Abstract
Sugar cane processing sites are characterised by high sugar/hemicellulose levels, available moisture and warm conditions, and are relatively unexplored unique microbial environments. The PhyloChip microarray was used to investigate bacterial diversity and community composition in three Australian sugar cane processing plants. These ecosystems were highly complex and dominated by four main Phyla, Firmicutes (the most dominant), followed by Proteobacteria, Bacteroidetes, and Chloroflexi. Significant variation (p < 0.05) in community structure occurred between samples collected from ‘floor dump sediment’, ‘cooling tower water’, and ‘bagasse leachate’. Many bacterial Classes contributed to these differences, however most were of low numerical abundance. Separation in community composition was also linked to Classes of Firmicutes, particularly Bacillales, Lactobacillales and Clostridiales, whose dominance is likely to be linked to their physiology as ‘lactic acid bacteria’, capable of fermenting the sugars present. This process may help displace other bacterial taxa, providing a competitive advantage for Firmicutes bacteria.
Sugar cane (Saccharum officinarum L.) is a perennial grass grown in many tropical countries. Globally, it is one of the most important staple crops, both in terms of total production (ranked #1 at 1,685 Million tonnes) and area planted (#13 at 23.8 Million Ha) (2010 data; http://faostat.fao.org). In many tropical countries, sugar cane production represents the most important land-use and agricultural commodity. The processing of sugar cane involves crushing of the cane stalks, with water added to enhance recovery of juices. The juices are then processed through to a clarified solution which is concentrated to syrup by evaporators. The syrup may be later crystallised to form solid sugar. A by-product of the process is bagasse, the fibrous residue of the cane stalk. This may itself be used in the sugar-cane processing, either to remove sediment from the juice clarification process, and/or burned to provide energy to operate the facility.
Sugar cane production sites offer an unusual habitat for microbial life. The inherent nature of sugar cane processing results in the accumulation of high levels of organic material, some of which contain a high percentage of labile C (sugars) and the rest of which is largely a mix of complex carbohydrates (bagasse/trash)1. The sites themselves are located in tropical to sub-tropical environments (alongside cane farms) and, as such, are warm and humid. Moisture availability is enhanced by the high use of water throughout the processing system.
A diverse range of ‘micro niches’ can be identified as common to most processing sites. However, most microbiological investigations have focussed on microbiota affecting the quality of the sugar product, or those which affect industrial operation of the plant. In the first instance, fungi such as Aspergillus, Penicillium, Saccharomyces, Torula, Pichia, and bacteria including Streptomyces, Pseudomonas, and Bacillus are commonly reported as responsible for the deterioration of sugar2,3. Cooling tower water (CTW) systems are often colonised by oligotrophic, thermophilic microbial communities with Caulobacteriaceae, Sphingomonadaceae, and Microbacteriaceae bacteria being the most abundant4,5. These bacteria can form biofilms which reduce the efficiency of the cooling towers and affect production at the cane-mills.
Furthermore, microbial micro-niches result from the bagasse processing. Typically, bagasse is stock-piled (“heaped”) and rainwater is passed through the piles to minimize the risk of spontaneous combustion and the associated generation of contaminated water and is captured in floor dump sumps (FDS) before being further processed. If contaminated storm water, leachate and windborne bagasse enters a waterway it has the potential to reduce dissolved oxygen levels and in extreme circumstances this can result in contaminated water with possible negative outcomes such as fish kills1. In comparison to the CTW, the bagasse leachate (BGL) represents a habitat highly enriched in nutrients (hemicellulose and others) but otherwise under a similar range of environmental conditions. In comparison to CTW, the microbial community with FDS is less well understood, yet also has important applied aspects relating to environmental (odor) and human health issues (exposure to microbial rich environments)1.
The primary objective of this study was to compare microbial diversity within hemi-cellulose enriched (FDS) and non-hemicellulose enriched habitats (CTW) in a tropical environment. As a secondary goal, we further aimed to understand the diversity and structures of microbial communities within the processing plants of one of the most important global commodities - sugar cane.
Results
Alpha diversity
Across all the samples, 34 different Phyla were found to be present (Table 1), which represents approximately half of those detectable by PhyloChip. The total OUT's present was 1367 – approximately 16% of those covered on the array. A table summarising the number of taxa present for each sample is given in the Supplementary Information. Richness of microbial communities was assessed using Margalefs index (d). Not surprisingly, richness increased with taxonomic level across all samples (Table 1). The levels of richness in the bacterial communities were similarly high across all sites and for all micro-niches examined. No clear impacts of sample type or location were evident.
Table 1. Richness (Margalefs index; d) of bacterial communities present at sugar-cane processing sites.
| Treatment | BGL | FDS | CTW | FDS | CTW | FDS | CTW |
|---|---|---|---|---|---|---|---|
| Location | Mackay | Maryborough | Prosperine | ||||
| Phylum | 1.91 | 1.88 | 1.85 | 1.88 | 1.82 | 1.86 | 1.85 |
| Class | 3.81 | 3.77 | 3.70 | 3.75 | 3.64 | 3.72 | 3.70 |
| Order | 7.28 | 7.19 | 7.07 | 7.17 | 6.96 | 7.10 | 7.06 |
| Family | 13.47 | 13.29 | 13.07 | 13.25 | 12.86 | 13.13 | 13.06 |
| Sub-family | 16.41 | 16.20 | 15.93 | 16.15 | 15.68 | 16.01 | 15.92 |
| OTU | 78.94 | 77.94 | 76.64 | 77.69 | 75.41 | 76.99 | 76.59 |
Similarly, the evenness of the bacterial communities was similar between sampling locations and micro-niches examined (Table 2). As such, the communities were not dominated by a few taxa and this evenness is balanced across the samples. Combined analysis of the data set (Tables 1 and 2) showed there was a high degree of correlation between richness and evenness in the samples (Spearman correlation ρ = 0.926; p < 0.001). Samples with high richness of taxa had greater evenness; were less inclined to be dominated by any particular taxa.
