Abstract
Background:
Escherichia coli have an optimum pH range of 6-7 for growth and survival that's why, called neutrophiles. The ΔpH across the cytoplasmic membrane is linked to cellular bioenergetics and metabolism of the body which is the major supplier of the proton motive force, so homeostasis of cellular pH is essential. When challenged by low pH, protons enter the cytoplasm; as a result, mechanisms are required to alleviate the effects of lowered cytoplasmic pH.
Materials and Methods:
The activities of Succinate dehydrogenase, isocitrate dehydrogenase, malate dehydrogenase and glucose-6-phosphate dehydrogenase in acid shocked cells of E. coli DH5 α and E. coli W3110 subjected to pH 3, 4, and 5 by two types of acidification, like external (using 0.1 N HCl), external along with the monensin (1 μM) and cytoplasmic acidification using the sodium benzoate as an acid permeant (20 mM) which is coupled to the electron transport chain by the reducing power, as yet another system possessed by E. coli as an armor against harsh acidic environments.
Result:
Results showed that an exposure to acidic environment (pH 3, 4 and 5) for a short period of time increased the activities of these dehydrogenases in all types of acidification except cytoplasmic acidification, which shows that higher recycling of reducing power results in pumping out of protons from the cytoplasm through the electron transport chain complexes, thereby restoring the cytoplasmic pH of the bacteria in the range of 7.4-7.8.
Conclusion:
Study indicates that acid shocked E. coli for a period of 2 h can survive for a sustained period.
Keywords: Acid tolerance, dehydrogenases, electron transport chain, external acidification, internal acidification and intracellular pH
INTRODUCTION
Under low pH conditions enteric bacteria have developed mechanisms to maintain pH homeostasis, which incorporates extreme gastric acidity and volatile fatty acids produced in the intestine by fermentation.[1] The fundamental property of all living organisms is the ability to harness energy and to channel it into the biological work along with it must have been acquired very early in cellular evolution.[2] These microorganisms contain membrane-bound proton pumps that generate a trans-membrane proton motive force (PMF) composed of a trans-membrane electrical potential and a trans-membrane chemical gradient, so if the influx of proton in the cytoplasm through the membrane bound F0F1 ATPase to produce Adenosine Tri-Phosphate ATP, is left unrestricted, it will rapidly destroy the internal pH away from neutral or near neutral to acidic.[3] In oxidative phosphorylation, electrons from Nicotinamide Adenine Dinucleotide – Hydrogen or 5,10-Methylenetetrahydrofolate Reductase (NADH or FADH) are passed onto O2 through the electron transport chain located in the plasma membrane of microorganism, which leads to the pumping of protons out of the cytoplasm.[2] Thus, the oxidation of fuels and the phosporylation of ADP are coupled by a proton gradient across the plasma membrane of microorganisms.[4] Bacteria have developed different ways to withstand stressful situations, such as a decrease in the external pH. Hommais et al. found that different induced acid resistant systems in E. coli depend on whether oxidative or fermentative metabolism is occurring. These systems are, change in the cellular envelope to decrease ionic permeability,[5,6] induction of DNA repair or chaperones,[7] development of ionic pumping system or proton extrusion system,[6] and majorly the amino acid decarboxylase system (glutamate decarboxylase, arginine decarboxylase, and lysine decarboxylase).[8]
Acetate is one of the major by-products of E. coli metabolism. The global gene expression changes seen after adaptation to external acetate, suggests regulation of carbon metabolism in E. coli to avoid further formation of acetate.[9]
E. coli W3110 is a mutant for phosphotransacetylase (PTA) an important enzyme in the acetate production pathway. The conversion between acetyl - Coenzyme ACoA and acetate is mediated by two pathways: (i) Acetate kinase and PTA, which rapidly convert acetyl-CoA via acetyl-phosphate to acetate as an overflow pathway, (ii) acetyl-CoA synthetase, a high-affinity, low-capacity uptake pathway for acetate, which produces acetyl-CoA via an enzyme-bound acetyl-adenylate intermediate.[10]
Sodium benzoate as acid permeant
