Abstract
The purpose of this paper is to describe pilot work on a semantic model of the pharmacogenomics information found in drug product labels. The model’s development is driven by a series of use cases that we have developed to demonstrate how structured pharmacogenomics information could be more effectively used to support clinical and translational efforts. Using an iterative process, the semantic model was field-tested by five pharmacists, who used it to manually annotate a subset of the sections that the Food and Drug Administration’s Table of Pharmacogenomic Biomarkers in Drug Labels cites as containing pharmacogenomics information. The five pharmacists identified a total of 213 pharmacogenomics statements in 29 sections. The model showed the potential to make the unstructured pharmacogenomic information currently written in product labeling more accessible and actionable through structured annotations of pharmacogenomics effects and clinical recommendations.
Introduction
Although pharmacogenomic information has been written into the product labels of many drugs marketed in the US, difficulties in the presentation and integration of this information limit its accessibility for supporting clinical and research oriented use cases. Genetic, clinical, and pharmacologic relations that might help both clinicians and translational researchers are spread across multiple unstructured data sources that use inconsistent and sometimes ambiguous terminology. The significant effort needed to overcome these barriers hinders the potential of this information to advance translational research and patient care.
The Food and Drug Administration (FDA) publishes a web page1 that indexes pharmacogenomic information present in the product labels of FDA-approved drug products. The table associates drug active ingredients with product label sections that contain pharmacogenomics information. Each entry in the table includes the name of the drug, a broad therapeutic area in which it is used, a common name for the relevant biomarker, and a list of one or more sections in the Structured Product Label (SPL) for the drug that contains pharmacogenomics information. At the time of this writing (fall 2012), the table listed 101 unique active ingredients and six active ingredient combinations. Although useful as an index, the information in this table is not sufficient for making informed decisions for either clinical or translational research applications. For example, a clinician or researcher who is using this table to seek pharmacogenomics information on a codeine drug product would be able to determine that three sections (Warnings and Precautions, Use in Specific Populations, and Clinical Pharmacology) contain information about the metabolic enzyme Cytochrome P-450 2D6 (CYP2D6). However, they would have to read each section closely to learn that the dose of codeine should be reduced in lactating mothers who are homozygous for the CYP2D6*2 allele because this genotype corresponds to the ultra-rapid metabolizer phenotype which is associated with an increased risk of neonatal toxicity.
The purpose of this paper is to describe pilot work on a semantic model of the pharmacogenomics information found in both the FDA table and the product label sections that it references. The model’s development is driven by a series of use cases we have developed to demonstrate how structured pharmacogenomics information could be more effectively used to support clinical and translational efforts. Using an iterative process, the semantic model was field tested by five pharmacists, who used it to manually annotate a subset of the sections that the FDA table cites as containing pharmacogenomics information.
Methods
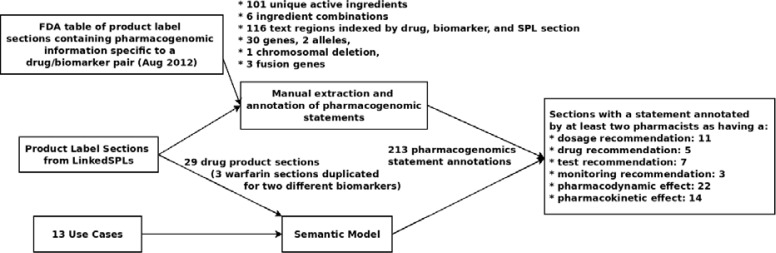
An overview of the methodology used is given in Figure 1. In summary, use cases were developed to guide the development of a semantic model, which was used to manually annotate a set of SPLs. The annotations were used to validate the semantic model and provide insight into range of pharmacogenomics effects and clinical recommendations reported in product labeling.
Figure 1.
Methodology and results overview
Use Cases
Drawing on both clinical background and translational research experience, we developed a set of use cases for pharmacogenomic information. Uses cases were defined from the perspective of professionals who are aware of the importance of pharmacogenomics, but need assistance in finding details relevant for specific clinical questions. These use cases were written and refined iteratively, without reference to any available information model. Once written, the queries were translated into an informal query notation, which will be used as the basis for eventual translation into SPARQL queries against an RDF knowledge base.
Mapping the FDA biomarker table
Data from the FDA’s pharmacogenomic biomarker table1 was downloaded and provided to a clinical pharmacologist (PEE) and a translational researcher with a background in pharmacogenomics (RRF) for review. Biomarkers in the table were mapped, where possible, to appropriate genes and proteins in the Protein Ontology2, official gene symbols maintained by the HUGO Gene Nomenclature Committee (HGNC)3, and biomarker identifiers in PharmGKB 4. Specific variant alleles, fused genes, and chromosomal deletions and translocations were noted but not mapped to any external data source in this iteration of development.
