Abstract
Several intersecting host, vector, and environmental factors have led to a re-emergence of rickettsial diseases such as Mediterranean Spotted Fever (MSF), and Dermacentor spp.-borne necrosis-erythema lymphadenopathy (DEBONEL). Some rickettsiae produce diffuse endothelial infection and systemic microvascular leakage leading in some cases to high morbidity and mortality. Unfortunately, little is known about the molecular pathways triggered by these diseases in humans. We therefore analyzed how candidate cytokines co-occur across acutely-ill patients with either a localized (DEBONEL), or a systemic (MSF) form of rickettsiosis, using bipartite visual analytics. The results revealed a network core consisting of a small set of MSF patients exhibiting high expressions of cytokines implicated in microvascular leakage, endothelial repair, and pro-inflammatory immune responses, and a network periphery consisting of a mixture of MSF and DEBONEL patients with relatively lower overall cytokine expressions. These results provide evidence of pathways triggered by rickettsiae in humans, and a testable hypothesis for the mechanisms in a rickettsia-induced cytokine storm with the translational goal of identifying therapeutic targets.
Introduction
Bacteria belonging to the genus Rickettsia cause a range of rickettsioses in humans including Mediterranean spotted fever (MSF), Rocky Mountain spotted fever (RMSF), and Dermacentor spp.-borne necrosis-erythema lymphadenopathy (DEBONEL). Transmitted through infected arthropods, most rickettsioses result in infection of the microvascular endothelium leading to increased microvascular permeability1. The diseases vary from localized forms such as DEBONEL, to diffuse infection of the microvascular endothelium resulting in mortality rates of 2–3% in MSF, and 20–25% in untreated cases of RMSF in the US.
Unfortunately, much remains to be discovered about the pathogenesis underlying rickettsial diseases in humans because most of the research has (1) been conducted in vitro and in mouse models, and (2) used univariate methods (e.g., t-test) to analyze potential biomarkers. For example, several mouse models have been developed for spotted fever and typhus group rickettsiae, leading to hypotheses about candidate biomarkers such as IFN-γ, TNF-α, and IL-1β2 in humans. However, to the best of our knowledge, no studies have used multivariate methods to analyze the pathogenetic role of cytokines and chemokines involved in the human immune response to rickettsiae. Such multivariate analysis could help to identify how multiple biomarkers act in concert, such as in a cytokine storm3.
Given the emergence and re-emergence of several rickettsial diseases due to multiple environmental and biological factors2, there is an urgent need to understand the complex multivariate nature of the disease with the goal of enabling the identification of molecular pathways and therapeutic targets. To address this gap, we used bipartite visual analytics to visualize and quantitatively analyze the multivariate co-occurrence of candidate cytokines across patients with a mild form (DEBONEL), and a systemic form (MSF) of rickettsiosis.
Methods
Our research began with the question: How do cytokines (implicated in rickettsial mouse models and in vitro models) co-occur across MSF and DEBONEL patients? To address our research question, we made critical decisions related to data selection, and data analysis as discussed below:
Data Selection
Our study was based on 49 DEBONEL, and 36 MSF patients that were diagnosed using serological assays (IFA) and/or PCR detection of rickettsial DNA amplicons from blood samples. Serum samples were collected between 0–20 days after symptoms first appeared, and a bioplex analysis was conducted to measure 26 candidate cytokines. The cytokine levels for both diseases were determined using a single standard curve.
Data Analysis
Our analysis consisted of two steps: (1) exploratory visual analysis to identify emergent bipartite relationships between patients and cytokines; and (2) quantitative analysis suggested by the emergent visual patterns. This two-step method was motivated by our earlier studies4–6, which have demonstrated that bipartite relationships can reveal different patterns each prompting the use of quantitative methods that make the appropriate assumptions about the underlying data.
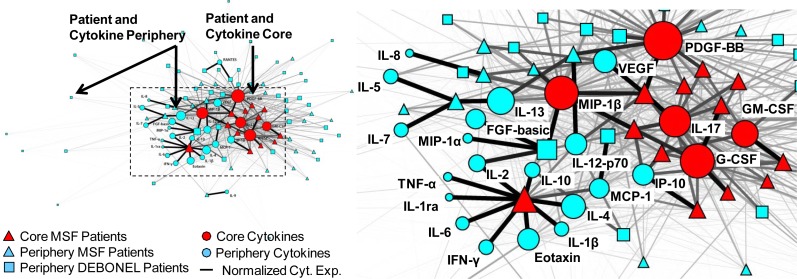
1. Exploratory Visual Analysis was conducted using network visualization and analysis7. Networks are increasingly being used to analyze a wide range of molecular phenomena such as gene and protein-protein interactions8–9, and to assess their relationships to diseases, symptoms, and syndromes. A network consists of nodes and edges; nodes represent one or more types of entities (e.g., patients or cytokines), and edges between the nodes represent a specific relationship between the entities. Figure 1 shows a bipartite network where edges exist only between patients and cytokines.