Generally used food preservatives are sodium benzoate, propionic acid and sorbitol which, retard the microbial growth thereby enhancing the shelf life of food. Sodium benzoate acts as a preservative of food and in packed food products by causing cytoplasmic acidification of bacterial cells at low pH. Addition of a permeant acid such as benzoate at high concentration depresses cytoplasmic pH with little or no recovery, without affecting external pH. On addition of sodium benzoate as an acid permeant to cell cultures which were suspended in media at pH adjusted to the acidic range, the cytoplasmic pH fell within seconds to that of the external medium.[11]
Monensin as an uncoupler
According to the chemiosmotic theory of Mitchell, bacteria use membrane-bound ATPases or electron transport system to translocate protons across the cell membrane. Proton extrusion establishes a pmf which is a summation of chemical (ΔpH) and electrical (ΔΨ) potential gradients. Ionophores, like monensin are highly lipophilic substances, which are toxic to many bacteria, protozoa, fungi, and higher organisms. The exterior of the molecule is hydrophobic, while the interior is hydrophilic and able to bind cations. Some ionophores bind only single cation (uniporters), but others are able to bind more than one cation (antiporters). Because cell membranes are composed of lipid bilayers, high activation energy is needed to translocate ions. Ionophores function as mobile carriers within the membrane and able to shield and delocalize the charge of ions thereby facilitating their movement across membranes. Monensin is an antiporter with a high selectivity for Na+ than K+.[12]
MATERIALS AND METHODS
Microorganism
The LAB used was from Department of Biochemistry and Biotechnology Institute of Science, Nirma University of Science and Technology, Ahmedabad. Pure cultures of different E. coli strains were obtained from the Microbial Type Culture Collection, Institute of Microbial Technology, Chandigarh, India, viz.
Microbial Type Culture CollectionMTCC 1652-E. coli DH5 α
MTCC 50-E. coli W3110
Media and cultivations
E. coli cells were grown in M9 minimal media and Nutrient media. All the cultures were obtained as lyophilized cultures from IMTECH, Chandigarh. Cultures were activated by incubating at 37°C for 10 min. All cultures were mixed with 300 μL broth and 100 μL culture was inoculated in 250 mL broth and then cultures were streaked on nutrient agar plates and slants.
Acid shock method
E. coli DH5 α and E. coli W3110 were given acid shock at different pH ranges for different time intervals by different types of acidification.
External acidification
Single cell colony of both bacteria was inoculated in 250 mL, M9 minimal media adjusted to pH 7 and was incubated at 37°C. When the optical density (OD) of the inoculated cultured broth reached A600 nm = 0.8-1.1, cells were collected by centrifuging at 10,000 rpm for 5 min, after that the collected cells were then subjected to acid shock in M9 minimal media with different pH of 3, 4, and 5 for incubation time periods of 1 h, 2 h, 3 h, and 4 h.
External acidification along with monensin
Single cell colony of both bacteria was inoculated in 250 mL M9 minimal media adjusted to pH 7 and was incubated at 37°C. When the OD of inoculated culture media reached A600 nm = 0.8-1.1, cells were collected by centrifuging at 10,000 rpm for 5 min, after that the collected cells were then subjected to acid shock in M9 minimal media along with 1 μM monensin at different pH of 3, 4 and 5 for incubation time periods of 1 h, 2 h, 3 h and 4 h.
Internal acidification
Single cell colony of both bacteria was inoculated in 250 mL M9 minimal media adjusted to pH 7 and was incubated at 37°C. When the OD of inoculated culture media reached A600 nm = 0.8-1.1, cells were collected by centrifuging at 10,000 rpm for 5 min, after that the collected cells were then subjected to acid shock in M9 minimal media along with 20 mM sodium benzoate at different pH of 3, 4 and 5 for incubation time periods of 1 h, 2 h, 3 h and 4 h.
Preparation of cell extract
Acid shocked cells were centrifuged at 10,000 rpm for 5 min and the pelleted cells were then resuspended in the phosphate buffer saline. Then the cells were sonicated for a total of 2 min with 30 s interval time and the sonicated sample was then centrifuged at 12,500 rpm for 25 min, after that the supernatant was collected for enzyme assays by continuous spectrophotometric rate determination.
