric, typically remaining in their natal groups for life, whereas males typically disperse around sexual maturity (Cheney and Seyfarth 1983). At birth, the male to female ratio is roughly 1:1 (with some variation [Cheney and Seyfarth 1987]), which drops as animals age to favor either sex depending on social rank and local ecology (e.g., Altmann and Alberts 2005; Cheney and Seyfarth 1987; Cheney et al. 1981) and in adults favoring females due to increased male mortality during dispersal (Isbell et al. 1991). Within their territories, vervets often return to known feeding and sleeping sites. This behavior provides an opportunity to capture the same individuals repeatedly in the same location. By marking each trapping site by GPS location, it has become possible to return to these sites for retrapping (enabling longitudinal investigations) and for geographic information system-based analysis of eco- and geographic covariables for biological samples (Figure 5).
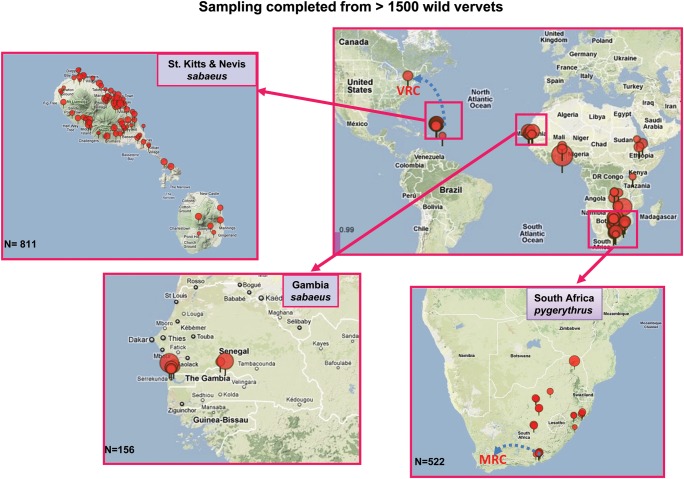
Figure 5.
Systems Biology Sample Repository sampling locations of wild vervets worldwide. To facilitate large-scale genetic analysis of various complex traits related to human diseases, biological material banked in the Systems Biology Sample Repository at the University of California–Los Angeles and multiple types of phenotypic data have been collected from more than 1500 wild vervets from South African pygerythrus, West African sabaeus, St. Kitts sabaeus, and Nevis sabaeus. Animals have been captured, assigned unique identifiers using a microchip injected subcutaneously to enable long-term follow-up studies of each monkey, sampled, and released in a trapping location. Red pins indicate GPS-defined geographic location, and pin size is proportional to the number of sampled animals at each location. MRC, Medical Research Council; VRC, Vervet Research Colony.
Wild Animal Capture Strategies for Research Purposes
Live trapping is the most efficient way of collecting biological samples, phenotype data, and health data from wild vervets (Brett et al. 1982). Using live trapping methods, our research collaboration has captured and sampled more than 1500 live vervet monkeys. This has been accomplished through two major trapping methods—individual drop traps and group traps.
Individual Drop Traps
Individual drop traps are used in Africa, based on a traditional African bird trap design (for details and diagrams, see Grobler and Turner 2010). Previously scouted vervet sleeping sites are regularly baited until the monkeys habituate to foraging under propped up traps. Once baited and set, trapping typically begins in the early morning as the monkeys leave the sleeping site and continues throughout the day for as long as the group forages in the area. Once trapped, monkeys are individually sedated through a mesh grate on top of the traps. Although the traps are meant for individual capture, younger animals sometimes are trapped together and up to five can fit in a trap. Using this method, most members of the group may be captured, with the majority (approximately 97% of total animals caught) being trapped in the first two days (Grobler and Turner 2010).
The composition of individuals trapped depends on the extent of sampling within the group; in heavily sampled groups, the sex ratio among trapped animals is often even, and adults predominate, whereas in less heavily sampled groups, older animals and females are predominate (Ma et al. 2013). Overall, in trapped immature monkeys, the sex ratio is skewed toward males, whereas in adults it is skewed toward females. A recapture rate of 44% was observed through longitudinal studies in South Africa (TR Turner, unpublished data); retraps are also typically higher in immature males. This increased trapping of immature males may be because they are more risk seeking than other age/sex classes (e.g., Fairbanks et al. 2004).
Using individual traps, within-trap behavior such as stereotypic responses to capture can also be reliably video- recorded and transformed into phenotypic measures using standardized protocols. Such in-trap behaviors show significant individual and regional variation in stereotypical behaviors in response to capture and human approach (CA Schmitt, unpublished data).
Group Traps
Group traps have been employed in National Institutes of Health-supported research in St. Kitts and Nevis through professional trappers that have a longstanding relationship with the St. Kitts Biomedical Research Foundation. Trappers maintain stable, GPS-marked large wire mesh cages that they provision (e.g., with sweet potatoes or corn) year-round to maintain a population of habituated and trappable monkeys. On trapping days, they set the wire mesh traps. Wire mesh traps are 10 feet × 5 feet ×10 feet wire mesh enclosures with a dropping door controlled by the trapper, with a detachable small 1-foot diameter mesh tube. Once as much of the group as possible is in the trap, the trapper closes the dropping door and then corrals the monkeys into the wire mesh tube. Once the desired number of monkeys is in the wire mesh tube, the animals are anesthetized through the tube and brought to the research area for sampling. Up to 50 animals may be trapped at a time using this method, although the number brought to researchers is modulated with work flow to maximize animal welfare. Sex ratios of trapped animals are consistently more even than in drop traps but are still, for the most part, biased toward males, including in adults. Animals do not become agitated in the traps unless a human approaches within 5 meters, which prevents within-trap monitoring comparable with drop traps. Animals not removed from traps immediately are monitored at a distance by staff remaining in the area.
Characteristics of Captured Animals
The sex, age, and size of the animal are estimated (by genital inspection, dental eruption patterns, and weight scale, respectively) once the animal is sedated. Animals are injected with a subdermal microchip that can be scanned to identify retrapped individuals. Parity status of females is assessed through observation of nipple length, milk expression, and abdominal palpation. Body weight (measured in kilograms using an electronic scale) is taken as data and to assess the amount of blood that can safely be subsequently withdrawn (up to 1.25% of body weight). Blood samples for DNA, RNA, viable cells, serum, and plasma are collected, depending on safe blood extraction allowances dictated by body weight and a skin punch. Microbiome sampling (of, for example, rectum, vagina, mouth) is also performed using sterile swabs on all individuals. Collection involves the use of sterile techniques to obtain fecal samples by digital insertion into the rectum to extract fecal matter. Extracted fecal matter is then placed in a sterile petri dish, divided, placed in a sterile container using a sterile scalpel, and frozen. After sample collection, morphometric measures are taken using a tape measure and sliding and digital calipers. No physical restraint is required because the animal is sedated throughout all procedures, which require approximately 30 minutes to complete per animal.
Characteristics of general health conditions comprise the presence of wounds and scars (new and old ones), body temperature, and body morphometric measurements of growth and obesity (e.g., body mass index and measures). Inscribed characterization is complemented by photographic documentation (facial and whole body pictures, hair and scrotal color, other individual features, such as wounds and scars) and by video recordings of in-trap behavior.
Collection of Vervet Population Samples and Creating the Systems Biology Sample Repository
A wide range of biological samples have been obtained and banked from vervets in Africa and the Caribbean (Figure 6). These samples include cerebrospinal fluid, hair, blood (for DNA, RNA, serum, plasma, mononuclear cells), skin biopsies (for fibroblast culture), samples from multiple body sites (for microbiome analysis), and morphometrics. All samples are submitted for long-term storage at the Systems Biology Sample Repository at UCLA. As of 2013, the Systems Biology Sample Repository includes samples from 1511 monkeys, including 833 sabaeus from St. Kitts and Nevis and 156 from the Gambia (the presumed ancestral population of the Caribbean monkeys) and 522 pygerythrus from South Africa, the most abundant population for SIV-related studies.