Figure 1.
A bipartite network (automatically laid out by the Kamada-Kawai algorithm10) shows how 26 cytokines (circular nodes) co-occur across 85 patients (square and triangular nodes). The size of the nodes is proportional to the sum of the edge weights (representing normalized cytokine values) that connect to them, and the thickness of edges is proportional to cytokine values. Therefore patients with high total cytokine values have large nodes, and higher cytokine values are represented by thicker edges. The network has a core-periphery topology (red=core, blue=periphery) for the patients, in addition to the cytokines.
Edge weights in the network were used to represent the strength of the cytokine expression values for each patient-cytokine pair. Because the cytokines had different ranges, we used the min-max normalization method (which does a linear mapping of each cytokine value to range from 0–1, and therefore preserves the relative distances between values to enable comparison). As shown in Figure 1, the edge thicknesses were drawn to be proportional to these normalized cytokine values. Node diameter was used to represent the sum of the edge weights connected to it (also referred to as the weighted degree centrality). This enabled a rapid visual inspection to determine for example, which patients have overall high aggregate cytokine values, and how such patients relate to the rest of the network. Finally, the node shape was used to represent phenotype (triangles=MSF, squares=DEBONEL, circles=cytokines), and node color was used to represent members of a cluster based on hierarchical cluster analysis.
Global patterns between patients and cytokines in the network were visualized and analyzed using the Kamada-Kawai layout algorithm10 in Pajek (version 3.02). As shown in Figure 1, the algorithm pulls together nodes that are strongly connected, and pushes apart nodes that are not. This algorithm is fast but approximate and is well-suited for medium sized networks consisting of between 100–1000 nodes. The result is that nodes with a similar pattern of connections (e.g., PDGF and MIP-1β in the middle of the network in Figure 1) are placed close to each other.
A key advantage of a network representation is the simultaneous visualization of multiple raw values (patient-cytokine associations, cytokine values), aggregated values (sum of cytokine values), and emergent global patterns (clusters) in a uniform visual representation. Such a representation enables the rapid generation of hypotheses based on complex multivariate relationships, which can be verified through appropriate quantitative methods.
2. Quantitative Analysis was conducted using three measures to verify the insights derived from the exploratory visual analysis. These methods were selected based on their appropriateness to the emergent patterns in the network.
Agglomerative Hierarchical Clustering. Because the network layout suggested a core-periphery topology (nodes with high overall edge weights in the core, and nodes with low overall edge weights in the periphery7) for patients and for cytokines, we used the agglomerative hierarchical clustering method. The clustering was done using the Manhattan dissimilarity measure with the Ward linkage function, and the number of clusters and their boundaries were determined based on natural breaks in the patient and cytokine dendrograms. The dendrograms were also combined with the heatmap to aid in the visual analysis of the results.
Clusteredness. To test whether the clusters in the network could have occurred by chance, we compared the variance, skewness, and kurtosis of the dissimilarities in the data, to 1000 random permutations of this data. For each network permutation, we preserved the size of the network, in addition to the edge weight distribution of each patient when analyzing the patient dendrogram, and the edge weight for each cytokine when analyzing the cytokine dendrogram. Significant breaks in the rickettsioses patient, or cytokine dendrograms would result in a significantly larger variance, skewness, and kurtosis of the dissimilarity measures, compared to the same measures generated from the random networks.
Weighted Degree Centrality. To test whether the patients in the network core had a higher overall cytokine expression, we calculated the weighted degree centrality7 for each patient node by adding its normalized cytokine expression across all cytokines. The Mann Whitney U test was then used to compare the weighted degree centrality of the patients in the core, to those in the periphery. The same measure was used to compare the weighted degree centrality of the cytokines in the core compared to cytokines outside the core. Furthermore, to characterize the continuous nature of the core-periphery topology, we plotted the relationship between the weighted degree centrality of the nodes (using 5 equal bins over the range 0–15), and the proportion of MSF (# MSF patients in each bin / # total MSF), or DEBONEL patients (# DEBONEL patients in each bin / # total DEBONEL) in each bin.