Enzyme assays
Isocitrate dehydrogenase assay: According to method describe by Garnak and Reeves 1979.[13]
Malate dehydrogenase assay: According to method describe by Manajit Hayer-Hartl 2000.[14]
Succinate dehydrogenase assay: According to method describe by Spencer and Guest, 1973.[15]
Glucose-6-phosphate dehydrogenase assay: According to method describe by Banerjee and Fraenkel, 1972.[16]
Calculating enzymes activity
μM/min/ml = (ΔA340 nm/min) × (3/6.2) × (1/extract volume) × (df)
where,
Formula for calculating enzyme activity[17]
ΔA340 nm/min = Change in absorbance per min
3 = Volume (in ml) of assay
df = Dilution factor
6.22 = Millimolar extinction coefficient for ß-NADPH at 340 nm
Extract volume = Volume (in ml) of enzyme used.
Note: For DCIP, 20.2 mM extinction coefficient was used.
Specific activity of enzymes
One unit of activity is the amount of enzyme required to catalyze the reduction of 1 μM of NADP+ per min at 25°C. Specific activity is expressed in μM/min per μg of protein.
μM/min/mg = (Enzyme activity) / (protein concentration)
Protein was estimated: By Folin Lowry method.[18]
Determination of viability
Viable population of acid shocked and control cultures were enumerated immediately after inoculation to M-9 minimal media adjusted to pH range of 3, 4, and 5 and after different shock period (1, 2, 3 and 4 h). Serial decimal dilutions in 0.1% peptone water were prepared. The viable population of cultures were then determined by plating 0.1 ml of the serially diluted samples on duplicate Sorbitol Mac Conkey Agar (SMAC, oxoid) plates. All plates were incubated at 37°C for 24-48 h, and then colorless colonies were enumerated.
Statistical analysis
The data presented are either from single experiments which were done at least 3 times or in the form of means accompanied by standard deviations. Statistical significance was accepted at the P < 0.05 level of probability by using Student's t-test of ANOVA Biostat software.
RESULT
When the external pH of E. coli cells in suspension was lowered from pH 7.5-5.5, the cytoplasmic pH fell within 10-20 s to pH 5.6-6.5. Rapid recovery occurred after 30 s of HCl addition and was followed by a slower recovery over the next 5 min. The pH of the periplasm equaled the external pH under all condition tested, including rapid acid shift. Addition of membrane permeant like sodium benzoate to the cell suspension has no effect on the periplasmic pH.[19] Uptake of a permeant acid can also dissipate the transmembrane pH, a component of the pmf. The permeant acids, such as sodium benzoate, can cross the bacterial membrane in the protonated and unprotonated forms and cyclically run down ΔpH, thereby collapsing pmf and uncoupling ATP synthesis.[20] The results show that an exposure to acidic environment (pH 3, 4 and 5) for a short period of time increased the activities of these dehydrogenases in all types of acidification except cytoplasmic acidification used in the current study. On cytoplasmic acidification, the activities of all dehydrogenases decreased at pH 3, 4, and 5. On external acidification along with monensin [Figures 1 and 2], activities of dehydrogenases increased further as compared to external acidification alone. Cells exposed to pH 3 for 2 h had the highest acid tolerance on external acidification with or without monensin, which again supports the work of Tosun and Gonul. It was also found that activity of G6PD remained unchanged at low pH. This result suggested that, in a low pH environment, metabolic flux in E. coli increases through Tricarboxylic acid cycleTCA cycle and remains unaffected through the pentose phosphate pathway. This increase in metabolic flux through TCA cycle, during oxidative phosphorylation, cause electrons from NADH or FADH2 to pass onto O2 through the ETC located in the plasma membrane of the microorganism, leading to the pumping of protons out of the cytoplasm and thus maintaining pH homeostasis.
Figure 1.

Comparison of specific activity of (a) succinate dehydrogenase (SDH), (b) isocitrate dehydrogenase(ICD), (c) MDH (malate dehydrogenase), (d) glucose-6-phosphate dehydrogenase (G6PD) from External acidification of E. coli DH5 α for different time periods upon exposure to different low pH (acidic shock) using monensin as an uncoupler (1 μM). pH 7 was used as control (Mean ± SD for triplicate values)
Figure 2.