Figure 6.
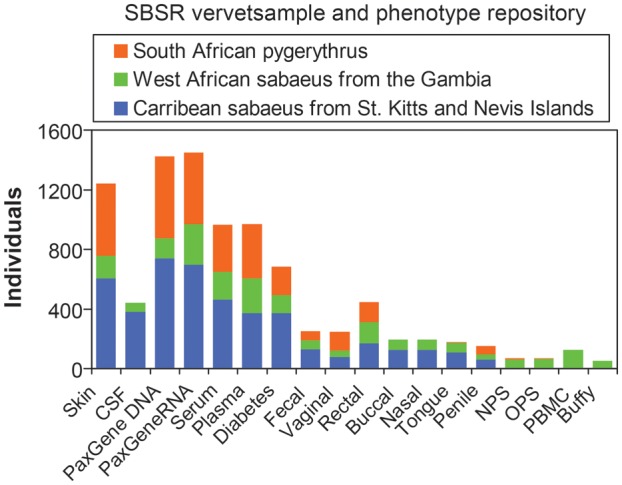
Vervet biomaterial and phenotypic resources managed by the Systems Biology Sample Repository. The histograms indicate the number of biological samples of different types that have been banked from the three most heavily sampled vervet populations, as shown in the key. The specific types of samples represented by the histograms are as follows: CSF, cisternal cerebrospinal fluid; diabetes, blood samples for diabetes-related measures; PaxGene DNA and RNA, blood samples collected for DNA and RNA extraction; serum and plasma, samples from which multiple aliquots have been established; skin, fibroblasts from punch skin biopsies. Samples for microbiome analyses were collected from feces, vagina, rectum, buccal swab, nose, tongue, penis, nasopharyngeal swab (NPS), and oropharyngeal swab (OPS). PBMC, peripheral blood mononuclear cells; Buffy, the buffy coat layer of blood cells.
Genetic and Genomic Investigations of Quantitative Traits in the Vervet
Although the collection of samples from large numbers of wild vervets will ultimately permit large-scale genetic association studies, current genetic and genomic investigations of the vervet focus mainly on the extended pedigree of the VRC; this is now the most thoroughly genomically characterized NHP resource worldwide. In this section, we describe the VRC and summarize the several related lines of research that combine to enable systems biology to be undertaken in the vervet.
History of the VRC Pedigree
The Wake Forest/UCLA Vervet Research Colony (VRC) is a large multigenerational pedigree of Caribbean-origin vervets, founded with 57 monkeys brought to UCLA in the 1970s and 1980s from several locations in St. Kitts and Nevis. From these founders, the pedigree expanded to include nearly 1300 individuals in the current genetically determined pedigree structure. As expected given the polygynic mating system in vervets, half-sib is the most common type of family relationship in the pedigree after grandparent–grandchild. In some branches, the pedigree is now 11 generations deep.
Establishment of the vervet pedigree was therefore preceded by two bottlenecks: a severe bottleneck during introduction from Africa to the Caribbean and a much milder bottleneck during founding of the VRC pedigree. Given the reduced genetic diversity present in the founder Caribbean populations, we anticipate that the large number of the VRC founders transferred to the VRC pedigree most of the genetic variation observed in the Caribbean populations and the same loci are contributing to traits in both populations. Because individuals in populations are separated from their most recent common ancestor by more meiotic steps (and therefore recombination events) than individuals in relatively recently established pedigree, shorter identical-by-descent segments carrying the functional variant contributing to a trait are present in the Caribbean populations. It gives us an opportunity of two-stage genetic mapping design consisting of initial mapping in the VRC followed by mapping of the most promising traits in the population, where higher resolution of the association signal is expected (Jasinska et al. 2012). This extraordinary history of the VRC pedigree (two expansions from population bottlenecks) is greatly facilitating genetic studies in this model, as we demonstrate further.
Investigation of Brain and Behavior in the VRC
Genetic Investigations of Brain Structure in the Vervet
Compared with apes and humans, OWMs show considerably less cross-species variability in the macroscopic and cytoarchitectonic anatomy of the brain. For example, macaques and vervets have very similar cortical sulcal and gyral anatomy (Woods et al. 2011) and very similar cytoarchitectonic maps (von Bonin and Bailey 1947). Despite more recent divergence than macaques from vervets, humans and chimpanzees are so anatomically dissimilar from one another that many uncertainties remain with regard to both macroscopic and microscopic brain homologies. Indeed, anatomic diversity in sulcal and gyral patterns and in the relationships between macroscopic anatomy and cytoarchitectonic boundaries within the human species alone complicates investigations of genetic contributions to human brain anatomy. The greater consistency of anatomic brain landmarks in OWMs simplifies parcellation of the brain into anatomic subregions but does not imply any lesser heritability of anatomic brain phenotypes than is observed in humans. Structural magnetic resonance imaging scans of 357 vervets from the VRC have shown that brain volume (h2 = 0.99), cerebral volume (with right and left hemispheres combined or analyzed separately, left h2 = 0.94, right = 0.97, combined h2 = 0.98), cerebellar volume (h2 = 0.86), hippocampal volume (h2 = 0.95), corpus callosum cross sectional area (h2 = 0.87), and right-left asymmetric transverse skewing of the brain (h2 = 0.29) are all heritable phenotypes in the vervet (Fears et al. 2009; Fears et al. 2011). These heritabilities are comparable with, and perhaps greater than, those reported in human studies (Alkan et al. 2011; Pfefferbaum et al. 2004; Posthuma et al. 2000; Scamvougeras et al. 2003; Sullivan et al. 2001), possibly reflecting reduced heterogeneity of environmental exposures in the VRC, compared with human study samples.
Behavioral and Cognitive Studies in the Vervet
NHPs have long played an indispensable role in mechanistic studies aimed at relating neurobiological processes to complex behavior, particularly cognition, motivation, and temperament. It is not surprising, therefore, that the first uses of Caribbean vervet monkeys in biomedical research were by neuroscientists and psychologists (Bacopoulos et al. 1980; McGuire 1974; Murphey et al. 1979; Raleigh et al. 1979; Roth et al. 1982; Young and Ervin 1984), and it was their biobehavioral research that ultimately led to the creation of the VRC.
From the beginning, the vervet monkey has been involved in research projects that have identified relationships between brain monoamine signaling and cognition and behavior. These studies include revealing a fundamental relationship between central serotonin signaling and social behavior and impulsivity (Brammer et al. 1991; Fairbanks et al. 2001; Manuck et al. 2003; McGuire and Raleigh 1985; Raleigh et al. 1980; Raleigh et al. 1984; Raleigh et al. 1985; Raleigh et al. 1986; Raleigh et al. 1991), between dopaminergic neurotransmission and motivated behavior and cognitive and executive functions (Elsworth et al. 1989; Groman et al. 2011; Jentsch et al. 1997; Melega et al. 2008; Palmour et al. 1998), and between norepinephrine and the pathological behaviors that characterize drug withdrawal (Elsworth et al. 1982; Grant et al. 1985; Roth et al. 1982). This earlier work focused mostly on the pharmacological manipulations, but subsequent work in the VRC has established similar relationships between markers in genes involved in monoamine neurotransmission and behavior (Bailey et al. 2007; Fairbanks et al. 2012; James et al. 2007).
Based upon the notion that certain dimensions of cognitive and executive function are reliable indicators of the pathophysiology of mental illness (e.g., working memory deficits in schizophrenia, problems with inhibitory control in attention defecit hyperactivity disorder, and addictions), ongoing efforts involve large-scale studies of individual differences in cognition in the VRC and attempts to relate these individual differences to objective biological markers (Groman et al. 2012; Groman et al. 2013) and, eventually, to genetic variation.