Results
Patient Clusters
As shown in Figure 1, the network layout revealed a core-periphery topology, where there were a few patients with high overall cytokine expression in the central core of the network (henceforth referred to as the patient core), and many patients with low overall cytokine expression in the periphery (henceforth referred to as the patient periphery).
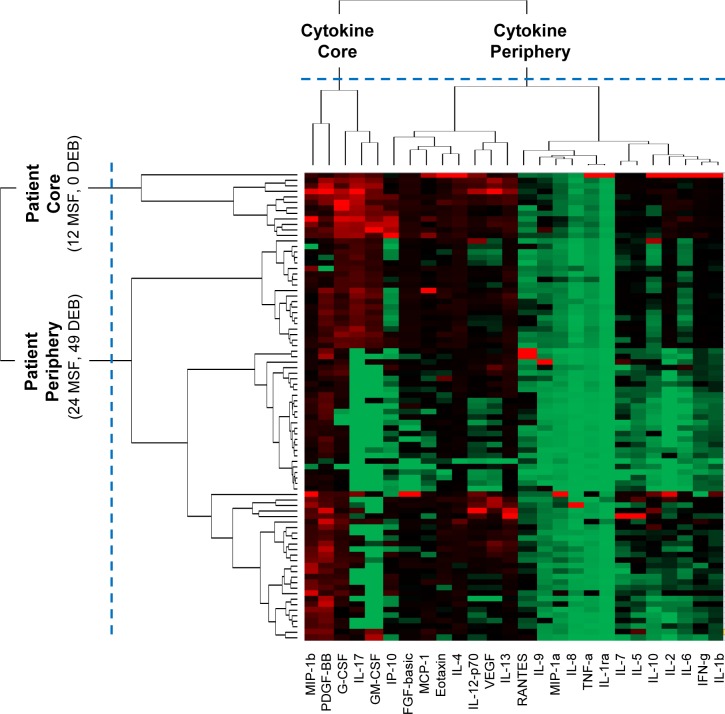
The above topology was quantitatively verified through agglomerative hierarchical clustering. The vertical dendrogram in Figure 2 shows that there exist mainly two clusters: a small cluster consisting of 12 (33.3%) MSF patients in the core, and the remaining 24 (66.7%) MSF and 49 (100%) DEBONEL patients were scattered at different distances outside the core. The proportion of the two types of patients in the core (12 MSFs, 0 DEBONELs) and periphery (24 MSFs, 49 DEBONELs) clusters was significantly different (χ2 Yates (1, N=85) = 16.368, p < .0001). Furthermore, the weighted degree centrality (sum of edge weights) of patient nodes in each cluster was also significantly different (U = 854.0, p < .001, two-tailed test), suggesting that the overall cytokine expression of the patients in the core (Median=5.1) was higher compared to those in the periphery (Median=1.9).
Figure 2.
A heat map where rows represent patients, columns represent cytokines, and colors represent normalized cytokine values (green = 0, red = 1). The rows and columns are ordered based on the results of the agglomerative hierarchical clustering, with dendrograms for the patient and cytokines shown on the vertical and horizontal axes respectively.
To test whether the above clusters could have occurred by chance, we measured their clusteredness with respect to random permutations of the data. The patient clustering in the rickettsia data was significant when compared to 1000 random networks based on variance of the dissimilarities (rickettsiae = 7.12, Random Mean = 6.46, p<.001 two-tailed test), skewness of the distribution of dissimilarities (rickettsiae = 3.27, Random Mean = 2.2, p<.001 two-tailed test), and kurtosis of the distribution of dissimilarities (rickettsiae = 15.26, Random Mean = 9.63, p<.001 two-tailed test).
Although the clustering result enabled the identification of a discrete set of patients that had a significantly higher overall cytokine expression value compared to patients in the periphery, the network topology reflected a continuous change from the periphery to core of the network. For example, moving from the periphery towards the core of the overall network, the proportion of MSF patients progressively decreases, whereas the node size (representing weighted degree centrality) progressively increases. This relationship was captured by plotting the proportion of MSF patients at increasing levels of weighted degree centrality, which was best fitted by a decreasing log curve (y = −0.372ln(x) + 0.5565, R2 = 0.92). In contrast, the equivalent curve for DEBONEL was best fitted by a steeper decreasing log curve (y = −0.561ln(x) + 0.7375, R2 = 0.74), which captured the critical difference that a higher proportion of DEBONEL patients (that had a steeper head in the curve) had lower total cytokine expression compared to MSF patients (that had a shallower head in the curve). We refer to these as the proportionate degree of cytokine expression (PDCE) curves useful for comparing the cytokine expression profiles across diseases with different numbers of patients. The visual and quantitative results therefore together provided evidence that a small percentage of MSF patients experienced an overall high cytokine expression, whereas a larger number of MSF and DEBONEL patients in the periphery experienced a lower overall cytokine expression.