Table 2. Evenness (Pielous index; J′) of bacterial communities isolated from sugar-cane processing sites.
| Treatment | BGL | FDS | CTW | FDS | CTW | FDS | CTW |
|---|---|---|---|---|---|---|---|
| Location | Mackay | Maryborough | Prosperine | ||||
| Phylum | 0.464 | 0.519 | 0.474 | 0.502 | 0.486 | 0.475 | 0.526 |
| Class | 0.658 | 0.672 | 0.677 | 0.679 | 0.680 | 0.678 | 0.699 |
| Order | 0.814 | 0.764 | 0.821 | 0.793 | 0.803 | 0.828 | 0.785 |
| Family | 0.867 | 0.838 | 0.867 | 0.856 | 0.855 | 0.873 | 0.854 |
| Sub-family | 0.856 | 0.828 | 0.858 | 0.846 | 0.845 | 0.865 | 0.844 |
| OTU | 0.986 | 0.959 | 0.990 | 0.976 | 0.981 | 0.967 | 0.962 |
Identification of dominant taxa
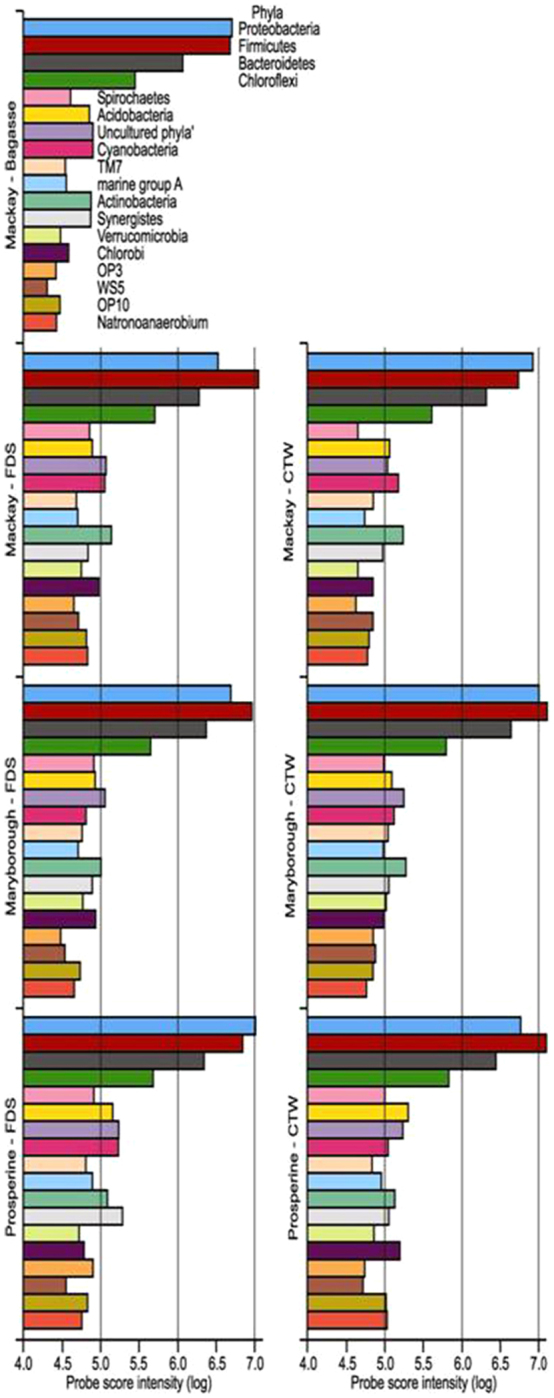
The ‘top100’ probe intensity score data was used to describe the dominant bacterial community of the sugar-cane processing mill. At Phylum level, the bacterial communities for all samples were dominated by Firmicutes, Proteobacteria, Bacteroidetes and Chloroflexi (Figure 1). However, even when looking within the top100 intensity data, a large range of phyla (18 of the 34 in total detected by PhyloChip) were present (Figure 1). These included a rage of common (e.g. Cyanobacteria, Acidobacteria, Chlorobi) and rarer (e.g. OP3, TM7, Natronoanaerobium) taxa.
Figure 1. Abundance of bacterial Phyla in samples of bagasse, floor dump sediment (FDS) and cooling tower water (CTW) from sugar-cane processing sites.