Vervet Model for Stress Studies
Psychological stress is a key risk factor for numerous chronic diseases, in particular cardiovascular, metabolic, and neuro-psychiatric disorders (Balodis et al. 2010; Hansel et al. 2010). Although susceptibility to stress-related disorders shows enormous interindividual variation, the genetic heterogeneity of most human study samples complicates efforts to uncover the genetic basis of such variation. More important, differential exposures among human subjects to a wide range of environmental factors contribute to variability in the magnitude of stress that individuals experience as well as in their response. These factors (including, for example, diet, lifestyles, medications, and substance use) may be difficult to account for and virtually impossible to control. In contrast, the possibility to regulate environmental exposures is a major advantage of NHP stress models. The vervet is an established model for investigations of response to acute psychosocial stress on gene expression and endocrine and behavioral phenotypic level. In 2008, the VRC was relocated from UCLA to Wake Forest, and gene expression and endocrine and behavioral (novelty-seeking) measures were collected pre- and postmove, with the following results: cortisol levels increased, novelty seeking behavior decreased, and 1184 genes showed deregulation in response to this stressor. Moreover, pedigree analysis indicated a significant genetic component to the magnitude of change in hair cortisol (h2 > 0.3) (Fairbanks, Jorgensen, et al. 2011), novelty-seeking behavior (h2 > 0.45) (Fairbanks, Bailey, et al. 2011), and abundances of specific transcripts.
Investigation of Cardiometabolic Traits in the VRC
Vervet Model for Metabolic Traits
The vervet, in comparison with other OWM models, more closely resembles human beings in lipoprotein metabolism and in the distribution and characteristics of atherosclerotic lesions that develop in response to a Western diet (Bullock et al. 1975, Fusegawa et al. 2001; Rudel et al. 1981). Moreover, visceral fat deposition, a major risk factor for cardiometabolic disorders, readily accumulates in vervet monkeys, even when they consume a low-fat diet.
Metabolism measures associated with obesity (e.g., body mass index, waist circumference, total cholesterol, triglyceride, high-density lipoprotein, and insulin) are moderately heritable among VRC monkeys (h2 = 0.18–0.44) (Kavanagh et al. 2007). Vervets also have proven useful in modeling the development of metabolic disorder, including obesity-related insulin resistance and type 2 diabetes (Kavanagh et al. 2007; Clarkson et al. 1985). Cann and colleagues (2010) observed two clinical forms of naturally occurring diabetes in vervet, and Kavanagh and colleagues (2011) showed that poor maternal glycemic control as measured by glycosylated A1C and low social ranking are associated with infant mortality.
Dietary Manipulation
Probably the best example of the use of the vervet model for investigations of cardiometabolic traits is a study that was conducted in the VRC to assess the impact of different diets on metabolic phenotypes, as evaluated by analysis of genotype (G) by diet (D) (G × D) interaction. The entire colony was initially fed with a standard low-fat primate diet (chow), and then was challenged over 8 weeks with a high-fat diet mimicking typical American diet. Although monkeys did not gain weight over this relatively short period of time, measures of most of their lipid and glycemic biomarkers showed a shift toward unhealthy values. These measures were moderately heritable in both environments (low- and high-fat diet). Evidence was observed for a significant G × D interaction for insulin, body weight, and lipid measures (Voruganti 2013). This study is an example of the type of longitudinal large-scale genetic investigation that is difficult to conduct in humans because of the infeasibility of obtaining a similar level of control of diet and other environmental exposures.
Investigation of the Vervet Microbiome
The microbiome is the microbial ecosystem comprising all the microorganisms that colonize an animal. The habitats of the animal body differ (e.g., oral, gut, skin, vagina) in terms of physical, chemical, nutrient, and other characteristics, and thus different types of microbes are found in different body sites. But each body site has a rich microbial community—mainly bacteria, but also viruses and eukaryotic microbes.
The function of the microbiome is only beginning to be unraveled, but its complexity and integration with the host suggests its impact is significant. The microbiome can be health promoting, for example, by providing additional metabolic capabilities over those in the host genome. There is evidence that the microbiome is required for normal development of tissues, such as the gastrointestinal tract. The microbiome is also protective against infection, possibly by competing with pathogens. Understanding causal effects of the microbiome allows insights into mechanisms and may lead to therapeutics. Likewise, understanding correlative effects on the microbiome can lead to new diagnostics.
The goal of microbiome analysis, then, is to describe these communities and determine how they differ with diet, health and disease, genotype, and other circumstances. This is done by two general methods. In one, the bacterial 16S rRNA gene is targeted for DNA sequencing. The 16S rRNA gene is different in sequence for each bacterial species, and thus determining the 16S rRNA sequences present in a microbial community is a way of taking a taxonomic census. Typically, one uses PCR primers that are slightly degenerate and can amplify many different versions of the 16S rRNA gene. The fragments resulting from amplification are sequenced (e.g., on the 454 FLX sequencing platform), and the resulting sequences are compared with a database of known 16S rRNA genes to identify the taxa that are present. The number of sequences found for a particular taxon reflects the abundance of that organism. In an alternative method, one does not rely on a database of known sequences but simply compares all the sequences found in all the samples. Sequences that are at least 97% identical are called operational taxonomic units and are a surrogate for species. In either method, at the end, for each sample, one obtains a table of taxa/operational taxonomic units and their abundances. These tables are used for the comparisons of different samples.
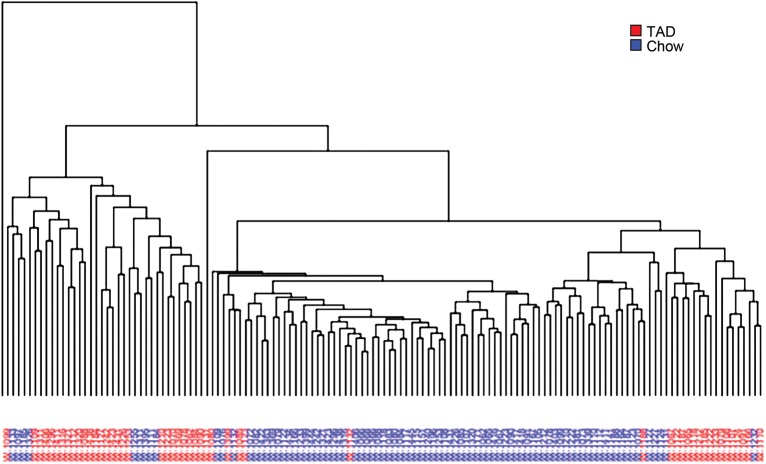
An example of this type of analysis is shown in Figure 7. In this experiment, 138 vervets were kept on extended diets of either monkey chow or the higher-fat typical American diet. For each animal, a rectal swab was obtained, DNA was extracted from the stool, and 16S rRNA gene sequencing was performed. The tables of taxa and abundances described above were then used to analyze the samples by hierarchical clustering. As can be seen, the samples from similar diets tended to cluster together, showing the different diets had distinct microbial communities.
Figure 7.
Dendrogram of the vervet fecal microbiome under two diets: monkey chow and a typical American Diet (TAD). Eighty-nine female vervets (aged 4.6–24.9 years) on a routine diet of monkey chow were switched to TAD (35% calories from fat) for 27 to 28 weeks, and samples were collected before and after the diet change. In all, 138 samples (rectal swabs) were obtained. DNA was extracted using the MOBIO PowerSoil kit, and the 16S rRNA genes were amplified (variable regions 3–5) and subjected to pyrosequencing using the 454 Life Sciences sequencing platform. Sequences were compared with the Ribosomal Database Project (RDP) database after quality filtering and the taxa, and their abundances found in each sample were tallied. The Bray–Curtis index was calculated for all pairings of samples, and this was used to construct the dendrogram shown by hierarchical clustering with complete linkage.
The second type of analysis does rely on a single (16S rRNA) gene but performs shotgun sequencing on the entire metagenomic sample. Because all DNA molecules are sequenced, this method provides a picture of the total gene content of a sample. Not only the genes of bacteria but also those from viruses and eukaryotic microbes as well as parasites are sequenced and are part of the dataset. This data can be analyzed to indicate which taxa are present and their abundances, as for the 16S rRNA gene method, by comparing the sequences to databases of known genomes or even databases of all known genes, such as GenBank at the National Center for Biotechnology Information. This analysis can focus on viruses by using viral genome databases. It is more challenging for eukaryotic microbes because there are fewer such sequences in databases and thus many of their metagenomic sequences cannot be identified with a particular organism.