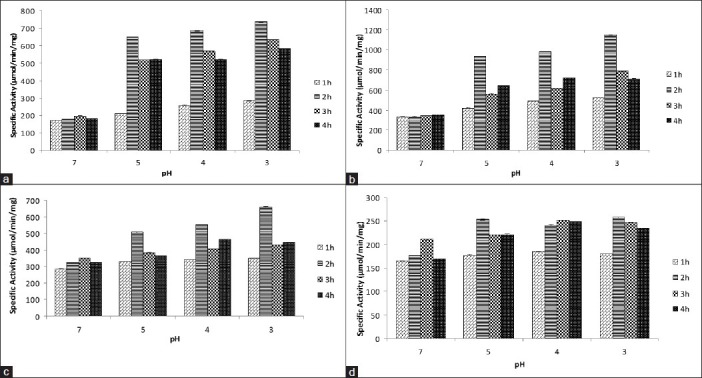
Comparison of specific activity of (a) SDH, (b) ICD, (c) MDH, (d) G6PD from External acidification of E. coli W3110 for different time periods upon exposure to different low pH (acidic shock) using monensin as an uncoupler (1 μM). pH 7 was used as control (Mean ± SD for triplicate values)
DISCUSSION
Most studies on pH homeostasis have been focused on those aspects where bacterial growth and survival have an impact on human health and economics, i.e., survival and growth in food, effect on the oral cavity, gastric transit, and intracellular survival. The pH surrounding cells or cell aggregates is the main environmental factor that strongly determines their growth and metabolism. In order to colonize their mammalian hosts, both commensal and pathogenic enteric bacteria must survive passage through the low pH environment of the stomach. Humans secrete approximately 2.5l of gastric juice each day, generating a fasting gastric pH of 1.5, which increases to pH between 3.0 and 5.0 during feeding.[21] Enteric pathogens must survive this level of pH for at least 2 h, the average emptying time for the stomach, before reaching the intestine.[22]
Bacteria have developed different ways to withstand stressful situations, such as a decrease in the external pH. Florence et al. found that different induced acid resistant systems in E. coli depend on whether oxidative or fermentative metabolism is occurring. These systems are, change in the cellular envelope to decrease ionic permeability, induction of DNA repair, chaperones, development of ionic pumping system and majorly the amino acid decarboxylase system (glutamate decarboxylase, arginine decarboxylase, and lysine decarboxylase). While our understanding of the mechanisms used by neutrophiles to grow or survive in low pH environments has increased, much remains to be determined. The present work is an investigation of the otherwise unexplored role of dehydrogenases in the acid adaptation of a neutrophile.
E. coli cells when subjected to acid shock environments were shown to undergo dramatic changes in specific activities of ICD, SDH, and MDH while the activity of G6PD remained unchanged. The extent of increased acid tolerance was affected by the time of adaptation and adaptation pH. Among the various adaptation times and adaptation pH tested, exposure of E. coli to acid at pH 3 for 2 h resulted in an increased acid tolerance for both the conditions including external acidification and external acidification along with monensin.
On external acidification
Much of the success of bacteria depends on their ability to survive and thrive in adverse conditions. External acidification of the environment of E. coli results in a net inflow of protons resulting in acidification of cytoplasm. PH homeostasis is an energy dependent process, which restores the cytoplasmic pH to near neutrality. The pH of the cell cytoplasm is a critical parameter controlling a variety of cellular processes.
The effect of a lowered extracellular pH on E. coli activates various pH homeostasis mechanisms, which restore the pH of the cytoplasm to near neutral. Pumping out protons from the cell is one of the many mechanisms that undo the effect of a lowered extracellular pH. Dehydrogenases when convert their substrate into its product, generate reducing power which on getting recycled at the ETC results in extrusion of protons out of the cell. In the present work, this expected result was confirmed by an increase in the specific activities of SDH, ICD and MDH on external acidification [Figures 3 and 4]. An increase in the specific activity suggests a higher flux of metabolites through the TCA cycle along with the recycling of the reducing power at the ETC. Moreover, the results in the present work are in accord with the work of Kannan et al. where these authors have, on a transcriptomic level, observed an up-regulation of succinate dehydrogenase on rapid external acidification of E. coli W3110.