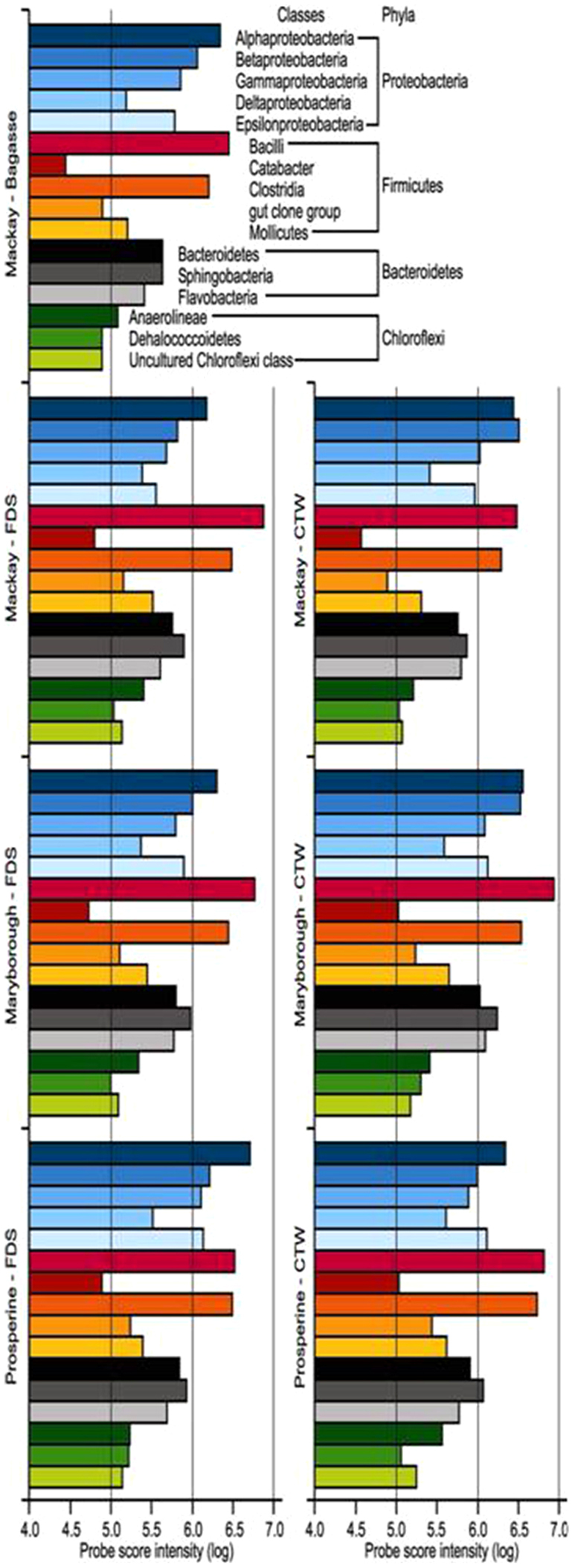
At the Phylum, a similar pattern of abundance occurred for the dominant Phyla for each of the sugar-cane communities. Of near-equal importance were always Firmicutes and Proteobacteria; Bacteriodetes were always third most abundant; Chloroflexi always fourth (Figure 1). The ‘minor placings’ were distributed reasonably across the remaining Phyla (Figure 1).
Analysis of the same data but at the Class level provides deeper information as to the composition of the dominant fraction of the bacterial communities. Within the Proteobacteria, the dominant classes were α-, β-Proteobacteria, followed by γ-, and ε-Proteobacteria (Figure 2). δ-Proteobacteria were present but at relatively lower abundance when compared to the other classes. The Firmicutes component of the community was dominated by Bacilli and Clostridia. In fact, Bacilli were the most dominant bacterial Class present in all but one sample (FDS at Prosperine; Figure 2). The Phylum Bacteroidetes were dominated by Classes Sphinogobacteria and Bacteroidetes. Many other classes were present but contributed only a minor component of the overall community composition (Figure 2).
Figure 2. Abundance of bacterial Classes in samples of bagasse, floor dump sediment (FDS) and cooling tower water (CTW) from sugar-cane processing sites.
Effects of treatment (niches) and sampling location on community composition
The effect of treatment type and sampling location on bacterial community was tested using the whole PhyloChip data set. CAP analysis could not partition effects of ‘location’ over any of the taxonomic levels (p > 0.05; Table 2); i.e. the Firmicutes-dominated community type was consistent across all sugar-cane processing mills examined. However, significant effects of ‘treatment’ or niches/habitats were evident at high-taxonomic levels, with Phyla > Class > Order (Table 3). For all three taxa levels, the bagasse sample always partitioned from CTW or FDS samples. However, single allocation errors were made between one of the CTW and FDS samples (they were more similar to each other than their respective treatment groups).
Table 3. Summary CAP analysis results testing for effects of treatment type (bagasse, FDS, and CTW) or location (Mackay, Maryborough, Prosperine) on bacterial community composition.
| Treatment | Location | |
|---|---|---|
| Phyla | 0.019 | 0.783 |
| Class | 0.019 | 0.784 |
| Order | 0.095 | 0.816 |
| Family | 0.168 | 0.674 |
| Sub-family | 0.168 | 0.637 |
| OTU | 0.133 | 0.488 |
Similarly, ordination by nMDS was used to visualise similarity in community composition between samples (at Class level). The bagasse sample was quite different to the FDS and CTW samples; however there is only a single observation for this treatment. A reasonable level of separation exists between the samples from CTW and FDS (Figure 4). Overall, the bacterial communities changed from bagasse-type, to FDS-type and then to CTW-type structures (Figure 3).
Figure 3. Non-metric multi-dimensional scaling ordination (NMDS) plot showing similarity in community composition between samples.
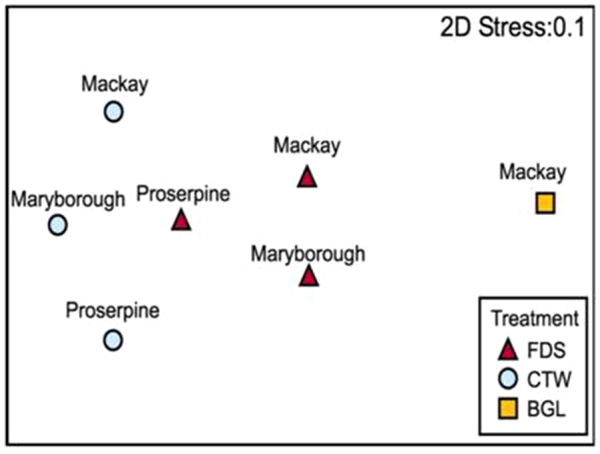
On the plot, increasing distance between sample points equates to more dissimilarity in community composition. Data is Class-level taxa and has been log(x + 1) transformed. Similarity is based on Gower's similarity method.