Alternatively, one may use shotgun data to analyze which pathways are present in the microbial community. One approach for this end is to compare the sequences to the Kyoto Encyclopedia of Genes and Genomes, which contains sequences of genes for most of the known metabolic pathways. In this way, one can identify which pathways are present and their relative abundances, independent of knowledge of the particular organism contributing the genes.
All of these analyses are of use in observing the changes in organisms, genes, pathways, and other genetic characteristics of the microbiome. The vervet is a powerful system to use in conjunction with microbiome analysis. For example, the ability to keep animals on controlled diets for long term allows the effects of diet on the microbiome to observed, as in Figure 7. This is much harder in humans because diet is harder to manipulate. Likewise, there are many other variables that are likely to affect the microbiome that can be observed. And in the case of a vervet colony whose genetics are well-defined, it should be possible to analyze the effect of host genotype on microbiome structure.
Genetic Trait Mapping in the VRC Pedigree
To facilitate genetic mapping studies in the VRC pedigree, before the vervet reference genome was available, a genetic linkage map was constructed based on human microsatellite markers, as already mentioned with respect to vervet chromosomal structure. This map took advantage of the low genomic sequence divergence between vervet and human. Through the testing of more than 5000 human microsatellites in vervet, 360 of these markers were chosen and then genotyped in in 434 members of the VRC pedigree. It was possible from these genotype data to construct a vervet genetic map, with markers at an average distance of 9.8 Kosambi centiMorgnans from each other, including 226 markers unequivocally mapped in a unique order (Jasinska et al. 2007). The maps covered all vervet autosomes except 18, 19, and 28. This map provided an initial framework for genetic mapping of QTL in the VRC.
Illustration of Initial Genetic Mapping in the VRC
QTL Mapping for Biochemical Markers (Monoamine Metabolites)
Monoamine metabolites are important regulators of primate behavior and show links to common human neuropsychatric phenotypes, but genetic regulation of their metabolism, which may underline these traits, is poorly known. Direct assessment of these neurotransmitters in brain is not practical; instead their levels in cerebrospinal fluid approximate their metabolism in brain. We measured levels of three monoamine metabolites—HVA from dopamine, 5-HIAA from serotonin, and MHPG from norepinephrine—in the VRC pedigree. All three metabolites showed moderate heritability with highest h2 of 0.52 for HVA (Freimer et al. 2007). The genomes scan using the first-generation vervet microsatellite linkage map yielded a suggestive linkage for two markers with 13 cM region on the vervet chromosome 9. We further fine-mapped this signal by high-resolution genotyping using a set of more than 100 local single nucleotide polymorphisms. Two single nucleotide polymorphisms reached genome-wide significance levels for HVA (logarithm of the odds, LOD = 4.23 and 3.96, respectively) and accounted for a high proportion of the heritable trait variance (56–60%) (Freimer et al. 2007). This relatively high proportion of trait variance explained by this signal (presumably representing a single functional locus) exemplifies the advantage for genetic studies of the founder vervet population, in which the genetic contribution to complex phenotypes may be simpler than in humans.
QTL Mapping of Brain Transcriptional Variation Using Peripheral Blood
As noted above, one of the key advantages of NHP models for systems biology investigations is the opportunity that they provide for analyzing functional genomic processes in tissues that would be impractical or unethical to collect in humans. One example of this possibility in the vervet was the identification of heritable transcripts whose profile in blood correlates with their profile in the brain (Jasinska et al. 2009). Linkage analysis of VRC pedigree-wide blood expression patterns of these transcripts led to discovery of 12 expression QTL, hypothesized to differentially regulate the levels of these transcripts in the brain (Jasinska et al. 2012). Furthermore, a subset of these expression QTL could be replicated in independent vervet samples from St. Kitts. This result provided an initial confirmation of the hypothesis that the VRC incorporates a substantial proportion of the genetic variability of the Caribbean vervet populations from which it derives and supports the idea that well-powered genetic investigations could be designed by integrating the use of both types of study sample.
Conclusions: Future Systems Biology Studies
In this article, we have described the features of the vervet that make it an ideal NHP model for systems biology, in particular its exceptional population genetic history and the availability of large, well-characterized pedigree and population samples for systems-level studies. Additionally, we have outlined the steps taken to enable such studies, including the assessment of a wide range of multisystem phenotypes, the establishment of the Systems Biology Sample Repository, and the use of next-generation sequencing to characterize vervet reference genomes and genome-wide genetic variation.
Over the next few years, we anticipate that a number of systems biology projects will be carried out, both by members of the collaborative team that has established these resources and by other scientists who are attracted to this model. In particular, we envision three broad types of studies using these resources:
Studies with a multistage experimental design: Although extended pedigrees such as the VRC are ideal for initial identification of QTL, the close relationship between the pedigreed animals makes it difficult to resolve such loci to the specific variant level. Here the possibility of following up such findings in large population samples (e.g., from St. Kitts) will be invaluable.
Intensive studies in captive populations of traits identified in wild populations: Although the large wild populations of vervets available offer exceptional opportunities for observational studies, they are not ideal for invasive or mechanistic investigations, which may be then designed in colony samples. For example, mechanistic studies of the ESN phenomenon discovered in African C. a. pygerythrus samples could be carried out in the South African Medical Research Council using monkeys derived from the population in which the phenomenon was discovered.
Studies using the vervet as a translational model: Interventional studies in captive vervet populations provide an opportunity to validate findings form human populations. Examples include controlled manipulation of diet (e.g. introduction of a typical Western diet to investigate cardiometabolic diseases).
Acknowledgments
This work was supported by the National Institutes of Health (grants R01RR016300, PL1NS062410, UL1DEO19580, P30NS062691, RL1MH083270, U54HG003079 and P40RR019963); Genome Quebec; and Genome Canada.