Figure 3.

Comparison of specific activity of (a) SDH, (b) ICD, (c) MDH, (d) G6PD from External acidification of E. coli DH5 α for different time periods upon exposure to different low pH (acidic shock). PH 7 is used as control (Mean ± SD for triplicate values)
Figure 4.
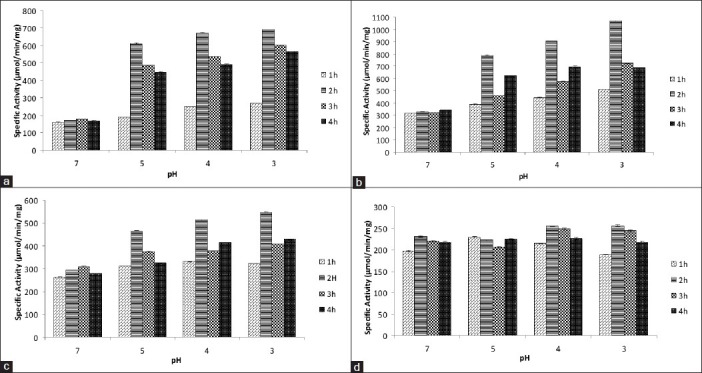
Comparison of specific activity of (a) SDH, (b) ICD, (c) MDH, (d) G6PD from External acidification of E. coli W3110 for different time periods upon exposure to different low pH (acidic shock). pH 7 was used as control (Mean ± SD for triplicate values)
G6PD is a key enzyme of the pentose phosphate pathway and its substrate G-6-P can be channeled along this pathway, or via glycolysis into the TCA cycle. Hence, under certain situations competition for the channeling of common substrate, G-6-P, between these pathways plays an important role. At low pH of 3, 4, and 5 channeling of G-6-P through glycolysis into TCA cycle was higher than when compared to pentose phosphate pathway in both E. coli DH5 α and E. coli W3100. Evidence of such a channeling came from the following observation-a rise in specific activities of SDH, ICD and MDH of the TCA cycle when compared to no noteworthy change in the specific activity of G6PD. This suggests that, at low pH, metabolites are not diverted from the central metabolic pathway to other sub-branching pathways. In this way nearly all the reducing power generated through the central metabolic pathway may be used for pumping out protons through the ETC, thereby helping in maintenance of pH homeostasis of the cytoplasm.
On internal acidification
In the gastrointestinal tract, enteric bacteria are subjected to acid stress from strong acid (HCl) as well as bacterial fermentation products such as acetic, propionic, and butyric acids, which are membrane-permeant weak acids.[9,23] The cytoplasm of E. coli may be acidified without changing the external pH, for example, by addition of membrane permeant weak acid like sodium benzoate, which is used as a food preservative. The action of sodium benzoate is dependent on the pH value which involves diffusion of the lipophilic, undissociated acid molecule through the plasma membrane into the cytoplasm where it dissociates into charged anions and protons resulting in cytoplasmic acidification. A fall by two to three folds in the specific activities of all four dehydrogenases was observed as a consequence of cytoplasmic acidification in E. coli, which suggesting that these enzymes may have an optimum pH range of near neutral to alkaline [Figures 5 and 6].
Figure 5.
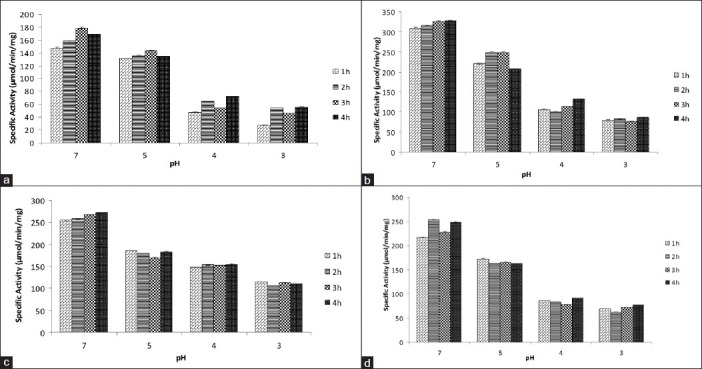
Comparison of specific activity of (a) SDH, (b) ICD, (c) MDH, (d) G6PD from internal acidification of E. coli DH5 α for different time periods upon exposure to different low pH (acidic shock) using sodium benzoate as an acid permeant (20 mM). pH 7 was used as control (Mean ± SD for triplicate values)
Figure 6.