Cytokine Clusters
As shown in Figure 1, most of the cytokines are pushed into the core of the overall network, but the network also suggests that the cytokines themselves have a core-periphery topology. As shown by the larger diameter of the cytokines in the center, some cytokines are more highly expressed compared to others.
Similar to the patient clusters, the above pattern was also quantitatively verified through hierarchical clustering. As shown by the horizontal dendrogram in Figure 2, the cytokines fell into two clusters: a small cluster of 5 cytokines: MIP-1β, PDGF, G-CSF, GMCSF, and IL-17 (henceforth referred to as the cytokine core), and a larger cluster of the remaining 21 cytokines (henceforth referred to as the cytokine periphery). In addition to the presence of the cytokine clusters, the bipartite network also revealed the inter-cluster relationships: the 12 MSF patients in the patient core had a significantly higher mean cytokine expression of the 5 cytokines in the core, compared to the patients in the periphery (U = 876, p < .0001, two-tailed test). This pattern can also be seen in the upper left hand corner of the heatmap in Figure 2 where the patient and cytokine cores intersect. One patient (large red triangle on the left in Figure 1, and the top patient row in Figure 2) within the patient core had a different profile of high cytokine expression compared to the rest in the core.
The clusteredness of the above cytokine clusters in the rickettsia data was significant when compared to 1000 random networks based on variance of the dissimilarities (rickettsiae = 80.08, Random Mean = 73.26, p<.001 two-tailed test), skewness of the distribution of dissimilarities (rickettsiae = 2.01, Random Mean = 1.65, p<.001 two-tailed test), and kurtosis of the distribution of dissimilarities (rickettsiae = 7.01, Random Mean = 5.8, p<.005 two-tailed test). Furthermore, the 5 cytokines in the core (Median=20.8) have a significantly higher weighted degree centrality compared to the cytokines in the periphery (Median=3.9) (U = 104.0, p < .0001, two-tailed test).
Discussion
The bipartite visualization and quantitative verifications revealed not only a core-periphery topology for the patients and the cytokines, but also a preferential connection between the 12 MSF core patients, and the 5 core cytokines. Among these 5 core cytokines, three (IL-17, GM-CSF, G-CSF) appear related to a pro-inflammatory pathway. IL-17 is considered an important pro-inflammatory cytokine that is produced in a broad range of diseases including infections and autoimmune diseases11. Furthermore, elevated levels of IL-17 have been associated with the production of the growth factors GM-CSF and G-CSF12, which in turn play a critical role in stimulating growth and differentiation of macrophages and granulocytes (two important cell effectors in the immune response). We hypothesize that IL-17 might also stimulate endothelial cells to produce chemokines, resulting in an amplification of the inflammatory response possibly leading to a cytokine storm.
Another potential mechanism suggested by the cytokine core involves PDGF (platelet-derived growth factor)-BB, MIP-1β (macrophage inflammatory protein also known as CCL4) and to a lesser degree VEGF (vascular endothelial growth factor) which appears outside of, but is similar in profile to members of the cytokine core. We hypothesize that PDGF-BB and VEGF are likely elevated due to diffused endothelial damage that would require “plugging” of denuded segments of microvessels with platelets, and subsequent repair by proliferation of endothelial cells to restore continuity of the endothelial monolayer in the microcirculation. However, both growth factors have also been implicated in increased microvascular permeability suggesting a potential important role in increased microvascular permeability seen in human rickettsioses and in their animal model counterparts13. While MIP-1β is produced by a large number of cells, the majority of MIP-1β in infections is produced by activated macrophages in response to LPS or other cytokines and growth factors such as GM-CSF13. The above two pathways might therefore be connected through their overlap with GM-CSF, in severe MSF patients.
While the 12 patients in the core are preferentially connected to the above 5 cytokines, it is important to note that the weighted degree centrality of those cytokines are not exclusively accounted for by the MSF patients, but are also expressed (although to a far lesser extent) by DEBONEL and MSF patients in the periphery. This suggests that the over-expression of the core cytokines in the 12 MSF patients could be caused by an immune response dysregulation. Supporting evidence is provided by platelet counts that were available for 8 of the 12 patients in the core. Of these 8 patients, 7 had evidence of thrombocytopenia, suggesting these patients had a more severe form of MSF with diffused endothelial injury and vascular leakage. The overall results therefore provide testable hypotheses that the core cytokines and associated pathways are important in severe MSF patients, especially because of their potential to cascade into a cytokine storm3.