Retrieval of product label sections referenced by the FDA biomarker table
The FDA requires industry to submit drug product labels using an Health Level Seven standard called Structured Product Labeling5. An SPL is an XML document written in the standard that specifically tags the content of each product label section with a unique code from the Logical Observation Identifiers Names and Codes (LOINC®) vocabulary6. The SPLs for all drug products marketed in the United States are available for download from the National Library of Medicine’s DailyMed resource7. As part of a separate ongoing project we have built a resource called LinkedSPLs that provides a Linked Open Data8 representation of the SPLs for all marketed prescription and over-the-counter drug products9. We have found that the LinkedSPLs resource makes it simple to retrieve content from specific product label sections along with a variety of SPL meta-data; a task that is more challenging to do by processing the SPL documents directly.
The FDA pharmacogenomics biomarker table lists one or more product label sections for each active ingredient/biomarker combination present in the table. Most active ingredients in the table are a component of more than one drug product and some are used in both prescription and over-the-counter products. The FDA’s description of the table makes it clear that the references are to sections in prescription drug products. However, no guidance is given on whether one can expect the same pharmacogenomics information across all sections for a given active ingredient. To address this issue, we randomly selected one representative section for each section listed in a row of the FDA’s table. Section retrieval was done using a Python10 script that queried the LinkedSPLs resource. The content for each section was stored in a text file, named so that we could identify both the product label from which it was extracted and the active ingredient/biomarker pair that is associated with it in the table. In accordance with the recommendations of a recent FDA draft guidance,11 some companies have included a “Pharmacogenomics” section in newly-submitted SPLs. Our script issued a separate query for the text of one representative Pharmacogenomics section for every drug product label in which this section was included.
Development of a semantic model of pharmacogenomic information
The use cases were used along with several of the representative product label pharmacogenomics statements to define a semantic model of the pharmacogenetic content found within SPLs (i.e., the focus of model development was on pharmacogenomics content present in the SPLs). Relevant sentences were examined to identify referenced entities and relationships, which were modeled using the Cmap concept mapping tool12. We chose this informal modeling approach to enable rapid development of the model which we plan to make more formal as iterative refinement provides input from perspectives relevant to both pharmacological and translational viewpoints.
Manual extraction and annotation of pharmacogenomics statements
Five pharmacist annotators independently extracted and annotated pharmacogenomics statements from a subset of the product label sections that were cited in the FDA’s table. The sections were chosen based on the anticipated clinical relevance of the pharmacogenomics statements in pharmacy practice. The annotators were trained according to guidelines developed for this effort which were provided for their reference. The primary units of annotation were pharmacogenomics statements, defined as a sentence or sequence of sentences (i.e., a phrase) published in an FDA-approved drug package insert that mentions a pharmacogenomics biomarker and (in most cases) provides some information on the importance of that biomarker for use of the drug. The annotators described pharmacogenomics statements using a variety of attributes from the semantic model, including the relevant biomarker; the medication of interest; the variant of interest; pharmacokinetic and pharmacodynamic effects; relevant patient state, medical conditions or side effects; and recommendations for monitoring, drug selection, dose selection, or testing. Annotation was done using the Knowtator plug-in13 for the Protégé modeling tool.
Results
Use Cases
A total of 13 clinical and translational use cases were identified (see Table 1 for a sample), covering medications, tests, genotypes, recommendations, alternatives, and additional fields.
Table 1.
A sample pharmacogenomics use case
| Use Case | Lauren is a physician in an outpatient clinic. She receives a pharmacogenomics test result from the Pathology lab for one of her patients. The result states that the patient has the genotype CYP2C19*2/*2. Lauren wants to know quickly what that means, and what the implications are for her patient, specifically the drugs the patient is currently taking. She would like to know if she should switch the patient to any new drugs, or change the dosage. |
| Abstract Query | For each drug ?d taken by patient with genotype ?g, identify recommendations for ?d, including suggested dosage changes ?c and/or recommended alternative medication ?e. |
Mapping the FDA biomarker table
We downloaded data from the August 3, 2012 version of the FDA’s pharmacogenomic biomarker table. The table indicated 116 unique drug/biomarker/SPL-section combinations. A total of 36 distinct biomarkers were listed in the table. Of these, 30 were listed by gene symbol and two by allele with three repeat entries. The remaining three biomarkers included three fusion genes and one chromosomal deletion. The 29 unique genes and alleles were mapped to unique HGNC symbols, Protein Ontology2 terms, and PharmGKB4 identifiers. The complete table is available for viewing at http://goo.gl/FN4D5.