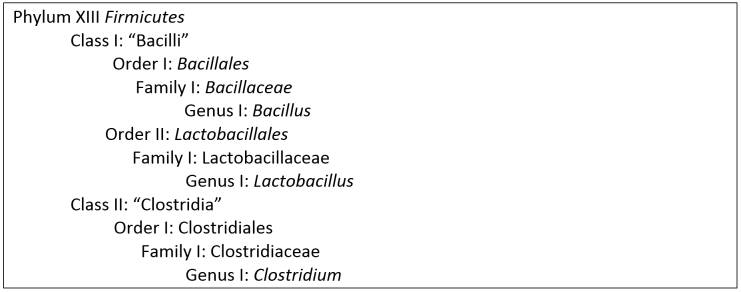
Figure 4. Taxonomy of selected genera of the Phylum Firmicutes (adapted from Whitman, 2009).
SIMPER analysis was used to determine which Classes of bacteria contributed towards defining differences in bacterial communities between the CTW and FDS treatments (Table 4). Most of the taxa contributing towards differences between the treatment groups were of minor relative abundance; as such, the rare bacterial groups were (generally) important in separating the two habitat types. A notable exception was the Firmicutes. Abundances of the Classes Bacillus, Lactobacillales, gut clone group, and Clostridia were all important in separating FDS and CTW communities; in all cases Firmicutes were of greater abundance in the FDS.
Table 4. SIMPER analysis identifying which bacterial Classes contribute most strongly towards differences in FDS and CTW community compositions. (UC) denotes an unclassified class within the Phyla identified. Average abundance data is log-transformed, average PhyloChip probe intensity data (standardised over samples). Average dissimilarity is the contribution of each taxa towards dissimilarity between the two treatment groups (FDS and CTW). Dissimilarity is the average contribution/standard deviation of the contribution over groups (i.e. consistency).
| Average abundance2 | ||||||
|---|---|---|---|---|---|---|
| Bacterial Class1 | FDS | CTW | Average Dissimilarity3 | Dissimilarity/standard deviation4 | Contrib-ution (%) | Cumulative contribution (%) |
| NC10-1 (UC) | 0.59 | 0.66 | 0.04 | 1.87 | 2.84 | 2.84 |
| Ellin6075/11-25 | 0.69 | 0.71 | 0.03 | 2.00 | 2.07 | 4.91 |
| Acetobacterales | 0.87 | 0.86 | 0.03 | 1.50 | 2.01 | 6.92 |
| MND1 clone group | 0.75 | 0.79 | 0.03 | 1.59 | 2.01 | 8.93 |
| Oscillatoriales | 0.59 | 0.58 | 0.03 | 1.14 | 1.97 | 10.90 |
| Methylophilales | 0.65 | 0.68 | 0.02 | 2.18 | 1.75 | 12.66 |
| Hydrogenophilales | 0.7 | 0.72 | 0.02 | 1.37 | 1.74 | 14.40 |
| Bacillales | 1.01 | 0.99 | 0.02 | 1.48 | 1.65 | 16.04 |
| Lactobacillales | 0.99 | 0.97 | 0.02 | 1.50 | 1.6 | 17.64 |
| TM7 (UC) | 0.67 | 0.68 | 0.02 | 1.66 | 1.53 | 19.17 |
| Xanthomonadales | 0.83 | 0.85 | 0.02 | 1.17 | 1.5 | 20.67 |
| Burkholderiales | 0.95 | 0.98 | 0.02 | 1.73 | 1.47 | 22.14 |
| gut clone group | 0.8 | 0.78 | 0.02 | 1.92 | 1.38 | 23.52 |
| Proteobacteria (UC) | 0.77 | 0.78 | 0.02 | 1.73 | 1.28 | 24.80 |
| Chloroflexi-4 (UC) | 0.71 | 0.68 | 0.02 | 1.30 | 1.22 | 26.03 |
| Nitrosomonadales | 0.79 | 0.81 | 0.02 | 1.71 | 1.19 | 27.22 |
| Clostridiales | 1.03 | 1.02 | 0.02 | 2.14 | 1.18 | 28.40 |
| Plectonema | 0.72 | 0.69 | 0.01 | 1.07 | 1.14 | 29.53 |
Discussion
Using high-density microarray tools (PhyloChip) a substantial level of bacterial diversity was uncovered in sugar-cane processing plants, a niche that has not been investigated widely to date. All of the samples studied were highly rich in taxa, which included not only the numerically dominant groups, but also rarer taxa, indicative of a very complex bacterial ecosystem. This data also revealed niche-partitioning among the treatments explored, from bagasse samples, floor dump sediments, through to cooling tower water. As discussed below, we propose that the habitat as a whole is remarkable in that it is highly selective for Firmicutes-type bacteria, and the shifts in the abundance of groups of Firmicutes (Classes, Orders etc) of these bacteria play a dominant role in defining the different habitat types.
This metagenomic study was designed to interrogate intact microbial communities, such as those found in the sugar mill samples, each of which comprises a separate micro-ecosystem. The intention of this research was to initiate a synecological approach to these communities, where a whole-of-community study would be used to determine such matters as Carbon degradation, Biofilm production, potential industrial processes to create useful end-products such as antibiotics or nutriceuticals, and other matters of interest. However, it is considered useful at this point to note some of the functional characteristics of the dominant Families in the majority of samples, in order to form a foundation for such community studies.
The presence of Firmicutes as a dominant group is quite rare for natural samples. The dominant Classes in the Firmicutes Phylum, according to the data shown in Figure 3, were the Bacilli, Catabacter, Clostridia, the “gut clone group” and the Mollicutes. In all of the samples (treatments), by far the most abundant were the Class Bacilli and the Class Clostridia. The taxonomical position of these classes is shown in Figure 4. Some functional physiological information about each Class, and about examples of selected Genus/species from each Class, follows.