References
- Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GM. Mammals of the West Indies. Bulletin of the Museum of Comparative Zoology. 1911;54:175–263. [Google Scholar]
- Altmann J, Alberts SC. Growth rates in a wild primate population: Ecological influences and maternal effects. Behav Ecol Sociobiol. 2005;57:490–501. [Google Scholar]
- Bacopoulos NG, Redmond DE, Baulu J, Roth RH. Chronic haloperidol or fluphenazine: Effects on dopamine metabolism in brain, cerebrospinal fluid and plasma of Cercopithecus aethiops (vervet monkey) J Pharmacol Exp Ther 212. 1980:1–5. [PubMed] [Google Scholar]
- Bailey JN, Breidenthal SE, Jorgensen MJ, McCracken JT, Fairbanks LA. The association of DRD4 and novelty seeking is found in a nonhuman primate model. Psychiatr Genet. 2007;17:23–27. doi: 10.1097/YPG.0b013e32801140f2. [DOI] [PubMed] [Google Scholar]
- Balodis IM, Wynne-Edwards KE, Olmstead MC. The other side of the curve: Examining the relationship between pre-stressor physiological responses and stress reactivity. Psychoneuroendocrinology. 2010;35:1363–1373. doi: 10.1016/j.psyneuen.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Baulu J, Evans G, Sutton C. Pathogenic agents found in Barbados Chlorocebus aethiops sabaeus and in Old World Monkeys commonly used in biomedical research. Laboratory Primate Newsletter. 2002;41:4–6. [Google Scholar]
- Bibollet-Ruche F, Bailes E, Gao F, Pourrut X, Barlow KL, Clewley JP, Mwenda JM, Langat DK, Chege GK, McClure HM, Mpoudi-Ngole E, Delaporte E, Peeters M, Shaw GM, Sharp PM, Hahn BH. New simian immunodeficiency virus infecting De Brazza's monkeys (Cercopithecus neglectus): Evidence for a cercopithecus monkey virus clade. J Virol. 2004;78:7748–7762. doi: 10.1128/JVI.78.14.7748-7762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brammer GL, Raleigh MJ, Ritvo ER, Geller E, McGuire MT, Yuwiler A. Fenfluramine effects on serotonergic measures in vervet monkeys. Pharmacol Biochem Behav. 1991;40:267–271. doi: 10.1016/0091-3057(91)90550-l. [DOI] [PubMed] [Google Scholar]
- Bjugstad KB, Redmond DE, Jr, Teng YD, Elsworth JD, Roth RH, Blanchard BC, Snyder EY, Sladek JR., Jr. Neural stem cells implanted into MPTP-treated monkeys increase the size of endogenous tyrosine hydroxylase-positive cells found in the striatum: A return to control measures. Cell Transplant. 2005;14:183–192. doi: 10.3727/000000005783983098. [DOI] [PubMed] [Google Scholar]
- Bosinger SE, Jacquelin B, Benecke A, Silvestri G, Muller-Trutwin M. Systems biology of natural simian immunodeficiency virus infections. Curr Opin HIV AIDS. 2012;7:71–78. doi: 10.1097/COH.0b013e32834dde01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett FL, Turner TR, Jolly CJ, Cauble R. Trapping baboons and vervet monkeys from wild, free-ranging populations. J Wildl Manag. 1982;46:164–174. [Google Scholar]
- Bullock BC, Lehner ND, Clarkson TB, Feldner MA, Wagner WD, Lofland HB. Comparative primate atherosclerosis. I. Tissue cholesterol concentration and pathologic anatomy. Exp Mol Pathol. 1975;22:151–175. doi: 10.1016/0014-4800(75)90060-x. [DOI] [PubMed] [Google Scholar]
- Cann JA, Kavanagh K, Jorgensen MJ, Mohanan S, Howard TD, Gray SB, Hawkins GA, Fairbanks LA, Wagner JD. Clinicopathologic characterization of naturally occurring diabetes mellitus in vervet monkeys. Vet Pathol. 2010;47:713–78. doi: 10.1177/0300985810370011. [DOI] [PubMed] [Google Scholar]
- Cheney DL, Lee PC, Seyfarth RM. Behavioral correlates of non-random mortality among free-ranging female vervet monkeys. Behav Ecol Sociobiol. 1981;9:153–161. [Google Scholar]
- Cheney DL, Seyfarth RM. Non-random dispersal in free-ranging vervet monkeys: social and genetic consequence. Am Nat. 1983;122:392–412. [Google Scholar]
- Cheney DL, Seyfarth RM. The influence of intergroup competition on the survival and reproduction of female vervet monkeys. Behav Ecol Sociobiol. 1987;21:375–386. [Google Scholar]
- Clarkson TB, Koritnik DR, Weingand KW, Miller LC. Nonhuman primate models of atherosclerosis: Potential for the study of diabetes mellitus and hyperinsulinemia. Metabolism. 1985;34:51–59. doi: 10.1016/s0026-0495(85)80010-x. [DOI] [PubMed] [Google Scholar]
- Coe CL. Psychosocial factors and immunity in nonhuman primates: A review. Psychosom Med. 1993;55:298–308. doi: 10.1097/00006842-199305000-00007. [DOI] [PubMed] [Google Scholar]
- Courgnaud V, Pourrut X, Bibollet-Ruche F, Mpoudi-Ngole E, Bourgeois A, Delaporte E, Peeters M. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: A new lineage in the nonhuman primate lentivirus family. J Virol. 2001;75:857–866. doi: 10.1128/JVI.75.2.857-866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denham W. Basel: Karger Press; 1987. West Indian Green Monkeys: Problems in Historical Biogeography. [Google Scholar]
- Detwiler KM, Burrell AS, Jolly CJ. Conservation Implications of hybridization in African cercopithecine monkeys. Int J Primatol. 2005;26:661–784. [Google Scholar]
- Disotell TR. Cambridge: Cambridge University Press; 2000. Molecular Systematics of the Cercopithecidae. [Google Scholar]
- Dutrillaux B. Chromosomal evolution in primates: Tentative phylogeny from Microcebus murinus (Prosimian) to man. Hum Genet. 1979;48:51–314. doi: 10.1007/BF00272830. [DOI] [PubMed] [Google Scholar]
- Emborg ME. Nonhuman primate models of Parkinson's disease. ILAR J. 2007;48:339–355. doi: 10.1093/ilar.48.4.339. [DOI] [PubMed] [Google Scholar]
- Elsworth JD, Deutch AY, Redmond DE, Jr, Taylor JR, Sladek JR, Jr, Roth RH. Symptomatic and asymptomatic 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-treated primates: biochemical changes in striatal regions. Neuroscience 33. 1989:323–331. doi: 10.1016/0306-4522(89)90212-1. [DOI] [PubMed] [Google Scholar]
- Elsworth JD, Redmond DE, Jr, Roth RH. Plasma and cerebrospinal fluid 3-methoxy-4-hydroxyphenylethylene glycol (MHPG) as indices of brain norepinephrine metabolism in primates. Brain Res. 1982;235:115–124. doi: 10.1016/0006-8993(82)90200-1. [DOI] [PubMed] [Google Scholar]
- Fairbanks LA, Bailey JN, Breidenthal SE, Laudenslager ML, Kaplan JR, Jorgensen MJ. Environmental stress alters genetic regulation of novelty seeking in vervet monkeys. Genes Brain Behav. 2011;10:683–688. doi: 10.1111/j.1601-183X.2011.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks LA, Jorgensen MJ, Bailey JN, Breidenthal SE, Grzywa R, Laudenslager ML. Heritability and genetic correlation of hair cortisol in vervet monkeys in low and higher stress environments. Psychoneuroendocrinology. 2011;36:1201–1208. doi: 10.1016/j.psyneuen.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks LA, Melega WP, Jorgensen MJ, Kaplan JR, McGuire MT. Social impulsivity inversely associated with CSF 5-HIAA and fluoxetine exposure in vervet monkeys. Neuropsychopharmacology. 2001;24:370–378. doi: 10.1016/S0893-133X(00)00211-6. [DOI] [PubMed] [Google Scholar]
- Fairbanks LA, Newman TK, Bailey JN, Jorgensen MJ, Breidenthal SE, Ophoff RA, Comuzzie AG, Martin LJ, Rogers J. Genetic contributions to social impulsivity and aggressiveness in vervet monkeys. Biol Psychiatry. 2004;55:642–647. doi: 10.1016/j.biopsych.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Fairbanks LA, Way BM, Breidenthal SE, Bailey JN, Jorgensen MJ. Maternal and offspring dopamine D4 receptor genotypes interact to influence juvenile impulsivity in vervet monkeys. Psychol Sci. 2012;23:1099–1104. doi: 10.1177/0956797612444905. [DOI] [PubMed] [Google Scholar]