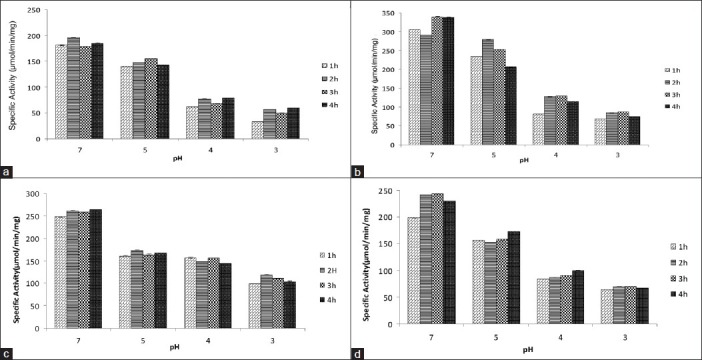
Comparison of specific activity of (a) SDH, (b) ICD, (c) MDH, (d) G6PD from internal acidification of E. coli W3100 for different time periods upon exposure to different low pH (acidic shock) using sodium benzoate as an acid permeant (20 mM). pH 7 was used as control (Mean ± SD for triplicate values)
On external acidification along with monensin
The pmf, generated across the inner membrane of E. coli, is a summation of electrical and chemical potential energies [pmf = Δψ + ΔpH]. Monensin, a monovalent ion-selective ionophore, facilitates the transmembrane exchange of principally sodium ions for protons.[24] The outer surface of the ionophore-ion complex is composed largely of nonpolar hydrocarbon, which imparts a high solubility to the complexes in nonpolar solvents. In biological systems, these complexes are freely soluble in the lipid components of membranes and, presumably, diffuse or shuttle through the membranes from one aqueous membrane interface to the other. Monensin binds to Na+ ions (from NaCl in the growth medium) in the extracellular space and shuttles them into the cytoplasm of the bacterium. In this way, it disrupts the electrical potential generated across its membrane and thus, acts as an uncoupler of oxidative phosphorylation. Since the electrical potential has been nullified by an uncoupler, to regain its actual pmf the bacterial cell enhances its chemical potential by increased expulsion of H+ ions through the ETC.[25] If, however, the external pH is lowered there will be a need to further increase the H+ ion expulsion. Reducing power generated within a cell with its recycling along the ETC expel H+ ions. Therefore, in both the scenarios (with or without acid shock), it is expected that the rate of recycling of the reducing power between the TCA cycle and the ETC may increase. In the present work, this expected result was confirmed by an increase in the specific activities of SDH, ICD and MDH on external acidification along with the use of monensin when compared to external acidification alone [Figures 1 and 2]. This further increase in specific activities suggests a higher rate of recycling of reducing power between the TCA cycle and the ETC and also an increment in the metabolic flux through the TCA cycle.
E. coli excretes acetate due to increased pyruvate flux from glucose transport by the phosphoenolpyruvate-dependent phosphotransferase system.[26] E. coli W3100 PTA mutant is defective in PTA, the first enzyme of the acetate pathway, sparing acetyl-CoA molecules to be fluxed through the TCA cycle which otherwise would have been excreted as acetate molecules in E. coli DH5 α.