Conclusions and Future Research
Although a range of cytokines have been implicated in rickettsioses, little is known about the underlying biological mechanisms in humans. Here we presented a multivariate analysis of cytokine expression in human data using bipartite visual analytics. We believe this study makes three biological and methodological contributions. First, we have shown evidence for two biological mechanisms that were known to be present in mouse models and in vitro (at least partially) but have never been shown comprehensively in human patients with MSF. Second, we believe there is evidence for a cytokine storm expressed in the patient core based on the hypothesized role of the identified pathways. Third, we believe the overall bipartite visual analytical methodology and the PDCE curve can be used to analyze the nature and degree of cytokine expression during and after a storm across diseases.
Cytokine storms are of course a time-related phenomenon, and a limitation of our study includes the lack of serum samples from several time-points during the disease progression in order to study the kinetics of cytokine expression. Furthermore, due to the policy hurdles of obtaining patient data from a foreign country, we have yet to receive complete clinical data for the patients to conduct inferential comparisons between the molecular and clinical variables. However, despite these limitations, the comparison of a mild and localized form of the disease to a more severe systemic form helped to identify which pathways could be activated in humans. Our future research aims to test these hypotheses in other datasets, and to use our visual analytical methodology to further define and use quantitative measures to characterize a cytokine storm over time. These advances should enable researchers to rapidly analyze the over-expression of cytokines in a wide range of infectious diseases, with the translational goal of identifying effective therapeutic targets.
Acknowledgments
This research was supported in part by a grant from IHII, UTMB, the Western Regional Center for Excellence in Biodefense and Emerging Infectious Diseases (WRCE; U54 AI057156-06 NIH/NIAID), and NIH 1U54RR02614 UTMB CTSA (ARB).
References
- 1.Olano JP. Rickettsial infections. Annals NY Academy of Sciences. 2005;1063:187–196. doi: 10.1196/annals.1355.031. [DOI] [PubMed] [Google Scholar]
- 2.Walker DH, Popov VL, Wen J, Feng HM. Rickettsia conorii infection of C3H/HeN mice. A model of endothelial-target rickettsiosis. Laboratory Investigation. 1994;70(3):358–68. [PubMed] [Google Scholar]
- 3.Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev Mar. 2012;76(1):16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhavnani SK, Bellala G, Victor S, et al. The Role of Complementary Bipartite Visual Analytical Representations in the Analysis of SNPs: A Case Study in Ancestral Informative Markers. JAMIA. 2012;19:e5–e12. doi: 10.1136/amiajnl-2011-000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhavnani SK, Victor S, Calhoun WJ, Busse WW, Bleecker E, Castro M, Ju H, Brasier AR. How Cytokines Co-occur across Asthma Patients: From Bipartite Network Analysis to a Molecular-Based Classification. Journal of Biomedical Informatics. 2011:44. S24–S30. doi: 10.1016/j.jbi.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhavnani SK, Bellala G, Ganesan A, et al. The Nested Structure of Cancer Symptoms: Implications for Analyzing Cooccurrence and Managing Symptoms. Methods of Information in Medicine. 2010;49:6. 581–591. doi: 10.3414/ME09-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman M. Networks: An Introduction. Oxford University Press; 2010. [Google Scholar]
- 8.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007 May 22;104(21):8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008 Apr;18(4):644–52. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamada T, Kawai S. An algorithm for drawing general undirected graphs. Information Processing Letters. 1989;31(1):7–15. [Google Scholar]
- 11.Joseph M, Reynolds P, Chen D. IL-17 family member cytokines: Regulation and function in innate immunity. Cytokine & Growth Factor Reviews. 2010;(21):413–423. doi: 10.1016/j.cytogfr.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawaguchi M, Kokubu F, et al. Induction of granulocyte-macrophage colony-stimulating factor by a new cytokine, ML-1 (IL-17F), via Raf I-MEK-ERK pathway. Journal of Allergy & Clinical Immunology. 2004;114(2):444–50. doi: 10.1016/j.jaci.2004.03.047. [DOI] [PubMed] [Google Scholar]
- 13.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine & Growth Factor Reviews. 2002;13(6):455–81. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]