Members of the class Bacilli generally form a Gram-positive cell wall, may or may not form endospores, and grow aerobically, microaerophilically or as facultative anaerobes. The order Bacillales is delineated on 16S sequence analysis, and includes the families Bacillaceae, Listeriaceae, Paenibacillaceae, Pasteuriaceae, Planococcaceae, Staphylococcaceae, Thermoactinomycetaceae, and others. Many of this order have in common the characteristic of producing endospores, and are Gram-positive, at least in young cultures. The type genus for the order is Bacillus17. The genus Bacillus is characterized by cells which are Gram-positive, mostly straight rod-shaped, occurring singly, in pairs, and in chains17. They are generally aerobic endospore formers, the heat resistant endospore being arguably their most important characteristic. Endospores also confer resistance to radiation, disinfectant action, UV and desiccation, making the genus one of nature's great survivors. The heat-resistant nature of the endospore accounts for the presence of Bacillus spp. in cooling tower waters, as the heat exchange process selects for heat resistant microorganisms, which become even more dominant as the water is recycled.
Using information from related research by this group, as yet unpublished, it was found by DGGE analysis and 16S rDNA sequencing (data not included), that the one of the major genera from the Phylum Firmicutes that were present in the liquid samples collected from sugar mills in Queensland, Australia, was Bacillus subtilis (found in all samples except one - CTW Maryborough). B. subtilis is known to use many sugars, its carbohydrate uptake and metabolism having been extensively reviewed by Deutscher et al. (2002)18. In fact, the majority of information regarding the metabolism and biochemistry of all Bacillus species relates specifically to Bacillus subtilis. An additional feature of B. subtilis is its ability to form biofilms, as a result of extracellular polysaccharide matrix production by the cells19. This capability is of great advantage to micro-communities, as the biofilm provides a matrix on which the community can develop.
The natural habitat of B. subtilis is soil, which contains a wide range of carbohydrates and polysaccharides from micro-organisms, plants and animals, and so it can utilize a wide range of such substrates and possesses a large number of enzymes which degrade polysaccharides. Interestingly, however, B. subtilis is also able to be isolated with ease from dried grass (“hay bacillus”). This may help to explain its major presence in the sugar mill samples, with sugar cane being a form of grass. The crushing and processing of sugar cane would release associated micro-flora, such as B. subtilis, into the mill environment, and the processing machinery would aid in the dissemination of aerosolized micro-flora throughout the mill, including into water sources such as the floor dump sumps and cooling towers. The common food-spoilage of sugar by “flat sours” refers to Bacillus species (Aerobic endospore formers), so its association with sugar processing is well recognized. The metabolic capabilities of B. subtilis make it an ideal organism to inhabit the sugar mill environment, as it is capable of metabolising the rich and diverse variety of sugars present.
The order Lactobacillales is distinguished by considerable gene loss, indicating early adaptation to nutritionally rich environments20. Members of the order do, however, retain the genes for production of various specific antimicrobials (bacteriocins), demonstrating the long-term survival of Lactobacillales in complex microbial communities, such as those reported here. The members of this order are known for their sugar metabolism and energy conversion systems, using various parts of the glycolysis pathway, the pentose-phosphate pathway and the mannose-specific phosphotransferase pathway. Further metabolic abilities seem to be specific to particular members of the order. Lactobacillales are able to change from sole production of lactic acid to the production of mixed end-products21,22. Lactobacilli commonly share the habitat when the content of sugar level is high in that habitat23. It has been mentioned earlier that bagasse leachate contains very high levels of hemi-cellulose and residual hexose sugars. According to Kleerebezem and Vaughan (2009)23, lactic acid bacteria are beneficial in the intestinal tract, producing lactic acid as an end-product of carbohydrate fermentation as well as bacteriocins20. This ability has been exploited in food production owing to the growth-inhibiting effect that both acidification and bacteriocins have on spoilage agents. Since the lactic acid bacteria are generally regarded as safe (GRAS) for specifically defined uses and they often produce narrow spectrum antibacterial peptides active against pathogenic bacteria, they have also been used for health-promoting purposes as probiotics24,25. Thus the presence of Lactic acid bacteria in sugar mill waste samples may have significant implications in the nutriceuticals and food industries. Like Bacillus species, many species of Lactobacillus are capable of producing biofilms. This capacity is advantageous in the probiotic industry, but is also of importance to the environmental survival of Lactobacillus species in microbial communities26.
The third member of the order Firmicutes to be examined is the genus Clostridium, type species Clostridium butyricum. Like other Firmicutes, Clostridium is characterised as being an endospore forming Gram-positive rod-shaped bacterium, but is distinguished by its anaerobic metabolism. C. butyricum is saccharolytic, producing butyric acid from glucose, as well as acetic acid, carbon dioxide and hydrogen gas. This species of Clostridium is “the dominating anaerobic spore-forming bacterium in certain silages”27 and ferments a wide variety of carbohydrates, including polysaccharides such as starch and pectin. C. butyricum is one of a number of Clostridia under investigation for their ability to produce industrial solvents27.
Summarizing the functional characteristics of the three dominant members of the Order Firmicutes, it can be seen that all three have wide-ranging metabolic capabilities, able to use various sugars as sole carbon sources. This ability is important in the niche environment being studied here. In addition, all three genera are able to produce useful end-products, particularly the Clostridium and Lactobacillus species. An interesting feature, given that these three genera are examples of the three dominant classes present in a single micro-environment, is the fact that one group is aerobic (Bacillus), one is microaerophilic (Lactobacillus) and the other is anaerobic (Clostridium). This is an ideal situation for a microbial community as the component genera are complimentary rather than competitive in their gaseous requirements. The ability of B. subtilis and the Lactobacillus sp. to form an extracellular matrix is also beneficial to the microbial community. Given the ability of Clostridium species to break down polysaccharides, it is interesting to note the potential dynamic – polysaccharide production and degradation occurring within the same community.