- Favre D, Lederer S, Kanwar B, Ma ZM, Proll S, Kasakow Z, Mold J, Swainson L, Barbour JD, Baskin CR, Palermo R, Pandrea I, Miller CJ, Katze MG, McCune JM. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000295. e1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fears SC, Melega WP, Service SK, Lee C, Chen K, Tu Z, Jorgensen MJ, Fairbanks LA, Cantor RM, Freimer NB, Woods RP. Identifying heritable brain phenotypes in an extended pedigree of vervet monkeys. J Neurosci. 2009;29:2867–2875. doi: 10.1523/JNEUROSCI.5153-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fears SC, Scheibel K, Abaryan Z, Lee C, Service SK, Jorgensen MJ, Fairbanks LA, Cantor RM, Freimer NB, Woods RP. Anatomic brain asymmetry in vervet monkeys. PLoS One. 2011;6 doi: 10.1371/journal.pone.0028243. e28243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finelli P, Stanyon R, Plesker R, Ferguson-Smith MA, O'Brien PC, Wienberg J. Reciprocal chromosome painting shows that the great difference in diploid number between human and African green monkey is mostly due to non-Robertsonian fissions. Mamm Genome. 1999;10:713–718. doi: 10.1007/s003359901077. [DOI] [PubMed] [Google Scholar]
- Fowke KR, Nagelkerke NJ, Kimani J, Simonsen JN, Anzala AO, Bwayo JJ, MacDonald KS, Ngugi EN, Plummer FA. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet. 1996;348:1347–1351. doi: 10.1016/S0140-6736(95)12269-2. [DOI] [PubMed] [Google Scholar]
- Freimer NB, Service SK, Ophoff RA, Jasinska AJ, McKee K, Villeneuve A, Belisle A, Bailey JN, Breidenthal SE, Jorgensen MJ, Mann JJ, Cantor RM, Dewar K, Fairbanks LA. A quantitative trait locus for variation in dopamine metabolism mapped in a primate model using reference sequences from related species. Proc Natl Acad Sci U S A. 2007;104:15811–15816. doi: 10.1073/pnas.0707640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusegawa Y, Kelley KL, Sawyer JK, Shah RN, Rudel LL. Influence of dietary fatty acid composition on the relationship between CETP activity and plasma lipoproteins in monkeys. J Lipid Res. 2001;42:1849–1857. [PubMed] [Google Scholar]
- Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Gordon S, Pandrea I, Dunham R, Apetrei C, Silvestri G. The call of the wild: What can be learned from studies of SIV infection of natural hosts? In. In: Leitner BF, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B, editors. HIV Sequence Compendium 2004. Los Alamos National Laboratory (NM): Theoretical Biology and Biophysics Group. 2005. pp. 2–29. [Google Scholar]
- Grant SJ, Galloway MP, Mayor R, Fenerty JP, Finkelstein MF, Roth RH, Redmond DE., Jr. Precipitated diazepam withdrawal elevates noradrenergic metabolism in primate brain. Eur J Pharmacol. 1985;107:127–132. doi: 10.1016/0014-2999(85)90050-0. [DOI] [PubMed] [Google Scholar]
- Grobler JP, Turner T. A novel trap design for the capture and sedation of vervet monkeys (Chlorocebus aethiops) S Afr J Wildli Res. 2010;40:163–168. [Google Scholar]
- Groman SM, James AS, Seu E, Crawford MA, Harpster SN, Jentsch JD. Monoamine levels within the orbitofrontal cortex and putamen interact to predict reversal learning performance. Biol Psychiatry. 2013;73:756–762. doi: 10.1016/j.biopsych.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groman SM, Lee B, London ED, Mandelkern MA, James AS, Feiler K, Rivera R, Dahlbom M, Sossi V, Vandervoort E, Jentsch JD. Dorsal striatal D2-like receptor availability covaries with sensitivity to positive reinforcement during discrimination learning. J Neurosci. 2011;31:7291–7299. doi: 10.1523/JNEUROSCI.0363-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groman SM, Lee B, Seu E, James AS, Feiler K, Mandelkern MA, London ED, Jentsch JD. Dysregulation of D(2)-mediated dopamine transmission in monkeys after chronic escalating methamphetamine exposure. J Neurosci. 2012;32:5843–5852. doi: 10.1523/JNEUROSCI.0029-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- Hansel A, Hong S, Camara RJ, von Kanel R. Inflammation as a psychophysiological biomarker in chronic psychosocial stress. Neurosci Biobehav Rev. 2010;35:115–21. doi: 10.1016/j.neubiorev.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Harrison MJS. Patterns of range use by the green monkey, Cercopithecus sabaeus, at Mt. Assirik, Senegal. Folia Primatol. 1983;41:157–179. [Google Scholar]
- Haus T, Akom E, Agwanda B, Hofreiter M, Roos C, Zinner D. Mitochondrial diversity and distribution of African green monkeys (chlorocebus gray, 1870) Am J Primatol. 2013;75:350–360. doi: 10.1002/ajp.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch VM, Dapolito G, Goeken R, Campbell BJ. Phylogeny and natural history of the primate lentiviruses, SIV and HIV. Curr Opin Genet Dev. 1995;5:798–806. doi: 10.1016/0959-437x(95)80014-v. [DOI] [PubMed] [Google Scholar]
- Horrocks J. Life-history characteristics of a wild population of vervets (Cercopithecus aethiops sabaeus) in Barbados, West Indies. Int J Primatol. 1986;15:31–47. [Google Scholar]
- Imagawa DT, Lee MH, Wolinsky SM, Sano K, Morales F, Kwok S, Sninsky JJ, Nishanian PG, Giorgi J, Fahey JL. Human immunodeficiency virus type 1 infection in homosexual men who remain seronegative for prolonged periods. N Engl J Med. 1989;320:1458–1462. doi: 10.1056/NEJM198906013202205. [DOI] [PubMed] [Google Scholar]
- Isbell LA, Cheney DL, Seyfarth RM. Are immigrant vervet monkeys, Cercopithecus aethiops, at greater risk of mortality than residents? Anim Behav. 1991;45:729–734. [Google Scholar]
- IUCN [International Union for Conservation of Nature and Natural Resources] 2013 IUCN Red List of Threatened Species. Version 2013.1. Available from www.iucnredlist.org . Accessed on September 2, 2013. [Google Scholar]
- Jacquelin B, Mayau V, Targat B, Liovat AS, Kunkel D, Petitjean G, Dillies MA, Roques P, Butor C, Silvestri G, Giavedoni LD, Lebon P, Barre-Sinoussi F, Benecke A, Muller-Trutwin MC. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119:3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AS, Groman SM, Seu E, Jorgensen M, Fairbanks LA, Jentsch JD. Dimensions of impulsivity are associated with poor spatial working memory performance in monkeys. J Neurosci. 2007;27:14358–14364. doi: 10.1523/JNEUROSCI.4508-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinska AJ, Lin MK, Service S, Choi OW, DeYoung J, Grujic O, Kong SY, Jung Y, Jorgensen MJ, Fairbanks LA, Turner T, Cantor RM, Wasserscheid J, Dewar K, Warren W, Wilson RK, Weinstock G, Jentsch JD, Freimer NB. A non-human primate system for large-scale genetic studies of complex traits. Hum Mol Genet. 2012;21:3307–3316. doi: 10.1093/hmg/dds160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinska AJ, Service S, Choi OW, DeYoung J, Grujic O, Kong SY, Jorgensen MJ, Bailey J, Breidenthal S, Fairbanks LA, Woods RP, Jentsch JD, Freimer NB. Identification of brain transcriptional variation reproduced in peripheral blood: an approach for mapping brain expression traits. Hum Mol Genet. 2009;18:4415–4427. doi: 10.1093/hmg/ddp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinska AJ, Service S, Levinson M, Slaten E, Lee O, Sobel E, Fairbanks LA, Bailey JN, Jorgensen MJ, Breidenthal SE, Dewar K, Hudson TJ, Palmour R, Freimer NB, Ophoff RA. A genetic linkage map of the vervet monkey (Chlorocebus aethiops sabaeus) Mamm Genome. 2007;18:347–360. doi: 10.1007/s00335-007-9026-4. [DOI] [PubMed] [Google Scholar]