Retrieval of product label sections referenced by the FDA biomarker table
Our query of LinkedSPLs for all relevant sections from every drug product label containing a drug in the biomarker table as an active ingredient returned 2,625 sections. These sections were from SPLs that were loaded into LinkedSPLs from DailyMed on August 23rd 2012. For all of the drugs listed in the table but two (galantamine and trimipramine), the query was able to return from one to many (>40) sections depending on the number of products containing a drug, and the number of SPLs for those products that provided a relevant section (some SPLs are missing sections). The failure to retrieve sections for galantamine and trimipramine might have been due to the use in the SPL of LOINC codes other than those intended for the cited sections.a Our query for “Pharmacogenomics” sections returned only three results; two for products containing the drug cisplatin (Platinol®) and Platinol-AQ), and one for a drug containing carisoprodol and aspirin. Co-authors PE and RF selected 9 high priority drugs from the FDA biomarker table and investigator RDB randomly selected twenty-nine representative prescription drug product label sections for manual annotation from the set of sections cited in the FDA’s table.
Pharmacogenomics knowledge model
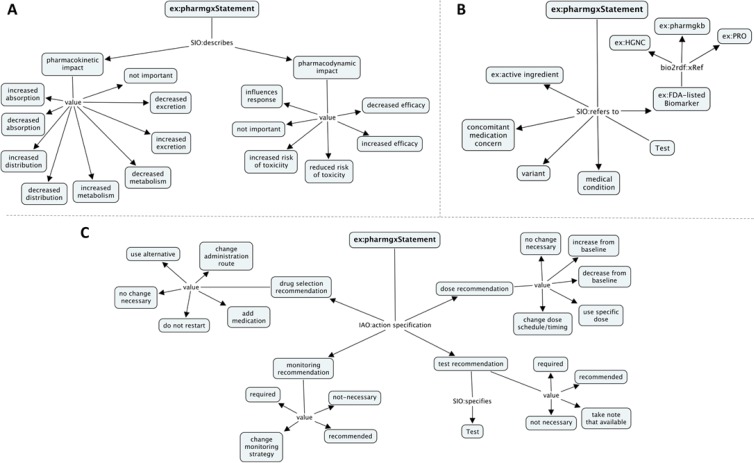
The current knowledge model (partially shown in Figure 2) is available at http://purl.org/net/linkedspls/pharmgx-fall-2012. The focus of the model is the SPL pharmacogenetic statement, which applies to a pharmacologic entity of interest, refers to a specific biomarker, describes pharmacodynamic and/or pharmacokinetic effects, and makes monitoring, dosage, and/or drug selection recommendations.
Figure 2.
Three parts of the semantic model with preliminary ontology mappings. A) what is described, B) what is referred to, and C) what action is specified. SIO: Semantic science Integrated Ontology, IAO: Information Artifact Ontology.
Manual extraction and annotation of pharmacogenomics statements
The five pharmacists identified a total of 213 pharmacogenomics statements in the 29 sections they were assigned to annotate. The results are summarized in the box to on the right in Figure 1. As the figure shows, at least two pharmacists agreed that 11 sections (five drug product labels) contained dosage recommendations, five sections seven sections (three drug product labels) provided recommendations for genetic testing, and three sections (in two drug product labels) provided specific monitoring recommendations. Additionally, pharmacological context was added regarding whether impact of the pharmacogenomic information involved altered pharmacokinetics (14 sections in six drug product labels) and/or pharmacodynamics (22 sections in nine drug product labels). Interestingly, some sections listed in the FDA biomarker table as containing pharmacogenomic recommendations received no annotations by any of the five pharmacists (e.g., the citalopram Drug Interactions section for CYP2C19 and CYP2D6).
Discussion
A semantic model of pharmacogenomics statements in product labels has the potential to make the information and recommendations that are currently buried within product labeling actionable through structured annotations. While our field test only focused on a small set of product label sections, the range of annotations identified suggest that the model has the potential to more directly answer questions regarding drug and dosage selection, genetic test recommendations, pharmacological consequences of genetic variation, and associated medical conditions. For example, clinicians and translational researchers seeking pharmacogenomics information from the product label on warfarin would have to integrate the text contained within the Dosage and Administration, Precautions, Clinical Pharmacology sections of the drug label regarding two biomarkers CYP2C9 and VKORC1 to gain an understanding of the potential impact of genetic variants on drug response. The annotations created in this study succinctly state that variants in CYP2C9 impact pharmacokinetics (decrease metabolism) and consequently drug response, while VKORC1 only impacts pharmacodynamics. For both biomarkers, specific dose levels and a change in monitoring are recommended, but no alternative drug therapy or specific testing recommendations are made. Similarly, annotations created for codeine identified that CYP2D6 variants alter pharmacokinetics (increasing metabolism), increase the risk of toxicity, and that dose reductions are suggested, as well as the additional specific monitoring recommendation in special populations (nursing mothers). Finally, in 56 instances within the 29 sections, annotators noted medical conditions that were either important for identifying subpopulations impacted by a pharmacogenomic biomarker, or that were a consequence of a variant. This allows for future mapping to medical ontologies or queries across different biomarkers for effects such as “which biomarkers increase the likelihood of drug hypersensitivity?”