Proteobacteria were also one of the dominant groups present in all samples. The Proteobacteria Phylum includes a wide range of pathogens, such as Escherichia coli, Salmonella sp., Vibrio sp., Helicobacter sp. etc.28. However, the majority of species in this Phylum are free-living, and also include a number of nitrogen fixing bacteria29,30. The presence of the dominating soil Phylum such as Proteobacteria, Bacteroidetes and Chloroflexi in the environment is very common31.
Although the ecosystems were dominated by four major Phyla, a large overall diversity of taxa was present. The bagasse leachate sample was the most different in terms of community composition. The subsequent analysis revealed that the bacterial community was more diverse in bagasse leachate than FDS and CTW. This may be because of the temperature and composition of nutrients in the habitat. This information revealed that bagasse leachate is naturally enriched in carbon and nitrogen sources such as hemicelluloses1. Lignocellulosic materials, such as sugar cane bagasse represent an abundant, inexpensive source of organic material which can be a carbon source for microbial growth1. Sugar cane bagasse wastes are also sources of microbial nutrients (nitrogen and phosphate source). The micro flora is very likely to have the ability to grow in lignocellulose type substrates. The spent compost/cane bagasse mix, besides being an important source of nitrogen and phosphate nutrients, also provides an appropriate support matrix for native microorganisms.
In addition to Firmicutes being important discriminators of CTW versus FDS samples, a number of other Classes of bacteria were also found to be important. For example, Acetobacterales, Oscillatoriales, Methylophilales, Hydrogenophilales, Bacillales, Lactobacillales, Xanthomonadales, Burkholderiales, Chloroflexi-4, Nitrosomonadales, Clostridiales and Plectonema. Some of these taxa have few cultured isolates; as such their wider ecology can only be inferred on the basis of environmental DNA sampling. Although these Classes were important in defining differences between the two habitat types, this information needs to be considered in light of the low relative occurrence of many of the taxa. For the main part, the ecology of these ecosystems is dominated by the Firmicutes.
In order to confirm the data obtained in this study, the authors performed denaturing gradient gel electrophoresis (DGGE) followed by 16S rDNA sequencing on the same samples analysed by PhyloChip (unpublished data). The DGGE and sequencing results obtained confirmed the dominant genera in the Firmicutes phylum to be B. subtilis, B. cereus and B. thuringiensis. Other genera identified by DGGE and 16S rDNA sequencing representing the Proteobacteria phylum were Brevibacillus brevis, Rhodospirillaceae species, Vibrio aestuarianus, V. vulnificus, Pseudomonas aeruginosa and P. fluorescens. These are very important genera in various ways due to their physiological properties. For example genera capable of degrading hemicellulose include Bacillus species and Pseudomonas species, and genera capable of degrading cellulose and lignin such as Pseudomonas species. In addition, Vibrio species often degrade chitin and hemicelluloses. On the whole, B. brevis, Rhodospirillaceae species, Bacillus species, Vibrio species and Pseudomonas species are decomposers which have the ability to recycle nutrients in their niches. Protein producing B. brevis was found in all of the samples, and Rhodospirillaceae species are known to be photosynthetic bacteria which are responsible for oxygenation of the microbial environment.
Approaches based on metagenomic level analysis are needed if the complexity of these ecosystems was to be successfully explored. Amann et al., (1995)32 and Craig et al., (2010)33 estimated that > 99% of microorganisms that were observable in nature typically were not able to be cultivated using standard culture techniques. The majority of the bacterial taxonomic divisions are poorly represented by cultured organisms34. The overall performance of 16S rDNA sequence analysis is considered to be excellent, since it is able to resolve and almost indentify the microbial species, when applied to a large collection of phenotypically unidentifiable bacterial isolates35. Although 16S rDNA databases such as the arb-silva (http:www.arb-silva.de/), greengenes (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi) and the ribosomal database project (RDP) database (http://rdp.cme.msu.edu/) have been developed to perform sequence comparisons and interpretation, further efforts should be made to expand 16S rDNA databases with high-quality sequences. This study has determined the general microbial community composition and potentially useful industrial micro-organisms in sugar mill cooling tower, bagasse leachate and floor dump sump liquid samples.
A high-density microarray system, PhyloChip used in this research to investigate the bacterial diversity and community composition of waste materials in three sugar cane processing plants in Australia. The samples investigated all harboured highly complex bacterial ecosystems. Four main Phyla dominated the ecosystems, with Firmicutes being the most dominant. This was followed by the Phyla Proteobacteria, Bacteroidetes, and then Chloroflexi according to the numbers. Within each sugar-cane mill, significant variation (p < 0.05) in community structure occurred between samples collected from the tested micro-niches, which were ‘floor dump sump’, ‘cooling tower water’, and ‘bagasse leachate’. These differences were due to variation in a large number of bacterial Classes; however most were of low numerical abundance. Importantly, separation in community composition between these habitat types was also linked to Classes of Firmicutes, in particular the Bacillales, Lactobacillales and Clostridiales. The dominance of Firmicutes bacteria in these systems is likely to be linked to their physiology. As ‘lactic acid bacteria’, they are capable of fermentation of sugars present in these samples. This ability may assist in the displacement of other bacterial taxa, so providing a competitive advantage for Firmicutes bacteria.
Methods
Sites and sample collection
Sugar-cane mills were sampled from Proserpine (P), Mackay (MK) and Maryborough (M), all of which are located in Queensland, Australia. At each site, samples were collected from floor dump sump (FDS) and cooling tower water (CTW). In addition, bagasse leachate (BGL) was collected from the Mackay mill, but this was not possible at the other two mills due to lack of rain. Samples of 250 mL were collected during spring (Oct – Dec), in sterile containers fitted with ice packs. All samples were kept at −20°C for short term use and long term storage was at −80°C.
DNA extraction and PhyloChip analysis of bacterial communities
DNA was extracted from all liquid samples including bagasse leachate using the ultra clean water DNA isolation kit (MOBIO laboratory Inc. USA) following the manufacturer's instructions. Extracted DNA was quantified by spectrophotometry (NanoDrop ND-100) and stored at −80°C until use.