PTA is a key enzyme of the acetate pathway and in a number of bacteria, its substrate is acetyl-CoA which can, in certain metabolic situations, be channeled along this pathway. In normal situations, acetyl-CoA is channeled preferably through the tricarboxylic acid cycle. Hence, in certain situations, a competition between the acetate pathway and the tricarboxylic acid cycle for the common substrate, acetyl-CoA may take place. At low pH conditions of 3, 4, and 5 along with an uncoupler-monensin, acetyl-CoA channeling through the TCA cycle was higher in E. coli W3110 when compared to E. coli DH5 α. The evidence of such a channeling came from the following observation-a rise in specific activities of SDH, ICD, and MDH of the TCA cycle in E. coli W3110 when compared to that in E. coli DH5 α. This is in agreement with the mutant nature of the strain E. coli W3110. This suggests, in E. coli W3110, a high metabolic flux through the TCA cycle results in a higher recycling of the reducing power between the TCA cycle and the ETC resulting in enhanced pumping of protons as compared to E. coli DH5 α. This manifests improved acid tolerance in E. coli W3110 than E. coli DH5 α.
The specific activity of G6PD remained unchanged in both E. coli W3100 and E. coli DH5 α stains indicating that there may be a normal flux of metabolites through the pentose phosphate pathway. Here again, results indicate that metabolites are preferably routed through the central metabolic pathway leading to higher recycling of reducing power at the ETC resulting in enhanced pumping of protons, thereby, contributing to the maintenance of pH homeostasis.
The human stomach produces gastric juice having a pH range of 1.8-2.5, which increases to between pH 3.0 and 5.0 during feeding and 2 h is the average emptying time for the stomach.[22] Documentation of highest activities of all tested dehydrogenases, viz. SDH, ICD and MDH in the 2nd h of time adaptation to external pH under both the conditions, including external acidification and external acidification along with monensin may point towards increased pH homeostasis as one of the many survival strategies possessed by E. coli, in its successful colonization of the human gastrointestinal tract. Proposed mechanism for acid shock generated acid tolerance in Escherichia coli is shown in Figure 7.
Figure 7.
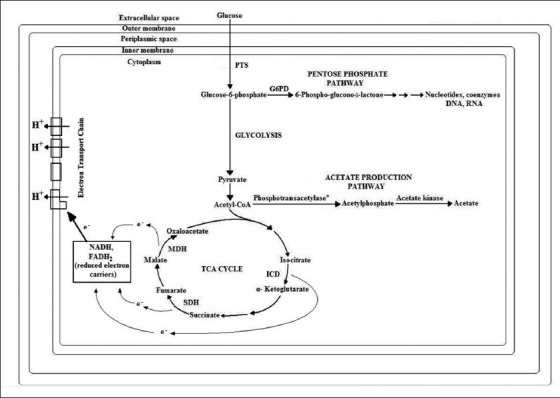
Proposed mechanism for acid shock generated acid tolerance in Escherichia coli
CONCLUSION
The results of this study may indicate that acid shocked E. coli for a period of 2 h can survive for a sustained period.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Hommais F, Krin E, Coppée JY, Lacroix C, Yeramian E, Danchin A, et al. GadE (YhiE): A novel activator involved in the response to acid environment in Escherichia coli. Microbiology. 2004;150:61–72. doi: 10.1099/mic.0.26659-0. [DOI] [PubMed] [Google Scholar]
- 2.Dawson AG. Oxidation of cytosolic NADH formed during aerobic metabolism in mammalian cells. Trends Biochem Sci. 1979;4:171–6. [Google Scholar]
- 3.Michels M, Bakker EP. Generation of a large, protonophore-sensitive proton motive force and pH difference in the acidophilic bacteria Thermoplasma acidophilum and Bacillus acidocaldarius. J Bacteriol. 1985;161:231–7. doi: 10.1128/jb.161.1.231-237.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–69. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 5.Benjamin MM, Datta AR. Acid tolerance of enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1995;61:1669–72. doi: 10.1128/aem.61.4.1669-1672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dilworth MJ, Glenn AR. Problems of adverse pH and bacterial strategies to combat it. Novartis Found Symp. 1999;221:4–14. doi: 10.1002/9780470515631.ch2. [DOI] [PubMed] [Google Scholar]