While not the focus of this study, the work also identified some data quality issues within the FDA table and the mapping of external resources. Several of the drug product label sections listed in the FDA table contained no pharmacogenomics information according to a consensus of our pharmacist annotators. Anecdotally, in attempting to reconcile these perceived errors, two reviewers also identified sections within drug product labels that were not listed in the FDA biomarker table. Similarly, mapping inconsistencies were uncovered while attempting to resolve linkages between biomarkers in the FDA table, HGNC gene symbols, and PharmGKB. For example, the HGNC gene symbol of CD20 antigen, a pharmacogenomic biomarker within the tositumomab label, is MS4A1. PharmGKB currently links this drug product label to the gene MS4A2 which, although an accepted alias for MS4A1, is an independent gene symbol on its own.
Conclusions
Through ongoing work, our team will further refine the semantic model in parallel to the use cases and will generate annotations for the remaining sections. We believe adding structure to the rich pharmacogenomics data currently scattered throughout the appropriate sections of the drug product labels will unlock the potential of using this rich information to advance translational research and patient care.
Acknowledgments
This work was supported by the NIH/NIGMS (U19 GM61388; the Pharmacogenomic Research Network), the Agency for Healthcare Research and Quality (K12HS019461), NIH/NCATS (KL2TR000146), NLM (T15 LM007059-24). The content is solely the responsibility of the authors and does not represent the official views of the Agency for Healthcare Research and Quality or any of the other funding sources.
Footnotes
For example, one of the Trimipramine SPLs (http://goo.gl/Zvw1Y) has the “Drug Interactions” section tagged with the LOINC code for an “SPL UNCLASSIFIED SECTION”
References
- 1.Center for Drug Evaluation and Research Genomics - Table of Pharmacogenomic Biomarkers in Drug Labels. 2012. Available at: http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm. Accessed September 14, 2012.
- 2.Natale DA, Arighi CN, Barker WC, et al. Framework for a protein ontology. BMC Bioinformatics. 2007;8(Suppl 9):S1. doi: 10.1186/1471-2105-8-S9-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2011. Nucleic Acids Research. 2010;39(Database):D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonagh EM, Whirl-Carrillo M, Garten Y, Altman RB, Klein TE. From pharmacogenomic knowledge acquisition to clinical applications: the PharmGKB as a clinical pharmacogenomic biomarker resource. Biomark Med. 2011;5(6):795–806. doi: 10.2217/bmm.11.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.FDA . Providing Regulatory Submissions in Electronic Format — Content of Labeling. Rockville, MD: Food and Drug Administration; 2005. Report UCM072331. [Google Scholar]
- 6.Regenstrief Institute Inc Logical Observation Identifiers Names Codes (LOINC®) 2012. Available at: http://loinc.org/.
- 7.NLM DailyMed. 2012. Available at: http://dailymed.nlm.nih.gov/dailymed/about.cfm. Accessed September 18, 2012.
- 8.Marshall MS, Boyce R, Deus HF, et al. Emerging practices for mapping and linking life sciences data using RDF — A case series. Web Semantics Science Services and Agents on the World Wide Web. 2012;14(null):1–12. [Google Scholar]
- 9.Boyce R, Horn J, Hassanzadeh O, et al. Dynamic Enhancement of Drug Product Labels to Support Drug Safety, Efficacy, and Effectiveness. Journal of Biomedical Semantics. Journal of Biomedical Semantics. 2013 doi: 10.1186/2041-1480-4-5. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Python Software Foundation Python Programming Language – Official Website. 2012. Available at: http://python.org/.
- 11.FDA . Clinical Pharmacogenomics: Premarketing Evaluation in Early Phase Clinical Studies. Rockville, MD: Federal Drug Administration; 2011. [Google Scholar]
- 12.Cañas AJ, Carff R, Hill G, et al. Knowledge and Information Visualization, LNCS Springer-Verlag. Heidelberg/NY: Springer; 2005. Concept Maps: integrating knowledge and information visualization; pp. 205–219. [Google Scholar]
- 13.Ogren PV. Knowtator: a protégé plug-in for annotated corpus construction. Proceedings of the 2006 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology: companion volume: demonstrations; Stroudsburg, PA, USA: Association for Computational Linguistics; 2006. pp. 273–275. NAACL-Demonstrations ’06. [Google Scholar]