A high-density oligonucleotide microarray system (PhyloChip G2) was used to taxonomically identify the bacterial community present in the liquid samples. The PhyloChip, based around the Affymetrix GeneChip platform (Affymetrix CA, USA), has probes covering 8,741 prokaryotic 16S rRNA OTU's PhyloChip6,7.
Bacterial 16S rRNA genes were PCR amplified using primers 27F 5′(AGAGTTTGATCCTGGCTCAG)3′ and 1492R 5′(GGTTACCTTGTTACGACTT)3′. PCR mixtures included primers at 0.3 μM each, dNTP's at 200 μM each, 1.2 U of Taq polymerase (Takara), 10 × reaction buffers, 10 ng of template DNA and water to 25 μL. Eight individual PCR reactions were set up over a primer annealing range of 48–58°C (reducing primer bias). After hot-start enzyme activation, PCR thermocycling consisted of 35 cycles of denaturation at 95°C for 30 s, annealing for 30 s and with an extension at 72°C for 90 s. A final elongation step was performed for 10 min at 72°C. PCR products from the separate 25 μL reactions were pooled, precipitated with isopropanol, washed with 80% ethanol and resuspended in water to concentrate the PCR products for PhyloChip analysis.
For each sample, 500 ng of PCR product was mixed with a control oligonucleotide spike and digested into 50–200 bp fragments with DNase I (Invitrogen) in One-Phor-All buffer (GE HealthCare). The 3′ ends of the fragments were labelled with biotin using terminal deoxynucleotidyl transferase (Promega) according to the GeneChip DNA labelling procedure (Affymetrix, CA). The biotinylated mixture was denatured (99°C for 5 min) and subsequently hybridised to a PhyloChip microarray at 48°C and 60 rpm for 16 h. The hybridised array was washed and stained (streptavidin-phycoerythrin) on an Affymetrix fluidics station according to protocols described previously6.
Identification of taxa
Raw PhyloChip array data (CEL data output files from the Affymetrix GeneChip Operating System) was imported into PhyloTrac for processing8. Pixel images (fluorescence intensities in well defined grid) were resolved as probe pairs (perfect match (PM) and mismatch (MM)), which were grouped into probe sets (OTUs). Each probe set contains an average of 24 probe-pairs per OTU and also contains a central 17-mer not found in other OTUs. For each PM probe, a MM probe, with a single nucleotide difference was present. For each probe set, the trimmed mean fluorescence intensity (highest and lowest probe values removed before averaging) were normalized to internal spike-in control intensities using a maximum likelihood method, then scaled to the mean overall array and finally, log transformed. Taxa were considered present in a sample when at least 90% of the probes in its probe set pass the following criteria: PM/MM ≥ 1.3 and PM ≥ 130 × background noise9.
A data file containing information on OTU's detected by the microarray, probe set intensity (mean value excluding the highest and lowest values), and hierarchical taxonomic classification was compiled. Manual reassignment of a few ambiguously-classified taxa was conducted (e.g. some Bradyrhizobiaceae were classified into the Bradyrhizobiales order and other Bradyrhizobiaceae into the Rhizobiales). A final tree, linking OTU intensity data in each sample to taxonomic rankings, was imported into the Primer 6 software package10 (PRIMER-E Ltd, UK) for analysis.
Microbial community analysis
Alpha-diversity was measured as both richness and evenness of the microbial communities, using selected indices as described by Kennedy & Smith (1995)11. Richness of bacteria OTU's was measured across the taxonomic levels using Margalefs index (d). Similarly, community evenness was measured using the Pielous index (J′); essentially the Shannon–Weiner index (H′) divided by the maximum value H′ can take.
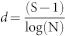 |
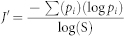 |
Where S = number of ‘species’ (at a given taxonomic ranking), N = number of individuals, pi is the proportion of intensity fluorescence for an individual OTU (at a given taxonomic level) relative to the sum of all the intensities detected in the sample.
When determining which taxa of bacteria are dominant in a system, the overall intensity data from the PhyloChip cannot be directly used past OTU (sub-family) level. Aggregation of this data to higher taxonomic levels imposes bias as probe coverage for bacteria such as Proteobacteria are much greater than those for ‘rare’ groups such as Phylum TM7. This can be overcome by using the ‘top 100’ taxa (probe intensity scores) for each treatment. The most abundant taxa are thereby selected and effects of differing coverage of probes across taxonomic groups are minimized. Accordingly, the top 100 probe intensity values were ranked for each treatment and the combined data merged into a single file containing the top 100 for all treatments (total of 351 unique OTU). This data was used to explore which taxa were dominant in abundance across the treatments. Taxonomic aggregation of OTUs was performed in Primer 610 as described previously.
Similarity in microbial community composition between samples was assessed to explore the effects the sample type and/or location effects. For each taxonomic level, OTU intensity values (full PhyloChip data set) were log-transformed and a resemblance matrix was generated using Gower's similarity method12,13. The influence of ‘sample type’ and ‘location’ were then tested at each level of taxonomy using CAP analysis – canonical analysis of principal components – with probability testing using permutation (999 repeats). Ordination by non-metric, multidimensional scaling (nMDS) was used to visualise similarity in community composition (Class aggregated taxa) between samples14. As before, the underlying resemblance matrix for the nMDS was Gower's similarity measure. SIMPER analysis (similarity percentages/species contributions; data standardised) was used to determine which taxa were important in describing differences in taxa composition between the CTW and FDS treatments15. All multivariate data analysis in the Primer 610/PERMANOVA software package (PRIMER-E Ltd., UK)16 using routines described previously10,14,16.