- Jentsch JD, Redmond DE, Jr, Elsworth JD, Taylor JR, Youngren KD, Roth RH. Enduring cognitive deficits and cortical dopamine dysfunction in monkeys after long-term administration of phencyclidine. Science. 1997;277:953–955. doi: 10.1126/science.277.5328.953. [DOI] [PubMed] [Google Scholar]
- Jin MJ, Hui H, Robertson DL, Muller MC, Barre-Sinoussi F, Hirsch VM, Allan JS, Shaw GM, Sharp PM, Hahn BH. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K, Dozier BL, Chavanne TJ, Fairbanks LA, Jorgensen MJ, Kaplan JR. Fetal and maternal factors associated with infant mortality in vervet monkeys. J Med Primatol. 2011;40:27–36. doi: 10.1111/j.1600-0684.2010.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K, Fairbanks LA, Bailey JN, Jorgensen MJ, Wilson M, Zhang L, Rudel LL, Wagner JD. Characterization and heritability of obesity and associated risk factors in vervet monkeys. Obesity (Silver Spring) 2007;15(7):1666–1674. doi: 10.1038/oby.2007.199. [DOI] [PubMed] [Google Scholar]
- Kavanagh M. Variable territoriality among tantalus monkeys in Cameroon. Folia Primatol (Basel) 1981;36:76–98. doi: 10.1159/000156009. [DOI] [PubMed] [Google Scholar]
- Kuhmann SE, Madani N, Diop OM, Platt EJ, Morvan J, Muller-Trutwin MC, Barre-Sinoussi F, Kabat D. Frequent substitution polymorphisms in African green monkey CCR5 cluster at critical sites for infections by simian immunodeficiency virus SIVagm, implying ancient virus-host coevolution. J Virol. 2001;75:8449–8460. doi: 10.1128/JVI.75.18.8449-8460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer S, Favre D, Walters KA, Proll S, Kanwar B, Kasakow Z, Baskin CR, Palermo R, McCune JM, Katze MG. Transcriptional profiling in pathogenic and non-pathogenic SIV infections reveals significant distinctions in kinetics and tissue compartmentalization. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000296. e1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemere CA, Beierschmitt A, Iglesias M, Spooner ET, Bloom JK, Leverone JF, Zheng JB, Seabrook TJ, Louard D, Li D, Selkoe DJ, Palmour RM, Ervin FR. Alzheimer's disease abeta vaccine reduces central nervous system abeta levels in a non-human primate, the Caribbean vervet. Am J Pathol. 2004;165:283–297. doi: 10.1016/s0002-9440(10)63296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Jasinska A, Kristoff J, Grobler JP, Turner T, Jung Y, Schmitt C, Raehtz K, Feyertag F, Martinez Sosa N, Wijewardana V, Burke DS, Robertson DL, Tracy R, Pandrea I, Freimer N, Apetrei C. SIVagm infection in Wild African green monkeys from South Africa: Epidemiology, natural history, and evolutionary considerations. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003011. e1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestripieri D, Hoffman CL. Chronic stress, allostatic load, and aging in nonhuman primates. Dev Psychopathol. 2011;23:1187–1195. doi: 10.1017/S0954579411000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manches O, Bhardwaj N. Resolution of immune activation defines nonpathogenic SIV infection. J Clin Invest. 2009;119:3512–3515. doi: 10.1172/JCI41509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuck SB, Kaplan JR, Rymeski BA, Fairbanks LA, Wilson ME. Approach to a social stranger is associated with low central nervous system serotonergic responsivity in female cynomolgus monkeys (Macaca fascicularis) Am J Primatol. 2003;61:187–194. doi: 10.1002/ajp.10118. [DOI] [PubMed] [Google Scholar]
- McGuire MT. The St. Kitts vervet (Cercopithecus aethiops) J Med Primatol. 1974;3:285–297. doi: 10.1159/000460030. [DOI] [PubMed] [Google Scholar]
- McGuire MT, Raleigh MJ. Serotonin-behavior interactions in vervet monkeys. Psychopharmacol Bull. 1985;21:458–463. [PubMed] [Google Scholar]
- Mekonnen A, Bekele A, Fashing PJ, Lernould JM, Atickem A, Stenseth NC. Newly discovered Bale monkey populations in forest fragments in southern Ethiopia: Evidence of crop raiding, hybridization with grivets, and other conservation threats. Am J Primatol. 2012;74:423–432. doi: 10.1002/ajp.21999. [DOI] [PubMed] [Google Scholar]
- Melega WP, Jorgensen MJ, Lacan G, Way BM, Pham J, Morton G, Cho AK, Fairbanks LA. Long-term methamphetamine administration in the vervet monkey models aspects of a human exposure: Brain neurotoxicity and behavioral profiles. Neuropsychopharmacology. 2008;33:1441–1452. doi: 10.1038/sj.npp.1301502. [DOI] [PubMed] [Google Scholar]
- Murphy DL, Redmond DE, Jr, Garrick N, Baulu J. Brain region differences and some characteristics of monoamine oxidase type A and B activities in the vervet monkey. Neurochem Res. 1979;4:53–62. doi: 10.1007/BF00963831. [DOI] [PubMed] [Google Scholar]
- Palmour RM, Ervin FR, Baker GB, Young SN. An amino acid mixture deficient in phenylalanine and tyrosine reduces cerebrospinal fluid catecholamine metabolites and alcohol consumption in vervet monkeys. Psychopharmacology (Berl) 1998;136:1–7. doi: 10.1007/s002130050532. [DOI] [PubMed] [Google Scholar]
- Pandrea I, Apetrei C. Where the wild things are: Pathogenesis of SIV infection in African nonhuman primate hosts. Curr HIV/AIDS Rep. 2010;7:28–36. doi: 10.1007/s11904-009-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandrea I, Apetrei C, Dufour J, Dillon N, Barbercheck J, Metzger M, Jacquelin B, Bohm R, Marx PA, Barre-Sinoussi F, Hirsch VM, Muller-Trutwin MC, Lackner AA, Veazey RS. Simian immunodeficiency virus SIVagm.sab infection of Caribbean African green monkeys: A new model for the study of SIV pathogenesis in natural hosts. J Virol. 2006;80:4858–4867. doi: 10.1128/JVI.80.10.4858-4867.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandrea I, Silvestri G, Apetrei C. AIDS in african nonhuman primate hosts of SIVs: A new paradigm of SIV infection. Curr HIV Res. 2009;7:57–72. doi: 10.2174/157016209787048456. [DOI] [PubMed] [Google Scholar]
- Pandrea I, Sodora DL, Silvestri G, Apetrei C. Into the wild: Simian immunodeficiency virus (SIV) infection in natural hosts. Trends Immunol. 2008;29:419–428. doi: 10.1016/j.it.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen L. Positional cloning of disease genes: Advantages of genetic isolates. Hum Hered. 2000;50:66–75. doi: 10.1159/000022892. [DOI] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MP, Silva A, O'Brien SJ, Pecon-Slattery J. A molecular phylogeny of living primates. 7. 2011;PLoS Genet doi: 10.1371/journal.pgen.1001342. e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Carmelli D. Morphological changes in aging brain structures are differentially affected by time-linked environmental influences despite strong genetic stability. Neurobiol Aging. 2004;25:175–183. doi: 10.1016/s0197-4580(03)00045-9. [DOI] [PubMed] [Google Scholar]
- Poirier FE. The St. Kitts green monkey (Cercopithecus aethiops sabaeus): ecology, population dynamics, and selected behavioral traits. Folia Primatol. 1972;17:20–55. doi: 10.1159/000155415. [DOI] [PubMed] [Google Scholar]
- Posthuma D, de Geus EJ, Neale MC, Hulshoff Pol HE, Baare WEC, Kahn RS, Boomsma D. Multivariate genetic analysis of brain structure in an extended twin design. Behav Genet. 2000;30:311–319. doi: 10.1023/a:1026501501434. [DOI] [PubMed] [Google Scholar]