- 7.Bearson S, Bearson B, Foster JW. Acid stress responses in enterobacteria. FEMS Microbiol Lett. 1997;147:173–80. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- 8.Castanie-Cornet MP, Foster JW. Escherichia coli acid resistance: CAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology. 2001;147:709–15. doi: 10.1099/00221287-147-3-709. [DOI] [PubMed] [Google Scholar]
- 9.Polen T, Rittmann D, Wendisch VF, Sahm H. DNA microarray analyses of the long-term adaptive response of Escherichia coli to acetate and propionate. Appl Environ Microbiol. 2003;69:1759–74. doi: 10.1128/AEM.69.3.1759-1774.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkpatrick C, Maurer LM, Oyelakin NE, Yoncheva YN, Maurer R, Slonczewski JL. Acetate and formate stress: Opposite responses in the proteome of Escherichia coli. J Bacteriol. 2001;183:6466–77. doi: 10.1128/JB.183.21.6466-6477.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannan G, Wilks JC, Fitzgerald DM, Jones BD, Bondurant SS, Slonczewski JL. Rapid acid treatment of Escherichia coli: Transcriptomic response and recovery. BMC Microbiol. 2008;8:37. doi: 10.1186/1471-2180-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pressman BC. Biological applications of ionophores. Annu Rev Biochem. 1976;45:501–30. doi: 10.1146/annurev.bi.45.070176.002441. [DOI] [PubMed] [Google Scholar]
- 13.Garnak M, Reeves HC. Purification and properties of phosphorylated isocitrate dehydrogenase of Escherichia coli. J Biol Chem. 1979;254:7915–20. [PubMed] [Google Scholar]
- 14.Hayer-Hartl M. Assay of malate dehydrogenase. A substrate for the E. coli chaperonins GroEL and GroES. Methods Mol Biol. 2000;140:127–32. doi: 10.1385/1-59259-061-6:127. [DOI] [PubMed] [Google Scholar]
- 15.Spencer ME, Guest JR. Isolation and properties of fumarate reductase mutants of Escherichia coli. J Bacteriol. 1973;114:563–70. doi: 10.1128/jb.114.2.563-570.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banerjee S, Fraenkel DG. Glucose-6-phosphate dehydrogenase from Escherichia coli and from a “high-level” mutant. J Bacteriol. 1972;110:155–60. doi: 10.1128/jb.110.1.155-160.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smeets EH, Muller H, de Wael J. A NADH-dependent transketolase assay in erythrocyte hemolysates. Clin Chim Acta. 1971;33:379–86. doi: 10.1016/0009-8981(71)90496-7. [DOI] [PubMed] [Google Scholar]
- 18.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- 19.Wilks JC, Slonczewski JL. pH of the cytoplasm and periplasm of Escherichia coli: Rapid measurement by green fluorescent protein fluorimetry. J Bacteriol. 2007;189:5601–7. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert LA, Abshire K, Blankenhorn D, Slonczewski JL. Proteins induced in Escherichia coli by benzoic acid. J Bacteriol. 1997;179:7595–9. doi: 10.1128/jb.179.23.7595-7599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill MJ. Factors controlling the microflora of the healthy upper gastrointestinal tract. In: Hill MJ, Marsh PD, editors. Human Microbial Ecology. Boca Raton, Fla: CRC Press, Inc; 2002. pp. 57–85. [Google Scholar]
- 22.Texter EC., Jr Pressure and transit in the small intestine. The concept of propulsion and peripheral resistance in the alimentary canal. Am J Dig Dis. 1968;13:443–54. doi: 10.1007/BF02233667. [DOI] [PubMed] [Google Scholar]
- 23.Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, Foster JW. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell JB, Strobel HJ. Effect of ionophores on ruminal fermentation. Appl Environ Microbiol. 1989;55:1–6. doi: 10.1128/aem.55.1.1-6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell JB. Effect of extracellular pH on growth and proton motive force of Bacteroides succinogenes, a cellulolytic ruminal bacterium. Appl Environ Microbiol. 1987;53:2379–83. doi: 10.1128/aem.53.10.2379-2383.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang DE, Shin S, Rhee JS, Pan JG. Acetate metabolism in a pta mutant of Escherichia coli W3110: Importance of maintaining acetyl coenzyme. A flux for growth and survival. J Bacteriol. 1999;181:6656–63. doi: 10.1128/jb.181.21.6656-6663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