Author Contributions
F.S. was responsible for sample collection, DNA extraction and co-writing the manuscript. S.W. performed the PhyloChip microarray experiment and assisted with data analysis. F.H., S.W. and M.H. also contributed to writing the manuscript and reviewed the manuscript.
Supplementary Material
Supplementary file
Acknowledgments
This research has been supported by grants from the Faculty of Science, Queensland University of Technology, Brisbane, Australia.
References
- Womersley J. Guideline: Managing impacts from the bulk storage of bagasse. Department of Environment and Resource Management publication, Queensland, Australia (2006).
- Bentham R. H. Routine sampling and the control of Legionella spp. in cooling tower water systems. Curr. Microbiol. 41, 271–275 (2000). [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan T. Role of cyanobacteria in sugar mill effluent. MPhilDissertation, Bharathidasan University, Tiruchirapalli, India (2007).
- Cheesman O. D. Environmental Impacts of Sugar Production: the Cultivation and Processing of Sugarcane and Sugar Beet. CAB International, Wallingford, UK (www.cabi-publishing.org)Date accessed 22 August 2012 (2004).
- World Wildlife Fund (WWF). Sugar and the environment, encouraging better management and practices in sugar protection. WWF Global Freshwater Programme, Netherlands (2004).
- Brodie E. L. et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microb. 72, 6288–6298 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie E. L. et al. Urban aerosols harbour diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U S A. 104, 299–304 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz M. C. et al. Integrated microbial survey analysis of prokaryotic communities for the PhyloChip microarray. Appl. Environ. Microb. 76, 5636–5638 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desantis T. et al. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microbial. Ecol. 53, 371–383 (2007). [DOI] [PubMed] [Google Scholar]
- Clarke K. R. & Gorley R. N. PRIMER v6: User manual/Tutorial.PRIMER-E Ltd, Plymouth, UK (2006).
- Kennedy A. C. & Smith K. L. Soil microbial diversity and the sustainability of agricultural soil. Plant Soil. 170, 75–86 (1995). [Google Scholar]
- Gower J. C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 53, 325–338 (1966). [Google Scholar]
- Kuczynski J. et al. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat. Methods. 7, 813–819 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K. R. & Warwick R. M. Change in marine communities: An approach to statistical analysis and interpretation.Plymouth, UK: second ed Primer-E Ltd. (2001).
- Clarke K. R. Non-parametric multivariate analysis of changes in community structure. Australian J. Ecol. 18, 117–143 (1993). [Google Scholar]
- Anderson M. J., Gorley R. N. & Clarke K. R. PERMANOVA+ for PRIMER: Guide to software and statistical methods.PRIMER-E, Plymouth, UK (2008).
- Whitman W. B. Ed., 2009. Bergey's Manual of Systematic Bacteriology Vol 3, 2nd Edition, Pub by Springer, New York.
- Deutscher J., Galinier A. & Martin-Verstraete I. Carbohydrate uptake and metabolism. Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC, 129–150 (2002).
- Aguilar C., Vlamakis H., Guzman A., Losick R. & Kolter R. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. mBio (mbio.asm.org). Date accessed 22 August 2012 (2010). [DOI] [PMC free article] [PubMed]
- Marakova K. et al. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U S A. 103, 15611–15616 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucaud C., Hemme D. & Desmazeaud M. C. Peptide utilization by Lactococcus lactis and Leuconostoc mesenteroides. Lett. Applied Microbiol. 32, 20–25 (2004). [DOI] [PubMed] [Google Scholar]
- Liu S. Practical implications of lactate and pyruvate metabolism by lactic acid bacteria in food and beverage fermentations. Int. J. Food Microbiol. 83, 115–131 (2003). [DOI] [PubMed] [Google Scholar]
- Kleerebezem M. & Vaughan E. E. Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Ann. Rev. Microbiol. 63, 269–290 (2009). [DOI] [PubMed] [Google Scholar]
- Teusink B., Walsh M. C., Van-Dam K. & Westerhoff H. V. The danger of metabolic pathways with turbo design. Trends Biochem. Sci. 23, 162–169 (1998). [DOI] [PubMed] [Google Scholar]
- Teusink B. & Smid E. J. Modelling strategies for the industrial exploitation of lactic acid bacteria. Nat. Rev. Microbiol. 4, 46–56 (2006). [DOI] [PubMed] [Google Scholar]
- Lebeer S., Verhoeven T., Velez M., Vanderleyden J. & De Keersmaecker J. Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 73, 6768 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya D., Arevalo C., Gonzales S., Aristizabal F. & Schwarz W. New solvent-producing Clostridium sp. strains, hydrolyzing a wide range of polysaccharides, are closely related to Clostridium butyricum. J. of Ind. Microbiol. Biotech. 27, 329–335 (2001). [DOI] [PubMed] [Google Scholar]
- O'Sullivan D. J. Methods for analysis of the intestinal microflora. Curr. Iss. Intes. Microbiol. 1, 39–50 (2000). [PubMed] [Google Scholar]
- Madigan M. & Martinko J. Brock's biology of microorganisms. Prentice-Hall (2005).
- Patel J. B. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol. Diagn. 6, 313–321 (2001). [DOI] [PubMed] [Google Scholar]
- Li H., Yu Y., Luo W., Zeng Y. & Chen B. Bacterial diversity in surface sediments from the Pacific Arctic Ocean. Extremophiles. 13, 233–246 (2009). [DOI] [PubMed] [Google Scholar]
- Amann R. I., Ludwig W. & Schleifer K. H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Mol. Biol. R. 59, 143–169 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J. W., Chang F. Y., Kim J. H., Obiajulu S. C. & Brady S. F. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl. Environ. Microb. 76, 1633–1641 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessitsch A. et al. Diagnostic microbial microarrays in soil ecology. New Phytol. 171, 719–736 (2006). [DOI] [PubMed] [Google Scholar]
- Huse S. M. et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genetics. 4, e1000255 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file