- Raleigh MJ, Brammer GL, McGuire MT, Pollack DB, Yuwiler A. Individual differences in basal cisternal cerebrospinal fluid 5-HIAA and HVA in monkeys. The effects of gender, age, physical characteristics, and matrilineal influences. Neuropsychopharmacology. 1992;7:295–304. [PubMed] [Google Scholar]
- Raleigh MJ, Brammer GL, McGuire MT, Yuwiler A. Dominant social status facilitates the behavioral effects of serotonergic agonists. Brain Res. 1985;348:274–282. doi: 10.1016/0006-8993(85)90445-7. [DOI] [PubMed] [Google Scholar]
- Raleigh MJ, Brammer GL, Ritvo ER, Geller E, McGuire MT, Yuwiler A. Effects of chronic fenfluramine on blood serotonin, cerebrospinal fluid metabolites, and behavior in monkeys. Psychopharmacology (Berl) 1986;90:503–508. doi: 10.1007/BF00174069. [DOI] [PubMed] [Google Scholar]
- Raleigh MJ, Brammer GL, Yuwiler A, Flannery JW, McGuire MT, Geller E. Serotonergic influences on the social behavior of vervet monkeys (Cercopithecus aethiops sabaeus) Exp Neurol. 1980;68:322–334. doi: 10.1016/0014-4886(80)90089-8. [DOI] [PubMed] [Google Scholar]
- Raleigh MJ, McGuire MT, Brammer GL, Pollack DB, Yuwiler A. Serotonergic mechanisms promote dominance acquisition in adult male vervet monkeys. Brain Res. 1991;559:181–190. doi: 10.1016/0006-8993(91)90001-c. [DOI] [PubMed] [Google Scholar]
- Raleigh MJ, McGuire MT, Brammer GL, Yuwiler A. Social and environmental influences on blood serotonin concentrations in monkeys. Arch Gen Psychiatry. 1984;41:405–410. doi: 10.1001/archpsyc.1984.01790150095013. [DOI] [PubMed] [Google Scholar]
- Raleigh MJ, Steklis HD, Ervin FR, Kling AS, McGuire MT. The effects of orbitofrontal lesions on the aggressive behavior of vervet monkeys (Cercopithecus aethiops sabaeus) Exp Neurol. 1979;66:158–168. doi: 10.1016/0014-4886(79)90071-2. [DOI] [PubMed] [Google Scholar]
- Redmond DE, Jr, Bjugstad KB, Teng YD, Ourednik V, Ourednik J, Wakeman DR, Parsons XH, Gonzalez R, Blanchard BC, Kim SU, Gu Z, Lipton SA, Markakis EA, Roth RH, Elsworth JD, Sladek JR, Jr, Sidman RL, Snyder EY. Behavioral improvement in a primate Parkinson's model is associated with multiple homeostatic effects of human neural stem cells. Proc Natl Acad Sci U S A. 2007;104:12175–12180. doi: 10.1073/pnas.0704091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth RH, Elsworth JD, Redmond DE., Jr. Clonidine suppression of noradrenergic hyperactivity during morphine withdrawal by clonidine: Biochemical studies in rodents and primates. J Clin Psychiatry. 1982;43:42–46. [PubMed] [Google Scholar]
- Rudel LL, Reynolds JA, Bullock BC. Nutritional effects on blood lipid and HDL cholesterol concentrations in two subspecies of African green monkeys (Cercopithecus aethiops) J Lipid Res. 1981;22:278–286. [PubMed] [Google Scholar]
- Sabatti C, Service SK, Hartikainen AL, Pouta A, Ripatti S, Brodsky J, Jones CG, Zaitlen NA, Varilo T, Kaakinen M, Sovio U, Ruokonen A, Laitinen J, Jakkula E, Coin L, Hoggart C, Collins A, Turunen H, Gabriel S, Elliot P, McCarthy MI, Daly MJ, Jarvelin MR, Freimer NB, Peltonen L. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scamvougeras A, Kigar DL, Jones D, Weinberger DR, Witelson SF. Size of the human corpus callosum is genetically determined: An MRI study in mono and dizygotic twins. Neurosci Lett. 2003;338:91–94. doi: 10.1016/s0304-3940(02)01333-2. [DOI] [PubMed] [Google Scholar]
- Service S, DeYoung J, Karayiorgou M, Roos JL, Pretorious H, Bedoya G, Ospina J, Ruiz-Linares A, Macedo A, Palha JA, Heutink P, Aulchenko Y, Oostra B, van Duijn C, Jarvelin MR, Varilo T, Peddle L, Rahman P, Piras G, Monne M, Murray S, Galver L, Peltonen L, Sabatti C, Collins A, Freimer N. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- Struhsaker TT. Social structure among vervet monkeys (Cercopithecus aethiops) Behaviour. 1967;29:6–121. [PubMed] [Google Scholar]
- Sullivan EV, Pfefferbaum A, Swan GE, Carmelli D. Heritability of hippocampal size in elderly twin men: equivalent influence from genes and environment. Hippocampus. 2001;11:754–762. doi: 10.1002/hipo.1091. [DOI] [PubMed] [Google Scholar]
- Tchitchek N, Jacquelin B, Wincker P, Dossat C, Silva CD, Weissenbach J, Blancher A, Muller-Trutwin M, Benecke A. Expression sequence tag library derived from peripheral blood mononuclear cells of the chlorocebus sabaeus. BMC Genomics. 2012;13 doi: 10.1186/1471-2164-13-279. 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi AJ, Detwiler KM, Disotell TR. X-chromosomal window into the evolutionary history of the guenons (Primates: Cercopithecini) Mol Phylogenet Evol. 2005;36:58–66. doi: 10.1016/j.ympev.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Turner TR, Anapol F, Jolly CJ. Growth, development, and sexual dimorphism in vervet monkeys (Cercopithecus aethiops) at four sites in Kenya. Am J Phys Anthropol. 1997;103:19–35. doi: 10.1002/(SICI)1096-8644(199705)103:1<19::AID-AJPA3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- van der Kuyl AC, Kuiken CL, Dekker JT, Goudsmit J. Phylogeny of African monkeys based upon mitochondrial 12S rRNA sequences. J Mol Evol. 1995;40:173–180. doi: 10.1007/BF00167111. [DOI] [PubMed] [Google Scholar]
- von Bonin G, Bailey P. Urbana: University of Illinois Press; 1947. The Neocortex of Macaca mulatta. [Google Scholar]
- Voruganti VS, Jorgensen MJ, Kaplan JR, Kavanagh K, Rudel LL, Temel R, Fairbanks LA, Comuzzie AG. Significant genotype by diet (G x D) interaction effects on cardiometabolic responses to a pedigree-wide, dietary challenge in vervet monkeys (Chlorocebus aethiops sabaeus) Am J Primatol. 2013;75:491–499. doi: 10.1002/ajp.22125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Worobey M. A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes. PLoS Pathog. 2007;3 doi: 10.1371/journal.ppat.0030095. e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NI, Berga SL, Cameron JL. Synergism between psychosocial and metabolic stressors: Impact on reproductive function in cynomolgus monkeys. Am J Physiol Endocrinol Metab. 2007;293:E270–E276. doi: 10.1152/ajpendo.00108.2007. [DOI] [PubMed] [Google Scholar]
- Woods RP, Fears SC, Jorgensen MJ, Fairbanks LA, Toga AW, Freimer NB. A Web-based brain atlas of the vervet monkey. Chlorocebus aethiops. Neuroimage. 2011;54:1872–1880. doi: 10.1016/j.neuroimage.2010.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M, Telfer P, Souquiere S, Hunter M, Coleman CA, Metzger MJ, Reed P, Makuwa M, Hearn G, Honarvar S, Roques P, Apetrei C, Kazanji M, Marx PA. Island biogeography reveals the deep history of SIV. Science. 2010;329:1487. doi: 10.1126/science.1193550. [DOI] [PubMed] [Google Scholar]
- Young JM, Turpin JA, Musib R, Sharma OK. Outcomes of a National Institute of Allergy and Infectious Diseases Workshop on understanding HIV-exposed but seronegative individuals. AIDS Res Hum Retroviruses. 2011;27:737–743. doi: 10.1089/aid.2010.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SN, Ervin FR. Cerebrospinal fluid measurements suggest precursor availability and sex are involved in the control of biogenic amine metabolism in a primate. J Neurochem. 1984;42:1570–1573. doi: 10.1111/j.1471-4159.1984.tb12743.x. [DOI] [PubMed] [Google Scholar]
- Ziegler TE, Bercovitch FB. New York: Wiley-Liss; 1990. Socioendrocrinology of Primate Reproduction. [Google Scholar]